Abstract
Mitotic movements of chromosomes are usually coupled to the elongation and shortening of the microtubules to which they are bound. The lengths of kinetochore-associated microtubules change by incorporation or loss of tubulin subunits, principally at their chromosome-bound ends. We have reproduced aspects of this phenomenon in vitro, using a real-time assay that displays directly the movements of individual chromosome-associated microtubules as they elongate and shorten. Chromosomes isolated from cultured Chinese hamster ovary cells were adhered to coverslips and then allowed to bind labeled microtubules. In the presence of tubulin and GTP, these microtubules could grow at their chromosome-bound ends, causing the labeled segments to move away from the chromosomes, even in the absence of ATP. Sometimes a microtubule would switch to shortening, causing the direction of movement to change abruptly. The link between a microtubule and a chromosome was mechanically strong; 15 pN of tension was generally insufficient to detach a microtubule, even though it could add subunits at the kinetochore–microtubule junction. The behavior of the microtubules in vitro was regulated by the chromosomes to which they were bound; the frequency of transitions from polymerization to depolymerization was decreased, and the speed of depolymerization-coupled movement toward chromosomes was only one-fifth the rate of shortening for microtubules free in solution. Our results are consistent with a model in which each microtubule interacts with an increasing number of chromosome-associated binding sites as it approaches the kinetochore.
INTRODUCTION
During prometaphase, chromosomes become attached to microtubules that emanate from the centrosomes; these microtubules form tracks along which the chromosomes move. Microtubules impinge on both the arms and the kinetochores of a chromosome, but it is the kinetochore-associated microtubules that contribute most to chromosome movement (reviewed by Nicklas, 1997). As a chromosome moves, its kinetochore-associated microtubules elongate or shorten. These length changes are primarily a result of addition or loss of tubulin subunits at the kinetochore-bound ends, but there is also some subunit exchange at or near the spindle poles (Mitchison et al., 1986; Gorbsky et al., 1987; Wise et al., 1991; Mitchison and Salmon, 1992).
The mechanisms underlying the tight linkage between chromosome movements and the incorporation or loss of kinetochore microtubule subunits remain mysterious, but progress has been made toward understanding the dynamic behavior of microtubules in solution. The energy that drives tubulin polymerization dynamics in vitro derives from GTP that binds to soluble tubulin. After tubulin is incorporated into a microtubule, its bound GTP is hydrolyzed to GDP, and much of the energy released during hydrolysis may be stored in the lattice of the microtubule (Caplow et al., 1994; Mickey and Howard, 1995). Microtubules use this energy to support “dynamic instability” (Mitchison and Kirschner, 1984), a process by which microtubule polymers spontaneously alternate between growth and rapid shortening (for review, see Desai and Mitchison, 1997). This behavior is qualitatively similar to the behavior of chromosome-bound microtubules during the oscillatory chromosome movements of prometaphase, and it has long been thought that the energy released by the disassembly of tubulin might be harnessed to help drive chromosome movements (for review, see Inoué and Salmon, 1995).
The complexity of the mitotic apparatus has deterred analyses of the mechanics and chemistry that underlie the links between microtubule polymerization and chromosome movement. In the cells of animals and higher plants, an uncounted number of motor enzymes and unknown cellular chemistries influence the behavior of chromosomes. Moreover, mechanical descriptions are confounded by the large number and complex arrangement of the microtubules that interact with a mitotic chromosome. To circumvent these difficulties, in vitro assays have been developed that allow study of microtubule–chromosome interactions. With these assays it has been shown that shortening microtubules can maintain their attachments with chromosomes at their depolymerizing ends (Koshland et al., 1988; Coue et al., 1991). The force exerted on a chromosome during this depolymerization-coupled movement exceeds 1 pN (Coue et al. 1991) and is therefore comparable to, or greater than, the 2–6 pN exerted on a filament by an ATP-driven motor protein, such as myosin or kinesin (Finer et al., 1994; Hunt et al., 1994; Meyhöfer and Howard, 1994; Svoboda and Block, 1994; Molloy et al., 1995). However, these assays too have been limited by their inability to reveal dynamic interactions between a single microtubule and a chromosome; such an interaction either cannot be resolved or has been inferred from the analysis of fixed samples. As a consequence, the literature includes apparently conflicting conclusions on several fundamental issues, such as the direction of microtubule movements in the presence of ATP (Mitchison and Kirschner, 1985b; Hyman and Mitchison, 1991), whether kinetochore-bound microtubules are stabilized or destabilized against catastrophe (Mitchison and Kirschner, 1985b; Hyman and Mitchison, 1990), the ability of depolymerizing microtubules to remain attached to chromosomes (Mitchison and Kirschner, 1985b; Koshland et al. 1988; Hyman and Mitchison, 1990; Coue et al. 1991; Lombillo et al. 1995a), the ability of microtubules bound laterally to kinetochores to translocate in the presence of ATP (Huitorel and Kirschner, 1988; Hyman and Mitchison, 1991), and whether Taxol-stabilized microtubules maintain their link with kinetochores in the presence of ATP (Mitchison and Kirschner, 1985b; Huitorel and Kirschner, 1988; Hyman and Mitchison, 1991). Also, in some of these assays the rapid shortening of microtubules was induced by tubulin dilution. This is quite different from the situation in vivo, in which microtubules continuously switch between periods of growth and shortening under conditions that approximate a steady state.
We have developed an assay wherein a labeled segment on a single microtubule is directly observed to move toward and/or away from a chromosome as the intervening microtubule depolymerizes or polymerizes at its chromosome-associated end. Conditions are such that the ends of microtubules in solution undergo dynamic instability, and we observe that chromosome-associated microtubules do so as well. Our results confirm that an isolated chromosome contains all of the structures necessary to maintain a link with both polymerizing (Mitchison and Kirschner, 1985b) and depolymerizing (Koshland et al., 1988; Coue et al., 1991) microtubules, but we also extend previously published work. By observing growth and shortening of kinetochore-associated microtubules in real time, we have learned that both of these processes will occur without ATP. By quantifying several of the polymerization–depolymerization parameters of kinetochore-associated microtubules we have shown that kinetochores regulate microtubule dynamics, decreasing both the rate of rapid shortening and the frequency of transition from growth to shortening. By measuring the force required to detach a dynamic microtubule from a kinetochore we have shown that this interaction is strong, even though it is labile to tubulin polymerization. These results are discussed in the context of a model originally proposed by Hill (1985).
MATERIALS AND METHODS
Preparation of the Assay Components
Mitotic Chinese hamster ovary (CHO) chromosomes were isolated essentially as described by Hyman and Mitchison (1993). We compared chromosomes prepared using their lysis buffers with or without 0.1 mM spermine and 0.2 mM spermidine. This addition had no apparent effect on the binding of microtubules to the chromosomes, and in later experiments was left out. It was impossible to know whether the resulting chromosomes were “normal,” that is, unmodified by the isolation procedure, because the components of these structures are not all known. We did, however, assess the presence of a few known kinetochore components: CENP-E, dynein, and MCAK (Yen et al. 1991; Lombillo, 1994; Lombillo et al. 1995a; Wordeman and Mitchison, 1995). All these motors were still associated with the kinetochores of chromosomes isolated by this procedure.
Phosphocellulose-purified tubulin was prepared from bovine or porcine brain (Weingarten et al., 1974; Howard et al., 1993). Tubulin was fluorescently labeled with rhodamine as described by Hyman et al. (1991). Stable, rhodamine-labeled microtubules were polymerized at 37°C for 15 min by incubating 2 mg/ml labeled and unlabeled tubulin with 0.5 mM guanylyl(a,b)methylene-diphosphonate (GMPCPP; Hyman et al., 1991) in 80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 1 mM EGTA, and 1 mM MgCl2 adjusted to pH 6.9 with KOH (BRB-80). These polymers were then diluted sixfold in standard buffer solution (80 mM PIPES, 1 mM EGTA, 4 mM MgCl2, and 5 mM DTT, adjusted to pH 6.9 with KOH and supplemented with protease inhibitors: 1 μg/ml leupeptin and pepstatin and 10 μg/ml chymostatin), pelleted, and resuspended in standard buffer to their original volume. GMPCPP was kindly provided by T.J. Mitchison (University of California, San Francisco, CA); all other reagents were obtained from Sigma Chemical (St. Louis, MO).
