Abstract
In yeast mitochondria, RNA degradation takes place through the coordinated activities of ySuv3 helicase and yDss1 exoribonuclease (mtEXO), whereas in bacteria, RNA is degraded via RNaseE, RhlB, PNPase, and enolase. Yeast lacking the Suv3 component of the mtEXO form petits and undergo a toxic accumulation of omega intron RNAs. Mammalian mitochondria resemble their prokaryotic origins by harboring a polyadenylation-dependent RNA degradation mechanism, but whether SUV3 participates in regulating RNA turnover in mammalian mitochondria is unclear. We found that lack of hSUV3 in mammalian cells subsequently yielded an accumulation of shortened polyadenylated mtRNA species and impaired mitochondrial protein synthesis. This suggests that SUV3 may serve in part as a component of an RNA degradosome, resembling its yeast ancestor. Reduction in the expression levels of oxidative phosphorylation components correlated with an increase in reactive oxygen species generation, whereas membrane potential and ATP production were decreased. These cumulative defects led to pleiotropic effects in mitochondria such as decreased mtDNA copy number and a shift in mitochondrial morphology from tubular to granular, which eventually manifests in cellular senescence or cell death. Thus, our results suggest that SUV3 is essential for maintaining proper mitochondrial function, likely through a conserved role in mitochondrial RNA regulation.
Prokaryotes and eukaryotes both utilize RNA processing and degradation, albeit via different pathways, as one mechanism for controlling gene expression (1–4). The RNA degradosome of Escherichia coli, yeast, and mammalian cells have all conserved the ability to turnover RNA; however, the assembly of their multicomponent machineries differ (5, 6). In E. coli, RNA degradation is conducted by the cooperation of four principal enzymes (7). The first enzyme is an endoribonuclease, RNase E, which initiates the turnover of many RNAs by cleaving single-stranded RNA internally and is essential for cell growth (2, 8, 9). The second enzyme is a 50-kDa DEAD-box helicase, RhlB, which unwinds and translocates RNA substrates (10). RhlB facilitates the degradation of structured mRNA decay intermediates by the third 85-kDa enzyme, polynucleotide polymerase (PNPase).3 This process, requiring ATP hydrolysis, is believed to involve the local unwinding of structures that block PNPase, thereby ensuring rapid degradation by PNPase (11–13). The fourth enzyme, a 48-kDa glycolytic enzyme enolase, associates with RNase E in response to stress caused by the overabundance of phosphosugars (14, 15). However, enolase is a unique and mysterious contributor to the degradosome, as it is not an RNA-binding protein nor does it appear to be directly involved in RNA turnover.
In Saccharomyces cerevisiae, mitochondrial RNA degradation also requires the activity of multifunctional components (mtEXO) (16), which consist mainly of two enzymes: ySuv3 and yDss1 (17–20). Yeast Suv3 is a nuclear gene-encoded protein that localizes to mitochondria in cells and displays NTP-dependent RNA helicase activity in vitro (20–22). Together with a 3′-5′ exoribonuclease (yDss1), it forms a multifunctional complex that is primarily involved in yeast mitochondrial RNA degradation (16–20). Genetic ablation of ySuv3 results in a respiratory negative phenotype and on the molecular level causes a total block of mitochondrial translation, accumulation of RNA by-products, and changes in stability and processing of mitochondrial RNAs (mtRNAs) (20).
Human mitochondria possess SUV3, PNPase, and mitochondrial polyadenylate polymerase but no yDss1 homolog (12, 19, 23, 24). Similar to ySuv3, human SUV3 (hSUV3) primarily localizes to the mitochondrial matrix and harbors an ATP-dependent unwinding activity on multiple substrates in vitro, although the precise in vivo substrate remains to be identified (26). SUV3 knock-out mice die in utero, supporting the idea that SUV3 is an essential gene (27). Recent data have further implicated SUV3 in maintaining mitochondrial DNA and cell viability (28–30). Although some reports stipulate a potential housekeeping role for SUV3, there is a dearth of significant data to authenticate that claim (31, 32). Thus, the precise cellular function of SUV3 remains largely unknown.
It has been suggested that in some mammalian mitochondrial mRNAs, internalppe polyadenylation generates functional mRNAs that can be translated in mitochondria, because polyadenylation is critical to creating UAA stop codons during mRNA maturation (33). In normal mammalian mitochondria, truncated heterodisperse polyadenylated transcripts do not accumulate and are rapidly degraded (33). The rapid turnover of truncated molecules, which are integral to the degradation process in mammalian mitochondria, highly resembles the intimate involvement of the mtEXO. This prompted us to investigate whether SUV3 participates in a human mitochondrial degradosome, and we found that indeed a significant accumulation of shortened polyadenylated mitochondrial RNA disrupts mitochondrial function and cell viability in human cells. Herein, we have elucidated the cellular role of SUV3 and highlighted a potential evolutionary convergence of the bacterial degradosome and yeast mtEXO, within mammalian mitochondria.
