Abstract
Pak1 (p21-activated kinase-1) and the dynein light chain, LC8, are overexpressed in breast cancer, and their direct interaction has been proposed to regulate tumor cell survival. These effects have been attributed in part to Pak1-mediated phosphorylation of LC8 at serine 88. However, LC8 is homodimeric, which renders Ser88 inaccessible. Moreover, Pak1 does not contain a canonical LC8 binding sequence compared with other characterized LC8 binding sequences. Together, these observations raise the question whether the Pak1/LC8 interaction is distinct (i.e. enabled by a unique interface independent of LC8 dimerization). Herein, we present results from biochemical, NMR, and crystallographic studies that show that Pak1 (residues 212-222) binds to LC8 along the same groove as canonical LC8 interaction partners (e.g. nNOS and BimL). Using LC8 point mutants K36P and T67A, we were able to differentiate Pak1 from canonical LC8 binding sequences and identify a key hydrogen bond network that compensates for the loss of the conserved glutamine in the consensus sequence. We also show that the target binding interface formed through LC8 dimerization is required to bind to Pak1 and precludes phosphorylation of LC8 at Ser88. Consistent with this observation, in vitro phosphorylation assays using activated Pak1 fail to phosphorylate LC8. Although these results define structural details of the Pak1/LC8 interaction and suggest a hierarchy of target binding affinities, they do not support the current model whereby Pak1 binds to and subsequently phosphorylates LC8 to promote anchorage-independent growth. Rather, they suggest that LC8 binding modulates Pak1 activity and/or nuclear localization.
The dynein light chain, LC8 (DYNLL1), is a highly conserved, small homodimeric protein that is associated with the dynein motor complex through its interaction with the dynein intermediate chain (IC)4 (1). In addition to dynein, LC8 binds a diverse set of proteins, including the signaling molecules nNOS and Pak1; the apoptosis regulator Bim/Bmf; the transcription factors TRPS1, NRF1, Swallow, and Ciz1; and viral products, including the rabies phosphoprotein, African swine virus, and herpes simplex virus (2, 3). Due to its association with the dynein complex, LC8 is frequently considered to be an adaptor protein linking these nondynein targets to the motor complex for retrograde transport along microtubules. However, recent structural and thermodynamic studies do not support this mechanism (4). Rather, LC8 may directly regulate functional characteristics of interacting proteins independently of its association with the dynein IC. In support of this contention, LC8 inhibits nitric oxide production when bound to nNOS (5, 6) and reduces the affinity of TRPS1 binding to GATA consensus sequences (7). LC8 is also necessary for early transcription of the rabies genome in N2A cells (8). Additional studies indicate that the interaction of LC8 with the rabies phosphoprotein facilitates its nuclear import (9). However, the molecular mechanisms by which LC8 interaction affects function of these diverse target proteins remain elusive.
Among most of the LC8 targets, two seemingly disparate sequences, GIQVD and KETQT, have been identified as a minimal region required for molecular interaction with LC8 (10). Structural characterization of these peptide-LC8 complexes show that each target peptide binds to a groove formed along the dimeric LC8 interface (4, 11, 12). Binding specificity is apparently achieved through a glutamine residue conserved in LC8-interacting peptides that caps the amide of Lys36 at the beginning of the α2-helix on one side of the groove. In addition, the residues flanking this glutamine interchelate hydrophobic pockets of the opposite LC8 monomer. Thus, the canonical LC8 binding sequence consists of a β-strand and a conserved glutamine flanked by hydrophobic residues (3).
A small number of putative LC8-interacting proteins do not contain the canonical recognition sequence. These include IκBα (13), estrogen receptor α (14), myosin Va (15), and Pak1 (16). Recently, the concurrent overexpression of LC8 and Pak1 as observed in breast cancer was shown to promote the tumorigenic phenotype (16). This earlier report also showed that LC8 bound Pak1 between residues 203 and 273. Further, it was reported that the kinase activity of Pak1 is necessary for matrix-independent cell survival and that Pak1 specifically phosphorylated LC8 at Ser88. Conversely, cells overexpressing an S88A LC8 mutant with active Pak1 failed to grow in soft agar assays. Based on these results, the overexpression of Pak1 and LC8 and the phosphorylation of LC8 by Pak1 have been implicated in cancer development and metastasis.
The reported Pak1 phosphorylation site on LC8, Ser88, is at the dimer interface, and recent studies show that the LC8 phosphomimic, S88E, produces a stable monomer (17, 18). Moreover, the monomeric LC8 cannot bind the dynein IC. The absence of an identifiable LC8 target sequence suggests that Pak1 may bind to a unique surface on LC8 independent of the dimeric state of LC8 and amenable to therapeutic targeting. A unique interface would also be consistent with the hypothesis that LC8 could bridge unique targets to the dynein motor complex for retrograde transport.
To address this possibility, we characterized the LC8-Pak1 interaction biochemically and structurally. We report that, although Pak1 represents a unique LC8 target sequence, it binds to dimeric LC8 in a manner similar to proteins containing canonical interaction sequences. Furthermore, Pak1 binding to the LC8 dimer renders LC8 S88 inaccessible to phosphokinases. Finally, we provide evidence that Pak1 does not phosphorylate LC8. Taken together, our data support a model where the LC8-Pak1 interaction functions in a dynein-independent manner to regulate its function and/or activation.
EXPERIMENTAL PROCEDURES
Materials—Wild type Drosophila LC8 (94% identity and 98% similarity to human LC8) (accession number NP 525075) was subcloned into the NcoI and EcoRI sites of the pet21D vector (Novagen). LC8-K36P was generated using the site-directed mutagenesis method by QuikChange (Stratagene). PCR was initially performed using a vector-specific T7 primer and appropriate reverse mutant primer to generate extended, intermediate mutagenesis primers. The purified, extended primers were then used for the site-directed mutagenesis PCR method by QuikChange. Human LC8 (accession number NP 001032584) and S88D-LC8 were subcloned into the NcoI and EcoRI sites of the pET-15b vector (Novagen). We initially subcloned a Pak1 (accession number NP 002567) peptide, residues 203-270, into the BamHI and XhoI sites of the SMT3 vector. Pak1-(204-226), Pak1-(240-224), Pak1-(204-221), Pak1-(212-226), T212E, T214E, R215A, D216A, A218Q, T219E, S220A, P221A, and I222A were generated using site-directed mutagenesis. Constructs were verified using automated sequencing. Positive clones were transformed into BL21(DE3)* cells for protein expression. The Pak1 peptides, SVI-EPLPVTPTRDVATSPISPTE (residues 204-226) and TPTRD-VATSP (residues 212-221), were synthesized and purified by the Kimmel Cancer Core facility.
Protein Expression and Purification—WT-LC8, K36P-LC8, T67A-LC8, and S88D-LC8 were expressed in Escherichia coli and purified as previously described (4). SMT3-Pak1 fusion proteins were purified using the protocol by Lima et al. (19). Proteins were first isolated using nickel-nitrilotriacetic acid-agarose (Qiagen) and subsequently purified using a Superdex g75 sizing column (GE Healthcare) preequilibrated in Buffer C (50 mm Tris, pH 8.0, 100 mm NaCl, 1 mm EDTA, and 1 mm DTT).
Preparation of Isotopically Enriched LC8 for NMR Experiments—Two isotopically enriched LC8 protein samples were prepared according to the expression protocol (20) and purified as described above. Both samples were dissolved in 500 μl of phosphate NMR buffer (20 mm Na2PO4, pH 6.0, 50 mm NaCl, 1 mm DTT, 3 mm NaN3) containing 10% D2O. The 15N-enriched sample (0.33 mm) was used for recording the 1H-15N HSQC spectra of apo-LC8 and for the LC8-Pak1 titration experiments. The 13C, 15N doubly enriched sample (0.52 mm) was used for triple resonance experiments for establishing the resonance assignments. The Pak1 solution was prepared by dissolving 2.6 mg of peptide in 30 μl of the NMR buffer described above. After adding Pak1 solution to LC8 solution, the solution was brought to a total volume of 500 μl. The molar concentration ratio of Pak1 to LC8 was 0, 0.3, 0.5, 0.7, 1, and 1.3, respectively.