Motility Assays
The motility assay described here is based on aspects of assays described by Lombillo et al. (1993) and by Hyman and Mitchison (1993). Most observations were made in perfusion chambers that were 75 μm deep, bounded at the bottom by a glass microscope slide and on top by a coverglass (Howard et al., 1993). For samples that were to be later examined by electron microscopy, the chambers were constructed by placing the coverglass onto two strips of cellophane tape and then securing the edges with wax. This allowed a coverglass to be easily removed for processing and did not expose the glass to grease, which tended to redistribute across the sample during critical point drying. Before use, chromosomes were washed by ninefold dilution into standard buffer, followed by centrifugation at 12,000 × g for 1 min in an Eppendorf (Hamburg, Germany) 5415c tabletop centrifuge. The pellet was then resuspended with standard buffer in two times the original volume.
The glass surfaces of the perfusion chamber were prepared to bind chromatin by introducing supernatant from a hybridoma cell line, 1D12, which produces an immunoglobulin G that binds DNA (Kotzin et al., 1984). Except where noted, 20 μl of solution (∼2 chamber vol) were introduced into the perfusion chamber at each solution exchange. The chamber surface was blocked by introducing 10% (wt/vol) BSA in 10 mM PIPES (pH 7.4) and washed with 40 μl of standard buffer. CHO chromosomes were introduced into the chamber, which was then inverted for 15 min on an aluminum block chilled with ice.
GMPCPP-stabilized microtubules were diluted 1:30 in standard buffer containing 1 mM GTP and 0.5 mg/ml unlabeled tubulin and then introduced into the perfusion chamber, which was transferred to a humidified chamber on a 37°C heating block. In some experiments the tubulin added in this step contained 50% rhodamine-labeled tubulin to stain the kinetochores (see Figures 2B and 4A). After 5 min, unbound microtubules were washed out with 40 μl of standard buffer; 1.0 mg/ml unlabeled tubulin, 0.13 mg/ml rhodamine-labeled tubulin, and 1.0 mM GTP were then introduced in standard buffer and incubated for 5 min to allow dim elongations to grow from the ends of the GMPCPP-stabilized microtubules. This solution was then washed out with 40 μl of standard buffer containing 1.0 mM GTP, 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), 0.75–2.0 mg/ml unlabeled tubulin, and an oxygen-scavenging mixture to inhibit photobleaching (0.6 mg/ml glucose oxidase, 0.12 mg/ml catalase, and 30 mM glucose). The ends of the perfusion chamber were sealed with immersion oil, and the chamber was immediately transferred to a prewarmed microscope stage for observation at 32°C.
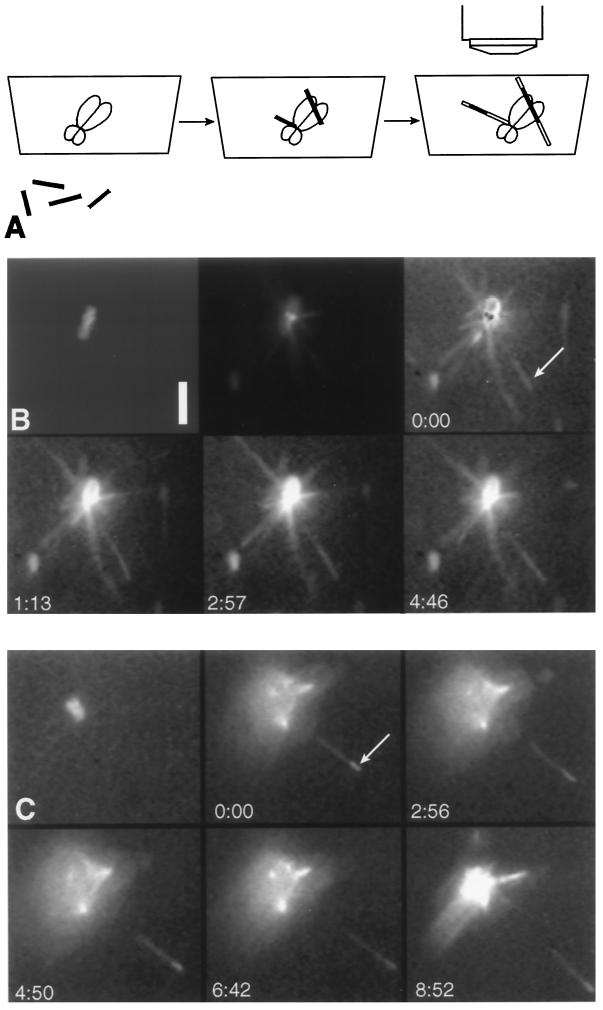
Figure 2.
Stable and brightly labeled microtubule segments that are tethered to a chromosome by labile elongations will undergo polymerization-coupled movement away from the chromosome. (A) Schematic representation of the procedure used to assay polymerization- and depolymerization-coupled movement. Microtubules that were brightly labeled and GMPCPP stabilized (filled thin rectangles) were incubated with chromosomes. Unbound microtubules were washed out, and dimly labeled, labile microtubules (unfilled thin rectangles) were grown from the ends of the bright segments. After the dimly labeled tubulin was exchanged with unlabeled tubulin, the samples were observed by epifluorescence microscopy. As the microtubules continued to elongate, the bright segments bound to the primary constriction of the chromosome moved away from the chromosome, but the bright segments bound to the arms did not move. (B and C) The top left panel in each series shows a chromosome visualized by DAPI fluorescence. The other panels are visualized by fluorescence of rhodamine-labeled tubulin. Arrows indicate the location of brightly labeled microtubule segments and time is in minutes and seconds. In the top center panel of B the gain of the camera is turned down, and the staining of the kinetochores by fluorescent tubulin can be seen. In the remaining panels the gain is turned up to reveal the microtubules; one brightly labeled microtubule segment (arrow) is tethered to the chromosome by a segment of dimly labeled and unlabeled microtubule. Because of the limited dynamic range of the camera, the chromosome now appears as a bright flare (bottom panels). In the top right panel the kinetochores are identified by black dots formed by subtracting the image of the kinetochore-bound rhodamine tubulin from an image at higher gain that reveals the microtubules. The microtubule whose bright segment moves extrapolates to the vicinity of the kinetochores. (C) Another example of polymerization-coupled movement. Bar, 5 μm.
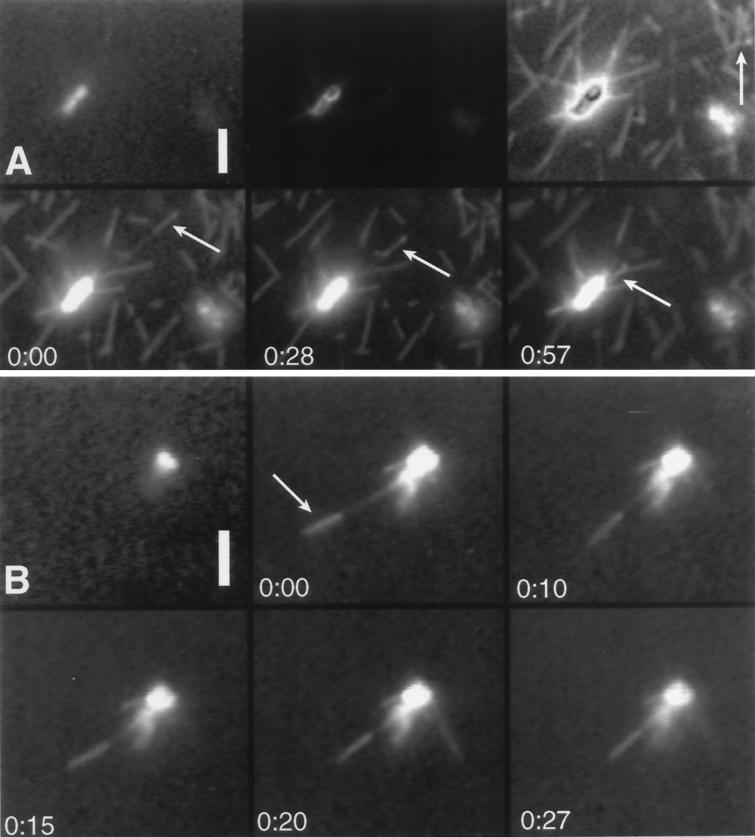
Figure 4.
Some of the stable and brightly labeled microtubule segments that are tethered to a chromosome by labile elongations undergo depolymerization-coupled movement toward the chromosome. Arrows indicate the location of the brightly labeled microtubule segments. Times are indicated in seconds. The top left panel in each series shows a chromosome visualized by DAPI fluorescence. The other panels are visualized by fluorescence of rhodamine-labeled tubulin. (A) In the top center and top right panels the kinetochores are visualized as described for Figure 2B. The microtubule indicated by the arrow in the top right panel swivels diffusively about its attachment to the chromosome so it projects more vertically in the bottom panels. (B) Another example of depolymerization-coupled movement. Bars, 5 μm.