EXPERIMENTAL PROCEDURES
Cell Culture, Cloning, and Reagents—U2OS, a human osteosarcoma cell line, was cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. 143B206 (ρ0), a human osteosarcoma cell line lacking mitochondrial DNA, was cultured similarly to U20S with the addition of pyruvate and uridine (34). U2OS stable clones were established for doxycycline-inducible expression of SUV3 shRNA. Briefly, SUV3 shRNA expression cassette was created in pTER (pTER-SUV3i). Four such cassettes from pTER-SUV3i were inserted in tandem into pPUR (pPUR-4xSUV3i). U2OS cells with inducible SUV3 shRNA were established by Lipofectamine 2000-mediated transfection of pPUR-4xSUV3iand a TetR-expressing construct, pCDNA6TR, followed by selection with 5.0 μg/ml blasticidin and 5.0 μg/ml of puromycin. One of the resultant clones was named LK6. The shRNA target sequence is 5′-GGG CCT GGA CCA GAG CAC TTC-3′. The synthetic SUV3 siRNA (ordered from Integrated DNA Technologies) is 5′-CCA GCA UAU UCC ACU AAG UCU GCG A-3′; control luciferase siRNA is 5′-UUA CGC UGA GUA CUU CGA-3′. LK6R, a LK6-derived cell line, was generated by retroviral integration of shRNA-resistant SUV3. To obtain shRNA-resistant SUV3 expression, three silent mutations were introduced into the shRNA targeting region of SUV3 (A942G, A943C, and A945T), which was subcloned into pQCXIG, a vector modified from pQCXIP (Clontech) by replacing the puromyocin resistance gene with green fluorescent protein for bicistronic expression of SUV3 and green fluorescent protein. 293GP2-packaging cells were used for virus production with a VSVG (vesicular stomatis virus G protein) expression plasmid and pQCXIG-SUV3RNAi.
Mouse monoclonal SUV3 antibodies (6B7 or 18C5) were raised using bacterially expressed recombinant full-length hSUV3 as the immunogen. The p48 and α-tubulin mouse antibodies were purchased from Genetex and Sigma, respectively. The oxidative phosphorylation (OXPHOS) antibody mixture (Mitosciences Inc.) contains the five antibodies against complex I subunits NDUFB8 (MS105), complex II-30 kDa (MS203), complex III-core protein 2 (MS304), complex IV subunit II (MS405), and complex V α-subunit (MS507), respectively. Other commercial antibodies used were γ-H2AX and ATM-S1981P (Genetex) and HP1-γ (Upstate).
Cell Proliferation Assay—U2OS and LK6 cells were seeded in 6-cm dishes in triplicate and induced with 5 μg/ml doxycycline. The viable cells at designated time points were counted by a trypan blue exclusion assay as described extensively elsewhere (35).
Cell Cycle Analysis by Flow Cytometry—Exponentially growing cells were treated with 5 μg/ml doxycycline for various periods of time. Cells were trypsinized, fixed with chilled 70% ethanol, and stained for 30 min with propidium iodide solution (50 μg/ml propidium iodide, 0.1% sodium citrate, 50 μg/ml of RNase A, and 0.03% Nonidet P-40 in PBS) at room temperature. Flow cytometric analysis was performed using a FACSCalibur flow cytometer, and the cell cycle distribution was analyzed using CellQuest software (BD Biosciences).
Immunofluorescent Microscopy—Immunostaining was performed essentially as described previously (36). In brief, cells were fixed with cold methanol for 10 min or 3.2% paraformaldehyde for 20 min. Cells were then permeated with 0.1% Triton X-100 in PBS for 10 min, blocked with 2% bovine serum albumin in PBS, 0.1% Triton X-100. Cells were incubated with primary antibodies for 1–2 h at room temperature, washed with PBS, 0.1% Triton X-100 for 5 min (three times), and then incubated with Alexa Fluor-conjugated secondary antibody (Invitrogen). After a 5-min wash with PBS/0.1% Triton X-100 (three times), DAPI was used to stain the DNA/nucleus before mounting the coverslip onto a coverglass with Prolong Gold antifade solution (Invitrogen). For mitochondrial staining, cells grown on coverslips were incubated with 25 nm MitoTracker Red CMXROS at 37 °C for 15 min and then fixed as instructed in the manual (Invitrogen) for subsequent immunostaining with antibodies as described above. For the senescence-associated β-galactosidase assay, the experiments were done as described previously (37). Microscopy was performed with a Carl Zeiss Axiovert 200 microscope equipped with a charge-coupled device Hamamatsu camera. Images were captured with AxioVision software and processed with Adobe Photoshop.
Electrochemical Membrane Potential Measurement— Changes in membrane potential were determined using a JC-1 mitochondrial membrane potential detection kit (Invitrogen). Briefly, 106 cells were washed, pelleted, and resuspended in 500 μl of JC-1 (5,5′-6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanineiodide) working solution. Cells were incubated at 37 °C for 15 min. Following staining, cells were washed twice in assay buffer and then resuspended in 200 μl of assay buffer and immediately analyzed using a BD FACSCalibur flow cytometer equipped with dedicated software.
Mitochondrial Isolation and OXPHOS Enzymology—Mitochondrial isolation and spectrophotometric studies of respiratory chain enzymes were carried out as described previously (38, 39) on U2OS, LK6, and induced LK6 cells. Mitochondrial protein concentration was measured using the Lowry method. Spectrophotometric analysis of the respiratory chain complexes was performed with mitochondrial preparations from the aforementioned cells using a spectrophotometer (model DU-650, Beckman Coulter). Mitochondria were freeze-thawed three times at 1 mg/ml and used immediately for enzymatic activity assays. All experiments were done at least in triplicates.
Mitochondrial ROS Determination—106 cells were harvested, washed in 1× Hanks balanced salt solution, and then resuspended in 1 ml of MitoSOX working solution for 10 min at 37 °C, protected from light. Following the incubation, the cells were spun down for 3 min at 2000 rpm. The cells are finally resuspended in 3 ml of Hanks balanced salt solution, placed in a glass cuvette, and read at wavelengths of 510/580 nm (ex/em) using an LS50B luminescence spectrophotometer (PerkinElmer Life Sciences) for a 30-min total reading. After allowing the base line to stabilize for several minutes, we induced superoxide production by adding 5 μl each of the complex I substrates 1 m sodium pyruvate and 1 m malic acid. To determine the rate of superoxide formation in the mitochondria, we calculated the ascending slope of the superoxide trace. The remaining pellet was resuspended in 10 μl of cell lysis buffer and kept at -80 °C until protein determination was performed by the Bradford colorimetric protein assay.
