Abstract
Homeodomain transcription factors serve important functions in organogenesis and tissue differentiation, particularly with respect to the positional identity of individual cells. The Nkx6 subfamily controls tissue differentiation in the developing central nervous system where they function as transcriptional repressor proteins. Recent work indicates that Nkx6.3 is expressed in hindbrain V2 interneurons that co-express Nkx6.1, suggesting the possibility of functional redundancy. Here, we report that Nkx6.3 expression is specific to Chx10+ V2a interneurons but not Gata3+ V2b interneurons of the hindbrain, and that Nkx6.3 expression appears to mark cells of the prospective medullary reticular formation. Molecular analysis of Nkx6.3 null embryonic mouse hindbrain did not reveal detectable defects in progenitor markers, motor neuron or V2 interneuron sub-types. Forced expression of Nkx6.3 and Nkx6.1 promote V2 interneuron differentiation in the developing chick hindbrain. These findings indicate Nkx6.3 function is dispensable for CNS development and lead to the proposal that absence of overt defects is due to functional compensation from a related homeodomain transcription factor.
Section: Nervous System Development, Regeneration and Aging
Keywords: Homeobox transcription factor, CNS development, V2 interneuron, Nkx6.3, Nkx6.1, rhombomere, medullary reticular formation
INTRODUCTION
The “reticular formation” is located in the central brainstem running through the mid-brain, pons and medulla. The ascending reticular activating system connects to areas in the thalamus, hypothalamus, and cortex, while the descending reticular activating system connects to the cerebellum and sensory nerves. The reticular formation is an important regulator in the autonomic nervous system for such processes as respiration rate, heart rate and gastrointestinal activity, and it is proposed to have further roles in sleep and consciousness, modulation of pain and other behaviors. Development of this system is poorly understood.
The Nkx6 subfamily is part of the Nkx class of transcription factors. Its 18 members have diverse functions in precise regional specification of precursor cells in a variety of organ systems including the central nervous system (CNS) (Kimura et al., 1996) (Lyons et al., 1995) (Pabst et al., 1999; Stanfel et al., 2005) (Sussel et al., 1998). Two Nkx6 subfamily members, Nkx6.2, previously called Gtx (Komuro et al., 1993), and Nkx6.1 (Rudnick et al., 1994) share DNA-binding preference, repress transcription, and show features of dynamic regulation in broad areas of the developing ventral hindbrain and spinal cord (Mirmira et al., 2000) (Muhr et al., 2001) (Qiu et al., 1998). Sonic hedgehog (Shh) signaling induces Nkx6.1 and Nkx6.2 expression in ventral neural tube progenitors where they exhibit redundant function in neuron and oligodendrocyte specification (Briscoe et al., 2000) (Cai et al., 2005) (Vallstedt et al., 2001).
Absence of Nkx6.1 results in a substantial decrease in the number of V2 interneurons and somatic motor neurons with significant cell loss along the murine CNS anterior-posterior (A–P) axis (Sander et al., 2000a); oligodendrocyte differentiation is delayed in the spinal cord but not in the hindbrain (Liu et al., 2003). Isolated Nkx6.2 loss causes an approximately 50% decrease in V1 interneurons and a corresponding increase in V0 neurons without affecting the number of somatic motor neurons or V2 neurons (Vallstedt et al., 2001). However, combined loss of Nkx6.1 and Nkx6.2 reduces the number of motor neuron by 90% throughout the spinal cord (Vallstedt et al., 2001) and interferes with proper differentiation, migration and projection of visceral motor neurons in the caudal hindbrain (Pattyn et al., 2003). Furthermore, Nkx6.1 induces V2 interneuron differentiation in the chick ventral hindbrain (Briscoe et al., 2000). These results reveal complex and overlapping Nkx6 gene functions that vary according to tissue position along the A–P axis of the CNS.
Nkx6.3 is more closely related to Drosophila Nk6 than other Nkx gene family members, which raises the possibility that it is the ancestral founder of the vertebrate Nkx6 subfamily (Pedersen et al., 2005). Like its paralogs, Nkx6.3 contains an Engrailed-homology domain that may mediate interaction with transcriptional co-repressors (Muhr et al., 2001); however, its physiologic functions have not been fully characterized. In E12.5 mouse embryos, Nkx6.3 expression is restricted to a subset of differentiating V2 interneurons in the caudal hindbrain that co-labels with Chx10 (Alanentalo et al., 2006) (Pedersen et al., 2005).
Here we show that Nkx6.3 expression is specifically associated with the Chx10+ subset of V2 interneurons but not the Gata3+ sub-type of V2 interneurons of the hindbrain. Nkx6.3 expression overlaps significantly with that of Nkx6.1 in the brain. To determine the in vivo requirements for Nkx6.3, we used gain- and loss-of-function approaches. First, we used homologous recombination in mouse embryonic stem cells to inactivate the mouse Nkx6.3 gene. Nkx6.3−/− animals developed normally without detectable defects in brain development. Second, we show that forced expression of Nkx6.3 and Nkx6.1 promote V2 interneuron differentiation in the developing chick hindbrain. Thus, the absence of overt defects in mice lacking Nkx6.3 is likely due to functional compensation from related homeodomain transcription factors, such as Nkx6.1.
