Abstract
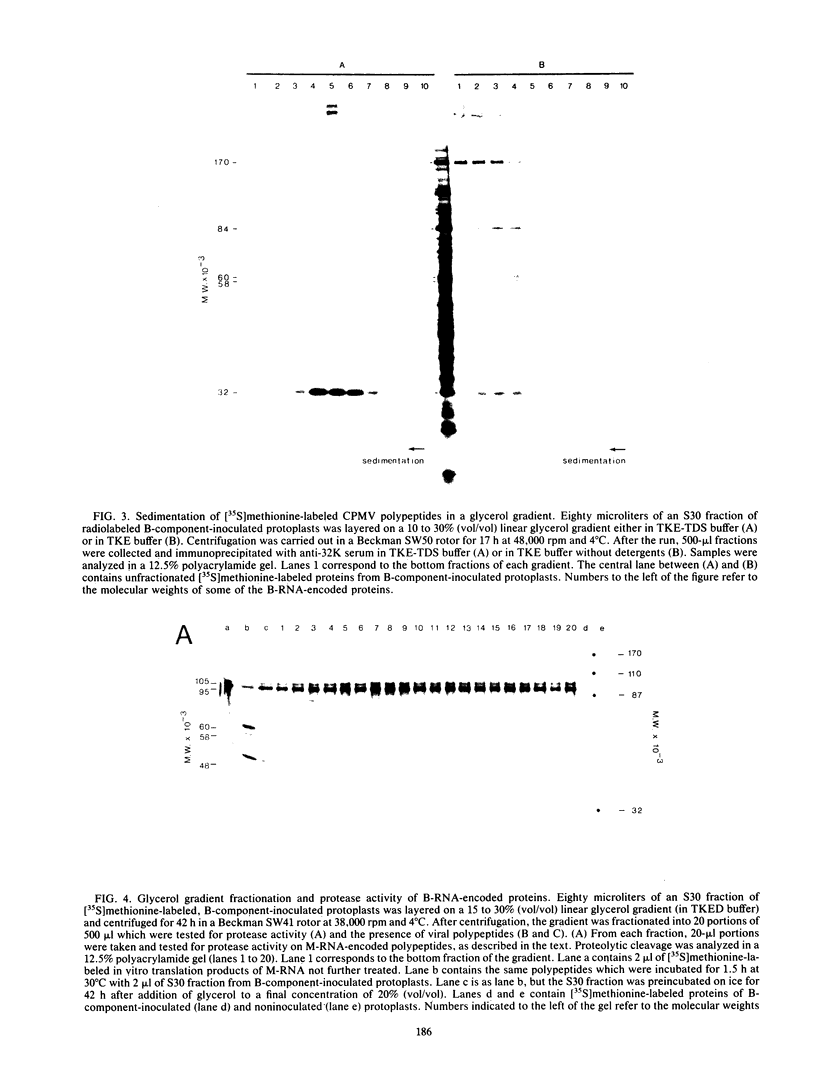
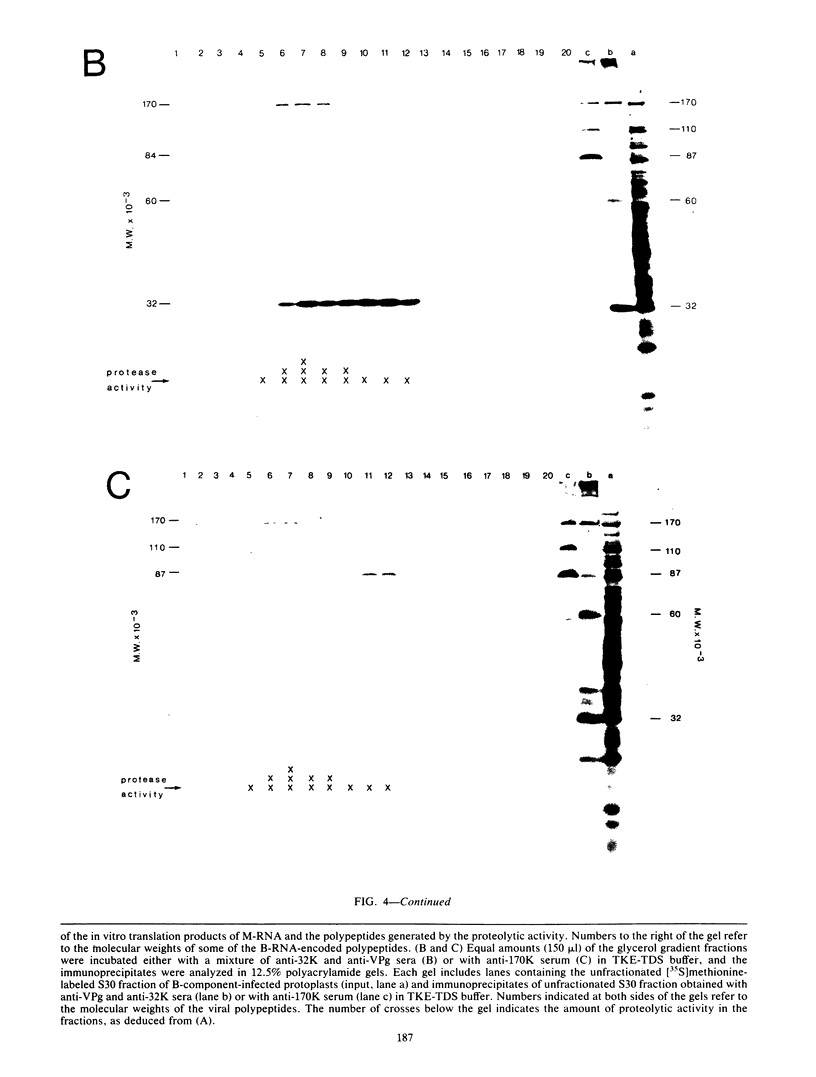
Translation of middle-component RNA of cowpea mosaic virus in vitro produced two polypeptides of 95 and 105 kilodaltons (95K and 105K, respectively) with overlapping amino acid sequences, which were specifically cleaved by a protease encoded by the bottom-component RNA. The proteolytic cleavage was studied by the addition of antibodies raised against various bottom-component RNA-encoded proteins to extracts prepared from bottom-component RNA-inoculated cowpea protoplasts. Since antiserum to the 32K polypeptide efficiently inhibited the proteolytic activity of such extracts, although antiserum to VPg or to the 170K polypeptide did not, evidence was obtained which indicates that the 32K polypeptide represents the protease involved. Fractionation of proteolytically active extract by glycerol gradient centrifugation demonstrated that 32K polypeptides do not exist as free proteins but are aggregated to the bottom-component RNA-encoded 170K, 84K, 60K, or 58K polypeptides. Maximal proteolytic activity was observed for 32K polypeptides associated with 170K polypeptides, suggesting that the activity was unstable and confined to newly synthesized molecules.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Kaesberg P. Determination of the length distribution of poly(A) at the 3' terminus of the virion RNAs of EMC virus, poliovirus, rhinovirus, RAV-61 and CPMV and of mouse globin mRNA. Nucleic Acids Res. 1979 Nov 10;7(5):1195–1204. doi: 10.1093/nar/7.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubert S. D., Bruening G., Najarian R. C. Protein bound to the genome RNAs of cowpea mosaic virus. Eur J Biochem. 1978 Dec 1;92(1):45–51. doi: 10.1111/j.1432-1033.1978.tb12721.x. [DOI] [PubMed] [Google Scholar]
- Franssen H., Goldbach R., Broekhuijsen M., Moerman M., van Kammen A. Expression of Middle-Component RNA of Cowpea Mosaic Virus: In Vitro Generation of a Precursor to Both Capsid Proteins by a Bottom-Component RNA-Encoded Protease from Infected Cells. J Virol. 1982 Jan;41(1):8–17. doi: 10.1128/jvi.41.1.8-17.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R. W., Schilthuis J. G., Rezelman G. Comparison of in vivo and in vitro translation of cowpea mosaic virus RNAs. Biochem Biophys Res Commun. 1981 Mar 16;99(1):89–94. doi: 10.1016/0006-291x(81)91716-2. [DOI] [PubMed] [Google Scholar]