ATP Contamination
In several experiments, we tried to deplete any contaminating ATP by supplementing the assay buffer with hexokinase (15 U/ml) and glucose (50 mM). These efforts produced no data because hexokinase induced complete depolymerization of dynamic microtubules within 3 min. Because this effect was seen with nominally pure hexokinase from two different sources, we suspected that the hexokinase was depleting the GTP, despite previous reports that it has a high specificity for ATP (Darrow and Colowick, 1962; Mitchison and Kirschner, 1985b). The GTPase activity of hexokinase was verified in two ways. The first was by observing kinesin-dependent microtubule gliding (Howard et al., 1993) in the presence of an initial concentration of 1 mM GTP, using Taxol-stabilized microtubules. In 15 U/ml hexokinase the speed of microtubule movement decreased to less than half the speed of the control within 5 min and was down to about 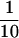 speed after 30 min. Second, we assayed GTP using a luciferin/luciferase luminescence assay (Molecular Probes, Eugene, OR). Although this assay is commonly used for measurements of ATP, it reveals GTP with reduced sensitivity. In the presence of the above concentrations of hexokinase and glucose, the GTP concentration dropped from 1 to <0.25 mM in <30 s and was down to ∼0.1 mM in 5 min. The GTP depletion was due to hexokinase and not a contaminating activity in the preparation, since the initial GTPase rate was decreased >50-fold in the absence of glucose.
speed after 30 min. Second, we assayed GTP using a luciferin/luciferase luminescence assay (Molecular Probes, Eugene, OR). Although this assay is commonly used for measurements of ATP, it reveals GTP with reduced sensitivity. In the presence of the above concentrations of hexokinase and glucose, the GTP concentration dropped from 1 to <0.25 mM in <30 s and was down to ∼0.1 mM in 5 min. The GTP depletion was due to hexokinase and not a contaminating activity in the preparation, since the initial GTPase rate was decreased >50-fold in the absence of glucose.
We have considered the remote possibility that both of the hexokinase preparations contained high levels of nucleoside diphosphate kinase, which catalyzes the conversion of GTP + ADP to ATP + GDP, and might thereby allow hexokinase to deplete GTP indirectly through hydrolysis of ATP. This possibility can be excluded for two reasons. First, at the trace ATP concentrations in these assays (see below), the enzymatic rate of hexokinase using glucose and ATP as substrates is at most 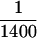 the rate needed to explain our results (Viola et al., 1982). Second, even if the hexokinase preparations were contaminated with as much as 10% nucleoside diphosphate kinase, the rate of phosphate transfer from GTP to ADP would be at most
the rate needed to explain our results (Viola et al., 1982). Second, even if the hexokinase preparations were contaminated with as much as 10% nucleoside diphosphate kinase, the rate of phosphate transfer from GTP to ADP would be at most 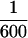 the rate necessary to explain these results (Garces and Cleland, 1969). The discrepancy between our observations and the initial characterization of hexokinase (Darrow and Colowick, 1962) is likely due to the different buffers that were used in these assays. The ability of hexokinase to cause depolymerization of microtubules is thus explained by its ability to decrease the GTP-to-GDP ratio and consequently the free energy of GTP hydrolysis. In light of this finding, interpretations of data that assumed ATP specificity for hexokinase will need to be reevaluated (Mitchison and Kirschner, 1985b).
the rate necessary to explain these results (Garces and Cleland, 1969). The discrepancy between our observations and the initial characterization of hexokinase (Darrow and Colowick, 1962) is likely due to the different buffers that were used in these assays. The ability of hexokinase to cause depolymerization of microtubules is thus explained by its ability to decrease the GTP-to-GDP ratio and consequently the free energy of GTP hydrolysis. In light of this finding, interpretations of data that assumed ATP specificity for hexokinase will need to be reevaluated (Mitchison and Kirschner, 1985b).
Our concerns about contaminating ATP were alleviated upon direct measurement of the ATP concentration using the luciferin/luciferase assay system. These enzymes are capable of using GTP or ATP as a substrate, but the Km is ∼200-fold larger and Vm ∼100 fold smaller for GTP then that for ATP (data not shown). We estimated the fraction of the luminescence that arose from ATP contamination by fitting the measured relation between luminescence and the GTP concentration with the equation:
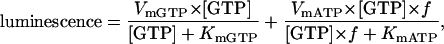 |
where f is the ATP concentration expressed as a fraction of the GTP concentration. Vm and Km for ATP were determined from separate experiments using ATP standards. The ATP concentration in our assays was 0.6 ± 1.1 nM.
Assay of Microtubule Growth from Axonemes
Axonemes isolated from Chlamydomonas (Gardner et al., 1994) were salt extracted by 10-fold dilution into 600 mM NaCl in BRB-80. After 15 min at 0°C, axonemes were further diluted 10-fold into standard buffer and introduced into a perfusion chamber at room temperature. After ∼1 min, axonemes not bound to the glass surface were washed out with standard buffer, and 40 μl of standard buffer containing 1.0 mM GTP and either unlabeled or a mixture of unlabeled and rhodamine-labeled tubulin was introduced. When rhodamine-labeled tubulin was used, oxygen scavengers were also added to the solution. The ends of the perfusion chamber were sealed with immersion oil, and the chamber was immediately transferred to the prewarmed microscope stage for observation at 32°C.
Extracted sea urchin axonemes (Gibbons and Fronk, 1972) were diluted 1:50 in standard buffer and introduced into a perfusion chamber. Thereafter, handling was the same as for Chlamydomonas axonemes.
Microscopy and Data Analysis
Preparations were viewed with a Zeiss Universal microscope (Carl Zeiss, Oberkochen, Germany) equipped with epifluorescence or differential interference contrast (DIC) optics. DIC microscopy was performed as previously described (Coue et al., 1991), and all video processing was done with an IMAGE-640 frame-grabber board (Matrox, Dorval, Canada) and MetaMorph software (Universal Imaging, West Chester, PA). When epifluorescence was used, samples were generally illuminated for only 76 ms every 1.3 s to slow photobleaching, although some observations were made continuously (i.e., at video rate). A Zeiss shutter placed in front of a 200-W Hg arc lamp was controlled using MetaMorph software and a custom-built interface. During the periods that the shutter was open, two successive video frames were acquired by a doubly intensified camera (DV2, Venus Scientific, Farmingdale, NY), averaged, and transferred on-line to a Super-VHS video cassette recorder (Panasonic AG-1970, Waxmans, Denver, CO).
The microscope stage temperature was maintained at 31.8 ± 0.2°C by an air curtain incubator (Sage Instruments, Cambridge, MA) regulated by a CN76000 microprocessor-based temperature and process controller (Omega Engineering, Stamford, CT), which monitored the temperature via a thermocouple wire inserted into the immersion oil between the microscope objective and the coverslip.
The position of objects was determined from video images that were digitized and displayed using MetaMorph software. Stationary objects, usually chromosomes, were used to align each series of images before they were analyzed. This corrected for stage drift on the microscope and time-base errors during digital frame capture. It was apparent that the errors introduced by this procedure (on the order of 200 nm, the light resolution limit) were small relative to the micrometers of movement that were being measured, because all objects on the glass surface aligned together. Coordinates of a specific object were determined from the position of a marker that was maneuvered across the image, using the computer’s mouse. In most cases the speed of polymerization-coupled movement was determined by measuring the distance between successive positions of one edge of a bright, GMPCPP-stabilized microtubule segment and the initial position of that edge. The mean speed and its SE were determined by linear regression. In some cases a segment underwent lateral motions as the entire microtubule swiveled about its attachment to a chromosome, apparently because of diffusive forces (e.g., Figure 4A). In these cases the position of a bright segment was measured relative to the point where the microtubule was bound to the chromosome.
Optical Tweezers
A single-beam optical gradient trap was constructed using a beam splitter to introduce a 1064-nm beam from the TEM00 mode of an Nd:YAG laser (C-95 YAGMAX, CVI Lasers or Series 700, Lee Laser) into the epifluorescence light path. Because the microscope does not contain infinity-corrected optics, the beam was set up to be divergent as it entered the back of the objective; the resulting trap was formed in the plane of focus of the microscope. The trapping force was calibrated by observing the displacement of a trapped 1.0-μm-diameter silica bead (Bangs Laboratories, Carmel, IN) subjected to viscous forces as the microscope stage was moved at known speeds, using computer-controlled motors attached to the stage translation lead screws (Figure 1). The displacement of a bead from one image to another was determined using MetaMorph software to calculate the centroid of the cross-correlation of the images (Gelles et al., 1988). The distance from the coverglass to the center of a trapped bead was 2.5 ± 0.5 μm during force measurements.