RESULTS
Identification of Nkx6.3 in a screen of the mouse transcriptome for transcription factors specifically expressed in the medullary reticular formation
In a recent study, Gray and coworkers used in situ hybridization to map the expression of ~1,100 transcription factor-encoding genes in the developing CNS (Gray et al., 2004), and found that 349 of these genes were sufficient to anatomically define subregions of the CNS. Of particular interest to us were genes with unique expression patterns in the medullary reticular formation, as the molecular mechanisms underlying early neuronal specialization in this region are not well understood. To address this question, we searched the brain atlas database for genes with spatially restricted expression patterns in this region at E13.5, the earliest time point in the study (Functional Genomic Atlas of the Mouse Brain; http://mahoney.chip.org/). Interestingly, we found a novel homeobox gene that is expressed exclusively in this region. This homeobox gene was most closely related to the homeobox sequence found in the Nkx gene family members, Nkx6.1 and Nkx6.2, and thus we called it Nkx6.3. This gene has been reported independently by several groups (Alanentalo et al., 2006; Henseleit et al., 2005; Nelson et al., 2005; Pedersen et al., 2005).
Nkx6.3 marks Chx10+ V2a neurons in the medullary reticular formation in the developing hindbrain
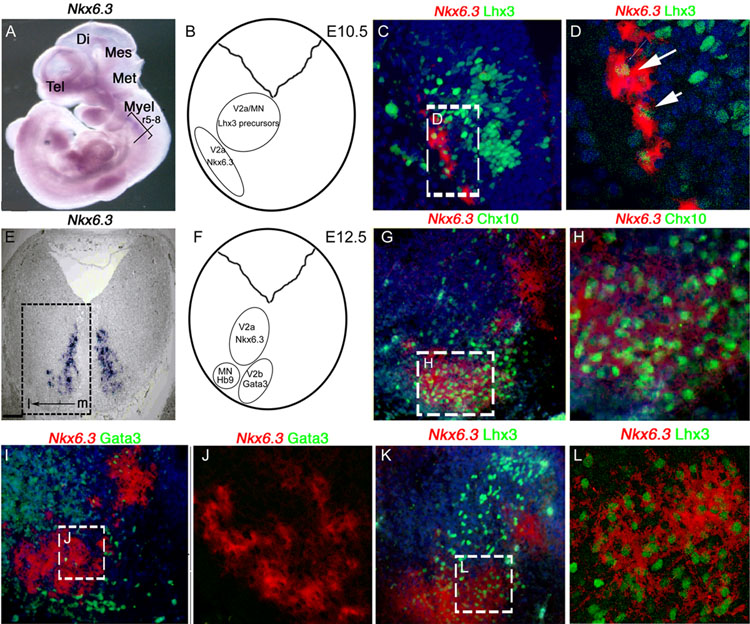
In the E10.5–12.5 embryonic CNS, Nkx6.3 mRNA expression is regionally restricted to the caudal hindbrain between rhombomeres (r) 5–8 (Fig. 1A). Analysis of transverse section at r7 shows Nkx6.3 expression in restricted region (Fig. 1B, C, E, F). We next examined Nkx6.3 mRNA relative to interneuron, and motor neuron markers. As shown (Fig. 1C, D), Nkx6.3 expression is detected at E10.5 in a subset of cells that co-express the V2 neuron marker Lhx3; co-expression is persistent at E12.5 (Fig. 1K, L). V2 interneurons are divided in V2a (Chx10+) and V2b (Gata3+) subclasses. Our analysis indicates that Nkx6.3 localizes to a subpopulation of Chx10+ V2a neurons but not Gata3+ V2b cells (Fig. 1G–J).
Figure 1. Expression of Nkx6.3 in V2a interneurons of the embryonic CNS.
(A–D) E10.5 embryo. (A) Whole mount in situ hybridization confirms Nkx6.3 expression between rhombomere 5 and rhombomere 8 (r5–8). A–P level of analysis (transverse black line at r7), telencephalon (tel), diencephalon (di), mesencephalon (mes), metencephalon (met), and myelencephalon (myel) are indicated. (B) Cartoon of transverse section at level of analysis showing prospective V2a/b and MN populations. (C) Combined double-label ISH for Nkx6.3 mRNA (pseudocolored red, cytoplasmic) and immunohistochemistry (IHC) for Lhx3 proteins (green, nuclear); (D) confocal photomicrograph showing overlap of Nkx6.3 and Lhx3 in some ventrolateral cells. (E–L) E12.5 embryo. (E) Nkx6.3 ISH in a transverse section from E12.5 caudal hindbrain. (F) Cartoon showing relative location of Nkx6.3 expression with V2a/b interneuron and motor neuron populations in the E12.5 caudal hindbrain at the level of analysis. (G–L) Double-label ISH for Nkx6.3 (red pseudocolor) and IHC with against Chx10, Gata3 and Lhx3 (green). (H, J, L) High-resolution confocal images from areas indicated within (G, I. K). (G. H) Note that (G, H) the V2a marker Chx10 and (K, L) Lhx3 are co-expressed in most Nkx6.3+ cells, whereas the V2b marker Gata3 is not co-expressed. In all panels except (A) dorsal is top, ventral is bottom, medial (m) is right and lateral (l) is to left. Scale bars: C, G, I, K 10 µM, F 100 µM, D, H, J, L 25 µM. Original magnification A x2.5, C, G, I, K x40, F x10, D, H, J, L x80.