- Goldbach R., Rezelman G. Orientation of the cleavage map of the 200-kilodalton polypeptide encoded by the bottom-component RNA of cowpea mosaic virus. J Virol. 1983 May;46(2):614–619. doi: 10.1128/jvi.46.2.614-619.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R., Rezelman G., Zabel P., van Kammen A. Expression of the Bottom-Component RNA of Cowpea Mosaic Virus: Evidence that the 60-Kilodalton VPg Precursor Is Cleaved into Single VPg and a 58-Kilodalton Polypeptide. J Virol. 1982 May;42(2):630–635. doi: 10.1128/jvi.42.2.630-635.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Svitkin Y. V., Kazachkov Y. A., Agol V. I. Encephalomyocarditis virus-specific polypeptide p22 is involved in the processing of the viral precursor polypeptides. FEBS Lett. 1979 Dec 1;108(1):1–5. doi: 10.1016/0014-5793(79)81164-3. [DOI] [PubMed] [Google Scholar]
- Hanecak R., Semler B. L., Anderson C. W., Wimmer E. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3973–3977. doi: 10.1073/pnas.79.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi T., Rezelman G., Van Kammen A. Infection of cowpea mesophyll protoplasts with cowpea mosaic virus. Virology. 1975 Apr;64(2):308–318. doi: 10.1016/0042-6822(75)90107-5. [DOI] [PubMed] [Google Scholar]
- Klootwijk J., Klein I., Zabel P., van Kammen A. Cowpea mosaic virus RNAs have neither m7GpppN ... nor mono-, di- or triphosphates at their 5' ends. Cell. 1977 May;11(1):73–82. doi: 10.1016/0092-8674(77)90318-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Palmenberg A. C., Pallansch M. A., Rueckert R. R. Protease required for processing picornaviral coat protein resides in the viral replicase gene. J Virol. 1979 Dec;32(3):770–778. doi: 10.1128/jvi.32.3.770-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C., Rueckert R. R. Evidence for intramolecular self-cleavage of picornaviral replicase precursors. J Virol. 1982 Jan;41(1):244–249. doi: 10.1128/jvi.41.1.244-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Synthesis and proteolytic processing of cowpea mosaic virus proteins in reticulocyte lysates. Virology. 1979 Jul 30;96(2):463–477. doi: 10.1016/0042-6822(79)90104-1. [DOI] [PubMed] [Google Scholar]