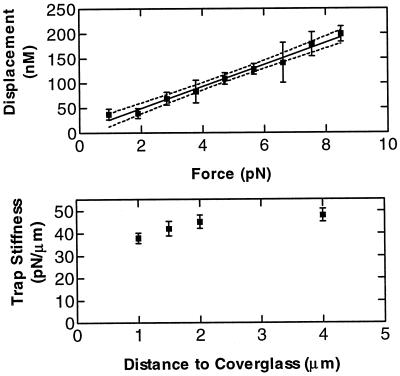
Figure 1.
Optical tweezers calibration curves. The top graph is the relation between the force on a 1.0-μm-diameter silica bead and the displacement of the bead from the center of the trap. Broken lines are 95% confidence intervals. The bottom graph shows how the trap stiffness varied with the distance from the coverglass that formed the top surface of the chamber. The laser power behind the objective was 38 mW.
Silica beads were attached to microtubules by means of a biotin–streptavidin link. The beads were coated by suspending them at 10 mg/ml in 20 μg/ml BSA-biotinamidocaproyl for 5 min, after which the beads were pelleted, washed two times in BRB-80, and resuspended at 1 mg/ml in 20 μg/ml streptavidin for 5 min. After pelleting and resuspending twice more in BRB-80, the beads were frozen in liquid nitrogen and stored at −70°C for later use.
GMPCPP microtubules were biotinylated by incubating them in BRB-80 with 1.7 mM biotin succinimydal ester (Molecular Probes) at 37°C for 15 min, after which 3 vol of 200 mM lysine were added. After 10 min microtubules were pelleted in an airfuge for 3 min at 28 psi and resuspended in standard buffer for immediate use.
To prevent the beads from sticking, the glass slides that formed the lower surface of the perfusion chambers were coated with tetrafluoroethylene telomer release agent (Miller-Stephenson, Sylmar, CA), BSA was not used, and casein was added at 10% of saturation. Despite these procedures the beads still tended to bind to the glass, so after ∼15 min no beads could be manipulated. This problem significantly increased the difficulty in performing these assays.
Electron Microscopy
In preparation for scanning electron microscopy, samples were fixed with BRB-80 supplemented with 0.1 M sucrose, 0.1% DMSO, 10 μM Taxol, 2% formaldehyde, and 0.8% glutaraldehyde. These were transferred into BRB-80 plus 1% osmium tetraoxide and transferred sequentially through solutions containing water with 0, 30, 50, 70, 95, and 100% ethanol before being subjected to critical point drying with CO2. Dry samples were coated with 3 nm of platinum using a BAF 60 freeze-fracture system (Bal-tec, Balzers, Liechtenstein) and viewed under a Zeiss DSM 940A scanning electron microscope. Sample chambers were prepared using coverslips on which a grid had been etched (Eppendorf Cellocate coverslips) to aid in reidentification of the same chromosome that had been monitored by light microscopy.
RESULTS
Brightly Labeled Microtubule Segments Undergo Polymerization-coupled Movements Toward and Away from Chromosomes In Vitro
Microtubules can attach to chromosomes in vitro (Mitchison and Kirschner, 1985; Koshland et al., 1988; Coue et al., 1991; Hyman and Mitchison, 1991), and a fraction of these are competent to undergo depolymerization-coupled movement (Koshland et al., 1988; Coue et al., 1991). To select for microtubules that had formed such associations, we used tubulin labeled with a low level of rhodamine to polymerize dim elongations from short, chromosome-associated microtubules that were brightly labeled and GMPCPP stabilized (Figure 2A). Such polymerization resulted in bright microtubule segments with dimly labeled tubulin elongations that were bound end-on to a chromosome (Figures 2–4).
Fluorescent tubulin in solution interferes with visualization of microtubules in the 75-μm-deep perfusion chambers, so it was exchanged with unlabeled tubulin before the chromosome–microtubule complexes were examined. In the presence of GTP and 0.75–2.0 mg/ml tubulin in standard buffer (no added ATP), ∼50% of the bright segments underwent movement away from or toward the chromosome to which they were tethered (Figures 2 and 4). The microtubules that displayed movement of their bright segments were associated with the chromosomes in the neighborhood of their primary constrictions (Figures 2–4), whereas microtubules associated with other parts of a chromosome never moved. Likewise, there was no movement by microtubules that bound laterally to a chromosome, such that both ends of the microtubule projected away from the chromosome. In some cases brightly labeled segments with unlabeled elongations were observed to undergo directed motion. If the association between such a segment and a chromosome could be established—either because it started at a chromosome and moved away, or because it moved toward a chromosome until it was clearly associated—the event was included in our analysis of polymerization-coupled motion. Chromosome-bound microtubules that did not contain bright segments were not followed in this study.
In light of the recent discovery of kinesin-like motor enzymes associated with chromosome arms (e.g., Wang and Adler, 1995), we were interested in learning whether the microtubules that moved in our assay were associated with kinetochores. We were able to visualize kinetochores on the chromosomes under study by using their affinity for tubulin to stain them with rhodamine (Figures 2B and 4A; see MATERIALS AND METHODS). Appropriate setting of the gain and offset of the camera displayed such labeled kinetochores as two bright dots at the primary constriction of a chromosome. Higher settings of camera gain revealed the microtubules elongated by polymerization of dimly labeled tubulin that were associated with the chromosome. We could then observe the brightly labeled segments moving away (Figure 2, B and C) or coming closer (Figure 4, A and B) to the chromosome. These dimly labeled microtubules extrapolate to one or the other of the kinetochores, identified in the third frames of Figures 2B and 4A by dark dots that were formed by subtracting the image of the kinetochore-bound rhodamine tubulin from the higher-gain image necessary to see the microtubules themselves. Although these data are highly suggestive of a direct kinetochore–microtubule association, CHO kinetochores are at the limits of the resolving power of the light microscope (Witt et al., 1980; Rieder, 1982), and microtubules are a factor of 10 smaller, so kinetochore association cannot be rigorously established by light microscopy. Microtubules that displayed polymerization-coupled movements also appeared to be bound at the kinetochores when examined by scanning electron microscopy (Figure 3). We conclude that the microtubules that display polymerization- and depolymerization-coupled movement on chromosomes are associated with kinetochores.
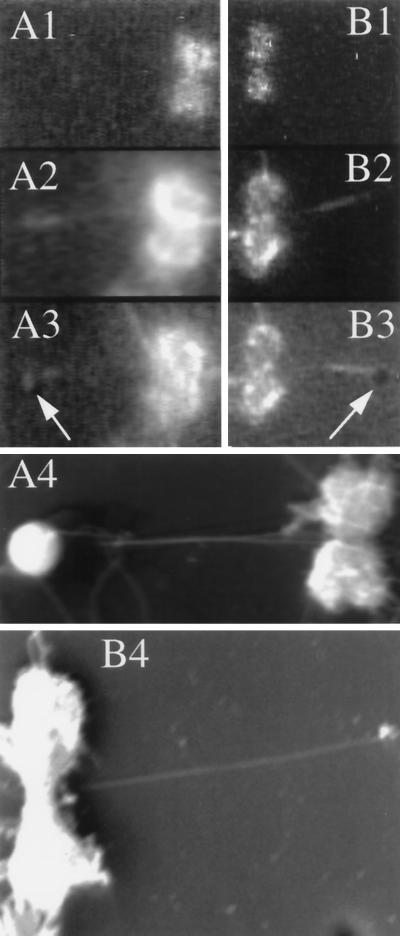
Figure 3.
Scanning electron micrographs of chromosome-bound microtubules. Two different chromosomes (A and B) and associated microtubules were viewed by light microscopy. The chromosomes were visualized first by DAPI fluorescence (A1 and B1) and then by rhodamine fluorescence to see the microtubules (A2 and B2). (A3 and B3) Silica beads placed on the microtubules by the optical tweezers were visible by bright-field illumination (arrows) in combination with rhodamine fluorescence. (A4) Chromosome shown in A1–A3 as seen by scanning electron microscopy. Note that two microtubules emanate from the primary constriction, one of which was formed from unlabeled tubulin and was thus not visible by light microscopy. (B4) Electron micrograph of the chromosome in B1–B3. The bead was lost during preparation of the sample for electron microscopy.