Pull-down Experiments—An N-terminal His-tagged Pak1 (∼75 μm) was incubated with LC8 (2 OD, 150 μm) at room temperature for 15 min. Samples (total volume 50 μl) were loaded on Handee centrifuge columns (Pierce) containing 50 μl of nickel-nitrilotriacetic acid-agarose beads (Qiagen). The flow-through was collected, and samples were washed with 3× 150 μl of 1× PBS with 10 mm imidazole. Samples were eluted with 1 m imidazole and analyzed using SDS-PAGE.
Analytical Size Exclusion Chromatography—WT-LC8, K36P-LC8, and T67A-LC8 were diluted to 37 μm (0.5 OD) in 1× PBS (137 mm NaCl, 10 mm phosphate, 2.7 mm KCl, pH 7.4). Proteins were incubated with ∼40 μm (0.1 mg/ml) dynein intermediate chain fragment (residues 106-240 (4)) for 15 minutes at 37 °C before being loaded onto an analytical Superdex g75 10/300 GL column (GE Healthcare). The column was preequilibrated in Buffer C. Column runs were performed with a flow rate of 0.5 ml/min and monitored at 230 and 280 nm. For LC8 and Pak1 assays, WT-LC8, K36P-LC8, T67A-LC8, or S88D-LC8 were diluted to 75 μm (1 OD) and incubated with ∼500 μm Pak1 for 15 min at 37 °C. Samples were analyzed via SEC as previously described. SDS-PAGE was used to verify protein content in the eluted fractions.
Circular Dichroism—CD Spectra were taken with 1 nm bandwidth over a spectral range of 205-250 nm with a 4-s response time and at 100 nm/min on a Jasco J-810 Spectropolarimeter (Jasco, Inc.). Melting scans were performed at 220 nm over a temperature range of 10-90 °C, collecting data in 0.5 °C increments at 40 °C/h. Total protein concentration was between 5 and 7 μm in 1× PBS, as determined by an absorbance scan at 280 nm.
Analytical Ultracentrifugation—Equilibrium sedimentation analysis was performed at 20 °C with a Beckman XLI Analytical Ultracentrifuge (Beckman Coulter) at three different speeds using an AN-50 rotor. WT-LC8, K36P-LC8, and S88D-LC8 were diluted in 1× PBS and 1 mm tris(2-carboxyethyl)phosphine to an A280 between 0.3 (23 μm) and 1 OD unit (75 μm). The optical density was measured at 230 and 280 nm. The total protein concentration was calculated using molar extinction coefficients of ε = 13,430 m-1 cm-1 at 280 nm and ε = 80,580 m-1 cm-1 at 230 nm. The partial specific volume of 0.7443 for LC8 was calculated as a weight average of amino acid residue values reported by Kharakoz (21). The value for solvent density of PBS was set at ρ = 1.0041 g/ml. Radial scans at 10 and 12 h were performed for each speed to verify that the sample reached equilibration. Data were analyzed using FastFitter and implemented in Igor Pro software (22).
Diffraction Data, Collection, and Processing—Purified WT-LC8 and K36P-LC8 were concentrated to ∼4 mm (∼50 mg/ml) in crystallization buffer (10 mm Tris, pH 8.0, 10 mm NaCl, 1 mm EDTA, 10 mm DTT), flash-frozen, and stored at -80 °C. A fresh aliquot was thawed and used for each crystallization trial. For the Pak1 peptide K36P-LC8 complex, K36P-LC8 (1 mm) was mixed with our Pak1 peptide, residues 212-221 (1.5 mm). Initial crystallization screens of WT-LC8, apo-K36P-LC8, and K36P-LC8/Pak1 peptide were performed using the Matrix Hydra II (Thermo Scientific) and Hampton Research crystallization screens. The apo-LC8 protein screens consisted of a 1:1 ratio of protein to crystallization solution. The K36P-LC8 peptide complex was screened alongside apo-K36P-LC8 using a 2:1 ratio of protein to crystallization solution. Conditions were optimized at 20 °C using the hanging drop/vapor diffusion method with a 1 μl drop volume.
Needles (0.4 × 0.1 × 0.1 mm3) of apo-WT-LC8 grew between 1 and 2 days in 50 mm MgCl2, 100 mm MES, pH 6.0, 32% (w/v) polyethylene glycol 4000, and 20% xylitol as a cryoprotectant. Diffraction data were collected from a single crystal at 100 K at wavelength 0.9793 Å (National Synchrotron Light Source X4C beamline). Data were collected at 1° oscillations on a MAR1800 and processed using HKL2000 (23).
Crystal clusters of apo-K36P-LC8 grew within 2 days in 50 mm CaCl2, 100 mm HEPES, pH 7.0, 28% (w/v) polyethylene glycol Mono Methylether (MME) 2000, 10% glycerol, and 10 mm DTT. Individual crystals (0.3 × 0.2 × 0.1 mm3) were generated via breaking large clusters of thick crystal sheets. Paratone was used to remove excess water from the crystal. Diffraction data were collected at 100 K with 1° oscillations using a Rigaku RU200 at wave-length 1.54 Å and an R-AXIS IV++ imaging plate. Data were indexed using MOSFLM and merged using SCALA (24).
Large crystals (1.0 × 0.8 × 0.7 mm3) of K36P-LC8-Pak1 peptide grew in 3-5 days in 100 mm MES, pH 6.0, 200 mm NaCl, 27% (w/v) polyethylene glycol 4000, and 10% 0.1 m (NH4)6CoCl3 as an additive. Crystals were broken (0.3 × 0.3 × 0.2 mm3), transferred to the same buffer with the addition of glycerol (20%), and directly cooled in the cryostream at 100 K. Diffraction data were collected with 1° oscillations using an XCalibur™ PX Ultra CCD (wavelength 1.54 Å; Oxford Diffraction). Data were processed and merged using the automated data processing system included in the CrysAlisPro software (version 171.32.5; Oxford Diffraction). The data were merged and reduced using SCALA.
Model Building and Refinement—The initial phases for each structure were obtained from molecular replacement using PHASER (25) and Protein Data Bank entry 2PG1 as a search model. The structure was refined by alternating rounds of manual model building in COOT (26) and refinement with REFMAC (27) with TLS. The Pak1 peptide (residues 212-221) was modeled manually using COOT and refined using REFMAC. The structures were refined through multiple iterations of Refmac, COOT, and Molprobity (28).
NMR Spectroscopy—All NMR experiments were performed at 298 K on a 14.1 T Bruker Avance instrument (1H frequency of 600.13 MHz, 15N frequency of 60.81 MHz). 1H-15N HSQC spectra were acquired as 2048 × 512 complex points with 32 acquisitions added up for each Free Induction Decay (FID) (except 4096 × 512 complex points and eight scans for apo-LC8). The States-TPPI method was used for phase-sensitive detection in the indirect dimension. 1H chemical shifts were referenced with respect to 2,2-dimethylsilapentene-5-sulfonic acid (0 ppm).
All spectra were processed in NMRpipe (29). HSQC spectra were processed using the sine bell square apodization function shifted by 30° in the direct dimension and 45° in the indirect dimension, followed by zero filling to twice the original number of points and the Fourier transformation in both dimensions. The acquisition and processing parameters of the three-dimensional HNCA, HNCO, HN(CO)CA, HNCACB, and CBCA(CO)NH experiments are summarized in supplemental Table 1.
The data analysis was performed in Sparky (30). The NMR titration curves were analyzed using the Origin software package (OriginLab Corp., Northampton, MA). The data were fit to several binding/exchange models in order to extract the apparent dissociation constants, as detailed in the supplemental materials.
Kinase Assays—GST-Cdc42 and Pak1 were produced and purified as previously described (31). GST-Cdc42 (100 μm) was incubated with 200 μm GTPγS in Cdc42 buffer (20 mm HEPES, pH 7.5, 50 mm NaCl) for 30 min at room temperature. The reaction was quenched with a 60 mm MgCl2 solution. For the kinase assay, human LC8 (75 μm) was mixed with 50 μm cold ATP, 2% bovine serum albumin, 10 μCi of [32P]ATP, and 2 μm activated Cdc42 in 250 mm HEPES (pH 7.5), 100 mm MgCl2, 20 mm MnCl2, and 5 mm DTT. Purified Pak1 (100 ng) was added last to the mixture to start the reaction. Samples were quenched after 30 min with SDS-sample buffer. Samples were separated on a 15% SDS-polyacrylamide gel, dried, and analyzed by autoradiography.