- Reijnders L., Aalbers A. M., van Kammen A., Thuring R. W. Molecular weights of plant viral RNAs determined by gel electrophoresis under denaturing conditions. Virology. 1974 Aug;60(2):515–521. doi: 10.1016/0042-6822(74)90345-6. [DOI] [PubMed] [Google Scholar]
- Rezelman G., Goldbach R., Van Kammen A. Expression of bottom component RNA of cowpea mosaic virus in cowpea protoplasts. J Virol. 1980 Nov;36(2):366–373. doi: 10.1128/jvi.36.2.366-373.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff R. D., Grandgenett D. P. Virus-coded origin of a 32,000-dalton protein from avian retrovirus cores: structural relatedness of p32 and the beta polypeptide of the avian retrovirus DNA polymerase. J Virol. 1978 Oct;28(1):279–291. doi: 10.1128/jvi.28.1.279-291.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Rottier P., Davies J. W., Zabel P., Van Kammen A. A protein linked to the 5' termini of both RNA components of the cowpea mosaic virus genome. Nucleic Acids Res. 1978 Dec;5(12):4505–4522. doi: 10.1093/nar/5.12.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R., Stinchcomb D., Losick R. Antibody directed against Bacillus subtilis rho factor purified by sodium dodecyl sulfate slab gel electrophoresis. Effect on transcription by RNA polymerase in crude extracts of vegetative and sporulating cells. J Biol Chem. 1975 Nov 25;250(22):8824–8828. [PubMed] [Google Scholar]
- Zabel P., Moerman M., van Straaten F., Goldbach R., van Kammen A. Antibodies Against the Genome-Linked Protein VPg of Cowpea Mosaic Virus Recognize a 60,000-Dalton Precursor Polypeptide. J Virol. 1982 Mar;41(3):1083–1088. doi: 10.1128/jvi.41.3.1083-1088.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Manna M. M., Bruening G. Polyadenylate sequences in the ribonucleic acids of cowpea mosaic virus. Virology. 1973 Nov;56(1):198–206. doi: 10.1016/0042-6822(73)90299-7. [DOI] [PubMed] [Google Scholar]
- van Kammen A. Purification and properties of the components of cowpea mosaic virus. Virology. 1967 Apr;31(4):633–642. doi: 10.1016/0042-6822(67)90192-4. [DOI] [PubMed] [Google Scholar]
- van Wezenbeek P., Verver J., Harmsen J., Vos P., van Kammen A. Primary structure and gene organization of the middle-component RNA of cowpea mosaic virus. EMBO J. 1983;2(6):941–946. doi: 10.1002/j.1460-2075.1983.tb01525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]