Polymerization-coupled Movement away from Chromosomes
The movement of brightly labeled microtubule segments away from the chromosome to which they were attached was the result of tubulin incorporation at the chromosome-bound end of a microtubule, as indicated by two observations. First, when examined after prolonged incubation in dimly labeled tubulin but immediately after replacement with unlabeled tubulin, the dim and labile elongations did not extend past a chromosome. Most probably they grew at the chromosome-bound end of a brightly labeled segment and pushed it away from the chromosome (remember that the unbound bright segments were washed away before the dimly labeled tubulin was added). Second, as a bright segment continued to move away from the chromosome after the addition of unlabeled tubulin, an unlabeled segment formed between the labeled microtubule and the chromosome; with time this segment grew longer (Figure 2, B and C).
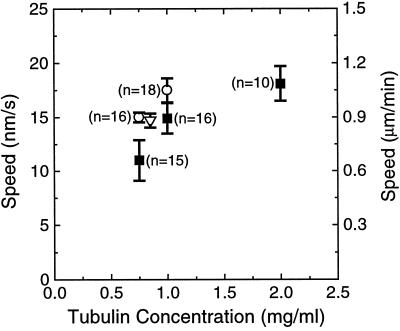
The speed of polymerization-coupled movement away from a chromosome depended on the concentration of soluble tubulin; the higher the concentration, the faster the movement (Figure 5). To compare these rates with the rate of polymerization at free microtubule ends in the same buffer and at the same temperature, we used DIC microscopy to observe microtubules grown from axonemes isolated from sea urchin sperm or Chlamydomonas flagella. The microtubule ends were initially identified as plus or minus by the morphological asymmetry of Chlamydomonas axonemes (Bergen and Borisy, 1982); thereafter they were distinguished by their rates of elongation (Walker et al., 1988). A small fraction of the microtubule ends may be misclassified by the latter method, because the tails of the distributions of the growth rates at the plus and minus ends of microtubules overlap (Kowalski and Williams, 1993). At 0.75 mg/ml tubulin, however, no microtubules were observed to grow from the minus ends of Chlamydomonas axonemes, and at 1.0 mg/ml tubulin only 10% (n = 20) of axonemes with microtubules growing from their plus ends had microtubules also at their minus ends. These frequencies are consistent with the observation that seeded nucleation from axonemes appears to be more frequent at the plus ends (Walker et al., 1988). To determine whether rhodamine-labeled tubulin altered microtubule growth, the elongation of axoneme-seeded microtubules at 0.75 mg/ml tubulin was also examined in 10% rhodamine-labeled tubulin. This fraction of labeled tubulin—the same as was used to polymerize dim microtubule elongations in the polymerization-coupled motility assays—had no effect on the rate of elongation (Figure 5).
Figure 5.
The speed of polymerization-coupled movement away from a chromosome is similar to the rate of growth at free microtubule plus ends. Bars are SEs, and the value in parentheses adjacent to each data point indicates the number of events used in calculating that point. Filled squares, Polymerization-coupled movement of brightly labeled microtubule segments away from the chromosome to which they are tethered; open circles, growth at the plus ends of axoneme-seeded microtubules; open triangle, growth at the plus ends of axoneme-seeded microtubules in the presence of 10% rhodamine-labeled tubulin (n = 4). Note that the open triangle was shifted to the right for clarity of display; the total concentration of labeled and unlabeled tubulin was 0.75 mg/ml.
At 0.75 and 1.0 mg/ml tubulin we observed very few microtubules that were not initiated from the ends of axonemes, but at 2.0 mg/ml tubulin our efforts to measure the elongation of free microtubule ends were frustrated because self-nucleated microtubules obscured the image. It is conceivable that the large number of unlabeled (and thus invisible) microtubules that spontaneously nucleate at 2 mg/ml might interfere with polymerization-coupled motility assays. High rates of spontaneous nucleation can also quickly reduce the concentration of free tubulin (Voter and Erickson, 1984). However, using data from Fygenson et al. (1995), and calculating tubulin consumption as,
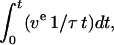 |
we estimate that during our assays the change in the tubulin concentration was negligible. In this equation ve is the rate of microtubule growth (at both ends) in subunits per second, 1/τ is the nucleation rate per unit volume, and t is time. This assumes that the catastrophe rate is zero and thus overestimates the tubulin depletion.
The speed of polymerization-coupled motility away from a chromosome was similar to the rate of elongation at the plus ends of axoneme-initiated microtubules (Figure 5). This suggests that the bright segments are tethered to the chromosomes by their plus ends. For several reasons we were unable to determine the polarity of the tethered microtubules from the relative lengths of the dim segments at opposing ends (Hyman and Mitchison, 1991). First, most bright segments had dim elongations at only one end. This was not surprising, because even at 37°C in 1.7 mg/ml tubulin we found that only 29 ± 8% of the brightly labeled GMPCPP-stabilized segments would grow elongations from both ends. Additionally, dim segments were lost over time as the labeled tubulin was turned over by dynamic instability. Finally, this type of assay for polarity would assume that the polymerization dynamics of chromosome-bound microtubules are the same as those of free microtubules. This assumption is untested for growing microtubules and, as will be discussed, is false for shrinking microtubules.
Depolymerization-coupled Movement toward Chromosomes
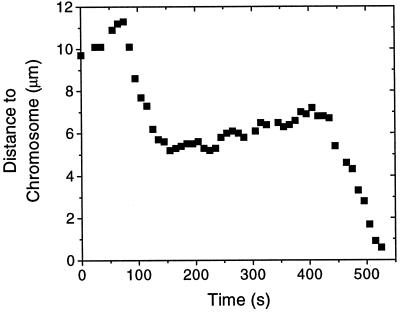
Like the microtubules bound to a kinetochore in vivo, an elongating microtubule would sometimes undergo a sudden transition from polymerization to depolymerization at its chromosome-bound end. Following the lexicon of microtubule dynamic instability, we will refer to such an event as a “catastrophe.” As a microtubule shortened, its brightly labeled segment moved toward the chromosome (Figure 4). This motion was coupled to depolymerization at the chromosome, as the labile, dimly labeled elongation of the microtubule was not extruded past the chromosome as the microtubule moved. Movement toward a chromosome was ∼10 times faster than movement away and in most cases continued until the bright segment contacted the chromosome. After contacting a chromosome, the bright segment then resumed slow movement away from the chromosome in all six cases that were observed for long enough to determine motion. In two cases shortening microtubules detached from a chromosome, and in two other cases the shortening microtubules underwent “rescue” (a transition from shortening to growth) before their bright segments reached a chromosome. After rescue the bright segments resumed slow motion away from the chromosome for several minutes before, in the first case, the bright segment stopped moving and, in the second case, another catastrophe occurred, whereupon the bright segment moved all the way to the chromosome (Figure 6). Interestingly, the rate at which these two microtubules grew before the catastrophes was different from the rate at which they grew after the rescue events. Before the catastrophes the speed of polymerization-coupled movement was, in the first case, 19.0 ± 0.5 nm/s (1.14 μm/min), and in the second case it was 23 ± 3 nm/s (1.38 μm/min). After the rescue events the speeds were, respectively, 13.0 ± 0.5 nm/s (0.78 μm/min) and 7 ± 1 nm/s (0.42 μm/min). For the microtubule that underwent two catastrophes the rates of shortening after each catastrophe were also different: 93 ± 6 nm/s (5.58 μm/min) and 67 ± 3 nm/s (4.02 μm/min). Both of these observations occurred at a soluble tubulin concentration of 1.0 mg/ml.
Figure 6.
A bright microtubule segment tethered to a chromosome can switch between polymerization-coupled movement away from the chromosome and depolymerization-coupled movement toward the chromosome. This trace shows two catastrophes and one rescue event. The tubulin concentration in this experiment was 1.0 mg/ml.
To determine whether the rhodamine-labeled microtubules shorten at the same rate as unlabeled microtubules, we measured the rate of rapid shortening of microtubules that were bound to the glass surface but not to the chromosomes. Because the rate of rapid shortening is only ∼16% different at the plus and minus ends of microtubules (Walker et al., 1988), we made no attempt to determine the polarity of these microtubules. To within the accuracy of measurement the microtubules exhibited the same rate of rapid shortening as unlabeled microtubules that were grown from the plus ends of axonemes.