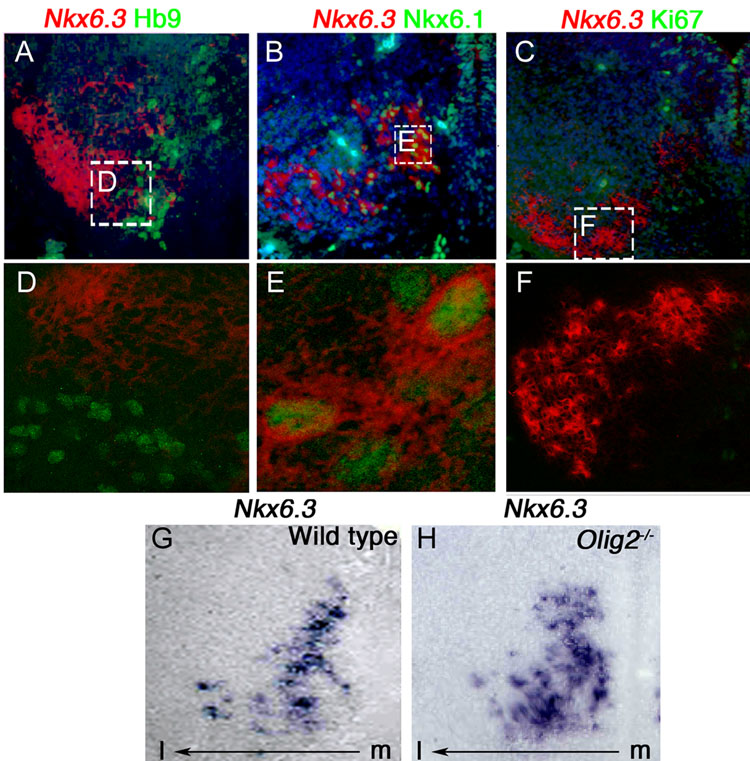
In E12.5 embryos Nkx6.3 is not expressed in Hb9+ motor neurons, indicating that it is specific to Chx10+ V2a neurons (Fig. 2A, D). Nkx6.1 and Nkx6.3 are co-expressed in post-mitotic V2 interneurons (Fig. 2B, E), raising the possibility that these two transcription factors share certain functions. At all stages examined between E10.5 to postnatal, Nkx6.3+ cells do not co-label with the proliferation marker Ki67, indicating that it is expressed in post-mitotic neurons (Fig. 2C, F, and data not shown). In the Olig2 null mutant embryo, which has a cell fate switch from motor neurons to V2 neurons (Lu et al., 2002; Zhou and Anderson, 2002), the population of Nkx6.3+ cells is expanded (Fig. 2G, H). This is consistent with our interpretation that Nkx6.3 expression is confined to V2 interneurons and also shows that Olig2 function is not required for regulation of Nkx6.3 expression. In summary, Nkx6.3 specifically marks V2a interneurons of the caudal hindbrain. Furthermore, our analysis confirms that Nkx6.3 expression localizes to cells of the prospective medullary reticular formation.
Figure 2. Further characterization of Nkx6.3 expression at E12.5 in wild type and Olig2 null mutant embryos.
(A–F) For orientation, see area indicated by the dashed box in Figure 1E. (A–C) Low resolution and (D–F) high resolution confocal images showing combined ISH for Nkx6.3 (pseudocolored red, cytoplasmic) with IHC for Hb9, Nkx6.1 and Ki67 (green, nuclear). Note that the motor neuron marker Hb9 and the proliferation marker Ki67 are not co-expressed in Nkx6.3+ cells. Nkx6.1 is co-expressed with Nkx6.3+ cells at E12.5. (G, H) Expression of Nkx6.3 is expanded in Olig2−/− mutant compared to wild type control. Scale bars: A–C, G–H 25 µM. Original magnification x40. D–F 10 µM original magnification x80.
Assessment of Nkx6.3 function in the embryonic CNS
To assess Nkx6.3 functions in vivo, we designed a targeting construct to delete exons 2 and 3, thus replacing the full homeodomain and flanking regions with the neomycin-resistance gene (Choi et al., 2008). The null allele segregated in the expected Mendelian proportions indicating viability (Choi et al., 2008).
