Abstract
Calmodulin plays a key role in the chemical gating of gap junction channels. Two calmodulin-binding regions have previously been identified in connexin32 gap junction protein, one in the N-terminal and another in the C-terminal cytoplasmic tail of the molecule. The aim of this study was to better understand how calmodulin interacts with the connexin32-binding domains. Lobe-specific interactions of calmodulin with connexin32 peptides were studied by stopped flow kinetics, using Ca2+ binding-deficient mutants. Peptides corresponding to the N-terminal tail (residues 1–22) of connexin32 engaged both the N- and C-terminal lobes (N- and C-lobes) of calmodulin, binding with higher affinity to the C-lobe of calmodulin (Ca2+ dissociation rate constants k3,4, 1.7 ± 0.5 s–1) than to the N-lobe (k1,2, 10.8 ± 1.3 s–1). In contrast, peptides representing the C-terminal tail domain (residues 208–227) of connexin32 bound either the C- or the N-lobe but only one calmodulin lobe at a time (k3,4, 2.6 ± 0.1 s–1 or k1, 13.8 ± 0.5 s–1 and k2, 1000 s–1). The calmodulin-binding domains of the N- and C-terminal tails of connexin32 were best defined as residues 1–21 and 216–227, respectively. Our data, showing separate functions of the N- and C-lobes of calmodulin in the interactions with connexin32, suggest trans-domain or trans-subunit bridging by calmodulin as a possible mechanism of gap junction gating.
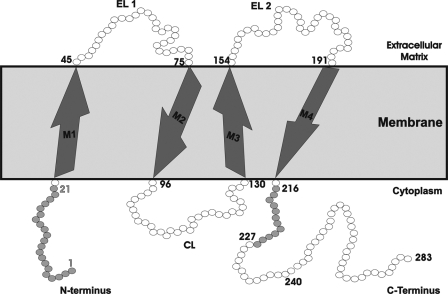
Gap junctions mediate direct intercellular communication by allowing the passage of ions and soluble molecules between cells. Gap junction channels are composed of two hemichannels (connexons); each connexon is composed of six connexins (Cx)2 (1). Each Cx polypeptide chain contains an N-terminal tail, four transmembrane domains (M1, M2, M3, and M4), two extracellular loops (EL1 and EL2), a cytoplasmic loop, and a C-terminal tail (Fig. 1).
FIGURE 1.
Schematic diagram of a Cx32 polypeptide chain spanning the membrane, data for residue numbers and positions obtained from NCBI data base search. The figure shows the N-terminal tail (residues 1–22), four transmembrane domains (M1, M2, M3, and M4), the intracellular loop (CL, residues 96–130), and the C-terminal tail (residues 215–283).
Gap junction channels are gated by transjunctional voltage (Vj), Ca2+, and H+ (2–5). Vj gradients activate two types of gates: fast and slow. The fast Vj gate flickers rapidly between the open and residual states, whereas the chemical/slow gate undergoes slow transitions between the open and closed states (6). Cytosolic acidification of small cells subjected to moderate Vj gradients allows the chemical/slow gate to be distinguished from the fast Vj-sensitive gate (6). Uncouplers activate a chemical gate that behaves identically to the slow Vj gate in terms of kinetics and efficiency. Thus, although chemical gate and slow Vj gate are usually referred to as separate gates, they may have closely related or identical mechanisms.
Ca2+ and H+ are pivotal to the operation of gap junctions because their concentrations within the cell dictate whether the junction is opened or closed (reviewed in Ref. 7). Increases in intracellular free Ca2+ concentration ([Ca2+]i) in the range of 200 to 1000 nm induce the opening of hemichannels (8). In contrast, [Ca2+]i in the high nanomolar to low micromolar range inhibits gap junctional communication (reviewed in Ref. 5), indicating that gap junction channels and hemichannels are both sensitive to [Ca2+]i but may be affected by Ca2+ in opposite ways.
The molecular mechanisms of gap junction gating are still poorly defined. Ca2+ may act via calmodulin (CaM) inducing an interaction between Ca2+-bound CaM and one or more intracellular domains of connexins (5, 9–12). CaM is a ubiquitous Ca2+-binding protein of 16.8 kDa (148 amino acids) with a sequence that is well conserved from plants to mammals. CaM has a high affinity for binding Ca2+, and in its Ca2+ bound form, CaM binds to a large number of target proteins regulating their function (13). The Ca2+-binding sites of CaM comprise four helix-loop-helix EF hands situated in two globular lobes; EF hands 1 and 2 are located in the N-terminal lobe (N-lobe), and EF hands 3 and 4 are found in the highly homologous C-terminal lobe (C-lobe). The N- and C-lobes are separated by a 30-amino acid linker, which, although appearing α-helical in crystals, is flexible in solution (14). The C-terminal lobe of Ca2+-free (apo) CaM has an about 5-fold higher affinity for binding Ca2+ because of the semi-open conformation of its two EF hands, differing significantly from the fully closed conformation of EF hands 1 and 2 in the N-lobe (15). This semi-open conformation adopted by the C-terminal lobe accounts for the ability of CaM to bind peptides in the absence of Ca2+ (15). Many target peptides, however, bind to CaM in a Ca2+-dependent manner, associating with the more open hydrophobic pockets exposed in the Ca2+-bound state (15). The bound peptide, once subjected to the hydrophobic pockets, tends to form an α-helical structure, often flanked by a Trp residue near the N terminus and a Phe, or other bulky hydrophobic residues, at the C terminus (16). CaM undergoes a number of structural changes upon binding of Ca2+. We have shown by Förster resonance energy transfer that apo-CaM exists in an extended conformation, with maximum distance between the N- and C-lobes, whereas in Ca2+·CaM·peptide complexes, the conformation of CaM is more compact as indicated by the significantly reduced distance between its N- and C-lobes (17). In the Ca2+-bound state (holo-CaM), CaM exists in a dynamic equilibrium of two major conformations, extended and semi-compact (17). This allows it enough flexibility to bind targets in variably extended conformations (17) and to form interdomain connections (18).
The possibility that CaM is involved in gap junction regulation was first proposed by Peracchia and co-workers (9, 10), suggesting a CaM-binding site at the C-terminal tail of connexin32 (Cx32, Gjb1). Previously, we have identified, by equilibrium binding studies, two CaM-binding domains in Cx32, a novel site in the N-terminal tail, and a site in the previously proposed C-terminal tail region (12). The peptide, consisting of residues 1–21, which represents the N-terminal tail CaM-binding domain of Cx32, bound to the fluorescent CaM derivative, TA-CaM with a high affinity (Kd, 27 nm) and in a Ca2+-dependent manner (12). The peptide representing residues 216–230 of the Cx32 C-terminal tail, bound TA-CaM with a Kd of 2.1 μm, also in a Ca2+-dependent manner (12). Interestingly, both the N-terminal and the C-terminal tail CaM-binding domains of Cx32 are located close to the membrane and are contiguous with hydrophobic membrane-spanning sequences (Fig. 1). Furthermore, although the C-terminal tail CaM-binding domain in Cx32 represents only 15% of the C-terminal tail of Cx32, the removal of the other 85% had little effect on junction permeability and chemical gating (19–22).