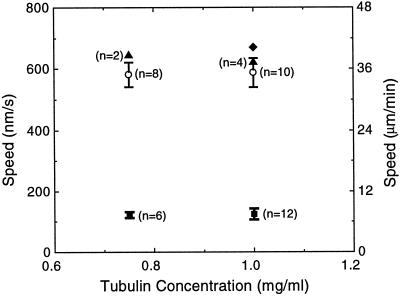
We compared the speed of depolymerization-coupled movement toward a chromosome with the rate of rapid shortening at free microtubule ends (Figure 7). The speed of microtubule depolymerization was significantly slowed by association with a chromosome. Bright microtubule segments proceeded toward a chromosome at about one-fifth the speed that free microtubule ends shrank during rapid shortening. There was no detectable change in the speed of movement toward a chromosome when the tubulin concentration was raised from 0.75 to 1.0 mg/ml. The rate of rapid shortening at free microtubule ends was also independent of the tubulin concentration, as has previously been observed by Walker et al. (1988).
Figure 7.
The speed of depolymerization-coupled movement toward a chromosome is slower than the rate of rapid-shortening at free microtubule ends. Open circles, Rapid shortening at the ends of axoneme-seeded microtubules. Bars are SEs, and the value in parentheses adjacent to a data point indicates the number of events used in calculating that point. Filled triangles, Rapid shortening at the ends of microtubules, dimly labeled with rhodamine, that were bound to the glass surface but not associated with chromosomes during polymerization-coupled motility assays; filled squares, depolymerization-coupled movement of brightly labeled microtubule segments toward the chromosome to which they were tethered. One outlier (filled diamond) was excluded from the calculations of the average speed of depolymerization-coupled movement at 1.0 mg/ml tubulin. Error bars associated with the filled triangles are not shown; the errors are, left and right, 32 and 106 nm/s, respectively.
Modulation of Microtubule Stability
We could observe the dynamic behavior of a tethered microtubule for only a limited duration (usually ∼15–20 min) before the microtubule became difficult to discern as a result of photobleaching. Consequently, we could only rarely measure the time between two catastrophes on a single microtubule. Instead, we calculated the mean time until catastrophe by dividing the total time that growing microtubules were observed by the number of catastrophes observed. This approach precluded calculating the SD of the estimate, because we do not know the form of the distribution of times until catastrophe for chromosome-bound microtubules, and a probabilistic analysis has shown that microtubules become more likely to undergo catastrophe over time (Odde et al., 1995). At 0.75 or 1.0 mg/ml tubulin the average time until catastrophe for tethered microtubules was more than double that at the plus ends of free microtubules (Table 1). This suggests that during polymerization-coupled movement an elongating microtubule is stabilized by its interactions with a chromosome. Although we did not measure the time until catastrophe at the minus end of microtubules, it has been observed to be about twice as long as at the plus end (Odde et al., 1995). Thus the average time until catastrophe at the minus ends will be similar to that for microtubules bound to chromosomes.
Table 1.
Average times until catastrophe (seconds)
| Tubulin concentration (mg/ml) | Chromosome bound (s) | Free ends (s) |
|---|---|---|
| 0.75 | 1470 (n = 5) | 627 (n = 12) |
| 1.00 | 1211 (n = 9) | 516 (n = 9) |
The average time until catastrophe at the chromosome-bound ends of microtubules was estimated by dividing the total time that the bright segments on these microtubules were observed moving away from chromosomes by the number (n) of times that these segments switched to movement toward a chromosome. The average time until catastrophe at the free ends of microtubules was calculated by dividing the total time that all microtubules grown from axonemes were observed by the number (n) of times these microtubules switched to rapid shortening.
A Strong Link Is Maintained between a Microtubule and a Chromosome during Polymerization-coupled Movement
During mitosis a microtubule must remain associated with a kinetochore, even when subjected to as much as 8–210 pN of tension (Nicklas, 1983). To examine the ability of chromosome-bound microtubules to bear tension in vitro, we constructed optical tweezers with which we could tug on a glass bead that had been attached to a microtubule as a handle. Using the optical tweezers, we placed streptavidin-coated beads on the bright, biotinylated segments of microtubules that were undergoing polymerization-coupled movement. A microtubule-bound bead was then held stationary by the optical tweezers while the microscope stage was moved at ∼2 μm/s, such that a surface-bound chromosome was pulled away from the bead-bound microtubule. At this speed the viscous drag on the bead was <20 fN, a force that is negligible in comparison with those developed by single, microtubule-dependent motor enzymes (reviewed by Howard, 1996). During polymerization-coupled movement microtubules could bear between 15 and 20 pN of tension without detaching from a chromosome. When the laser power was set so that the maximum trapping force was <15 pN, no microtubules could be detached from the chromosomes. At 15 pN, only one of six microtubules could be detached, and when the maximum trapping force was 20 pN, six of six microtubules were detached. During the course of these experiments we found that the force generated during polymerization-coupled movement was <2 pN (n = 3), which is the lower limit for force detection at the trap stiffness used in these studies. The uncertainty in the calibration of the optical tweezers was 10%.
DISCUSSION
We have developed an in vitro assay with which to study the mechanical coupling of a chromosome to the polymerizing or depolymerizing end of a microtubule. Through observing the movements of a stable and brightly labeled microtubule segment tethered to a chromosome by a dim and labile microtubule elongation, we have found that an isolated chromosome can maintain an attachment to a growing or shortening microtubule end, even at concentrations of ATP that are too low to produce appreciable movement from a microtubule-dependent motor enzyme. The distance between a chromosome and the chromosome-distal end of an active microtubule oscillated as the microtubule switched between polymerization and depolymerization at its chromosome-associated end, analogous to the movements of chromosomes during prometaphase. With this assay we have determined some fundamental properties of the interaction between a chromosome and a dynamic microtubule. Kinetochores form mechanically strong links with dynamic microtubules but generate little force against these microtubules as they polymerize. Kinetochores regulate the dynamic behavior of bound microtubules by slowing their rate of depolymerization and decreasing their frequency of catastrophe. These observations help explain how the links between kinetochores and microtubules are maintained and allow us to test models describing how chromosomes and microtubules interact.
The speed of depolymerization-coupled movements in the effective absence of ATP in vitro is similar to the speed of movements immediately after a chromosome attaches to the spindle (reviewed by Rieder and Salmon, 1995) but is ∼5- to 40-fold faster than poleward chromosome movements during later prometaphase and anaphase (Brinkley and Cartwright, 1971; Cande and Wolniak, 1978). The speed of movement in our assays showed no tendency to decrease with the duration of attachment, suggesting that factors other than time are important for the changes seen in vivo. Prometaphase movements may be slowed because the many spindle microtubules that contact a mitotic chromosome mechanically impede chromosome movement. This impediment might derive from “polar ejection forces” that sweep objects away from the spindle poles (Rieder et al., 1986; Ault et al., 1991) or from a subset of kinetochore microtubules that are in a state that resists chromosome movement. This idea is supported by the observation that when nocodazole or colcemid is used to induce simultaneous shortening of both kinetochore and nonkinetochore microtubules in newt lung cells, the rate that kinetochore microtubules depolymerize at their chromosome-bound ends increases 4- to 15-fold to ∼60 nm/sec (3.6 μm/min) (Washio and Sato, 1982; Cassimeris and Salmon, 1991; Skibbens et al., 1993). This approaches the ∼120 nm/sec (7.2 μm/min) that we observe in vitro. Also consistent with our in vitro observations, 60 nm/s is approximately one-fifth the rate that free microtubule ends shorten in vivo (Cassimeris and Salmon, 1991).
Our results, when taken together with those of Coue et al. (1991), indicate that depolymerization-coupled movement is supported by energy from GTP hydrolysis that is stored in the lattice of a microtubule during the polymerization process. Coue et al. (1991) found that chromosomes can maintain attachments with the depolymerizing ends of microtubules grown from immobilized Tetrahymena pellicles when the tubulin is reduced to low levels (≤0.023 mg/ml) and the nucleotides are removed by both dilution and enzymatic degradation. This demonstrates that the energy for depolymerization-coupled movement is not directly derived from nucleotides in solution. The speeds of these movements, although more variable, were not statistically different from those observed in the present work. Our experiments show that these movements also do not require free energy introduced by tubulin dilution, leaving only the microtubule lattice itself as the source for energy to support these movements.
The movements and forces described in this article have all been observed in the absence of added ATP and are interpreted as being independent of this nucleotide. We can dismiss the concern that ATP-dependent motor proteins were confounding our attempts to study polymerization-coupled movements in isolation, because the concentration of contaminating ATP (0.6 ± 1.1 nM; see MATERIALS AND METHODS) was more than an order of magnitude too low to support even the slowest movements seen (Vale and Toyoshima, 1988; Howard et al., 1989). Clearly, the roles of ATP-dependent enzymes, both motor proteins and kinases, are important issues for chromosome–microtubule interactions. They will be the explicit subject of future work.