RESULTS
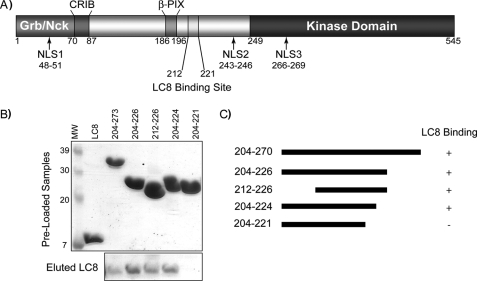
Refinement of LC8 Binding Site on Pak1—The initial reports of the Pak1-LC8 interaction identified a region spanning residues 203-273 (16) and 210-232 (32) of Pak1. Protein prediction algorithms predict that this region of Pak1 is primarily disordered (e.g. no β-strand (33)), whereas canonical LC8 binding sequences are frequently predicted to have β-strand character. To refine the sequence of the LC8 binding site of Pak1, we generated truncation mutants and fused these to the C terminus of a His-tagged SMT3 protein (19). A peptide fragment spanning residues 212-224 of Pak1 was sufficient to account for LC8 interaction (Fig. 1). The interaction of this peptide with LC8 was further confirmed using size exclusion chromatography (Fig. 2).
FIGURE 1.
LC8 binding site on Pak1. A, schematic of the domain structure and nuclear localization sequences of Pak1. The LC8 binding domain, residues 212-222, is C-terminal of the β-PIX binding site and N-terminal to the Pak1 kinase domain. B, SDS-PAGE of nickel pull-down assays with His-tagged SMT3-Pak1 constructs and WT-LC8. The initial components are shown before mixing at 1:1 ratio. WT-LC8 protein eluting with each respective Pak1 construct is shown in the bottom lane. Each experiment was repeated three times. C, graphical representation of nickel pull-down assays. Of note, each experiment was repeated three times, and some variation in intensity of the bound LC8 band was observed. This variation probably stems from the weak affinity of a monovalent interaction and slight differences in the binding affinity for each construct (see “Results & Discussion”). MW, molecular weight.
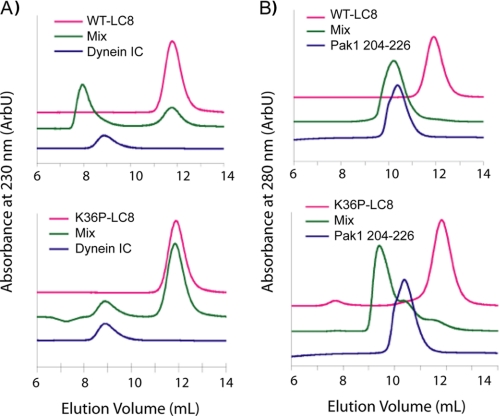
FIGURE 2.
Pak1 is a noncanonical LC8 binding partner. A, analytical SEC assays demonstrating that WT-LC8, but not K36P-LC8, binds the canonical dynein IC. B, analytical SEC assays demonstrating SMT3-Pak1, residues 204-226, bind both WT-LC8 and the noncanonical point mutant K36P-LC8. Note that the SMT3-Pak1/WT-LC8 peak elutes slightly earlier than the SMT3-Pak1 trial but nearly at the same volume fraction as the individual SMT3-Pak1 construct. However, WT-LC8 is absent at elution volumes consistent with the individual protein and co-migrates with the SMT3-Pak1 fusion as confirmed by SDS-PAGE. Also note that a much more dramatic shift is observed for the SMT3-Pak1/K36P-LC8 combination. Based on the structural data presented below, we suggest there is a slight difference in the peptide conformation due to the point mutant (e.g. the side chain of Ser220 may not form a backbone hydrogen bond the amide backbone of Lys36 in wild type).
Pak1 Represents a Novel LC8 Binding Sequence—To test whether Pak1 represents a novel LC8 binding sequence, we generated a point mutation in LC8, K36P, which would cap the α2-helix and sterically occlude the conserved glutamine side chain. The K36P mutant LC8 protein is a homodimer as determined by SEC and sedimentation equilibrium studies (Fig. 2 and data not shown). Since the introduction of a proline into the protein could destabilize the fold, we measured the thermal denaturation point of the K36P point mutant and wild type LC8 by circular dichroism. The melting point of the K36P mutant, Tm = 74 ± 0.4 °C, is comparable with wild type, Tm = 76 ± 0.5 °C) (supplemental Fig. 1). Neither protein reversibly folds to a native state after thermal denaturation.
To test whether the K36P point mutation blocks interaction with proteins containing a canonical LC8 binding sequence, we assessed the binding to a recombinant dimeric dynein IC that contains a canonical LC8 site (4). As determined by native gel electrophoresis and SEC, the K36P mutation abrogates LC8-IC complex formation (Fig. 2A). Next, we mixed the SMT3-Pak1-(204-226) construct with the K36P mutant and WT-LC8 and showed that the Pak1 peptide bound to both forms of LC8 (Fig. 2B). This suggests that the Pak1 peptide interacts with LC8 in a previously unrecognized way.
Mutational Analysis of the LC8 Binding Site Confirms Unique Sequence—In order to identify critical residues in Pak1 necessary for the Pak1-LC8 interaction, we mutated a number of residues in the minimal sequence described above and tested their ability to bind LC8 using nickel pull-down assays and analytical SEC. In Fig. 3, each trace (LC8, Pak1 mutant, and the combination) was normalized to 1 to show the shift and/or absence of the apo forms. Also, the molar extinction coefficients of the Pak1 and LC8 constructs differ significantly (e.g. the Pak1 constructs do not contain a tryptophan). For an admixture of wild type SMT3-Pak1 and LC8, we observe the absence of the LC8 peak eluting at 11.9 ml (Fig. 3A). SDS-PAGE of each peak shows that LC8 shifted to an earlier elution volume of 10.1 ml and coincides with the Pak1 peak. Please note that the SMT3-Pak1 construct has a mass of 15.3 kDa and elutes well before LC8 with a molecular mass of 20.6 kDa. We attribute this anomalous hydrodynamic behavior to the combination of high content of prolines in the Pak1 peptide and the SMT3 fusion (SMT3 without the C-terminal Pak1 fusion has a molecular mass of 16.5 kDa, elutes at 11.6 ml, and is monomeric by sedimentation equilibrium experiments; data not shown). Thus, qualitatively we judge binding based on the absence of a peak eluting at the LC8 and the presence of LC8 (by SDS-PAGE) at earlier elution volumes.
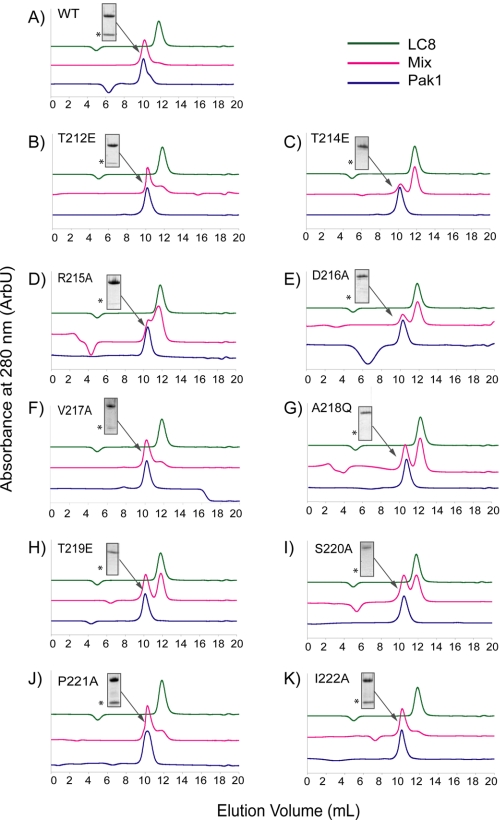
FIGURE 3.