Recently, CaM has also been shown to bind peptides matching a cytoplasmic loop domain of Cx43 (Gja1) (23) and C-terminal tail domains of Cx36 and Cx35 (24), suggesting that CaM plays a role in the function of several connexins. Peracchia and coworkers (19, 20), using Cx chimera Cx32/38IL, have shown that a domain in the second half of the cytoplasmic loop may be involved in the chemical gating mechanism of Cx32. Because this region in Cx32 does not contain a CaM-binding motif (12), in contrast with Cx43 (23), the mechanism of this involvement is yet to be determined.
Here, we explore the binding of CaM to Cx32 domains to better understand how CaM participates in the chemical gating mechanism of gap junction channels. To assess the binding potential of all four Ca2+ sites of CaM two Ca2+ binding-deficient CaM mutants were used: CaM12, in which a single-point mutation was made in each of the two N-lobe EF hands, and CaM34, in which single-point mutation was applied to each of the two C-lobe EF hands of CaM. CaM mutants were combined with Cx32-derived peptides and using stopped flow kinetics; the Ca2+ dissociation rate constants (koff) were measured. In addition, a resonance energy transfer CaM derivative was used to explore CaM conformation in the Cx32 peptide complexes and helped establish a more exact definition of the two CaM-binding regions of Cx32.
MATERIALS AND METHODS
Vectors—Human liver wild type CaM was subcloned between restriction sites NdeI for the 5′ and PstI for the 3′ end in the Escherichia coli expression vector pAED4. CaM and the T34C/T110C double mutant CaM were generated as previously described (17). CaM12 (D22A/D58A CaM) and CaM34 (D95A/D131A CaM) cDNA-s, kindly provided by Dr. J. P. Adelman (Vollum Institute, Portland, OR), were subcloned in the BamHI (5′) and EcoRI (3′) restriction sites in the E. coli expression vector pET-21b by Dr. Nael Nadif Kasri (Katholic University of Leuven, Leuven, Belgium). DNA sequencing confirmed that the mutations were in the desired positions.
Protein Expression and Purification—Wild type and mutant CaM-s were expressed and purified by previously described procedures (17). Final purification to homogeneity was performed by HPLC and the purity, and identity of the proteins is confirmed by mass spectrometry as in Ref. 17. The concentrations of CaM, its mutants, and fluorescent derivatives were determined as previously described (17).
CD Spectroscopy—CD spectra were collected using a Chirascan spectropolarimeter in the wavelength ranges of 185–260 and 230–400 nm. Protein samples were desalted, freeze-dried, and dissolved in phosphate-buffered saline and 10 mm EGTA. The instrument was flushed continuously with pure N2 gas throughout the experiment to improve performance below 200 nm. The path length was 0.5 mm for far UV and 1 mm for near UV measurements. All of the spectra were acquired at room temperature and were buffer base line-subtracted. The far UV CD spectra were corrected for concentration and path length and expressed in terms of molar differential extinction coefficient, Δε (m–1 cm–1). Secondary structure estimation was calculated using the Principle Component Analysis method (33).
DA-CaM—The synthesis, characterization, and properties of DA-CaM are described in Ref. 17. In short, cysteine residues in the double mutant T34C/T110C-CaM were labeled with the fluorophore, IAEDANS and DDP, a quencher compound. The two probes IAEDANS and DDP form a Förster resonance energy transfer pair with Ro of 2.6 nm (17); increased quenching of the donor AEDANS fluorescence by the acceptor DDP indicates close proximity of the probes and hence of the N- and C-CaM lobes. Following random labeling of the two sites, T34C and T110C, the labeled mixture was fractionated by HPLC to separate the CaM fractions labeled with different fluorophores at each lobe, termed DA-CaM (17). DA-CaM fractions were identified by maximum donor quenching displayed upon binding target peptides, e.g. CaM-binding domain peptides of αCaMKII (17) or myosin light chain kinase (25). These peptides cause Ca2+/CaM, which exists in an equilibrium of extended and semi-compact conformations to bind in a compact conformation.
Peptides—Cx32 (Gjb1) peptides were synthesized at the University of Rochester (Rochester, NY) and were purified to homogeneity by HPLC using previously described procedures (17). The concentrations of the two Cx32 N-terminal tail peptides representing residues 1–19 (MNWTGLYTLLSGVNRHSTA, mass 2121.4 Da) and 1–22 (MNWTGLYTLLSGVNRHSTAIGR, mass 2447.8 Da) were determined spectroscopically using a molar extinction coefficient εo of 7100 m–1 cm–1. The concentrations of the two Cx32 C-terminal tail peptides representing residues 208–226 and 208–227 (EVVYLIIRACARRAQRRSN and Ac-EVVYLIIRACARRAQRRSNP-NH2, masses 2274.7 and 2413.9 Da, respectively) were determined using a molar extinction coefficient εo of 1400 m–1 cm–1.
Fluorescence excitation was set to 320 nm with 1-nm slit width and fluorescence emission from quin 2 was collected using a 530-nm cutoff filter. The assay solution contained 50 mm K+-PIPES, pH 7.0, 100 mm KCl, 2 mm MgCl2. 90 μm quin 2 in assay solution with no added Ca2+ was mixed with 3 μm CaM or CaM·peptide complexes in 50 μm Ca2+-containing buffer solution (mixing chamber concentrations). Care was taken that all of the protein components were free of EGTA.
Conformation Studies and Equilibrium Binding Measurements of Cx32 Peptides with DA-CaM—Equilibrium fluorescence titrations of DA-CaM and Cx32 peptide binding were carried out using an ISS-SLM spectrofluorimeter as previously described (17) to assess the degree of compactness of CaM in Cx32 peptide complexes and to measure the dissociation constant (Kd) for CaM binding by Cx32 peptides.
Software—Stopped flow kinetic data were fitted using the KinetAsyst software program (Hi-Tech Scientific). Equilibrium binding fluorescence data were analyzed using GraFit software program, version 4.0. CaM binding propensity prediction was obtained using software provided by the Department of Medical Biophysics, University of Toronto.
Statistical Analysis—For each data set the stopped flow kinetic experiments produced five to nine records; these records were averaged and can be seen in Figs. 2, 3, 4; for the averaged records an “S.D. fit” was determined that indicates the standard deviation of the data from the fit. From independent averages a mean was produced for each type of experiment; the number of independent averages included in the mean is displayed in the format n = number of experiments and can be found in Tables 1, 2, 3. The S.D. associated with all data under “Results” and in the tables is a measurement of the standard deviation of the mean from all the averages and is denoted by either S.D. or the ± symbol. Typically, data are presented in the format: koff value ± S.D. of mean (n = the number of independent experiments).
FIGURE 2.
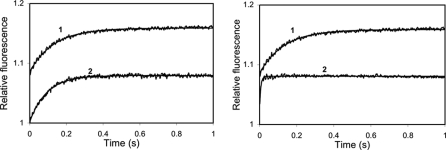
Ca2+ dissociation kinetics of wild type and Ca2+ binding-deficient mutant CaMs. 3 μm CaM or mutant CaM in the presence of 50 μm CaCl2 was rapidly mixed with 90 μm quin-2 in the same solution (see “Materials and Methods”) without added Ca2+ (concentrations in mixing chamber are given) at 21 °C. A, record 1, CaM, koff 10.14 ± 0.09 (S.D. of fit) s–1, ΔRF 0.076; record 2, CaM12, 10.74 ± 0.13 s–1, ΔRF 0.076. B, record 1, CaM, 10.14 ± 0.09 s–1, ΔRF 0.076; record 2, CaM34, koff 195.54 ± 7.10 s–1, ΔRF 0.045.