Several published studies have inferred polymerization- and depolymerization-coupled motility from the average lengths of microtubules in chemically fixed microtubule–chromosome complexes (Mitchison and Kirschner, 1985b; Koshland et al., 1988; Hyman and Mitchison, 1990). In these studies microtubules with a marked segment were bound to isolated chromosomes and then diluted into buffers containing different concentrations of tubulin and nucleotides. At various times after dilution, aliquots of the preparations were fixed, centrifuged onto coverslips, and examined by fluorescence microscopy. Changes over time in the average lengths between the chromosome-bound ends and the marked segments of microtubules were interpreted as the result of depolymerization-coupled motility, a conclusion that is consistent with our study. Our results do not, however, support the conclusions that polymerization-coupled movement requires added ATP (Mitchison and Kirschner, 1985b), and that kinetochores markedly increase the catastrophe rate of attached microtubules (Hyman and Mitchison, 1990). We suggest that these discrepancies might result from the indirectness of the earlier assays and hence their inability to study the dynamic behavior of single microtubules directly. It is problematic to infer changes in the lengths of individual microtubules from measurements of the average length of microtubule populations, because each population will probably contain microtubules that are exhibiting diverse behaviors. For example, some may be growing while others are shrinking at their chromosome-associated ends. Interpretation is further complicated if the microtubule populations vary over time because of microtubules detaching or attaching to the chromosomes (Huitorel and Kirschner, 1988). This is especially likely to be a problem when the tubulin concentration is above ∼1.5 mg/ml; at these concentrations many microtubules will spontaneously form in solution, and if these were to bind to a chromosome and one of the many marked segments in solution, perhaps by lateral association, the marked segment could mistakenly be interpreted as having been continuously tethered to the chromosome. Alternatively, the kinetochores could have nucleated microtubules (Mitchison and Kirschner, 1985a), which could then become associated with a marked segment. Hyman and Mitchison (1990, their Figure 4) observed that the average number of marked microtubules attached to a chromosome increased in the presence of a high concentration of tubulin, supporting these possibilities. Although Mitchison and Kirschner (1985b) found that the addition of ATP had a significant effect on the population distribution of fixed chromosome-bound microtubules, this does not demonstrate that ATP is required for movement away from chromosomes. A number of ATP-dependent processes could have altered the behavior of some population of microtubules and/or shifted the population of microtubules that remain attached to the chromosomes and/or shifted the population of microtubules that exhibited the criteria that were used for selection of the chromosomes that were considered. Examples of such processes include changes in the microtubule detachment rate, the corresponding attachment rate, the speed of microtubule movements toward or away from a chromosome, the catastrophe rate, or the rate of microtubule nucleation at the kinetochores.
By using optical tweezers to tug on beads attached to chromosome-bound microtubules, we have established that during polymerization-coupled movement a kinetochore forms a mechanically strong bond with a microtubule and can bear tensions equivalent to those that could be generated by several motor proteins acting in parallel. Some care must be taken in interpreting these measurements, because it is possible that the force required to detach a microtubule may depend on how the stress is applied. For example, it has previously been observed that tension can stabilize the interaction between a microtubule and chromosome in vivo (Nicklas and Koch, 1969). Because this stabilization may be accomplished by strengthening the bonds between the microtubule and the chromosome, it is plausible that we might observe an increase in the tension required to detach a microtubule if it were first subjected to a preload to stabilize its attachment. It is also possible that dynamic and mechanical features of the bonds between a chromosome and a microtubule might result in directional or temporal variations in the strength of the attachments. We are currently constructing a new optical trapping device that can be manipulated with finer resolution to address these issues in future studies.
A major motivation for our developing this in vitro assay was the possibility of characterizing microtubule–chromosome interactions by simple and direct observation while the conditions of the interaction were subject to experimental control. Isolated from the confounding influences of spindle structure and the many unknown properties of cytoplasm, the movements that we observe reflect only the mechanochemistry of the link between a microtubule and a chromosome. Our results are well described by a model proposed by Hill (1985), in which a microtubule interacts with binding sites on a chromosome that are arranged so an increasing number can bind to the microtubule as its tip moves toward the chromosome (Figure 8A). Following Hill’s description, we refer to this arrangement of binding sites as a “sleeve” into which a microtubule inserts, but note that this geometry is not required by the model. The free energy of the interaction between a microtubule and the sleeve will decrease as more sites bind, an effect that will tend to pull the microtubule into the sleeve. This is appealing because it describes a simple mechanism that a chromosome could use to maintain its association with a depolymerizing microtubule tip, deriving the forces necessary for movement from the energy released by the depolymerizing microtubule.
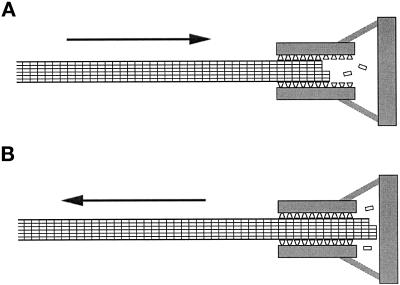
Figure 8.
Schematic representation of the model of Hill (1985) for how chromosome movements might be coupled to microtubule depolymerization (A) and polymerization (B). (A) A shortening microtubule interacts with a series of binding sites (represented as trapezoids) at the kinetochore. The microtubule is drawn toward the chromosome to interact with more binding sites. As tubulin subunits are lost from the tip of the microtubule, new binding sites are exposed, allowing the microtubule to move continuously. (B) A growing microtubule extends past the binding sites and encounters a barrier. The force exerted against this barrier pushes the microtubule away from the chromosome. This movement is not opposed by the force that draws the shortening microtubule in A toward the chromosome, because the number of binding sites that can interact with the growing microtubule in B is unchanged by its movements.
Hill’s model can accommodate our observation that microtubule depolymerization is slowed by association with a chromosome. For a microtubule to undergo sustained movement into the sleeve, tubulin subunits must be lost from its tip. This requires the breaking of bonds not only within the microtubule but also between tubulin and the sleeve. If, alternatively, tubulin subunits in the polymerized and unpolymerized conformations had the same affinity for the binding sites in the sleeve, the unpolymerized tubulin would prevent the microtubule from undergoing directed motion by competing for the same binding sites. The energy requirements for breaking interactions with the sleeve could significantly decrease the off rate for tubulin subunits, as was assumed in the formulation of Hill’s (1985) model.
The model can also be modified to describe how a chromosome might maintain a link with the growing end of a microtubule by adding the assumption that if an elongating microtubule extends far enough through the sleeve toward the chromosome, it will encounter a barrier (Figure 8B). If the polymerizing end of a microtubule exerts force against this barrier (e.g., by inhibiting its thermal motions in one direction), the rest of the microtubule will be pushed back through the sleeve and away from the chromosome. Forces exerted by polymerizing microtubules have been observed in several studies (Miyamoto and Hotani, 1988; Waterman-Storer et al., 1995; Dogterom and Yurke, 1997) and have been the subject of theoretical treatments (Hill, 1981; Peskin et al., 1993). An appealing aspect of this arrangement is that it explains the apparently paradoxical result that the chromosome–microtubule interaction can bear tensions >15 pN yet still allow a microtubule subjected to relatively small forces (i.e., <2 pN observed here or on the order of 4 pN against a glass wall, as observed by Dogterom and Yurke, 1997) to move away from a chromosome during polymerization-coupled movement. There is no free-energy change associated with the movement of a microtubule that has extended past the sleeve of binding sites (Figure 8B), so even a small force will move the microtubule, provided the force is applied for long enough to allow the binding sites to rearrange. However, if a tension is sufficient to pull the tip of the microtubule into the sleeve, the free-energy gradient will be reestablished to oppose movement away from the chromosome (Figure 8A).
A recent study by Lombillo et al. (1995b) has shown that beads coated with kinesin-like proteins can maintain a link with, and thereby follow, the end of a depolymerizing microtubule. This evokes the supposition that a kinesin-like microtubule-binding protein might play a role in the link between chromosomes and microtubules that supports depolymerization-coupled movement (Wordeman and Mitchison, 1995; Walczak et al., 1996). In the context of the model of Hill (1985), the kinesin-like protein might provide the microtubule binding sites. To fulfill this role the kinesin-like protein would need a higher affinity for microtubules than for tubulin.
In future studies, the ability to study polymerization-coupled motility during the interactions of a chromosome with a single microtubule should continue to provide valuable information about the mechanisms underlying this form of motility. We can now make quantitative physical measurements in a controlled chemical environment. This is critical for understanding mitosis, which is as much a mechanical process as it is a biochemical one. The in vitro assay presented here should aid the study of polymerization-coupled motility, much as assays for ATP-driven motility in vitro have shed light on the workings of the motor proteins myosin, dynein, and kinesin.