Mutational analysis of Pak1 binding sequence. Analytical SEC of WT-LC8 (A) and individual point mutants (B-K) in SMT3-Pak1-(204-226) was used to qualitatively assess the role of each residue in the LC8-binding region of Pak1. SDS-PAGE of fractions eluting between 10.1 and 10.5 ml is shown for each experiment. The expected position of LC8 through SDS-PAGE is indicated with an asterisk. Each experiment was performed three times.
We first mutated the central segment of the LC8 binding site on Pak1, TPTRDVATSPI, since it partially resembles the canonical site KETQT. Specifically, we mutated Ala218 to a glutamine to test whether the point mutant would bind to WT-LC8 but abrogate the interaction with the K36P mutant. Both nickel pull-down and SEC assays using WT-LC8 show that the A218Q mutant blocks the interaction with WT-LC8 (Fig. 3G). Mutation of Arg215 to alanine reduced the affinity for LC8, whereas the mutation of Asp216 and Ser220 failed to bind at a detectable level.
In addition, Thr212 of Pak1 is phosphorylated in vivo by Cdk5 (34), Erk2 (35), and Cdc2 (36), and phosphorylation at this site abrogates some Pak1 interactions (36). Therefore, we generated the phosphomimic, T212E, to see if this was consistent with the Pak1-LC8 interaction. Both nickel chelation pull-down assays and SEC showed that the phosphomimic reduces but does not fully abrogate the interaction relative to the nonmutated peptide (Fig. 3B). In contrast, both T214E and T219E point mutants blocked the interaction. Finally, the mutation of Val217, Pro221, or Ile222 to alanine did not strongly affect the interaction. These results further confirm that the Pak1 sequence is distinct from canonical targets and suggest that phosphorylation of Thr212 does not fully abrogate LC8 binding of the peptide.
Pak1-LC8 Interaction Requires Dimeric LC8—Recently, Pak1 has been implicated not only in binding LC8 but also in phosphorylating LC8 at Ser88. Since Ser88 is located at the dimer interface, we and others (17) suspect that phosphorylation of this residue would prevent the dimerization of LC8. We generated the phosphomimic S88D in LC8 and used SEC, analytical ultracentrifugation, CD, and NMR to show that this phosphomimic produces a stable, folded monomer but does not permit dimerization (Fig. 4 and supplemental Fig. 2). To test whether Pak1 can bind to the monomeric LC8, we mixed the SMT-Pak1 peptide fusion with the phosphomimic and used native gel electrophoresis, nickel pull-down assays, and SEC. No interaction between monomeric LC8 and Pak1 was observed despite multiple attempts at different protein concentrations and buffer conditions (Fig. 4D). Similarly, the dimeric dynein IC fragment also fails to bind the phosphomimic S88D-LC8, consistent with recently published results (17).
FIGURE 4.
Pak1-LC8 interaction requires dimeric LC8. Multiple sedimentation equilibrium experiments were performed on WT-LC8 and S88D. A representative series of radial concentration profiles (points) at three different speeds (40,000, 45,000, and 50,000 rpm), together with fits (lines) and residuals (top axes), are shown for WT-LC8 measured at 230 nm (wavelength chosen for maximum sensitivity) (A) and S88D measured at 280 nm (B). C, reduced chi-squared, χ 2r, is shown as a function of the difference between a fixed dissociation constant (pK fixD) and that of the best fit, pK BestD. Here, χ 2r is the sum of squared deviations between measured and fit-calculated values divided by the fit degrees of freedom, and the dissociation constant, pKD, is expressed as the negative log of the dimerization constant. The nearly symmetric minimum seen for the S88D indicates a well defined best fit value for the pKD. By contrast, the plateau seen for WT-LC8 at higher pKD values indicates a pKD well bounded only at lower affinities. Thus, we can only place a upper limit on the KD of 500 nm. D, S88D (top trace; pink) elutes later than WT-LC8, in agreement with the centrifugation experiments (elution volume, 13.2 ml). The mixture of S88D and the Pak1 peptide fragment (middle trace; green) did not produce a shift or reduce concentration of either component at their individual elution volumes (as judged by the absorbance). Analysis of the 10.6-ml fraction by SDS-PAGE did not indicate the presence of S88D.
LC8 Is Predominantly Dimeric at Low Concentrations—Pak1-dependent phosphorylation of Ser88 of LC8 requires access to the hydroxyl side chain (37). We used the Pak1 kinase structure and superposed other ligand-bound kinase structures to artificially dock LC8 on Pak1 and noted that both the dimeric and monomeric forms of LC8 produced severe steric clashes with the kinase domain (see supplemental Fig. 3). This crude modeling suggests that the β5-strand that contains Ser88 in LC8 must have access to the solvent (e.g. partially unfold) to be phosphorylated. Minimally, this would require a monomeric state of LC8.
The dimerization constant of LC8 has been reported to be relatively weak (KD = 12 μm) (38). However, we observed that LC8 is invariably dimeric in SEC experiments even at concentrations lower than the KD. To resolve this difference, we reevaluated the dimerization constant of LC8 by analytical ultracentrifugation. Sedimentation equilibrium data at multiple speeds and concentrations at 20 °C were collected for WT-LC8 (Fig. 4A) and analyzed using a monomer-dimer equilibrium model (22). Sequencing and mass spectrometry confirmed that the molecular weight of the LC8 construct is the same as the calculated value. The base-line and dimer dissociation constant were the only global parameters allowed to vary in the analysis. Interestingly, values for the dissociation constant (KD) of >1 μm produced poor fits to the data, whereas values at higher affinity than the best fit, KD = 160 nm, produced reasonable fits with only a slight increase in the χ2 values (Fig. 4C). Consequently, our data place a lower limit on the dimer dissociation constant of the wild type species, KD < 500 nm. This limitation is due simply to the detection limit of the instrument and the molar extinction coefficient of the sample. In contrast, similar χ2 analysis shows that the dimer dissociation constant of the S88D mutant, KD = 512 μm ± 10%, is bounded and thus well defined (Fig. 4C). Taken together, these data indicate a substantially greater pool of dimeric LC8 in the cell than previously implied (see supplemental materials).
Structural Analysis of the Pak1-LC8 Interaction—We used diffraction and NMR methods to obtain structural data of the Pak1 peptide bound to LC8. Based on the mutagenesis and binding studies, the Pak1 peptide synthesized for the diffraction studies spanned residues 212-221. We confirmed that this fragment bound to the LC8-K36P mutant (see supplemental Fig. 4). Crystals of the K36P-LC8 and this Pak1 peptide were obtained and diffracted with Bragg spacings that extend beyond 2.5 Å. The unit cell constants were 47.40, 57.38, and 64.80 Å in the P212121 space group (Table 1). We generated a monomeric LC8 search model (Protein Data Bank entry 2PG1) (4) for molecular replacement and obtained a satisfactory solution with two monomers in the asymmetric unit. The initial electron density maps clearly indicated that the Pak1 peptide bound to LC8 along the same groove as other canonical targets (Fig. 5B). The Pak1 peptide was built into the electron density using the dynein IC peptide as a template, resulting in a 2:2 Pak1 peptide/LC8monomer ratio. We noted a drop in the R and Rfree of ∼5% upon the addition of the peptide in refinement of the model.
TABLE 1.
Diffraction data and refinement statistics
| WT-LC8 | K36P-LC8 | K36P-LC8-Pak1 peptide | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 | C2 | P212121 |
| Unit cell (Å) | 36.48, 44.87, 84.83, 79.62, 77.54, 88.03 | 163.03, 37.95, 44.87, β = 100.92 | 47.40, 57.38, 64.80 |
| Bragg spacings (Å) | 30.0-2.3 | 28.9-2.0 | 11.65-2.5 |
| Wavelength (Å) | 0.9793 | 1.54 | 1.54 |
| Rmerge (last shell) (%) | 0.078 (22.8) | 0.077 (30.4) | 0.069 (19.8) |
| I/σ(I) (last shell) (%) | 10.7 (3.7) | 5.8 (2.2) | 9.8 (3.5) |
| Reflections | |||
| Measured | 42,597 | 55,345 | 39,783 |
| Unique | 22,187 | 18,462 | 6394 |
| Completeness (%) | 97.6 | 100 | 91.6 |
| Refinement | |||
| Resolution (Å) | 28.7-2.3 | 27.2-2.0 | 11.65-2.5 |
| Reflections (% complete) | 21,034 (95.2) | 17,514 (99.5) | 6095 (91.6) |
| R/Rfree (%) | 19.3/25.5 | 19.3/23.1 | 20.8/25.3 |
| No. of atoms protein/water | 4265/159 | 1078/105 | 1516/62 |
| r.m.s. deviations length (Å)/angle (degrees) | 0.013/1.415 | 0.022/1.68 | 0.028/2.162 |
| Ramachandran angles favored/outliersa (%) | 95.4/1.4 | 96.4/0.9 | 93.3/1.1 |
The ϕ/ψ angles of Gln51 are 70 ± 10°/160 ± 10° in each structure, in good agreement with LC8 models deposited in the Protein Data Bank.