FIGURE 3.
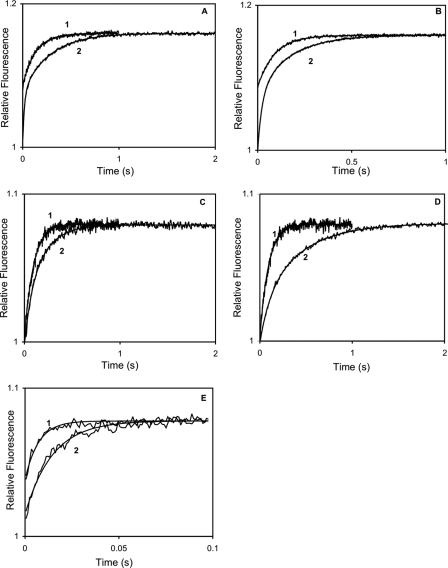
Ca2+ dissociation kinetics of wild type and Ca2+ binding-deficient mutant CaM complexes with Cx32 N-terminal peptides. 3 μm CaM or mutant CaM and 10 μm peptide (unless otherwise specified) in the presence of 50 μm CaCl2 was rapidly mixed with 90 μm quin-2 in the same solution (see “Materials and Methods”) without added Ca2+ (concentrations in mixing chamber are given) at 21 °C. A, record 1, CaM, koff 10.14 ± 0.09 (S.D. of fit) s–1, ΔRF 0.076; record 2, CaM with Cx32 1–19, koff1 56.04 ± 1.80 s–1, ΔRF1 0.063, koff2 3.61 ± 0.03 s–1, ΔRF2 0.077. B, record 1, CaM, koff 10.14 ± 0.09 s–1 ΔRF 0.076; record 2, CaM with Cx32 1–22, koff1 12.3 ± 1.06 s–1, ΔRF1 0.082, koff2 1.70 ± 0.03 s–1, ΔRF2 0.078. C, record 1, CaM12, koff 10.74 ± 0.13 s–1, ΔRF 0.076; record 2, CaM12 with Cx32 1–19, koff1 20.57 ± 3.30 s–1, ΔRF1 0.024, koff2 5.45 ± 0.19 s–1, ΔRF2 0.056. D, record 1, CaM12, koff 10.74 ± 0.13 s–1, ΔRF 0.076; record 2, CaM12 and Cx32 1–22, koff1 9.18 ± 0.91 s–1, ΔRF1 0.024, koff2 5.35 ± 0.04 s–1, ΔRF2 0.046. E, record 1, CaM34, koff 195.54 ± 7.10 s–1, ΔRF 0.045; record 2, CaM34 and Cx32 1–22, koff1 13.16 ± 1.63 s–1, ΔRF1 0.009, koff2 97.18 ± 5.00 s–1, ΔRF2 0.041.
FIGURE 4.
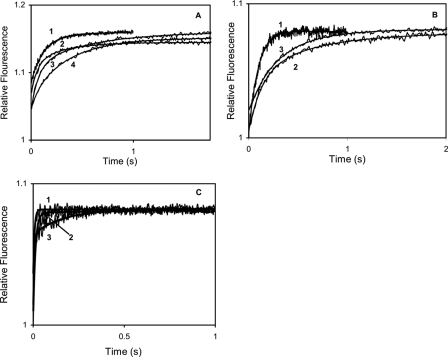
Ca2+ dissociation kinetics of wild type and Ca2+ binding-deficient mutant CaM complexes with Cx32 C-terminal peptides. 3 μm CaM or mutant CaM and 10 μm peptide (unless otherwise specified) in the presence of 50 μm CaCl2 was rapidly mixed with 90 μm quin-2 in the same solution (see “Materials and Methods”) without added Ca2+ (concentrations in mixing chamber are given) at 21 °C. A, record 1, CaM; record 2, CaM with Cx32 208–226, koff 6.10 ± 0.12 s–1, ΔRF 0.070; record 3, CaM with Cx32 208–226 (20 μm), koff1 11.45 ± 0.42 s–1, ΔRF1 0.069, koff2 1.66 ± 0.05 s–1, ΔRF2 0.041; record 4, CaM with Cx32 208–227, koff1 13.78 ± 1.90 s–1, ΔRF1 0.020, koff2 2.70 ± 0.04 s–1, ΔRF2 0.080. B, record 1, CaM12; record 2, CaM12 with Cx32 208–226, koff1 7.64 ± 0.12 s–1, ΔRF1 0.040, koff2 1.69 ± 0.53 s–1, ΔRF2 0.030; record 3, CaM12 with Cx32 208–227, koff 3.1 ± 0.08 s–1, ΔRF 0.060. C, record 1, CaM34, koff 195.54 ± 7.10 s–1 ΔRF 0.045; record 2, CaM34 with Cx32 208–226, koff1 19.85 ± 2.50 s–1, ΔRF1 0.060, koff2 177.44 ± 19.00 s–1, ΔRF2 0.020; record 3, CaM34 with Cx32 208–227, koff1 5.85 ± 0.31 s–1, ΔRF1 0.014, koff2 165.05 ± 11.00 s–1, ΔRF2 0.046.
TABLE 1.
Ca2+ dissociation kinetics of CaM and mutant CaMs
The mean dissociation rate constants (koff) and the standard deviation are shown for each EF hand in wild type CaM, CaM12, and CaM34, respectively; n refers to the number of independent experiments from which the data was obtained. The relative amplitude (A) is also shown for each EF hand; this represents the involvement of the particular hand in the binding of Ca2+, with an A of 1 being equal to 100% binding capability. The CaM N-lobe EF hands are represented in the left four columns, and the C-lobe EF hands are represented in the right four columns. A rate constant of 1000 s–1 represents a rate that was too fast to measure.
|
N-terminal CaM lobe
|
C-terminal CaM lobe
|
n
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
EF1a
|
EF2a
|
EF3a
|
EF4a
|
||||||
| k1 | A1 | k2 | A2 | k3 | A3 | k4 | A4 | ||
| s-1 | s-1 | s-1 | s-1 | ||||||
| CaM | 1000 | 1 | 1000 | 1 | 10.9 ± 1.2 | 1 | 10.9 ± 1.2 | 1 | 4 |
| CaM12 | 11.2 ± 0.7 | 1 | 11.2 ± 0.7 | 1 | 4 | ||||
| CaM34 | 1000 | 1 | 190 ± 15 | 1 | 3 | ||||
Note that the denotations given to the EF hands are for reference purposes only and do not correlate to the actual hand involved in the reaction.
TABLE 2.