Mitochondrial Protein Labeling—Twenty-four hours prior to labeling, cell growth medium was changed to methionine-free Dulbecco's modified Eagle's medium plus 10% dialyzed fetal bovine serum. On the day of labeling, medium was again changed to methionine-free Dulbecco's modified Eagle's medium plus 10% dialyzed fetal bovine serum and incubated for 5 min at 37 °C in 5% CO2. The medium was then replaced with methionine-free medium plus emetine (100 mg/ml working concentration, Sigma) and incubated for 5 min at 37 °C. As a negative control, one dish was simultaneously treated with chloramphenicol (100 μg/ml working concentration, Sigma). Then, cells were labeled with 200 μCi of [35S]methionine (GE Healthcare) in the presence of emetine and incubated for 2 h at 37 °C. Cells were then washed and harvested for lysis. Lysed proteins were quantified by performing the colorimetric Bradford assay, and additionally 50 μl (at least 15 μg of mitochondria) of crude lysate was used for citrate synthase activity to normalize the number of mitochondria in each sample. Mitochondrial proteins were separated on a 5–15% gradient SDS-PAGE. The gel was then fixed in acetic acid and methanol for 30 min at room temperature, dehydrated in glacial acetic acid for 20 min, and then treated with 20% diphenyloxozole in glacial acetic acid for 10 min. The gel was dried at 85 °C for 1 h on a Bio-Rad gel dryer, model 583. The dried gel was then subject to phosphorimaging using a Storm 820 PhosphorImager (GE Healthcare).
ATP Quantification—Cellular ATP was quantified using an ATP determination kit according to the manufacturer's instructions (Molecular Probes). Briefly, LK6 cells treated with 5 μg/ml doxycycline at various time points, and control U2OS, untreated LK6 cells, and 143B-206 (ρ0) cells plated at equal densities were lysed in passive lysis buffer (Promega). Equal volumes of cell lysate were added to the standard reaction solution, and luminescence was measured and normalized to protein amounts in each lysate. The values used fell in the linear range of the assay as determined by a standard curve.
mtDNA Analysis—DNA was extracted from control LK6 and doxycycline-induced LK6 cells using standard procedures. MtDNA was amplified by quantitative PCR using gene-specific primers targeting ND2 and ND6 and compared with 18 S nuclear DNA under the following cycling parameters: 94 °C for 10 min, 94 °C for 1 s, 53 °C for 10 s, and 72 °C for 10 s for 40 cycles. Primer sequences used were: 18 S forward, ACG GAC CAG AGC GAA AGC AT; 18 S reverse, GGA CAT CTA AGG GCA TCA CAG AC; ND2 forward, GCC CTA GAA ATA AAC ATG CTA; ND2 reverse, GGG CTA TTC CTA GTT TTA TT; ND6 forward, ATC CTC CCG AAT GAA CCC TG; ND6 reverse, GAT TGT TAG CGG TGT GGT CG. Quantification of mtDNA was performed as described previously (40, 41). Briefly, this method uses a double-stranded DNA dye, SYBR Green I, to continuously monitor product formation and quantify samples ranging up to 4 log units in concentration. The values found in each sample for the two genes (representative of mtDNA, ND2 and ND6; nuclear DNA, 18 S rDNA) allowed the calculation of a ratio of mtDNA/nDNA to make comparisons between the different groups. Real-time PCR was performed in a fluorescence temperature LightCycler (Roche Applied Science). The fluorescent product was detected at the last step of each cycle by single acquisition. After amplification, a melting curve was acquired by heating the product with continuous fluorescence collection. Melting curves were used to determine the specificity of the PCR products. Variations in sample loading or due to PCR tube-to-tube efficiency were corrected by performing triplicates for each standard and unknown sample. The mean value of the triplicates for each sample was used to determine the values of mtDNA and 18 S amount. The mtND2/18 S mean ratios of each group were used for comparisons between them.
Oligo(dT)-primed Reverse Transcription-PCR—Total RNA was isolated by using a Versagene kit (Gentra Inc.) and quantified by UV absorbance. Oligo(dT)17 with adaptor sequence 5′-GAC TCG AGT CGA CAT CGA T-3′ was used to prime the reverse transcription reaction as described (42), and the resulting cDNA was PCR-amplified using the adaptor and one ND2 gene-specific primer (sequence 5′-GGC CCA ACC CGT CAT CTA CTC T-3′). PCR products were visualized by 2% agarose gel or 6% polyacrylamide gel depending on the size of the DNA fragments.
RESULTS
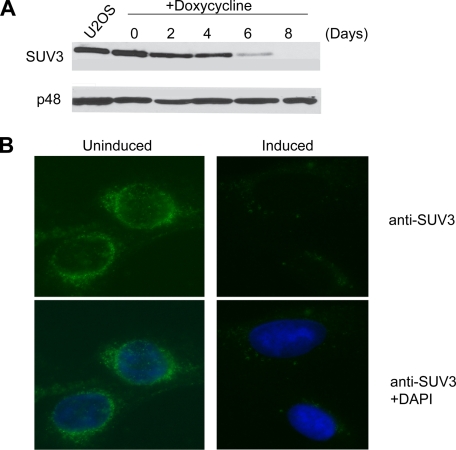
Recent studies have reported that HeLa cells lacking 75–80% of their SUV3 protein undergo apoptosis when transient transfection is used with its corresponding siRNA (27). However, the cellular and mitochondrial phenotypes associated with SUV3 deficiency and the underlying mechanisms therein have not been systematically examined in this transient knockdown system. SUV3 knock-out in mice leads to early embryonic lethality, making the availability of SUV3-null cells a formidable challenge.4 Therefore, to examine the cellular function of SUV3 in mammalian cells, an inducible shRNA system specifically targeting SUV3 was generated in an osteosarcoma cell line (U2OS). LK6 denotes one of several such clones generated in this matter, which was able to markedly reduce the expression of SUV3 by day 6 and nearly completely knock down expression by day 8 (>90% reduction) upon the addition of 5 μg/ml doxycycline, as shown by Western blotting and further validated by immunostaining (Fig. 1).