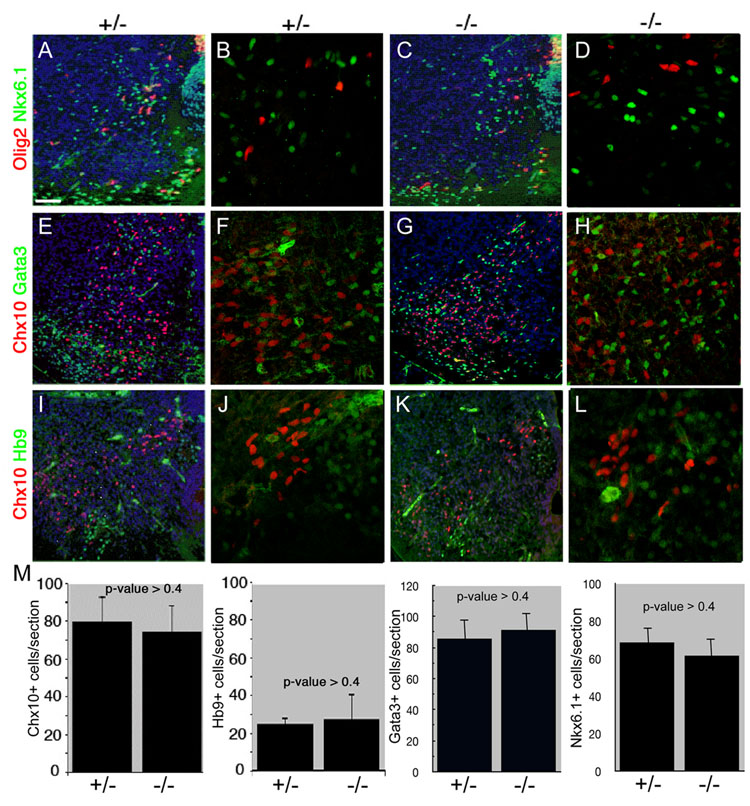
In situ hybridization on transverse hindbrain sections from E12.5 Nkx6.3 mutant embryos at r7 confirmed absence of Nkx6.3 expression, compared to Nkx6.3+/− embryos (Supplemental Figure 1). To determine the impact of Nkx6.3 function on mouse CNS development, heterozygous Nkx6.3+/− mice were crossed to generate Nkx6.3−/− E14.5 embryos. As shown in Figure 3, using a variety of molecular markers for neurons and oligodendrocytes, we failed to detect a significant phenotype. First, the neural specification factors Nkx6.1 and Olig2 are expressed normally in Nkx6.3 null mutant embryos at E14.5 (Fig. 3A–D). Second, the number and distribution of Chx10+ V2a interneurons was unaffected in the caudal hindbrain of E14.5 mutant mice. On average, 74 ±12.4 post-mitotic Chx10+ cells were identified in each section from E12.5 Nkx6.3−/− embryos compared to 81 ±14.0 such cells per section from Nkx6.3+/− embryos (Fig. 3 E–M), which was statistically insignificant (p >0.4). Numbers of cells expressing the V2b interneuron marker Gata3 and the motor neuron marker Hb9 were also unaffected (Fig. 3 E–M). Nkx6.3-null mice lack overt neurobehavioral abnormalities, and as the physiologic role of V2 interneurons is unknown, it is unclear how the animals could be challenged to elicit other defects. Our results indicate that Nkx6.3 function is dispensable for development of motor neurons or V2 interneurons in the embryonic brainstem. The normal domain of Nkx6.1 expression encompasses Chx10+ V2 interneurons (Alanentalo et al., 2006) (Fig. 2), and numbers of Nkx6.1 cells were not significantly diminished in Nkx6.3 null embryos (Fig. 3M), consistent with the possibility of functional overlap with between these to factors in V2a interneuron development.
Figure 3. Nkx6.3 function is dispensable for V2a interneuron development.
Analysis of expression of Olig2 and Nkx6.1 (A–D); Chx10 and Gata3 (E–H); and Chx10 and Hb9 (I–L) in the caudal hindbrain of heterozygote (A, B, E, F, I, J) and Nkx6.3-null (C, D, G, H, K, L) mouse embryos at E14.5. (M) Quantification of numbers of Chx10+, Hb9+, Gata3+ and Nkx6.1+ cells per section in heterozygote (n=10) and Nkx6.3-null (n=5) embryos. Error bars indicate SEM. (B, D, F, H, J, L) are high resolution confocal photomicrographs from adjacent (A, C, E, G, I, K) photomicrographs. Scale bars: A, C, E, G, I, K 25 µM. B, D, F, H, J, L 10 µM. Original magnification A, C, E, G, I, K x40. B, D, F, H, J, L x80.