Ca2+ dissociation kinetics of CaM and mutant CaM complexes with Cx32 N-terminal tail peptides
The rate constants of Ca2+ dissociation of CaM and CaM mutant complexes with Cx32 N-terminal tail peptides Cx32 1–19 and Cx32 1–22 (see “Materials and Methods” for sequence).
|
N-terminal CaM lobe
|
C-terminal CaM lobe
|
n
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
EF1a
|
EF2a
|
EF3a
|
EF4a
|
||||||
| k1 | A1 | k2 | A2 | k3 | A3 | k4 | A4 | ||
| s-1 | s-1 | s-1 | s-1 | ||||||
| CaM + 1-19 | 56.9 ± 2.3 | 1.5 | 1000 | 0.5 | 5.1 ± 1.3 | 1 | 5.1 ± 1.3 | 1 | 3 |
| CaM12 + 1-22 | 10.8 ± 1.3 | 1 | 10.8 ± 1.3 | 1 | 1.7 ± 0.5 | 1 | 1.7 ± 0.5 | 1 | 3 |
| CaM12 + 1-19 | 18.6 ± 2.9 | 0.5 | 4.9 ± 0.5 | 1.5 | 3 | ||||
| CaM12 + 1-22 | 10.5 ± 1.3 | 0.5 | 3.4 ± 1.7 | 1.5 | 3 | ||||
| CaM34 + 1-22 | 13.2 ± 0.3 | 0.5 | 97.2 ± 0.5 | 1.5 | 2 | ||||
Note that the denotations given to the EF hands are for reference purposes only and do not correlate to the actual hand involved in the reaction.
TABLE 3.
Ca2+ dissociation kinetics of CaM and mutant CaM complexes with Cx32 C-terminal tail peptides
The rate constants of Ca2+ dissociation of CaM and CaM mutant complexes with Cx32 C-terminal tail peptides Cx32 208–226 and 208–227 (see “Materials and Methods” for sequence).
|
N-terminal CaM lobe
|
C-terminal CaM lobe
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
EF1a
|
EF2a
|
EF3a
|
EF4a
|
||||||
| k1 | A1 | k2 | A2 | k3 | A3 | k4 | A4 | n | |
| s-1 | s-1 | s-1 | s-1 | ||||||
| CaM + 208-226 | 1000 | 1 | 1000 | 1 | 6.1 ± 0.9 | 1 | 6.1 ± 0.9 | 1 | 3 |
| CaM12 + 208-226 | 7.6 ± 0.1 | 1 | 1.6 ± 0.1 | 1 | 1 | ||||
| CaM34 + 208-226 | 17.1 ± 0.9 | 1 | 173 ± 0.4 | 1 | 2 | ||||
| CaM + 208-227 | 13.5 ± 0.4 | 0.5 | 1000 | 1.5 | 2.6 ± 0.1 | 1 | 2.6 ± 0.1 | 1 | 2 |
| CaM12 + 208-227 | 3.1 ± 0.1 | 0.5 | 3.1 ± 0.1 | 1 | 1 | ||||
| CaM34 + 208-227 | 6.3 ± 0.7 | 0.5 | 169 ± 5.6 | 1 | 2 | ||||
Note that the denotations given to the EF hands are for reference purposes only and do not correlate to the actual hand involved in the reaction.
RESULTS
Measurement and Interpretation of Ca2+ Dissociation Kinetics of CaM and Its Target Complexes
The interaction of CaM with Cx32 N-terminal and C-terminal tail peptides is Ca2+-dependent (12); thus it was expected that Ca2+ dissociation rate constants of CaM Ca2+-binding sites were affected by peptide target binding. The fluorescence intensity of the fluorescent Ca2+ chelator compound quin 2 increases upon Ca2+ binding, and the rate of the quin 2 fluorescence intensity increase is limited by the dissociation of Ca2+ ions from CaM or its peptide complexes. Quin 2 is used in a large excess rendering Ca2+ rebinding to CaM insignificant and thus allowing the observed rates to be interpreted as the rate constants of dissociation. The amplitude of the quin 2 fluorescence increase, which corresponds to the increase in [Ca2+.quin 2], as seen in Figs. 2, 3, 4, is converted to relative fluorescence (RF). The time courses of the change in RF (ΔRF) are fitted to exponentials to give the rate constants of Ca2+ dissociation. ΔRF provides a measure of the binding sites involved in each reaction, in our conditions, a ΔRF of 0.04 corresponded to one Ca2+-binding site. This value was obtained using the data from the experiments with CaM, which showed only two measurable binding sites with a ΔRF of 0.08; these binding sites correspond to the two sites on the C-terminal lobe. Ca2+ dissociation from the two sites in the N-terminal lobe are too fast to measure by stopped flow kinetics and are estimated to be ∼1000 s–1 (27).
Secondary Structure Analysis of CaM Mutants
CaM and mutant CaMs were characterized by CD spectroscopy to assess the effect of the single-point mutations on the structural integrity of the protein. Two mutant preparations were measured; the means and S.D. of the measurements were as follows: in the apo form, in the presence of 10 mm EGTA, the α-helix content of wild type CaM was 46.4 ± 0.1%. In CaM12, this was reduced to 37.3 ± 0.1%, and the β-sheet content decreased from 17.9 ± 0.1% for wild type to 16.5 ± 0.1% in CaM12. For CaM34, the α-helix content was reduced to 36.5 ± 0.1%, whereas the β-sheet content was increased to 29.6 ± 0.1%. These data show that the single point mutations that disable Ca2+ binding in the EF hands of one CaM lobe resulted in some structural differences in the mutated lobe. The Ca2+ dissociation kinetic experiments presented below were carried out to see whether the functionality of the unmutated lobe was affected.
Comparison of the Ca2+ Dissociation Kinetics of Wild Type and Mutant CaMs
Before using the CaM mutants to determine lobe-specific interactions with Cx32 peptides, the Ca2+ dissociation kinetics of CaM12 and CaM34 were first characterized to see whether the mutations of the EF hands of one lobe affected the functionality of the EF hands of the unmutated lobe. Fig. 2 shows the ΔRF of quin 2 on Ca2+ dissociation from CaM (Fig. 2, record 1) in comparison with that of our two mutants, CaM12 (Fig. 2A, record 2) and CaM34 (Fig. 2B, record 2), respectively. The average dissociation rate constant (koff) for CaM12 of 11.2 ± S.D. 0.7 s–1 (n = 4) was similar to that of CaM at 10.9 ± 1.1 s–1 (n = 4). In contrast, although dissociation from the N-terminal lobe Ca2+-binding sites of CaM was too fast to measure, a koff of 190.4 ± 15 s–1 (n = 3) was measured for one of the N-terminal lobe EF hands of CaM34, whereas dissociation remained too fast to measure from the other. These data support the understanding that the C-terminal EF hands (binding sites 3 and 4) have a higher affinity for binding Ca2+ than the EF hands of the N-terminal (27). As summarized in Table 1, when the average of all data obtained for each of the CaM mutants is compared against the control CaM data, it is apparent that CaM12 and CaM34 EF hands have largely preserved the functional integrity of CaM.