FIGURE 1.
Depletion of SUV3 by RNAi in human cells. A, Western blot probing SUV3 expression in parental U2OS and LK6 cells treated with 5 μg/ml doxycycline over a time course of 8 days (top) compared with p48 (an Rb-interacting partner) as a loading control. B, immunostaining of SUV3 in LK6 cells induced with 5 μg/ml doxycycline at day 8. DAPI staining in merged images (bottom panels) shows the DNA/nucleus.
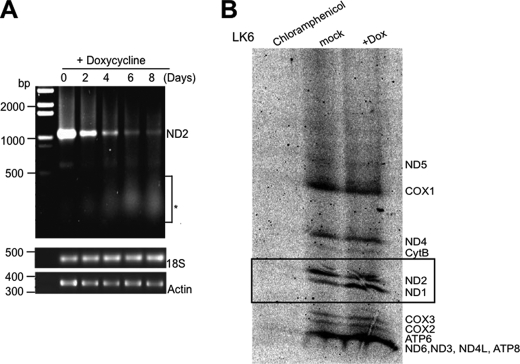
RNA degradation plays a key role in RNA metabolism by preventing the overabundance of RNA maturation by-products and aberrant transcripts, an essential process in gene regulation. SUV3 has an RNA helicase activity, and thus the major consequence upon SUV3 depletion is most likely a defect in mtRNA transactions. In yeast, Suv3 deficiency inhibits mitochondrial translation. If SUV3 engages in a mitochondrial degradosome-like activity, it is plausible that its depletion would lead to the accumulation of truncated RNAs that might adversely affect translation. To test this possibility, we determined the levels of polyadenylated mtRNA transcripts by reverse transcription-PCR, because mtRNAs in humans are known to be polyadenylated either at the 3′-tail or internally (43). Interestingly, we found an accumulation of polyadenylated, truncated RNA species with a concomitant diminution in the level of full-length transcript of ND2, as an example (Fig. 2A), suggesting defective clearance of the transiently and internally polyadenylated mtRNA species, which is consistent with the role of hSUV3 in RNA degradation.
FIGURE 2.
SUV3 knockdown leads to an accumulation of shortened polyadenylated mtRNA species and defective mitochondrial protein synthesis. A, oligo(dT)-primed reverse transcription-PCR results of mitochondrial ND2 mRNA. *, depicts the accumulation of polyadenylated and truncated RNA transcripts within 100 to 500 bases upon SUV3 knockdown. Lower panel (control PCR), shows the unchanged levels of 18 S rRNA or actin mRNA. B, defective mitochondrial protein translation in SUV3 shRNA-induced LK6 cells after 8 days of treatment with doxycycline in comparison with mock-treated control or the negative control with chloramphenicol treatment abrogating total protein synthesis. When the translation level of ND1 was normalized by citrate synthase activity (in lanes 2 and 3), all other mitochondrial proteins including ND4, cytB, and others were reduced in translational level. Boxed proteins indicate the quantified altered ratio of ND1/ND2 from 1:3 in controls to 2:1 in SUV3 knockdown cells as an example.
Because mtRNA transcription and translation have been shown to be coupled physically (43), the accumulation of truncated mtRNA transcripts may serve to suppress translational levels and consequently lead to a reduction in levels of mitochondrially encoded protein. To test this idea, we performed an in vitro S35 labeling assay in SUV3-depleted LK6 cells. Notably, ND1 is the first polypeptide on the polycistron to be translated by the mitoribosome (44) in which the transcript level is unchanged upon transient inhibition of mitochondrial translation in normal cells (44, 45). Using ND1 as an internal control, we compared doxycycline-treated LK6 cells with mock-treated controls. By normalizing the number of mitochondria, using citrate synthase activity as a marker (data not shown), the ND1 levels of doxycycline-treated LK6 cells become comparable with the level of ND1 in mock treated cells. Based on this comparison, the remaining mitochondrial encoded proteins in doxycycline-treated LK6 cells were reduced by at least 2–3 fold (Fig. 2B), suggesting a defect in mitochondrial protein synthesis. Collectively, these results imply that inefficient protein translation associated with accumulation of shortened mtRNAs may directly lead to reduction of mitochondrially encoded proteins.
Using this inducible knockdown system, by straight Western analysis we found a consistent decrease in the expression of mitochondrial-encoded components of complex I (20-kDa subunit) and complex IV (COX II), in addition to nuclear-encoded complex III (core-2 protein) in SUV3-depleted LK6 cells, but not in parental U2OS cells treated with doxycycline (Fig. 3A). The observed defect in complex III expands the dysfunction to an entire metabolon comprising complexes I, III, and IV. It has been reported that complexes I, III, and IV form a supercomplex that participates in NADH oxidation by direct substrate channeling in vivo (40–43).
FIGURE 3.