Nkx6.3 is sufficient to induce V2a interneurons in the chick CNS
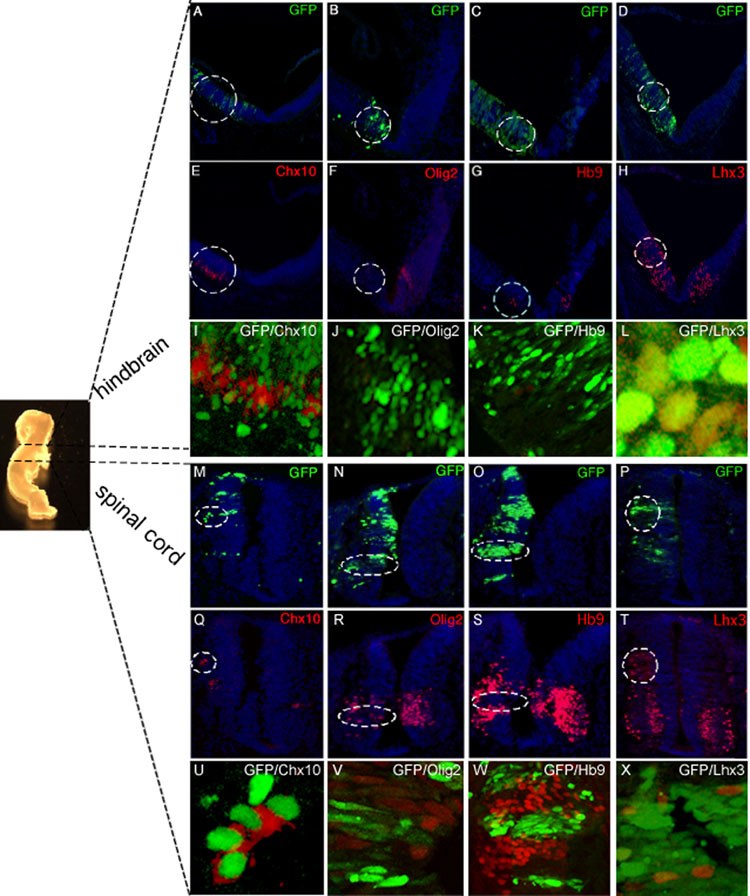
Nkx6.1 is well known to ectopically promote the generation of Chx10+ V2 interneurons and Hb9+ motor neurons in the chick neural tube (Briscoe et al., 2000, Liu et al., 2003). To investigate potential redundant functions between Nkx6.3 and Nkx6.1, we examined Nkx6.3 function through forced expression of Nkx6.3 in the spinal cord and brainstem of chick embryos. We cloned mouse Nkx6.3 cDNA into the expression vector pCIG, which contains an internal ribosome entry site-green fluorescent protein (GFP) cassette, electroporated the plasmid into the chick neural tube between Hamburger-Hamilton stages 9 and 11, and evaluated GFP-expressing embryos by immunostaining after 45–54 hours. In contrast to the empty pCIG vector (data not shown), Nkx6.3 misexpression promoted ectopic Chx10+ Lhx3+ V2a interneuron development in the hindbrain (Fig. 4 A, D, E, H, I, L) and spinal cord (Fig. 4 M, P, Q, T, U, X). Ectopic V2a cells were detected only on the electroporated side of the neural tube and were absent in embryos treated with control vector (n = 5); expression of Pax6 mRNA was unaltered (data not shown). Further, Nkx6.3 repressed Olig2 expression (Fig. 4 B, F, J, N, R, V), and development of Hb9+ motor neurons (Fig. 4 C, G, K, O, S, W). Misexpression of Nkx6.1 in the chick spinal cord and hindbrain produced ectopic Chx10+ Lhx3+ V2 interneurons and Hb9+ motor neurons (n=6) (Tab. 1, Suppl. Fig. 2), as previously described (Briscoe et al., 2000). Together, these observations indicate that Nkx6.3 is sufficient to induce the V2a interneuron determinants Chx10 and Lhx3, and repress the motor neuron marker Hb9 in the chick neural tube. The data suggests that Nkx6.3 and Nkx6.1 may possess similar biological functions in respect to induction of V2a interneurons.
Figure 4. Nkx6.3 is sufficient to induce specification of V2 interneurons in the chick CNS.
Chick embryos were electroporated unilaterally with the pCIG-Nkx6.3 plasmid at HH stages 9–11, and collected 44–54 h later. Whole mount embryo (left) indicates hindbrain and spinal cord A–P levels at which the analysis was performed. (A–D, M–P) GFP expression is readily detected on the electroporated side of the neural tube in green; the contralateral side lacks GFP signals and provides an internal control. Nuclei are stained blue with DAPI. Unlike the empty pCIG vector (data not shown), Nkx6.3 misexpression induced Chx10+ Lhx3+ V2 interneurons in the chick hindbrain (A, D, E, H, I, L) and spinal cord (M, P, Q, T, U, X) (areas marked by dotted circle). (I, L, U) are high resolution confocal photomicrographs showing overlap of GFP signal with Chx10, and Lhx3. Nkx6.3 misexpression further repressed appearance of Hb9+ motor neurons in the chick spinal cord (O, S, W) and hindbrain (C, G, K). (K, W) High resolution confocal photomicrographs showing lack of overlap of GFP signal with Hb9. Olig2+ pMN progenitors were also repressed in the spinal cord (N, R, V) and hindbrain (B, F, J) (areas marked by dotted circle). Forced Nkx6.3 expression did not alter the domain of Pax6 expression, which served as an internal control, as Pax6 is known to be upstream of Nkx6 factors (data not shown). Results are representative of at least 5 embyos. Scale bars: A–H and M–T 25 µM. Original magnification x40.