Kinetics of Ca2+ Dissociation of CaM Complexes with Cx32-derived Peptides
N-terminal Cx32 Peptide—To assess the mechanisms of CaM binding to Cx32-derived peptides, we examined the Ca2+ dissociation rate constant (koff) values of wild type CaM (CaM) and CaM mutants (CaM12 and CaM34) and their complexes with Cx32-derived peptides by stopped flow kinetics. Previously, we have shown that Cx32 1–21 peptide binds CaM with high affinity (12). Here, two homologous peptides, representing the N-terminal tail CaM-binding domain, were examined to determine the mechanism of their interaction with CaM in a lobe-specific manner. Two sequences, corresponding to residues 1–19 and 1–22, were studied to further probe the boundaries of the Cx32 N-terminal tail CaM-binding domain. The kinetic parameters for CaM complexes with Cx32NT peptides 1–19 and Cx32 1–22 are shown in Fig. 3 (A and B, respectively), and in Table 2.
The amplitude of ΔRF on Ca2+ dissociation from the CaM·Cx32 1–19 peptide complex (Fig. 3A, record 2) was 1.5-fold greater than that in the absence of the peptide (Fig. 3A, record 1), indicating that the binding of Cx32 1–19 engaged three Ca2+-binding sites, a slower rate constant of 5.1 ± 1.3 s–1 (n = 3) representing two sites presumed to correspond to the C-lobe EF hands (k3,4), and a faster rate constant of 56.9 ± 2.3 s–1 (n = 3), thought to correspond to the arbitrarily assigned EF1 of the N-lobe sites (k1). Both represented a marked rate constant reduction compared with those of CaM in the absence of peptide, consistent with Ca2+-dependent peptide binding. A further increase in the ΔRF value from 0.12 to 0.16 was seen for the dissociation of Ca2+ from CaM·Cx32 1–22 complex (Fig. 3B, record 2) compared with that from the CaM·Cx32 1–19 complex, showing that all four Ca2+-binding sites of CaM were involved in the CaM·Cx32 1–22 peptide complex; the two N-lobe EF hands were involved in the binding of the 1–22 peptide with a rate constant of 10.8 ± 2.5 s–1 (n = 3) and the CaM C-lobe with a lower rate constant of 1.7 ± 0.5 s–1 (n = 3) (Table 2). Thus, all four CaM EF hands bound Ca2+ with a higher affinity in the 1–22 complex than when associated with the Cx32 1–19 peptide.
When studying the interactions specific to the CaM C-lobe with the CaM12 mutant, Ca2+ dissociation from the CaM12·Cx32 1–19 peptide complex (Fig. 3C, record 2) was biphasic, with one EF hand showing a slow rate of dissociation at 4.9 ± 0.5 s–1 (n = 3) and the other a faster rate at 18.6 ± 2.9 s–1 (n = 3). Compared with the koff values for CaM12 without peptide (Fig. 3C, record 1), 11.2 ± 0.7 s–1, the koff values of CaM12 with the peptide have thus doubled and halved for each EF hand, respectively (Tables 1 and 2), Similarly, when CaM12 was in complex with Cx32 1–22 (Fig. 3D, record 2), a biphasic Ca2+ dissociation occurred, with a rate constant of 3.4 ± 1.7 s–1 (n = 2) from EF4 and a value of 10.5 ± 1.3 s–1 (n = 2) from EF3. Thus, although there was evidence of Ca2+-dependent peptide binding to the CaM C-lobe only, cooperativity between the two C-lobe Ca2+-binding sites, seen in CaM, was reduced or lost in the interaction of the Cx32 1–19 or the 1–22 peptide with the CaM C-lobe in the absence of N-lobe Ca2+ binding.
In the interaction of the CaM N-lobe with Cx32 1–22, studied by CaM34, both binding sites became involved: EF1 exhibited a koff value of 13.2 ± 0.2 s–1 (n = 2) (Fig. 3E, record 2), similar to that for EF1 in CaM·Cx32 1–22 complex, whereas the rate constant for EF2 decreased from 190 ± 15 s–1 to 97.2 ± 0.5 s–1 (n = 2). These data showed that both peptides could bind to the N-lobe of CaM in the absence of C-lobe Ca2+ binding but substantially more weakly than to CaM. The ∼0.5 binding site fitted to the data may indicate partial engagement of one of the EF hands in the peptide binding.
C-terminal Cx32 Peptides—Previously, we have shown that the Cx32 C-terminal tail 216–230 peptide binds CaM in a Ca2+-dependent manner but more weakly than the N-terminal tail peptide (12). Here, we investigated whether the Cx32 C-terminal tail CaM-binding domain extends further at the N-terminal end by including residues 208–215. Two peptides were studied: 208–226 and the terminally blocked Ac-208–227-NH2. The rate constant of Ca2+ dissociation from the CaM·Cx32 208–226 complex (Fig. 4A, record 2) was 6.0 ± 0.9 s–1 (n = 3), reduced from that of unbound CaM, indicating peptide binding to the C-lobe of CaM; the two Ca2+ sites of the N-lobe, however, remained too fast to measure by stopped flow kinetics, indicating that they were not involved in peptide binding. Increasing the peptide concentration to 20 μm (Fig. 4A, record 3) recruited more Ca2+-binding sites compared with that of 208–226 at 10 μm as shown by the greater ΔRF. An interpretation is that the increase in peptide concentration could have resulted in multiple peptides binding to CaM, as previously suggested (25).
The dissociation of Ca2+ from the CaM Cx32 208–227 complex occurred with a rate constant of 13.4 ± 0.4 s–1 (n = 2) (Fig. 4A, record 4) for one of the N-lobe EF hands; this, however, was only representing involvement of 50% of the EF hand binding. Dissociation from the other N-lobe EF hand remained too fast to measure, indicating that the N-lobe, much like in the case of the Cx32 208–226 peptide, had very little interaction with the peptide. This was corroborated by data on the CaM34 complex with Cx32 208–227; dissociation from one of the EF hands, arbitrarily assigned EF1 (Table 1), was slowed down to 6.3 ± 0.7 s–1 (n = 2) (Fig. 4C, record 3) and again only seemed to commit half of its binding potential; the dissociation rate from the second site assigned EF2 was 173.1 ± 0.4 s–1 (n = 2), little affected by the peptide.
Ca2+ dissociation rates for the C-lobe EF hands were reduced substantially to 2.6 ± 0.1 s–1 (n = 2) (Fig. 4A, record 4), indicating a strong affinity of the CaM C-lobe for the Cx32 208–227 peptide. The rate constant of 3.1 ± 0.1 s–1 (n = 1) for both C-lobe EF hands of CaM12 (Fig. 4B, record 3), similar to those for CaM with Cx32 208–227, was consistent with high affinity binding between the CaM C-lobe and the Cx32 C-terminal tail CaM-binding domain. These data showed that CaM binding the Cx32 C-terminal tail CaM-binding domain involves one CaM lobe at a time and demonstrated a marked preference for the CaM C-lobe over the N-lobe.