Impaired OXPHOS can be restored by the expression of RNAi-resistant SUV3. A, immunoblot showing a decrease in the expression of complex I, III, and IV proteins using 30 μg of whole cell lysate from LK6 cells treated with 5 μg/ml doxycycline (+dox) but not in parental or mock-treated cells. B, measurement of complex IV activity (left bar graph) and citrate synthase activity (middle) and their relative ratio (right) in SUV3 shRNA-induced LK6 cells. C, immunoblot of SUV3 from U2OS cells targeted by SUV3 siRNA. D, immunoblot showing a decrease in the expression of complex I, III, and IV proteins in SUV3 siRNA-treated U2OS cells after 2 and 5 days of transfection. E, immunoblot showing the level of SUV3 in control LK6 or in LK6R expressing the shRNA-resistant SUV3 version by retroviral infection. Both cells were induced (+) or uninduced (-) with doxycycline (5 μg/ml for 8 days). F, immunoblot showing only a partial diminution in the expression of complex I, III, and IV proteins in LK6R cells treated similarly to those described in E, with doxycycline compared with control. u, untreated; Luc, luciferase siRNA.
To extend our observation of decline in OXPHOS expression, we examined the electron transport chain by measuring complex activity, specifically COX activity, a major regulatory component for OXPHOS and the final step in electron transport. Mitochondria isolated from induced LK6 cells were examined for defects in complex IV activity, which was normalized against citrate synthase activity (COX/citrate synthase ratio). The COX/citrate synthase ratio revealed a decline in activity by 4-fold, with a concomitant 45% increase in the number of mitochondria/cell as determined by citrate synthase activity (Fig. 3B). Consistently, by immunostaining with MitoTracker Green FM, the number of mitochondria/cell also increased in doxycycline-induced LK6 cells as measured by flow cytometry (data not shown). This suggests that depleting SUV3 induces downstream retrograde signals from the mitochondria to the nucleus, which in turn up-regulates mitochondria proliferation to compensate for stress-induced conditions (46–48).
To exclude the possibility of an RNAi off-target effect, a siRNA that targets a different SUV3 region from the shRNA was used (Fig. 3C). Consistent with the observed defect in complexes I, III, and IV, SUV3 siRNA-treated cells also exhibited the same decrease in expression, whereas luciferase control siRNA-treated cells did not (Fig. 3D). To further confirm that the mitochondrial phenotype observed was specific to SUV3, we designed and generated a RNAi-resistant SUV3 version with wild type coding capacity in a retroviral system, which was used to infect LK6 cells to generate a stable clone (LK6R). Upon the addition of doxycycline, LK6R cells maintained the level of SUV3, whereas LK6 cells did not (Fig. 3E). The expression levels of I, III, and IV supercomplexes to near control amounts suggests that the expression of RNAi-resistant SUV3 can restore the mitochondrial phenotype of LK6 cells upon doxycycline treatment (Fig. 3F). Collectively, these data indicate that the defects observed are due to specific SUV3 ablation.
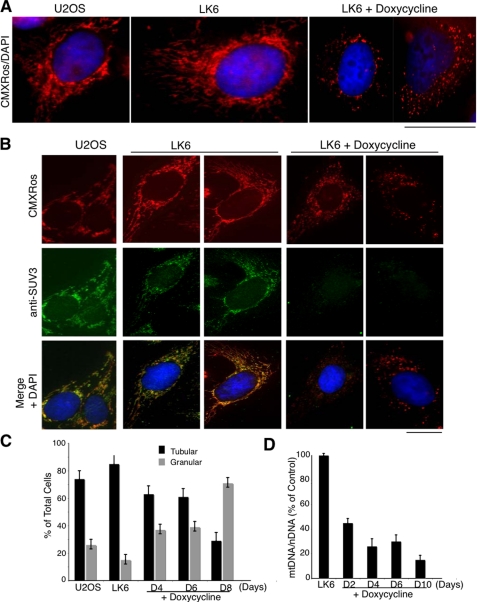
Destabilization of OXPHOS upon SUV3 depletion is bound to compromise the integrity of the mitochondria. Maintaining proper mitochondrial morphology is critical for its function. The basic morphology of mitochondria in cells is a dynamic tubulovesicular reticulum. The mitochondrial reticular network forms in the perinuclear region and extends outward toward the periphery of the cell. The balance of fission and fusion determines mitochondrial morphology (49). One of the consequences of active cycling of mitochondrial fusion and fission may be to permit an individual nucleoid to have access to a larger pool of diffusible proteins required for replication or transcription than would be possible if the nucleoid were trapped in a single mitochondrion (28, 30). A high rate of mitochondrial fusion appears to be essential for the maintenance of mtDNA in nucleoids (29). Bogenhagen et al. (28) recently reported that SUV3 is associated with the formation of mtDNA nucleoid complexes in the mitochondria. Therefore, we determined the effect of SUV3 depletion on the morphology of the mitochondria. In control cells, SUV3 was shown to colocalize with tubulovesicular mitochondria. On the contrary, the tubular, elongated, mitochondrial organization shifted to a punctuated, granular shape concomitant with the loss of SUV3 (Fig. 4). This result suggests the presence of SUV3 is essential for maintaining the tubulovesicular morphology of healthy mitochondria.
FIGURE 4.
Mitochondrial morphology is altered in SUV3 knockdown cells. A, fragmentation of the filamentous mitochondrial network upon SUV3 knockdown. CMXRos MitoTracker Red-stained parental control, LK6, and doxycycline-induced LK6 cells at day 8 showed a change in mitochondrial morphology from tubular and elongated to granular and punctate-shaped structures. Nuclei were stained with DAPI. B, immunofluorescence staining of CMXRos and SUV3 on parental, LK6, and doxycycline-induced LK6 cells (day 8). Nuclei were stained with DAPI. C, quantification of mitochondrial morphology changes in parental, uninduced LK6 and doxycycline-induced LK6 cells for 4, 6, or 8 days. D, decline in mtDNA copy number, quantified by the mean ratio of mtND2/18 S, is shown for LK6 cells induced for 2, 4, 6, and 10 days (5 μg/ml doxycycline). Scale bars, 10 μm.