Table 1. Summary of Nkx6.3 and Nkx6.1 misexpression analysis.
Summary of the results of the misexpression analysis in the chick spinal cord and brainstem of Nkx6.3 and Nkx6.1. Nkx6.3 misexpression in the chick spinal cord and brainstem induced ectopic expression of V2 neuron determinants Chx10+ and Lhx3+ (upward arrow). Nkx6.3 misexpression repressed motor neuron marker Hb9 and progenitor marker Olig2 (downward arrow). Nkx6.1 misexpression resulted in induction of Chx10+ and Lhx3+ V2 neurons, Hb9+ motor neurons and limited Olig2 progenitors (upward arrow). Nkx6.3 and Nkx6.1 misexpression resulted in no effect on Pax6, which was used as an internal control (horizontal arrow). Nkx6.3 results were representative of 5/5 embryos, and Nkx6.1 of 6/6 embryos.
| Markers: | |||||||
|---|---|---|---|---|---|---|---|
| Gene: | Region: | Chox10 | Lhx3 | Hb9 | Pax6 | n: | |
| Nkx6.3 |  |
Hindbrain | ↑ | ↑ | ↓ | +/− | 5/5 |
| Spinal cord | ↑ | ↑ | ↓ | +/− | 5/5 | ||
| Nkx6.1 |  |
Hindbrain | ↑ | ↑ | ↑ | +/− | 6/6 |
| Spinal cord | ↑ | ↑ | ↑ | +/− | 6/6 | ||
DISCUSSION
The transcription factors underlying development of the medullary reticular formation are not well understood. In this study, we searched the Mahoney brain atlas database for transcription factors specifically expressed in this region, and found a novel homeobox gene, Nkx6.3 specifically expressed within this region. This is, to our knowledge, the first transcription factor that specifically marks the medullary reticular formation.
It was previously shown that Nkx6.3 marks V2 neurons (Alanentalo et al., 2006). We extended this work by using a panel of markers and show that Nkx6.3 is selectively expressed in the V2a –but not V2b--subtype of V2 neurons. Loss of Nkx6.3 function does not result in a detectable CNS phenotype. The most likely explanation for this is functional redundancy with homologous factors. Candidates include other members of the Nkx6 subfamily, which share over 90% homology in the homeodomain sequence and common expression domains in the brain; Nkx6.3 expression overlaps significantly with that of Nkx6.1 in hindbrain V2a interneurons. To test the validity of this hypothesis, we misexpressed Nkx6.3 and Nkx6.1 in the developing chick CNS, and showed that they both promoted V2a interneuron differentiation. This finding is consistent with previous reports in the literature, where forced expression of Nkx6.1 resulted in ectopic induction of V2a interneurons (Briscoe et al., 2000). Thus Nkx6.3 and Nkx6.1 may function in a complementary manner at initial stages of V2 interneuron development.
We did find differences in the gain-of-function effects of Nkx6.3 and Nkx6.1 in motor neuron development. Although Nkx6.1 induces ectopic cells expressing the motor neuron marker Hb9 and progenitor marker Olig2 (Briscoe et al., 2000, Liu et al., 2003), Nkx6.3 forced expression potently repressed Hb9 and Olig2 expression. However, the functional significance of this effect in normal development is unclear because (1) Nkx6.3 is not normally expressed in motor neuron precursor pools and (2) loss of Nkx6.3 function did not result in ectopic dorsal activation of motor neuron markers. One interesting possibility is that a mechanism might exist to repress Nkx6.3 expression in motor neuron precursors.
Our findings lead us to propose that the absence of overt defects in mice lacking Nkx6.3 is due to functional compensation from related homeodomain transcription factors of the Nkx6 family, most likely Nkx6.1. Further work is needed to determine essential functions of Nkx6.1/6.2/6.3 functions in double and triple mutant mice, where one might predict specific defects in hindbrain neurogenesis. Other Nkx sub-families also share expression domains with Nkx6 genes in the CNS (Briscoe et al., 2000; Pabst et al., 2003) (Sander et al., 2000b) and influence Nkx6.3 function indirectly. In this regard, it is worth noting one basis for functional Nkx6 redundancy in ventral neurogenesis. Although Nkx6.2 expression increases in the absence of Nkx6.1 and compensates for the latter, it is not itself normally expressed in motor neuron progenitors. Rather, its expression is normally inhibited by Nkx6.1 and appears when Nkx6.1 transcriptional repression is lacking (Vallstedt et al., 2001). Thus, in principle, Nkx6.3 function could be substituted by a variety of equivalent proteins that it otherwise represses.