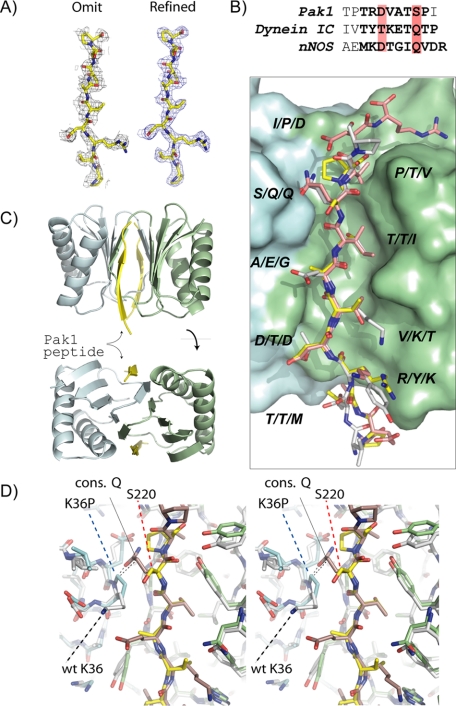
FIGURE 5.
Structural analysis of the Pak1-LC8 interaction. A, the initial maps provided clear evidence that the peptide was present in these crystals. Shown is an omitted map of the peptide after randomizing the atomic positions of final model by 0.1 Å and refinement of the model with REFMAC in the absence of the peptide. The second map shows the electron density of the peptide after the final refinement. B, superposition and structure-based sequence alignment of the Pak1 (yellow carbon), dynein IC (white), and nNOS (pink) peptides indicate that these peptides bind to the same groove on LC8. Note that the binding groove is at the dimeric interface (cyan and green surfaces indicate the individual monomers comprising the structure). A structure-based alignment of LC8 binding sequences is shown above. C, ribbon diagram of the Pak1-LC8 model. Note two Pak1 peptides (yellow) bind along the LC8 dimeric interface. D, stereoview of Pak1 (yellow) and the IC (pink) peptide near the N terminus of the α2-helix of LC8. Also shown is the stick figure of K36P (cyan for one monomer and green for the other) and WT-LC8 (white). Note that the hydroxyl group of Ser220 is pointing away from the proline ring and that the side chain of the conserved glutamine (cons. Q) of the IC is sterically blocked by the proline ring.
Superposition of the Pak1-LC8 complex with the dynein IC-LC8 and nNOS-LC8 structures shows the same pattern of backbone hydrogen bonds to the swapped β4-strand and similar side chain rotamers with the exception of Ser220 and Pro221 (Fig. 5B). Side chains of residues Arg215, Asp216, and Val217 of Pak1 match closely the nNOS residues Lys234, Asp235, and Thr236, and the side chain of Thr219 is nearly identical to Thr134 of the dynein IC, including the formation of a hydrogen bond to the side chain of Ser64 in LC8. The hydroxyl group of Ser220 is slightly skewered from the α2-helix, probably due to the substitution of proline at Lys36. Finally, Pro221 occupies a unique position compared with the other structures. This difference probably stems from the truncation of peptide at this position. Overall, the structure of the complex is consistent with the mutagenesis data and with the hypothesis that only dimeric LC8 can bind Pak1.
In addition, we collected and solved the structures of WT-LC8 and K36P-LC8 by molecular replacement (Table 1). The WT-LC8 crystal diffracted beyond 2.2 Å and belongs to the P1 space group (verified by Xtriage (39)). The K36P-LC8 crystal diffracted beyond 2.0 Å and belongs to the C2 space group. The resulting models showed little or no significant perturbation in overall tertiary structure of K36P-LC8 compared with WT-LC8 (supplemental Figs. 1G and 5A). The mean deviation between any monomer within WT-LC8 asymmetric unit or compared with any monomer in the K36P-LC8 mutant was less than 0.43 Å2 (85 calcium atoms). Likewise, the mean deviation was less than 0.53 Å2 between the apo and Pak1-LC8 complex. Overall, there are no significant perturbations in stereochemical values. The diffraction and refinement values are reported in Table 1.
NMR Data and Analysis—Since we could not obtain crystals of the WT-LC8 and Pak1 peptide, we collected HSQC NMR spectra of 15N-labeled WT-LC8 to confirm the diffraction data using the K36P-LC8. The spectrum of the apo-WT-LC8 was well dispersed, and a number of peaks could be correlated with published results. Since several resonances could not be correlated to the published spectra, we collected and assigned the heteronuclear three-dimensional correlation spectra using 13C/15N-labeled sample and following the standard protocols (see supplemental materials). Next, we titrated the 15N-labeled WT-LC8 with an unlabeled Pak1 peptide, spanning residues 204-226. A handful of resonances shifted as the peptide concentration increased, indicating fast exchange. These include Glu35, Lys36, Asp37, Ile38, Arg60, Asn61, Phe62, Gly63, Ser64, Thr70, Tyr75, Phe76, Tyr77, Ile83, Leu84, and Gly89. The residues map to the same site region as the Pak1 peptide and are similar to those found in other studies using the Bim (40) or nNOS peptides (41).
In addition, closer examination of these perturbed resonances as a function of increasing concentrations of the Pak1 peptide indicated perturbations of two types, following either fast or slow exchange behavior. The residues undergoing fast exchange on the chemical shift scale are Glu35, Arg60, Asn61, and Phe62; their resonances undergo monotonic shift upon Pak1 titration (Fig. 6 and supplemental Fig. 5). The residues undergoing slow exchange are Ala6, Asn10, Met17, Asn33, Lys36, Asp47, Tyr75, and Leu85 and exhibit two distinct chemical shifts corresponding to the free and bound LC8 in the presence of Pak1. The peak intensities of the free form decrease, whereas those of the Pak1-bound form increase upon the addition of Pak1. Residues Asp37 and Ser64 exhibit more complex exchange behavior, not consistent with either fast or slow exchange in a single binding site. These residues exhibit two distinct peaks corresponding to the free and the peptide-bound protein; both peaks shift, and their intensities change upon the addition of the varying Pak1 concentrations. These results suggest that peptide binding potentially induces conformational change in the peptide and LC8. Additional studies are needed to address potential cooperativity.
FIGURE 6.
NMR titration of WT-LC8 with unlabeled Pak1 peptide. A, 15N-labeled WT-LC8 was titrated with a synthetic Pak1 peptide, spanning residues 204-226. The 15N/1H HSQC spectra are coded for the LC8/Pak1 molar ratio as red for apo-LC8, orange for 1:0.3, yellow for 1:0.5, green for 1:0.7, blue for 1:1, and purple for 1:1.3. Select resonances of residues that line the Pak1 binding site are highlighted. Two resonances that are strongly affected in the titration experiment, also highlighted, could not be assigned. Asn10, Asn51, and Gln80 are near the peptide binding groove, and their side chain amides may correspond to these resonances. B, the residues that shifted significantly in the titration experiments are colored purple in the Pak1-LC8 model. Residues shown undergo fast exchange. Many of these shifts correspond to similar HSQC experiments with canonical peptides. C and D. Titration curves of fast exchange residues Glu35 and Asn61 plotted as a change in chemical shift (ppm) with respect to an increase in Pak1 levels.
Finally, we used the NMR titration data to estimate the binding constant of the Pak1 peptide to LC8. To extract the apparent dissociation constants, we fitted the titration curves to two binding models (one- and two-site binding), following the standard procedures for the residues undergoing fast and slow exchange (supplemental Figs. 5 and 6). These apparent dissociation constants are on the order of 10-8 to 10-7 m2, assuming a two-site binding (both fast and slow exchange residues), and on the order of 10-5 to 10-4 m, assuming a single binding site model. These data are a rough estimate, since we did not reach full saturation (partially due to limiting amounts of the peptide).