CaM Conformation in the Peptide Complexes
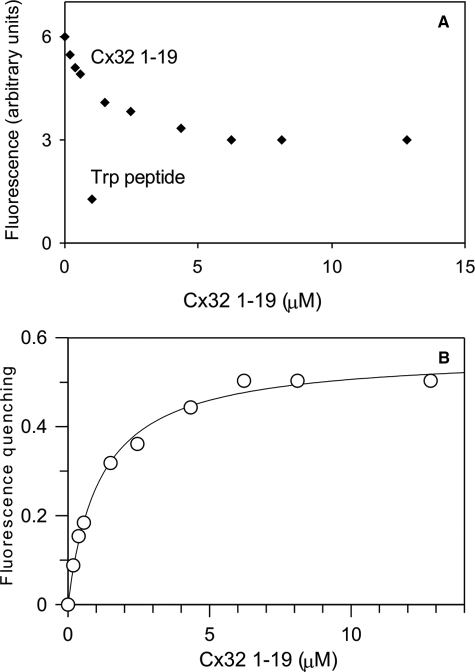
N-terminal Cx32 peptide—The Förster resonance energy transfer probe DA-CaM (Ref. 17 and see “Materials and Methods”) was used to explore the conformation of CaM in the Cx32 peptide complexes as explained under “Materials and Methods.” The smooth muscle myosin light chain kinase-derived Trp peptide (25) with a known compact structure in complex with CaM (16) induced a 79% quenching of DA-CaM fluorescence (17) (Fig. 5A). The degree of DA-CaM fluorescence donor quenching upon increasing concentrations of the Cx32 1–19 peptide is shown in Fig. 5. Maximal donor quenching was 56%, indicating that CaM conformation remained partially extended in complex with the Cx32 1–19 peptide. A weaker complex was also indicated by the dissociation constant (Kd) of DA-CaM for the Cx32 1–19 peptide, which was 1.14 ± 0.10 μm (n = 1), higher than the value of 27 nm, previously measured for the related Cx32 1–21 peptide using a Lys75-modified fluorescent CaM, TA-CaM (12, 25). Thus, residues 20–21 form an essential part of the Cx32 N-terminal tail CaM-binding domain.
FIGURE 5.
Equilibrium binding of Cx32 N-terminal peptide with DA-CaM. A, 433 nm DA-CaM in assay solution (see “Materials and Methods”) containing 0.5 mm CaCl2 is titrated at 21 °C with Cx32 1–19 peptide. B,a Kd of 1.14 ± 0.10 μm was obtained for DA-CaM.
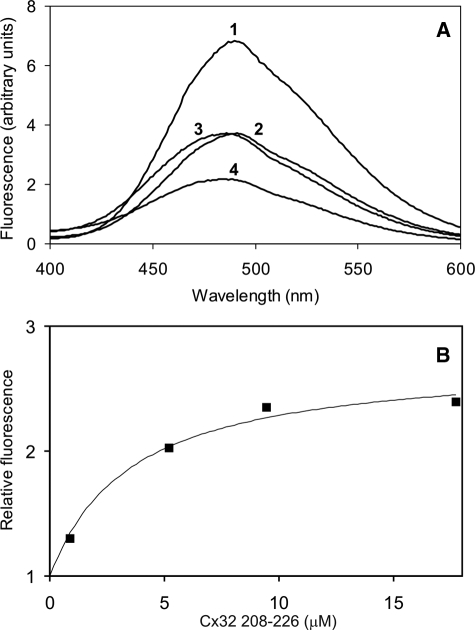
C-terminal Cx32 Peptides—CaM conformation was assessed by examining the maximum donor quenching of DA-CaM induced by Cx32 208–226 peptide binding. As shown in Fig. 6A, the binding of the Cx32 208–226 peptide to DA-CaM was complex. Donor quenching was maximal at 50% (Fig. 6A, record 2). This was consistent with the binding of peptide to one CaM lobe only as shown above by Ca2+ dissociation kinetic experiments. On increasing the peptide concentration, however, a blue shift was seen in the donor AEDANS fluorescence (Fig. 6A, record 3). This was likely to indicate binding of a second peptide molecule to the second CaM lobe. Interestingly, as seen in Fig. 6A (record 4), Trp peptide, even at a large excess, did not fully compete with Cx32 208–226 for CaM. In the light of the high affinity of Trp peptide for CaM (6 pm) (25) in comparison with the relatively low affinity of the Cx32 208–226 peptide, this indicates an unorthodox binding mode between the Cx32 C-terminal tail region and CaM.
FIGURE 6.
Equilibrium binding of Cx32 C-terminal peptide with DA-CaM. A, aliquots of Cx32 208–226 peptide are added to 433 nm DA-CaM in assay solution (see “Materials and Methods”) containing 0.5 mm CaCl2. Record 1, 433 nm DA-CaM; record 2, addition of 0.9 μm Cx32 208–226 to DA-CaM; record 3, further addition of 4.4 μm Cx32 208–226; record 4, addition of 6 μm Trp peptide. B, 150 nm AEDANS-T34C/T110C-CaM in assay solution (see “Materials and Methods”) containing 0.5 mm CaCl2 was titrated at 21 °C with Cx32 208–226 peptide. A Kd of 3.45 ± 1.09 μm was obtained for DA-CaM.
The binding affinity of the peptide to CaM was measured taking advantage of the sensitivity of donor fluorescence in DA-CaM to Cx32 208–226 binding. The donor-only labeled probe, AEDANS-T34C/T110C-CaM showed a 2.5-fold increase in fluorescence on 208–226 peptide binding and gave a Kd of 3.45 ± 1.09 μm (n = 1) (Fig. 6B), a value consistent with previously measured 2.1 μm for TA-CaM for a related peptide representing residues 216–230 (12). These data indicated that the inclusion of residues 208–215 did not increase the CaM binding affinity of the Cx32 C-terminal tail CaM-binding domain.
DISCUSSION
CaM association with two CaM-binding domains of Cx32 was characterized by fluorescence stopped flow and equilibrium measurements and by the use of Ca2+ binding-deficient CaM mutants with the aim to gain an insight into the binding of CaM to gap junctions in vivo.
First, the viability of the CaM mutants was tested. Far UV CD spectroscopy revealed changes in the helical, β-sheet, and loop contents of the CaM12 and CaM34 mutants compared with CaM. The CD results indicated that the EF hand mutations resulted in some changes in the secondary structures of the mutated lobe of CaM12 and CaM34. The functional integrity of CaM12 was, however, shown by the essentially unchanged Ca2+ dissociation rate constants of the CaM12 compared with those of the C-lobe of CaM (Table 1); this also indicated that C-lobe Ca2+ binding is independent of Ca2+ binding to the N-lobe and that the N-lobe mutations had no significant effect on Ca2+ binding by the C-lobe.
In contrast, whereas mutation of the EF3 and EF4 hands in CaM34 did not affect the EF1 site (Table 1), a significant (∼6-fold) rate reduction was seen for EF2 when compared with that in CaM. Higher affinity Ca2+ binding by the N-lobe in the absence of the C-lobe is likely to have unmasked the existence of negative cooperativity in Ca2+ binding exerted on the N-lobe by the C-lobe. Previous work showing that Ca2+ binding by the EF3 and EF4 sites destabilizes Ca2+ binding to EF2 via the 76–80 linker region (28) is consistent with this interpretation.