An alteration in the mitochondrial reticulum of induced LK6 cells is highly reminiscent of cells depleted of their mitochondrial DNA (ρ0), which show a characteristic lack of mitochondrially encoded protein subunits resulting in a disruption of respiratory function and prompting cellular energy production by glycolysis (50). Considering the localization of SUV3 (Fig. 4B), we examined the effect of SUV3 knockdown on mtDNA copy number and noted a decline of ∼3.5 fold as compared with nuclear DNA (Fig. 4D). Taken together, the abnormality in mitochondrial morphology and the decline in mtDNA copy number support the finding that SUV3 may impart a role in mtDNA nucleoid maintenance consistent with the findings of Bogenhagen et al. (28).
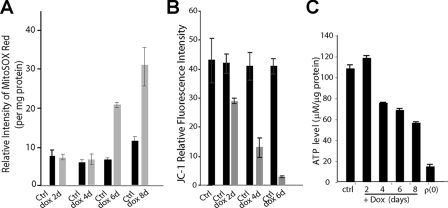
The observed defects in RNA degradation and translation, as well as mitochondrial morphology and OXPHOS, prompted us to examine the mitochondrial physiology in LK6 cells upon depletion of SUV3. Given the dominant subcellular localization of SUV3 to the mitochondria and the fact that mtDNA damage is closely interrelated with mitochondrial ROS production, we assessed the cellular levels of mitochondrial superoxide by measuring mitoSOX Red fluorescence and found a 4-fold increase in the rate of ROS production (Fig. 5A). ROS production is intimately linked to the electrochemical gradient generated by the OXPHOS complexes in mitochondria. Thus, we tested LK6 and induced LK6 cells with JC-1 dye to measure the mitochondrial electrochemical membrane potential by fluorescence-activated cell sorter analysis and found up to an 8-fold decrease in the mitochondrial electrochemical membrane potential in induced LK6 cells (Fig. 5B). Because mitochondria are cellular powerhouses, used to drive energy production, we reasoned that induced LK6 cells with dysfunctional mitochondria would have an abrogated production of ATP, which we calculated to decline by ∼50% in comparison with controls (Fig. 5C). Together, these results maintain a role for SUV3 in mitochondrial physiology, likely because of its apparent role in RNA turnover, which is critical to cell viability.
FIGURE 5.
SUV3 knockdown alters mitochondrial physiology. A, measurement of superoxide formation by mitoSOX Red staining upon SUV3 knockdown in control (Ctrl) and SUV3-ablated LK6 cells (SUV3 induced for 2, 4, 6, 8 days). B, detection of mitochondria membrane potential by JC-1 staining in the induced LK6 cells (2, 4, or 6 days with doxycycline (dox)). C, histogram to show the ATP levels measured in parental, LK6, SUV3-depleted LK6, and 143B-ρ0 cells.
To assess the overall cellular effect of SUV3 ablation on cell viability, we closely examined the growth rate of both parental and LK6 cells treated with doxycycline over a time course of 8 days. Even though there was no apparent change in doubling time in doxycycline-treated parental U2OS cells, a striking growth retardation was observed in doxycycline-induced LK6 cells, as shown in Fig. 6A, which correlated with depletion of SUV3 (Fig. 1). This suggests that the slowed proliferation observed in LK6 cells is an inherent characteristic of SUV3 deficiency, independently of doxycycline treatment. To explore the nature of the slowed proliferation due to SUV3 depletion, we examined the cell cycle profile of doxycycline-induced LK6 cells. The data indicate that the sub-G1 population exhibited an increase from 2% in control to 17% upon SUV3 depletion, suggesting increased cell death. In the meantime, there was a gradual decline in the G1 population, from 50 to 15%, and an increase in the multinucleated population, from 3 to 18% (Fig. 6B). Accordingly, an increase in the multinucleated population with a concomitant increase in G2/M strongly suggests that the slowed proliferation in SUV3-depleted cells may be in part attributed to a defect in cytokinesis. Moreover, upon SUV3 depletion, the remaining induced LK6 cells appeared to be enlarged and flattened, a sign of cell cycle exit or cellular senescence.
FIGURE 6.
SUV3 knockdown leads to cell senescence and cell death. A, parental U2OS and LK6 cells were treated with 5 μg/ml doxycycline over a time course of 8 days. Viable cells were counted via trypan blue exclusion, and the numbers were plotted in the graph. B, fluorescence-activated cell sorter analysis to show the cell cycle profile of LK6 cells induced with 5 μg/ml doxycycline over a time course of 8 days. Single arrows denotes 2N, and double arrows denotes 4N. C, senescence-associated β-galactosidase (SA β-gal) activity of LK6 cells treated with 5 μg/ml doxycycline for 8 days (left panels); the percentage of positive cells was quantified and is shown in thehistogram on the right. D, detection of senescence-associated heterochromatin foci in uninduced or SUV3 shRNA-induced LK6 cells (5 μg/ml doxycycline for 8 days). γ-H2AX, ATM, and HP1-γ were stained together with DAPI (nucleus). Bar graphs (bottom), quantification of the number of cells with positively stained foci.