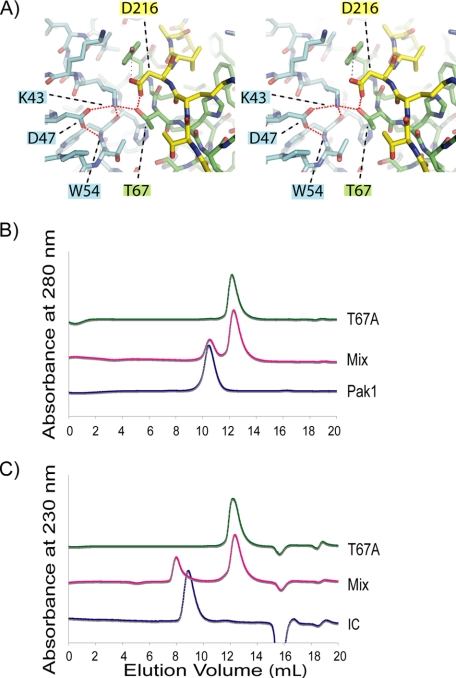
Thr67 in LC8 Selects for Asp in Target Sequence—Based on the similarity of the Pak1-LC8 interface to the canonical target-LC8 interface, we sought to identify residues in LC8 that compensate for the lack of the conserved glutamine in the Pak1 peptide. The crystal structure of Pak1 and LC8 indicates that Asp216 in the Pak1 peptide makes a short hydrogen bond to the completely conserved Thr67 (2.4 Å) (Fig. 7) and subsequently mutated Thr67 in LC8 to alanine. We show by SEC, CD, and analytical ultracentrifugation that the purified protein, T67A-LC8, is folded and dimeric (supplemental Fig. 1 and Fig. 7B). Under the same conditions used to show that the Pak1 peptide binds to WT-LC8 and K36P-LC8, we show by SEC that the Pak1 peptide fails to bind the T67A mutant. Moreover, we note that the equivalent residue to Asp216 in the dynein intermediate chain is a threonine (Thr131; rat IC2C numbering) and does not make a significant hydrogen bond (based on Protein Data Bank entry 2PG1 or 2P2T). Since the dynein intermediate chain encodes the conserved glutamine, we hypothesized that the T67A mutant would bind to the dynein intermediate chain. This was confirmed by SEC (Fig. 7B).
FIGURE 7.
Role of Thr67 on LC8-Pak1 interaction. A, stereoview of the LC8-Pak1 hydrogen bond network. A conserved hydrogen bond network in LC8 is formed through the side chains of Lys43, Asp47, and Thr67′ and the backbone of Trp54 in LC8 and interacts with Pak1 through the side chain of Asp216 through a short hydrogen bond (dotted red lines). B and C, analysis of T67A mutant by analytical SEC. Analytical SEC assays demonstrate that mutation of LC8-Thr67 to alanine abrogates the Pak1-LC8 interaction. The dynein intermediate chain, on the other hand, encodes threonine at the same position (Asp216 in Pak1) and does not form a significant hydrogen bond (distance is 5.54 Å based on Protein Data Bank entry 2PG1). Analytical SEC assays show that the dynein IC binds to the mutant, T67A-LC8.
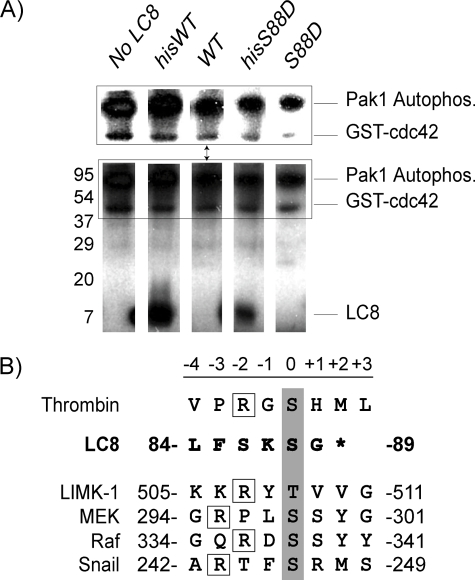
Pak1 Does Not Phosphorylate LC8 in Vitro—Our structural and sedimentation equilibrium studies also suggest that Ser88, which is at the dimer interface, is not readily accessible to the kinase active site of Pak1. Moreover, the interaction with Pak1 would necessarily reduce the exposure of Ser88 to the kinase active site. Since these observations are in contrast to recently published phosphorylation data, we tested whether purified Pak1, activated by GTP-loaded Cdc42, could phosphorylate LC8 in vitro. In these assays, we observe a 32P signal for Pak1 and the GST-Cdc42 but no signal at or near the expected weight for LC8 (Fig. 8A). Although the former observation is most likely due to the autophosphorylation of Thr423 in the kinase activation loop and was expected, we did not expect a signal from the GST-Cdc42 or the absence of a signal from LC8. To resolve this discrepancy, we scanned the GST-Cdc42 protein sequence for a Pak1 phosphorylation consensus sequence. Surprisingly, we observed that the thrombin site associated with the GST fusion could be a potential Pak1 phosphorylation site (Fig. 8B). To test this, we used an N-terminal His-tagged LC8 construct that contains an intervening thrombin site. Similar kinase assays using a His-tagged LC8 protein as well as the same protein after being completely cleaved with thrombin (as judged by SDS-PAGE). We observed phosphorylation of the His-tagged LC8 (as well as Pak1 and GST-Cdc42) but not of the cleaved LC8. Moreover, we repeated these experiments on the LC8 S88D mutant that must prevent phosphorylation at Ser88. We obtained similar results; the His-tagged S88D LC8 was phosphorylated but not the thrombin-cleaved LC8. These experiments were repeated three times.
FIGURE 8.
Pak1 does not phosphorylate LC8 in vitro. A, in vitro Pak1 kinase assay. Activated Pak1 kinase assays were performed using [32P]ATP, separated by SDS-PAGE, and detected by autoradiography. Individual lanes are shown for each reaction. Pak1 phosphorylates His-tagged WT-LC8 and the phosphomimic His-tagged LC8-S88D. These proteins contain a thrombin cleavage site C-terminal to the His tag and N-terminal to LC8. The same LC8 proteins in which the His tag was removed are not phosphorylated by activated Pak1. The inset above shows phosphorylation of Pak1 and GST-Cdc42 at different contrast settings. GST-Cdc42, which also contains a thrombin site C-terminal to the GST tag, is also phosphorylated in these assays. These experiments were repeated three times. B, the thrombin cleavage site contains a Pak1 phosphorylation consensus sequence. Residues that are phosphorylated by Pak1 are highlighted in gray. Boxed are the upstream arginine residues important for Pak1 target phosphorylation. As shown, LC8 does not contain the Pak1 consensus phosphorylation sequence.
DISCUSSION
The results presented here address two separate but related issues. First, this study presents the first structural characterization of a noncanonical LC8 binding peptide. The structure shows that the Pak1-LC8 interaction is similar to other LC8 binding proteins but, together with our point mutants, establishes a hierarchy of LC8 targets based on binding affinities. Second, the interaction between Pak1 and the LC8 dimer precludes phosphorylation of Ser88 at the dimeric interface, and furthermore, our in vitro data indicate that LC8 is not phosphorylated by Pak1.
On the first point, previous biochemical and structural efforts revealed that all LC8 interaction partners encode a conserved glutamine flanked by hydrophobic residues, thus suggesting a “canonical” sequence for LC8 targets (Fig. 5) (3, 40). The only exception being myosin Va (discussed below). The fact that the reported region of Pak1 did not contain a glutamine suggested that the LC8-Pak1 interface could be different from the canonical LC8 peptide-LC8 interface. Moreover, a tripartite complex between BimL, LC8, and Pak1 using a GST pull-down assay was also reported (16). Thus, we generated and demonstrated that the point mutant LC8-K36P blocks canonical targets (dynein IC) but retained its ability to bind Pak1. The structural data presented here, however, show in fact that the Pak1 peptide binds to the same groove as canonical targets. The backbone of Ser220 of Pak1 occupies the equivalent position as the conserved glutamine. The hydroxyl group of serine is 4.2 Å from the amide backbone of K36P, but simple modeling suggests that it could also cap the α2-helix with minor rearrangement of the peptide backbone. Mutation of Ser220 to alanine in the Pak1 peptide and its inability to bind to LC8 provides indirect support of this model.