EXPERIMENTAL PROCEDURES
Mouse procedures
Details of targeting strategy used to generate Nkx6.3 targeted mutant mice and genotyping procedures are published (see Choi et al., 2008). Embryos from wild type c57Bl6 or mutant mice were collected at E10.5, E12.5 or E14.5 days counted from the time of vaginal plug formation (E 0.5). Embryos were washed in PBS and processed as described below.
In situ hybridization on tissue sections and whole mount embryos
For section and whole mount in situ hybridization, E10.5 embryos and E12.5 brains were fixed in 4% paraformaldehyde at 4°C overnight. Tissue sections were prepared as described below in the immunohistochemistry protocol (Ma et al., 1997) (Gray et al., 2004) (Cheng et al., 2005). Embryos and sections were hybridized overnight at 65°C with digoxigenin-labeled riboprobe transcribed from Nkx6.3 sequence cloned into Topo II (Invitrogen); cloned constructs were confirmed by DNA sequencing. Slides were washed with 2X and 0.2X SSC, blocked with 10% fetal bovine serum, and incubated with 1:1000 alkaline phosphatase-conjugated digoxigenin antibody (Roche, Indianapolis). Staining was visualized with NBT/BCIP. Images were captured with an Olympus BX41 compound microscope with attached CCD camera and Zeiss LSM 510 laser scanning confocal microscope and processed using Photoshop 7.0 (Adobe) to match the microscopic view.
Immunohistochemistry and tissue staining
Mouse embryos were fixed by perfusion of 4% paraformaldehyde in phosphate-buffered saline (pH 7.0), embedded in OCT compound, and cryosectioned at 12–14 µm thickness. Tissue specimens were blocked with 10% fetal bovine serum for 1 h at ambient temperature. Primary antibody sources were as follows: Lhx3 and Hb9, Developmental Studies Hybridoma Bank, University of Iowa; Chx10, a gift from C. Cepko, Harvard Medical School; Ki67, Novocastra Laboratories, Newcastle-upon-Tyne, UK; Nkx6.1 (Pedersen et al., 2006), Beta Cell Biology Consortium; Gata3 (Santa Cruz Biotechnology, Santa Cruz, CA). These antibodies were applied overnight at 4°C at the following dilutions: Lhx3, 1:3000; Hb9, 1:500; Chx10, 1:2000; Ki67, 1:100; Olig2, 1:40,000; Nkx6.1, 1:1000; and Gata3, 1:50. Secondary antibodies were FITC-conjugated anti-mouse IgG (1:100, Jackson Immunoresearch, West Grove, PA) or Cy3-conjugated anti-rabbit IgG (1:200, Jackson). After washing in PBS supplemented with 0.05% Tween-20, fluorescent stains were visualized on Olympus BX41 compound and Zeiss LSM 510 laser scanning confocal microscopes using Photoshop 7.0 software.
In ovo electroporation of chick embryonic neural tube
Full-length mouse Nkx6.3 cDNA, amplified from a stomach cDNA library, was subcloned into the expression vector pCIG (Megason and McMahon, 2002). Chick neural tubes at Hamburger and Hamilton (HH) stages 9–11 were flushed with 3 µg/µl plasmid DNA and electroporated unilaterally (three 25-millisecond pulses at 25 mV) using an ECM830 square-wave electroporator (BTX Molecular Delivery Systems). GFP+ embryos were harvested at Hamburger and Hamilton stages 21–23, after 44–54 hrs incubation, and processed for immunostaining. At least 5 electroporated embryos were studied for each DNA construct and the results documented are representative of at least 5 embryos.
Supplementary Material
In situ hybridization analysis of E12.5 caudal hindbrain showing Nkx6.3 RNA expression in wild type (A) but not mutant (B) embryos. Original magnification x40.
Nkx6.1 is sufficient to induce specification of V2a interneurons in the chick CNS. Chick embryos were electroporated unilaterally with the CMV-Nkx6.1 plasmid at HH stages 9–11, and collected 44–54 h later. (A–C) GFP expression is detected on the electroporated side of the neural tube in green; the contralateral side lacks GFP signals and provides an internal control. (E, F) Nkx6.1 misexpression induced Chx10+ Lhx3+ V2 interneurons in the chick neural tube in regions of ectopic transgene expression (compare with A–C), and (D) Hb9+ motor neurons. Results are representative of at least 5 embryos. Scale bars: A–H 25 µM. Original magnification x40.