To test whether these cells underwent senescence, several prominent cellular senescence markers, such as senescence-associated β-galactosidase activity and senescence-activated heterochromatin-positive foci formation, were examined. Nearly 35% of the cells were positive for senescence-associated β-galactosidase activity upon SUV3 knockdown, whereas the control LK6 cells remained negative (Fig. 6C). As determined by indirect immunostaining, 70–80% of the nuclei of the doxycycline-induced LK6 cells showed positive foci formation for γ-H2AX and HP1-γ antibodies (Fig. 6D). Taken together, these results suggest that depletion of SUV3 leads to multinucleation, cellular senescence, and cell death.
DISCUSSION
In this study, we characterized human SUV3 function in mammalian mitochondria using both transcription-mediated shRNA and siRNA systems (Fig. 1). As summarized in Fig. 7, depletion of SUV3 leads to the accumulation of shortened mtRNA species, which is likely due to impaired mtRNA degradation. It is plausible that these truncated internally polyadenylated RNA species serve to impair mitochondrial protein synthesis, giving rise to defective OXPHOS complexes containing mitochondrially-encoded components. As a result, the function of OXPHOS is crippled, triggering a cascade of dysfunction in membrane potential, ATP production, and ROS generation, yielding disrupted mitochondria tubular network and ultimately the cessation of cell growth. Eventually, the cells undergo either cell death or senescence (Figs. 2, 3, 4, 5, 6).
FIGURE 7.
Summary diagram. This is a depiction of how SUV3 deficiency may give rise to the pleiotropic phenotypes at the cellular and molecular level.
The aforementioned accumulation of shortened mitochondrial RNA is similar to observations made in yeast Suv3-null cells, in which omega RNA and truncated mtRNA cannot be degraded (20). Disruption of the yeast mtEXO component, Dss1, has also been shown to lead to an accumulation of truncated mitochondrial omega RNA species (20). However, the alleged mammalian mitochondria degradosome does not contain Dss1, but perhaps it contains PNPase, the conserved component from the bacterial degradosome. It was reported that depletion of PNPase generates a plethora of defects such as impaired mitochondrial electrochemical membrane potential and ATP generation, decreased respiratory chain activity, and slowed cell proliferation (23). These defects bear a great resemblance to the phenotype we observed in SUV3-depleted cells. However, the detection of an accumulation of truncated mitochondrial RNA species was not reported. If PNPase is indeed a component of the mammalian mitochondria degradosome, the accumulation of the shortened polyadenylated RNA is very likely to be present in PNPase-depleted cells.
Human mtRNAs of both the heavy and light strands are internally polyadenylated, which strongly suggests that a prokaryotic polyadenylation-dependent degradation mechanism exists in human mitochondria (42). Given this notion, the key observation that an inhibition in mitochondrial translation occurs in SUV3-deficient human cells (Fig. 2B) may result in part from the accumulation of truncated polyadenylated mitochondrial transcripts (Fig. 2A). It is possible that the excessive shortened poly(A) mtRNAs may compete for and lodge ribosome binding sites, thus lowering the pool of available ribosomes for proper translation. Alternatively, the shortened RNAs may function in an antisense manner to block mtRNA translation of the complementary strand. The failure to eliminate these truncated mtRNA polyadenylated transcripts in SUV3-depleted cells strongly implicates a role for SUV3 as an evolutionarily conserved degradosome component in human mitochondria.
As a result of defective mitochondrial translation, SUV3 depletion led to the collapse in mitochondrial structure and function. Accordingly, the expression and activity of mitochondrially-encoded OXPHOS complexes were markedly defective upon SUV3 depletion (Fig. 3). The protein subunits of complex I and IV, encoded by the mitochondrial genome, declined. However, ATP-synthase (F0 subunit) and complex II (30-kDa unit), which are encoded by the nuclear genome, were expressed at normal levels. Surprisingly, the core-2 protein subunit of complex III, which is also nuclear encoded, declined. Because complexes I, III, and IV are associated functionally and physically in the inner membrane as part of a supercomplex, or respirasome (51–54), it is possible that SUV3 depletion dynamically destabilizes this supercomplex as a whole. Whether other mitochondrial protein complexes are also affected by SUV3 depletion remains to be tested systematically.
The breakdown in OXPHOS, at the physiological level, led to an increase in ROS production and a decline in membrane potential and energy (ATP) production (Figs. 3 and 5). Mitochondrial ROS production left unquenched leads to a plethora of damaging processes; perhaps one of the most damaging is to its own naked mtDNA nucleoids, thus amplifying mitochondrial defects (55, 56). Within a certain level, ROS may induce stress responses by altering the expression of specific nuclear genes to uphold the energy metabolism to rescue the cell. Once beyond the threshold, ROS may cause oxidative damage to mtDNA and other components of the affected cell and eventually elicit apoptosis.
The morphology of mitochondria in SUV3-depleted cells shift from the interconnected tubulovesicular reticulum to a mainly granular morphology that appears circular, donut-shaped, and swollen (Fig. 4). These changes in mitochondrial morphology strikingly resemble ρ(0) cells (57). Consistently in SUV3-depleted cells a decline in mtDNA copy number was detected (Fig. 5). During fission and fusion, damaged DNA is equally likely to be redistributed in newly biosynthesized mitochondria; a decline in mtDNA copy number suggests that this distribution is abrogated, evident in the predominantly granular shape of mitochondria in SUV3-depleted cells. At least two possibilities can explain SUV3-depleted cells devoid of mtDNA: an increase in ROS production from a faulty respiratory chain that severely damages mtDNA leads to errors and consequently may block mitochondrial DNA replication; alternatively, SUV3 may exert a role in mtDNA replication, although no data at the present time support this possibility. Interestingly, SUV3 together with a group of proteins was found to associate with mtDNA nucleoids (28). The association of mtDNA nucleoids may be important for maintaining mtDNA copy number and morphology. For example, inactivation of the first identified mitochondrial nucleoid-associated protein, mitochondrial transcription factor A (TFAM), reduced mtDNA copy number and altered mitochondrial morphology (58–60).