To compensate for the loss of the conserved glutamine, we identified a completely conserved hydrogen bond network in the structure centered at about Thr67 in LC8 that makes a short hydrogen bond to Asp216 of the Pak1 peptide. Complementary mutations, Thr67 to alanine in LC8 or Asp216 to alanine in Pak1, disrupt the interaction. Likewise, we note that the dynein IC does not encode an asparate at the equivalent site in Pak1 and was not affected by the T67A mutation in LC8. We also note that the binding affinity, roughly determined from the NMR titration, indicates that the Pak1 peptide has a weaker affinity for LC8 than peptides containing the glutamine, ∼10 μm for nNOS-LC8 compared with ∼100 μm for Pak1-LC8 (4).
These new data corroborate a previous study that identified a preference for an aspartate in LC8 binding sequences at residue i - 4 (3). Moreover, they are consistent with recent binding and NMR studies of a fragment from myosin Va isoform that binds to LC8 (42). Similar to Pak1, the myosin Va peptide (residues 1284DDKNTMTD1291) does not contain a canonical glutamine but rather a methionine. It also encodes an aspartate at the i - 4 position. Although the structure of the myosin Va peptide bound to LC8 was not determined, similar chemical shifts in the LC8 backbone were observed upon titration of the myosin Va peptide as the Pak1 peptide. In addition, it was shown using the pepscan method that substitution of Asp1285 in the myosin Va peptide (e.g. the i - 4 position) to any other residue significantly decreased LC8 binding (3).
Taken together, our findings and previous studies suggest a hierarchy in the binding affinity of LC8 targets to LC8. Specifically, we suggest that targets that contain both glutamine (position 0) and asparate (position i - 4) have the highest affinity for LC8 (e.g. nNOS). These are followed by LC8 targets encoding a glutamine but no aspartate at i - 4 (e.g. dynein intermediate chain and Bim). Finally, targets with an aspartate at the i - 4 position but no glutamine at the i = 0 position would be the weakest (e.g. Pak1 and myosin Va).
It is important to note, however, that the overall affinity of a single peptide to a half-site on LC8 from any of these targets is very weak (∼10-100 μm based on NMR data and unpublished results).5 Thus, to enable this interaction, a protein would have to be present at very high concentrations not readily achieved within a cell (43). LC8 itself, as well as all LC8 interaction partners characterized thus far, are homodimeric, consistent with the formation of a molecular complex consisting of a dimer of dimers. This type of interaction gives rise to energy additivity and produces a higher apparent affinity (44). Moreover, structural analysis of LC8 target peptides bound to LC8 indicates that they bind to each half-site in a parallel manner, further enhancing specificity due to geometric constraints. The role of geometric constraints and bivalent interactions as it relates to LC8 binding specificity is supported by our recent structural and thermodynamic data of a complex of LC8 and TcTex1 bound to a fragment of the dynein IC (4). Of direct relevance to these considerations, Pak1 has been demonstrated to be dimeric in the inactive state (45) and potentially in the active state (46).
On the second point, these results raise questions about the role of Pak1-mediated Ser88 phosphorylation of LC8 and the relevance of this phenomenon to the malignant phenotype (16). Based on simple modeling, we observed severe steric clashes between LC8 and the Pak1 kinase domain, even when using a monomeric model of LC8 (supplemental Fig. 3). The structure of the complex (Fig. 5) clearly demonstrates that the Pak1/LC8 interaction further precludes accessibility of Ser88. Moreover, the sedimentation equilibrium data of LC8 (Fig. 4) suggest that the dissociation constant of dimeric LC8 is significantly lower than previously reported and is consistent with a largely dimeric pool of LC8 in cells in vivo.
In addition, we were able to activate Pak1 using GTP-loaded Cdc42 but unable to detect any phosphorylation of LC8 in in vitro studies using autoradiography (Fig. 8A). Consistent with this finding, Pak1 substrates contain an arginine at the -3- and/or -2-positions of the serine phospho-acceptor and a hydrophobic group (e.g. Trp, Tyr, and Phe) at the +1-, +2-, and +3-positions (47). However, the flanking sites of LC8 S88 are Leu85 (-3), Phe86 (-2), and Gly89 (+1), and the sequence terminates after Gly89 (Fig. 8B). Our observation that the thrombin cleavage site contains elements of the consensus sequence and is phosphorylated by Pak1 suggests the possibility that the initial report of LC8 phosphorylation by Pak1 is due to an unintended introduction of a consensus sequence. Alternatively, it is possible that one or more intermediary factors are necessary to account for the mechanism of Pak1-mediated, posttranslational modification of LC8 in cells.
Although it is clear that phosphorylation of Ser88 will affect LC8 binding to its targets, including Pak1, the functional implications of the Pak1-LC8 interaction remain poorly defined. The LC8 binding site is unique to Pak1 and not found in other members of the Pak family. It is located in an intrinsically disordered region of Pak1 and lies between the kinase domain and its N-terminal regulatory domain. Such disordered regions have recently been identified as critical regulatory sites (37, 48, 49), consistent with LC8-mediated modulation of TRPS1 (7) and nNOS (5) activities. The Pak1-LC8 complex implies a spatial constraint that could manifest itself in a number of scenarios, including the regulation of kinase activity (e.g. orienting the kinase domains for trans-autophosphorylation), the regulation of binding targets (e.g. either facilitating or hindering activation by Cdc42 or β-PIX/Cool1), and/or the regulation of post-translational modifications (e.g. occlusion of Thr212 to block phosphorylation by Erk2, Cdk5, or Cdc2 kinases). Alternatively, LC8 was recently shown to facilitate nuclear import of the rabies phosphoprotein and may similarly affect intracellular trafficking of Pak1 (9). Of note, Pak1 contains three nuclear localization signals (Fig. 1A), of which one (residues 243-246) appears to be required for nuclear import (50). Similar to the rabies phosphoprotein, this second site is located immediately to the C-terminal region of the LC8 binding site. Thus, by analogy, the Pak1-LC8 interaction may facilitate Pak1 nuclear import. Studies are under way to address these possibilities.
CONCLUSIONS
The detailed biochemical and structural characterization of the Pak1-LC8 interaction reveals that Pak1 binds LC8 through the same interface as other LC8 targets and further expands the repertoire of its protein interaction partners. In addition, the results highlight the importance of LC8 dimerization for Pak1 interaction to occur, precluding access of the Pak1 kinase to Ser88 of LC8. Finally, because Pak1 shares the same binding site with all known LC8 interaction partners, a small molecule antagonist to disrupt this molecular interface may have limited specificity in therapeutic applications.
Supplementary Material
Acknowledgments
We thank Dr. Jonathan Chernoff (Fox Chase Cancer Center) for supplying the human Pak1 and Cdc42 cDNA and full-length, active Pak1 protein. We thank Christopher Lima (Memorial Sloan Kettering Cancer Center) for the SMT3 expression system. We thank present and former members of the Williams laboratory for general support and advice. We also acknowledge insightful discussions with Drs. John Pascal, Michael Root, and Charles Scott (Thomas Jefferson University) and Dr. Chernoff (Fox Chase Cancer Center).
The atomic coordinates and structure factors (code 3DVP, 3DVH, and 3DVT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health (National Center for Research Resources) Grant S10-RR022316 (to J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1-3 and Figs. 1-6.
Footnotes
The abbreviations used are: IC, intermediate chain; WT-LC8, wild type LC8; SEC, size exclusion chromatography; HSQC, heteronuclear single quantum correlation; His tag, polyhistidine purification sequence; DTT, dithiothreitol; PBS, phosphate-buffered saline; MES, 4-morpholineethanesulfonic acid; GTPγS, guanosine 5′-3-O-(thio)triphosphate.