ACKNOWLEDGMENTS
We would like to thank Sovann Kaing, Dong-in Yuk, and Anthony Romer for expert technical assistance. BPH is supported by an F30 training grant from the NINDS and the Harvard Center for Neurodegeneration and Repair. MYC is supported by the F32 training grant from the NIH. We are grateful to the Beta Cell Biology Consortium and C. Cepko. Lhx3 and Hb9 antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. This work was supported by grants from the NINDS (to DHR) and NIDDK (to RAS) and through the generosity of the Caring for Carcinoid Foundation (to RAS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE REFERENCES
- Alanentalo T, Chatonnet F, Karlen M, Sulniute R, Ericson J, Andersson E, Ahlgren U. Cloning and analysis of Nkx6.3 during CNS and gastrointestinal development. Gene Expr Patterns. 2006;6:162–170. doi: 10.1016/j.modgep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Choi MY, Romer AI, Wang Y, Wu MP, Ito S, Leiter AB, Shivdasani RA. Requirement of the tissue-restricted homeodomain transcription factor Nkx6.3 in differentiation of gastrin-producing G-cells in the stomach antrum. Mol Cell Biol. 2008 doi: 10.1128/MCB.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Henseleit KD, Nelson SB, Kuhlbrodt K, Hennings JC, Ericson J, Sander M. NKX6 transcription factor activity is required for alpha- and beta-cell development in the pancreas. Development. 2005;132:3139–3149. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Komuro I, Schalling M, Jahn L, Bodmer R, Jenkins NA, Copeland NG, Izumo S. Gtx: a novel murine homeobox-containing gene, expressed specifically in glial cells of the brain and germ cells of testis, has a transcriptional repressor activity in vitro for a serum-inducible promoter. Embo J. 1993;12:1387–1401. doi: 10.1002/j.1460-2075.1993.tb05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Cai J, Hu X, Tan M, Qi Y, German M, Rubenstein J, Sander M, Qiu M. Region-specific and stage-dependent regulation of Olig gene expression and oligodendrogenesis by Nkx6.1 homeodomain transcription factor. Development. 2003;130:6221–6231. doi: 10.1242/dev.00868. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Mirmira RG, Watada H, German MS. Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J Biol Chem. 2000;275:14743–14751. doi: 10.1074/jbc.275.19.14743. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Janiesch C, Sander M. Expression of Nkx6 genes in the hindbrain and gut of the developing mouse. J Histochem Cytochem. 2005;53:787–790. doi: 10.1369/jhc.5B6619.2005. [DOI] [PubMed] [Google Scholar]
- Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- Pabst O, Rummelies J, Winter B, Arnold HH. Targeted disruption of the homeobox gene Nkx2.9 reveals a role in development of the spinal accessory nerve. Development. 2003;130:1193–1202. doi: 10.1242/dev.00346. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Sander M, Ericson J. Complementary roles for Nkx6 and Nkx2 class proteins in the establishment of motoneuron identity in the hindbrain. Development. 2003;130:4149–4159. doi: 10.1242/dev.00641. [DOI] [PubMed] [Google Scholar]
- Pedersen IL, Klinck R, Hecksher-Sorensen J, Zahn S, Madsen OD, Serup P, Jorgensen MC. Generation and Characterization of Monoclonal Antibodies against the Transcription Factor Nkx6.1. J Histochem Cytochem. 2006;54:567–574. doi: 10.1369/jhc.5A6827.2006. [DOI] [PubMed] [Google Scholar]
- Pedersen JK, Nelson SB, Jorgensen MC, Henseleit KD, Fujitani Y, Wright CV, Sander M, Serup P. Endodermal expression of Nkx6 genes depends differentially on Pdx1. Dev Biol. 2005;288:487–501. doi: 10.1016/j.ydbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Qiu M, Shimamura K, Sussel L, Chen S, Rubenstein JL. Control of anteroposterior and dorsoventral domains of Nkx-6.1 gene expression relative to other Nkx genes during vertebrate CNS development. Mech Dev. 1998;72:77–88. doi: 10.1016/s0925-4773(98)00018-5. [DOI] [PubMed] [Google Scholar]
- Rudnick A, Ling TY, Odagiri H, Rutter WJ, German MS. Pancreatic beta cells express a diverse set of homeobox genes. Proc Natl Acad Sci U S A. 1994;91:12203–12207. doi: 10.1073/pnas.91.25.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell TM, Rubenstein JL. Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 2000a;14:2134–2139. doi: 10.1101/gad.820400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000b;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Stanfel MN, Moses KA, Schwartz RJ, Zimmer WE. Regulation of organ development by the NKX-homeodomain factors: an NKX code. Cell Mol Biol (Noisy-le-grand) 2005 Suppl 51:OL785–OL799. [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Muhr J, Pattyn A, Pierani A, Mendelsohn M, Sander M, Jessell TM, Ericson J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron. 2001;31:743–755. doi: 10.1016/s0896-6273(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In situ hybridization analysis of E12.5 caudal hindbrain showing Nkx6.3 RNA expression in wild type (A) but not mutant (B) embryos. Original magnification x40.
Nkx6.1 is sufficient to induce specification of V2a interneurons in the chick CNS. Chick embryos were electroporated unilaterally with the CMV-Nkx6.1 plasmid at HH stages 9–11, and collected 44–54 h later. (A–C) GFP expression is detected on the electroporated side of the neural tube in green; the contralateral side lacks GFP signals and provides an internal control. (E, F) Nkx6.1 misexpression induced Chx10+ Lhx3+ V2 interneurons in the chick neural tube in regions of ectopic transgene expression (compare with A–C), and (D) Hb9+ motor neurons. Results are representative of at least 5 embryos. Scale bars: A–H 25 µM. Original magnification x40.