ACKNOWLEDGMENTS
We thank S.J. King for supplying Chlamydomonas axonemes, P. Grissom for supplying sea urchin axonemes, T.J. Mitchison for GMPCPP, and B.L. Kotzin for 1D12 supernatant. We also thank S. Hird, C. Rogers, E.A. Vaisberg, and C.L. Troxell for their comments on the manuscript. This work was supported by Office of Naval Research grant N00014-94-1-0621, and the National Institutes of Health grant GM33787, and a Burroughs Wellcome career award to A.J.H. J.R.M. is a research professor of the American Cancer Society.
REFERENCES
- Ault JG, DeMarco AJ, Salmon ED, Rieder CL. Studies on the ejection properties of asters: astral microtubule turnover influences the oscillatory behavior and positioning of monooriented chromosomes. J Cell Sci. 1991;99:701–710. doi: 10.1242/jcs.99.4.701. [DOI] [PubMed] [Google Scholar]
- Bergen LG, Borisy GG. A direct method for analyzing the polymerization kinetics at the two ends of a microtubule. Methods Cell Biol. 1982;24:171–187. doi: 10.1016/s0091-679x(08)60654-8. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Cartwright J. Ultrastructural analysis of mitotic spindle elongation in mammalian cells in vitro. J Cell Biol. 1971;50:416–431. doi: 10.1083/jcb.50.2.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande WZ, Wolniak SM. Chromosome movement in lysed mitotic cells is inhibited by vanadate. J Cell Biol. 1978;79:573–580. doi: 10.1083/jcb.79.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplow M, Ruhlen RL, Shanks J. The free energy for hydrolysis of a microtubule-bound nucleotide triphosphate is near zero: all of the free energy for hydrolysis is stored in the microtubule lattice. J Cell Biol. 1994;127:779–788. doi: 10.1083/jcb.127.3.779. (erratum [1995] 129, 549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Salmon ED. Kinetochore microtubules shorten by loss of subunits at the kinetochores of prometaphase chromosomes. J Cell Sci. 1991;98:151–158. doi: 10.1242/jcs.98.2.151. [DOI] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow RA, Colowick SP. Hexokinase from baker’s yeast. Methods Enzymol. 1962;5:226–235. [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometer steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Fygenson DK, Flyvbjerg H, Sneppen K, Libchaber A, Leibler S. Spontaneous nucleation of microtubules. Phys Rev E. 1995;51:5058–5063. doi: 10.1103/physreve.51.5058. [DOI] [PubMed] [Google Scholar]
- Garces E, Cleland WW. Kinetic studies of yeast nucleoside diphosphate kinase. Biochemistry. 1969;8:633–640. doi: 10.1021/bi00830a026. [DOI] [PubMed] [Google Scholar]
- Gelfand VI, Bershadsky AD. Microtubule dynamics: mechanism, regulation, and function. Annu Rev Cell Biol. 1991;7:93–116. doi: 10.1146/annurev.cb.07.110191.000521. [DOI] [PubMed] [Google Scholar]
- Gelles J, Schnapp BJ, Sheetz MP. Tracking kinesin-driven movements with nanometer-scale precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Fronk E. Some properties of bound and soluble dynein from sea urchin sperm flagella. J Cell Biol. 1972;54:365–381. doi: 10.1083/jcb.54.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. Microfilament or microtubule assembly or disassembly against a force. Proc Natl Acad Sci USA. 1981;82:4404–4408. doi: 10.1073/pnas.78.9.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. The movement of kinesin along microtubules. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- Howard J, Hunt AJ, Baek S. Assay of microtubule movement driven by single kinesin molecules. Methods Cell Biol. 1993;39:137–147. doi: 10.1016/s0091-679x(08)60167-3. [DOI] [PubMed] [Google Scholar]
- Huitorel P, Kirschner MW. The polarity and stability of microtubule capture by the kinetochore. J Cell Biol. 1988;106:151–159. doi: 10.1083/jcb.106.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AJ, Gittes F, Howard J. The force exerted by a single kinesin molecule against a viscous load. Biophys J. 1994;67:766–781. doi: 10.1016/S0006-3495(94)80537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Hyman A, Mitchison T. Modulation of microtubule stability by kinetochores in vitro. J Cell Biol. 1990;110:1607–1616. doi: 10.1083/jcb.110.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Mitchison T. Two different microtubule-based motor activities with opposite polarities in kinetochores. Nature. 1991;351:206–211. doi: 10.1038/351206a0. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Mitchison TJ. An assay for the activity of microtubule-based motors on the kinetochores of isolated Chinese hamster ovary chromosomes. Methods Cell Biol. 1993;39:267–277. doi: 10.1016/s0091-679x(08)60176-4. [DOI] [PubMed] [Google Scholar]
- Inoué S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Kotzin BL, Lafferty JA, Portanova JP, Rubin RL, Tan EM. Monoclonal anti-histone autoantibodies derived from murine models of lupus. J Immunol. 1984;133:2554–2559. [PubMed] [Google Scholar]
- Kowalski RJ, Williams RC. Unambiguous classification of microtubule-ends in vitro: dynamic properties of the plus- and minus-ends. Cell Motil Cytoskeleton. 1993;26:282–290. doi: 10.1002/cm.970260403. [DOI] [PubMed] [Google Scholar]
- Lombillo VA. An in vitro study of motility driven by microtubule depolymerization. Doctoral Dissertation. University of Colorado; 1994. [Google Scholar]
- Lombillo VA, Coue M, McIntosh JR. In vitro motility assays using microtubules tethered to Tetrahymena pellicles. Methods Cell Biol. 1993;39:149–165. doi: 10.1016/s0091-679x(08)60168-5. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995a;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995b;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Meyhöfer E, Howard J. The force generated by a single kinsin molecule against an elastic load. Proc Natl Acad Sci USA. 1994;92:574–578. doi: 10.1073/pnas.92.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickey B, Howard J. Rigidity of microtubules is increased by stabilizing agents. J Cell Biol. 1995;130:909–917. doi: 10.1083/jcb.130.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding. J Cell Biol. 1985a;101:755–765. doi: 10.1083/jcb.101.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985b;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Salmon ED. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992;119:569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H, Hotani H. Polymerization of microtubules within liposomes produces morphological change of their shapes. Proc Tanaguchi Int Symp. 1988;14:220–242. [Google Scholar]
- Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DCS. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome micromanipulation III. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odde DJ, Cassimeris L, Buettner HM. Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys J. 1995;69:796–802. doi: 10.1016/S0006-3495(95)79953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the brownian ratchet. Biophys J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. The formation, structure and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LCW, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position in the vertebrate mitotic spindle. J Cell Biol. 1995;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Vale RD, Toyoshima YY. Rotation and translocation of microtubules in vitro induced by dyneins from tetrahymena cilia. Cell. 1988;52:459–469. doi: 10.1016/s0092-8674(88)80038-2. [DOI] [PubMed] [Google Scholar]
- Viola RE, Raushel M, Rendina AR, Cleland WW. Substrate synergism and the kinetic mechanism of yeast hexokinase. Biochemistry. 1982;21:1295–1302. doi: 10.1021/bi00535a029. [DOI] [PubMed] [Google Scholar]
- Voter WA, Erickson HP. The kinetics of microtubule assembly. J Biol Chem. 1984;259:10430–10438. [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MJ, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Adler R. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol. 1995;128:761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio H, Sato H. Differential effects of mitotic poisons on spindles and chromosomes in dividing newt lung epithelia. Cell Struct Funct. 1982;7:263–273. [Google Scholar]
- Waterman-Storer CM, Gregory J, Parsons SF, Salmon ED. Membrane/microtubule tip attachment complexes (TACs) allow the assembly dynamics of plus ends to push and pull membranes into tubulovesicular networks in interphase Xenopus egg extracts. J Cell Biol. 1995;130:1161–1169. doi: 10.1083/jcb.130.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise D, Cassimeris L, Rieder CL, Wadsworth P, Salmon ED. Chromosome fiber dynamics and congression oscillations in metaphase PtK2 cells at 23 degrees celsius. Cell Motil Cytoskeleton. 1991;18:1–12. doi: 10.1002/cm.970180208. [DOI] [PubMed] [Google Scholar]
- Witt PL, Ris H, Borisy GG. Origin of kinetochore microtubules in Chinese hamster ovary cells. Chromosoma. 1980;81:483–505. doi: 10.1007/BF00368158. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel centromer associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]