C. M. Lightcap, A. Dawn, and J. C. Williams, unpublished data.
References
- 1.King, S. M. (2000) Biochim. Biophys. Acta 1496 60-75 [DOI] [PubMed] [Google Scholar]
- 2.Vallee, R. B., Williams, J. C., Varma, D., and Barnhart, L. E. (2004) J. Neurobiol. 58 189-200 [DOI] [PubMed] [Google Scholar]
- 3.Lajoix, A. D., Gross, R., Aknin, C., Dietz, S., Granier, C., and Laune, D. (2004) Mol. Divers. 8 281-290 [DOI] [PubMed] [Google Scholar]
- 4.Williams, J. C., Roulhac, P. L., Roy, A. G., Vallee, R. B., Fitzgerald, M. C., and Hendrickson, W. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10028-10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCauley, S. D., Gilchrist, M., and Befus, A. D. (2007) Life Sci. 80 959-964 [DOI] [PubMed] [Google Scholar]
- 6.Jaffrey, S. R., and Snyder, S. H. (1996) Science 274 774-777 [DOI] [PubMed] [Google Scholar]
- 7.Kaiser, F. J., Tavassoli, K., Van den Bemd, G. J., Chang, G. T. G., Horsthemke, B., Moroy, T., and Ludecke, H. J. (2003) Hum. Mol. Genet. 12 1349-1358 [DOI] [PubMed] [Google Scholar]
- 8.Tan, G. S., Preuss, M. A., Williams, J. C., and Schnell, M. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7229-7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moseley, G. W., Roth, D. M., Dejesus, M. A., Leyton, D. L., Filmer, R. P., Pouton, C. W., and Jans, D. A. (2007) Mol. Biol. Cell 18 3204-3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro-Lerida, I., Martinez Moreno, M., Roncal, F., Gavilanes, F., Albar, J. P., and Rodriguez-Crespo, I. (2004) Proteomics 4 339-346 [DOI] [PubMed] [Google Scholar]
- 11.Liang, J., Jaffrey, S. R., Guo, W., Snyder, S. H., and Clardy, J. (1999) Nat. Struct. Biol. 6 735-740 [DOI] [PubMed] [Google Scholar]
- 12.Fan, J., Zhang, Q., Tochio, H., Li, M., and Zhang, M. (2001) J. Mol. Biol. 306 97-108 [DOI] [PubMed] [Google Scholar]
- 13.Crepieux, P., Kwon, H., Leclerc, N., Spencer, W., Richard, S., Lin, R., and Hiscott, J. (1997) Mol. Cell Biol. 17 7375-7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayala, S. K., den Hollander, P., Balasenthil, S., Yang, Z., Broaddus, R. R., and Kumar, R. (2005) EMBO Rep. 6 538-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckwith, S. M., Roghi, C. H., Liu, B., and Morris, R. N. (1998) J. Cell Biol. 143 1239-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vadlamudi, R. K., Bagheri-Yarmand, R., Yang, Z., Balasenthil, S., Nguyen, D., Sahin, A. A., den Hollander, P., and Kumar, R. (2004) Cancer Cell 5 575-585 [DOI] [PubMed] [Google Scholar]
- 17.Song, Y., Benison, G., Nyarko, A., Hays, T. S., and Barbar, E. (2007) J. Biol. Chem. 282 17272-17279 [DOI] [PubMed] [Google Scholar]
- 18.Song, C., Wen, W., Rayala, S. K., Chen, M., Ma, J., Zhang, M., and Kumar, R. (2008) J. Biol. Chem. 283 4004-4013 [DOI] [PubMed] [Google Scholar]
- 19.Mossessova, E., and Lima, C. D. (2000) Mol. Cell 5 865-876 [DOI] [PubMed] [Google Scholar]
- 20.Marley, J., Lu, M., and Bracken, C. (2001) J. Biomol. NMR 20 71-75 [DOI] [PubMed] [Google Scholar]
- 21.Kharakoz, D. P. (1997) Biochemistry 36 10276-10285 [DOI] [PubMed] [Google Scholar]
- 22.Arkin, M., and Lear, J. D. (2001) Anal. Biochem. 299 98-107 [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and Minor, W. (1997) Macromol. Crystallogr. A 276 307-326 [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computing Project 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 25.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 28.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S., and Richardson, D. C. (2003) Proteins 50 437-450 [DOI] [PubMed] [Google Scholar]
- 29.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 30.Goddard, T., and Kneller, D. (2006) Sparky3, San Francisco, CA
- 31.Sells, M. A., Pfaff, A., and Chernoff, J. (2000) J. Cell Biol. 151 1449-1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, J., Sun, Q., Chen, X., Wang, H., Hu, Y., and Gu, J. (2005) Biochem. Biophys. Res. Commun. 331 153-158 [DOI] [PubMed] [Google Scholar]
- 33.Rost, B., and Liu, J. (2003) Nucleic Acids Res. 31 3300-3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rashid, T., Banerjee, M., and Nikolic, M. (2001) J. Biol. Chem. 276 49043-49052 [DOI] [PubMed] [Google Scholar]
- 35.Sundberg-Smith, L. J., Doherty, J. T., Mack, C. P., and Taylor, J. M. (2005) J. Biol. Chem. 280 2055-2064 [DOI] [PubMed] [Google Scholar]
- 36.Thiel, D. A., Reeder, M. K., Pfaff, A., Coleman, T. R., Sells, M. A., and Chernoff, J. (2002) Curr. Biol. 12 1227-1232 [DOI] [PubMed] [Google Scholar]
- 37.Iakoucheva, L. M., Radivojac, P., Brown, C. J., O'Connor, T. R., Sikes, J. G., Obradovic, Z., and Dunker, A. K. (2004) Nucleic Acids Res. 32 1037-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbar, E., Kleinman, B., Imhoff, D., Li, M., Hays, T. S., and Hare, M. (2001) Biochemistry 40 1596-1605 [DOI] [PubMed] [Google Scholar]
- 39.Zwart, P. H., Grosse-Kunstleve, R. W., and Adams, P. D. CCP4 Newsletter; Winter 2007, Contribution 7
- 40.Day, C. L., Puthalakath, H., Skea, G., Strasser, A., Barsukov, I., Lian, L. Y., Huang, D. C., and Hinds, M. G. (2004) Biochem. J. 377 597-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan, J. S., Zhang, Q., Li, M., Tochio, H., Yamazaki, T., Shimizu, M., and Zhang, M. (1998) J. Biol. Chem. 273 33472-33481 [DOI] [PubMed] [Google Scholar]
- 42.Hodi, Z., Nemeth, A. L., Radnai, L., Hetenyi, C., Schlett, K., Bodor, A., Perczel, A., and Nyitray, L. (2006) Biochemistry 45 12582-12595 [DOI] [PubMed] [Google Scholar]
- 43.Wu, J. Q., and Pollard, T. D. (2005) Science 310 310-314 [DOI] [PubMed] [Google Scholar]
- 44.Mammen, M., Choi, S. K., and Whitesides, G. M. (1998) Angew. Chem. Int. Ed. Eng. 37 2755-2794 [DOI] [PubMed] [Google Scholar]
- 45.Lei, M., Lu, W., Meng, W., Parrini, M. C., Eck, M. J., Mayer, B. J., and Harrison, S. C. (2000) Cell 102 387-397 [DOI] [PubMed] [Google Scholar]
- 46.Pirruccello, M., Sondermann, H., Pelton, J. G., Pellicena, P., Hoelz, A., Chernoff, J., Wemmer, D. E., and Kuriyan, J. (2006) J. Mol. Biol. 361 312-326 [DOI] [PubMed] [Google Scholar]
- 47.Rennefahrt, U. E., Deacon, S. W., Parker, S. A., Devarajan, K., Beeser, A., Chernoff, J., Knapp, S., Turk, B. E., and Peterson, J. R. (2007) J. Biol. Chem. 282 15667-15678 [DOI] [PubMed] [Google Scholar]
- 48.Uversky, V. N., Oldfield, C. J., and Dunker, A. K. (2005) J. Mol. Recognit. 18 343-384 [DOI] [PubMed] [Google Scholar]
- 49.Dyson, H. J., and Wright, P. E. (2005) Nat. Rev. Mol. Cell Biol. 6 197-208 [DOI] [PubMed] [Google Scholar]
- 50.Singh, R. R., Song, C., Yang, Z., and Kumar, R. (2005) J. Biol. Chem. 280 18130-18137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.