Subtle differences were seen in the functioning of the EF hands of CaM12 in peptide complexes when compared with wild type CaM. Positive cooperativity was lost between EF3 and EF4, as shown by the heterogeneity of the Ca2+ dissociation rate constants of the N-terminal peptides and C-terminal 208–226 bound to CaM12 (Table 2). Cooperativity of Ca2+ binding between EF3 and EF4 was, however, observed again when CaM12 was bound to the Cx32 C-terminal tail 208–227 peptide. These data demonstrate a high level of adaptability in CaM target binding (13).
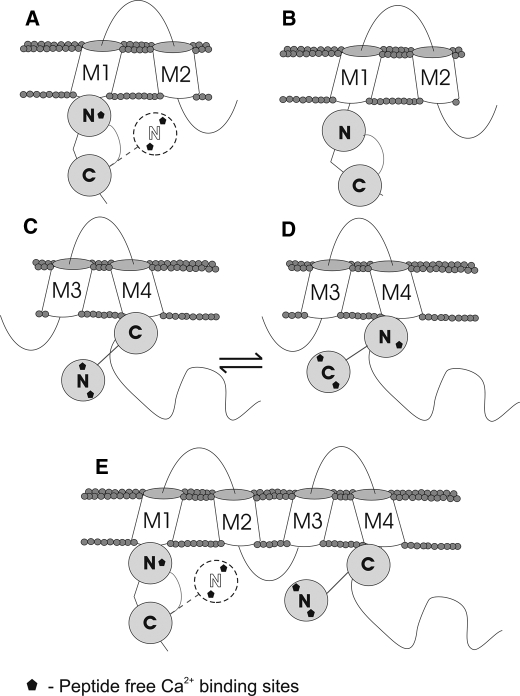
Our data suggest some possible binding modes of CaM to Cx32 gap junctions, which are illustrated in Fig. 7. Fig. 7 (A and B) depicts possible arrangements between the N-terminal tail of Cx32 and CaM. If the CaM-binding domain corresponded to residues 1–19 (Fig. 7A), that would cause a weak, dynamic interaction of the CaM N-lobe with the Cx32 N-terminal tail. In contrast, if the 1–22 region of the Cx32 N-terminal tail were accessible for CaM binding, CaM with all four Ca2+-binding sites engaged in the interaction, would have a firm grip on the Cx32 N-terminal CaM-binding domain (Fig. 7B). Previous data suggest that residues 1–21 would have a similarly high affinity interaction to that of the 1–22 peptide (12). The N-terminal tail CaM-binding domain is essential for the trafficking and assembly of functional Cx32 gap junctions (29), it remains to be determined whether and how CaM binding to this region may play a role in the regulation of pore permeability.
FIGURE 7.
Schematic representation of the binding of CaM to each of the four individual Cx32-derived peptides examined in this study. For dissociation kinetic results, refer to Tables 1, 2, 3. A, the binding of CaM to Cx32 N-terminal tail 1–19 peptide. The 1–19 N-terminal tail may not adopt a fully helical conformation because the lack of residues 20–21 results in a weak, dynamic interaction of the N-lobe, with only EF1 involved in peptide binding (Table 2). With a 1–19 as N-terminal CaM-binding domain, only the CaM C-lobe would be anchored. B, CaM binding to a Cx32 N-terminal 1–22 tail. A high affinity interaction with all four Ca2+-binding sites of CaM is involved in peptide binding (Table 2). C and D, CaM binding to Cx32 C-terminal tail 208–226/208–227 peptides. Binding site for only one CaM lobe is present in these peptides; the CaM C-lobe exhibits a higher affinity for the binding site (Table 3); however, in the absence of the C-lobe, the CaM N-lobe shows moderate affinity for binding via the EF1 site (Table 3), making trans-domain binding feasible. E, a representation of how CaM may bind to a whole connexin subunit. Should the Cx32 N-terminal tail be available, it is hypothesized that both lobes of CaM will bind; however, inaccessibility of residues 21 and 22 would result in the destabilization of CaM N-lobe binding and may result in its release. The N- and C-lobes of CaM compete for the C-terminal Cx32 tail-binding site, and engagement of the N-lobe would be especially favorable at high intracellular [Ca2+].
Fig. 7 (C and D) illustrates the possible binding modes of CaM to the C-terminal tail domain of Cx32. Our data showed that CaM binds to C-terminal Cx32 tail peptides with one lobe at a time. The Cx32 C-terminal tail 208–227 peptide showed a higher affinity for CaM than the 208–226 peptide. The differing results for 208–227 compared with those for 208–226 can be attributed to the acetylation of the N-terminal of the peptide or the addition of the Pro residue and amidation at the C terminus. Both of these extensions to the peptide help it mimic the real sequence in a connexin molecule. In previous work (12), Cx32 C-terminal tail 216–230 peptide was shown to bind CaM with a higher affinity than the 208–226 peptide. This indicates that residues 208–215 do not form part of the CaM-binding domain, and because the VVYLII motif is highly hydrophobic, these residues most likely form part of a transmembrane domain (M4). The Cx32 C-terminal tail CaM-binding domain therefore best corresponds to residues 216–227, and the CaM binding site is likely to be terminated by the 227–228 Pro-Pro sequence.
Fig. 7E summarizes the CaM binding modes to a connexin32 molecule, determined by our data. The CaM C-lobe had a substantially higher affinity for the Cx32 C-terminal tail peptides than the N-lobe (Table 3). An unbound CaM lobe has been suggested to act as a “cork” to gate gap junction conductivity (5). At high peptide concentrations, however, a second peptide molecule could attach to the N-lobe, resulting in one CaM molecule binding two separate Cx32 C-terminal peptides. In a gap junction, the local concentration of CaM-binding domains may be high enough for trans-domain or trans-subunit bridging to occur by the two lobes of a CaM molecule.
When considering possible mechanisms by which CaM could regulate gap junction conductance, the architecture (30) and properties of the gap junction channel need to be taken into account. Gap junctions show charge selectivity, which is not explained by a simple open pore model (31, 32) but which suggests similarities with ion channels. The open probability of the gap junction channel can decrease without selective decrease in large solute permeability (31). A dynamic interaction model of CaM with gap junctions as outlined above would be consistent with a reduced open probability but unaffected large solute permeability. The significance of CaM binding to the N- and C-terminal binding domains of Cx32 in the regulation of pore permeability and open probability, however, requires further investigation.
Several questions remain to be answered to obtain a clear and definitive a model of how CaM may function as a gate for the gap junction channel. First, the stoichiometry of CaM binding to gap junctions is important in understanding the gating mechanisms but has not been determined. Second, it is not clear whether, in the case of connexin32, the N-terminal tail is accessible for CaM binding in assembled junctions (29). Third, CaM appears to interact with different connexins at different regions and by different mechanisms, raising the question of how heteromeric gap junctions may interact with and be regulated by CaM.
When considering the proximity of CaM-binding domains in connexins to the transmembrane region, an intriguing possibility of regulation emerges: CaM may affect channel open probability without reducing large solute permeability by blocking conformational changes that would involve the rearrangement of transmembrane regions; conformational changes necessary for channel function may not take place.
It is clear that peptide studies alone are not sufficient to produce a clear understanding of how CaM controls the gating of gap junctions or hemichannels; however, the studies presented here have produced strong evidence toward CaM binding to Cx32 and show that mutants CaM12 and CaM34 can be used in target binding experiments to help assess the role that each individual lobe plays in the binding process. Further investigation is required to achieve a definitive understanding of gap junction gating mechanisms by CaM.
Acknowledgments
We are thankful to Dr. Nael Nadif Kasri (Katholic University of Leuven, Belgium) for providing us with the E. coli expression vectors for CaM12 and CaM34. Zawahir (Jack) Chowdhury is thanked for expression and purification of the mutant CaMs, and Dr. Gerald Connolly assisted with HPLC purification of the Cx32 peptides and CaMs. We thank the Pharmaceutical Optical & Chiroptical Spectroscopy Centre of King's College London for performing the CD spectroscopic conformational analysis of CaM and its mutants. We thank Dr. Alice Warley (Centre for Ultrastructural Imaging, King's College London) for reading the manuscript.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grant GM20113. This work was also supported by long term support of research in the laboratory on CaM probes and stopped flow kinetics from the Wellcome Trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Cx, connexin; IAEDANS, 5-[[2-[(iodoacetyl)amino]ethyl]amino]naphthalene-1-sulfonic acid; AEDANS, 5-[[2-[amino]ethyl]amino]naphthalene-1-sulfonic acid; HPLC, high pressure liquid chromatography; CaM, calmodulin; CaM-s, calmodulins; N-lobe, N-terminal lobe; C-lobe, C-terminal lobe; PIPES, 1,4-piperazinediethanesulfonic acid; RF, relative fluorescence.
References
- 1.Bruzzone, R., White, T. W., and Paul. D. L. (1996) Eur. J. Biochem. 238 1–27 [DOI] [PubMed] [Google Scholar]
- 2.Bennett, M. V., and Verselis, V. K. (1992) Semin. Cell Biol. 3 29–47 [DOI] [PubMed] [Google Scholar]
- 3.Harris, A. L. (2001) Q. Rev. Biophys. 34 325–472 [DOI] [PubMed] [Google Scholar]
- 4.Bukauskas, F. F., and Verselis, V. K. (2004) Biochim. Biophys. Acta 1662 42–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peracchia, C. (2004) Biochim. Biophys. Acta 1662 61–80 [DOI] [PubMed] [Google Scholar]
- 6.Bukauskas, F. F., and Peracchia, C. (1997) Biophys J. 72 2137–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spray, D. C., White, R. L., Mazet, F., and Bennett, M. V. (1985) Am. J. Physiol. 248 H753–H764 [DOI] [PubMed] [Google Scholar]
- 8.De Vuyst, E., Decrock, E., Cabooter, L., Dubyak, G. R., Naus, C. C., Evans, W. H., and Leybaert, L. (2006) EMBO J. 25 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peracchia, C., Bernardini, G., and Peracchia, L. L. (1983) Pflugers Arch. Eur. J. Physiol. 399 152–154 [DOI] [PubMed] [Google Scholar]
- 10.Peracchia, C. (1988) in Gap Junctions (Hertzberg, E. L., and Johnson, R. G., eds) pp. 267–282, Alan R. Liss, New York
- 11.Peracchia, C., Sotkis, A., Wang, X. G., Peracchia, L. L., and Persechini, A. (2000) J. Biol. Chem. 275 26220–26224 [DOI] [PubMed] [Google Scholar]
- 12.Török, K., Stauffer, K., and Evans, W. H. (1997) Biochem. J. 326 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoeflich, K. P., and Ikura, M. (2002) Cell 108 739–742 [DOI] [PubMed] [Google Scholar]
- 14.Chin, D., and Means, A. R. (2000) Trends Cell Biol. 10 322–328 [DOI] [PubMed] [Google Scholar]
- 15.Grabarek, Z. (2006) J. Mol. Biol. 359 509–525 [DOI] [PubMed] [Google Scholar]
- 16.Ikura, M., Clore, G. M., Gronenborn, A. M., Zhu, G., Klee, C. B., and Bax, A. (1992) Science 256 632–638 [DOI] [PubMed] [Google Scholar]
- 17.Török, K., Tzortzopoulos, A., Grabarek, Z., Best, S. L., and Thorogate, R. (2001) Biochemistry 40 14878–14890 [DOI] [PubMed] [Google Scholar]
- 18.Schumacher, M. A., Rivard, A. F., Bachinger, H. P., and Adelman, J. P. (2001) Nature 410 1120–1124 [DOI] [PubMed] [Google Scholar]
- 19.Wang, X., Li, L., Peracchia, L. L., and Peracchia, C. (1996) Pflugers Arch. Eur. J. Physiol. 431 844–852 [DOI] [PubMed] [Google Scholar]
- 20.Peracchia, C., Wang, X. G., and Peracchia, L. M. In: Peracchia, C. (eds) (2000) Gap Junctions: Molecular Basis of Cell Communication in Health and Disease, Academic Press, Inc., pp. 271–295, San Diego, CA
- 21.Werner, R., Levine, E., Rabadan-Diehl, C., and Dahl, G. (1991) Proc. Biol. Sci. 243 5–11 [DOI] [PubMed] [Google Scholar]
- 22.Wang, X., and Peracchia, C. (1997) Biophys. J. 271 C1743–C1749 [DOI] [PubMed] [Google Scholar]
- 23.Zhou, Y., Yang, W., Lurtz, M. M., Ye, Y., Huang, Y., Lee, H. W., Chen, Y., Louis, C. F., and Yang, J. J. (2007) J. Biol. Chem. 282 35005–35017 [DOI] [PubMed] [Google Scholar]
- 24.Burr, G. S., Mitchell, C. K., Keflemariam, Y. J., Heidelberger, R., and O'Brien, J. (2005) Biochem. Biophys. Res. Commun. 335 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Török, K., and Trentham, D. R. (1994) Biochemistry 33 12807–12820 [DOI] [PubMed] [Google Scholar]
- 26.Tzortzopoulos, A., Best, S. L., Kalamida, D., and Török, K. (2004) Biochemistry 43 6270–6280 [DOI] [PubMed] [Google Scholar]
- 27.Martin, S. R., Andersson, T. A., Bayley, P. M., Drakenberg, T., and Forsen, S. (1985) Eur. J. Biochem. 151 543–550 [DOI] [PubMed] [Google Scholar]
- 28.Sorensen, B. R., Faga, L. A., Hultman, R., and Shea, M. A. (2002) Biochemistry 41 15–20 [DOI] [PubMed] [Google Scholar]
- 29.Evans, W. H., and Martin, P. E. M. (2002) Mol. Mem. Biol. 19 121–136 [DOI] [PubMed] [Google Scholar]
- 30.Yeager, M., and Harris, A. L. (2007) Curr. Op. Cell Biol. 19 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veenstra, R. D., Wang, H. Z., Beblo, D. A, Chilton, M. G., Harris, A. L., Beyer, E. C., and Brink, P. R. (1995) Circ. Res. 77 1156–1165 [DOI] [PubMed] [Google Scholar]
- 32.Harris, A. L. (2007) Prog. Biophys. Mol. Biol. 94 120–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venyaminov, S. Y. and Yang, J. T. (Fasman, G. D., ed) (1996) Circular Dichroism and the Conformational Analysis of Biomolecules, pp. 69–108, Springer