One effect of cells devoid of mtDNA is slowed proliferation (61). At the cellular level, using our inducible knockdown system, we monitored the gradual reduction of SUV3 expression (with a robust knockdown efficiency of >90%) and observed multinucleation and senescence (Fig. 6). Multinucleation is the result of defects in cytokinesis, as the energetics of ATP as well as specialized proteins are required to facilitate this process. It is plausible that damaged mitochondria, devoid of mtDNA with a defective respiratory chain, generate low levels of ATP in SUV3-depleted cells and account for the apparent increase in multinucleation and, consequently, reduced growth rate.
Growth retardation was also associated with severe changes in cell morphology, such as cell flattening, increased granularity, and positive β-galactosidase staining (Fig. 6), hallmarks of an activated senescence program (62). We detected senescence-activated heterochromatin-positive foci formation and also found that cells depleted of SUV3 exhibited an up-regulation of p53 (data not shown). p53 up-regulation mediates activation of p21, which has been shown to be the major regulator of the cellular senescence program (25, 63–65). Thus, a link between a mitochondrial RNA helicase and cellular senescence can be established. Indeed our animal studies faithfully reflect this link bridging mitochondrial homeostasis with aging and cancer.5
Acknowledgments
We thank Dr. Phang-Lang Chen and Chi-Fen Chen for invaluable help. We also thank Dr. Zhanyong Shu, Bella Haroutyunyan, and Maria Lvova for technical assistance and Dr. Kyoko Yokomori for providing some of the reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AG027877 (to W.-H. L.) and AG24373, AG13154, NS41850, DK73691, and NS21328 (to D. W.). It was also supported by the Donald Bren Endowment (to W.-H. L. and D. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PNPase, polynucleotide polymerase; shRNA, small hairpin RNA; RNAi, RNA interference; siRNA, small interfering RNA; OXPHOS, oxidative phosphorylation; PBS, phosphate-buffered saline; DAPI, 4′,6-diamidino-2-phenylindole; ROS, reactive oxygen species; mtEXO, mitochondrial RNA degradosome.
L. Khidr, G. Wu, A. Davila, V. Procaccio, D. Wallace, and W.-H. Lee, unpublished data.
P. L, Chen, C. F, Chen, and W. H, Lee, manuscript in preparation.
References
- 1.Carpousis, A. J. (2002) Biochem. Soc. Trans. 30 150-155 [PubMed] [Google Scholar]
- 2.Ehretsmann, C. P., Carpousis, A. J., and Krisch, H. M. (1992) FASEB J. 6 3186-3192 [DOI] [PubMed] [Google Scholar]
- 3.Houseley, J., LaCava, J., and Tollervey, D. (2006) Nat. Rev. Mol. Cell. Biol. 7 529-539 [DOI] [PubMed] [Google Scholar]
- 4.Carpousis, A. J., Leroy, A., Vanzo, N., and Khemici, V. (2001) Methods Enzymol. 342 333-345 [DOI] [PubMed] [Google Scholar]
- 5.Bessarab, D. A., Kaberdin, V. R., Wei, C. L., Liou, G. G., and Lin-Chao, S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3157-3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coburn, G. A., Miao, X., Briant, D. J., and Mackie, G. A. (1999) Genes Dev. 13 2594-2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miczak, A., Kaberdin, V. R., Wei, C. L., and Lin-Chao, S. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 3865-3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babitzke, P., and Kushner, S. R. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callaghan, A. J., Marcaida, M. J., Stead, J. A., McDowall, K. J., Scott, W. G., and Luisi, B. F. (2005) Nature 437 1187-1191 [DOI] [PubMed] [Google Scholar]
- 10.Gebauer, D., Mais, C., Zinger, K., Hock, R., Lieb, B., and Scheer, U. (1996) Int. J. Dev. Biol. 40 239-244 [PubMed] [Google Scholar]
- 11.Regonesi, M. E., Briani, F., Ghetta, A., Zangrossi, S., Ghisotti, D., Tortora, P., and Deho, G. (2004) Nucleic Acids Res. 32 1006-1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar, D., and Fisher, P. B. (2006) Pharmacol. Ther. 112 243-263 [DOI] [PubMed] [Google Scholar]
- 13.Yehudai-Resheff, S., and Schuster, G. (2000) Nucleic Acids Res. 28 1139-1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita, T., Kawamoto, H., Mizota, T., Inada, T., and Aiba, H. (2004) Mol. Microbiol. 54 1063-1075 [DOI] [PubMed] [Google Scholar]
- 15.Chandran, V., and Luisi, B. F. (2006) J. Mol. Biol. 358 8-15 [DOI] [PubMed] [Google Scholar]
- 16.Dziembowski, A., Piwowarski, J., Hoser, R., Minczuk, M., Dmochowska, A., Siep, M., van der Spek, H., Grivell, L., and Stepien, P. P. (2003) J. Biol. Chem. 278 1603-1611 [DOI] [PubMed] [Google Scholar]
- 17.Golik, P., Szczepanek., T., Bartnik, E., Stepien, P. P., and Lazowska, J. (1995) Curr. Genet. 28 217-224 [DOI] [PubMed] [Google Scholar]
- 18.Margossian, S. P., and Butow, R. A. (1996) Trends Biochem. Sci. 21 392-396 [PubMed] [Google Scholar]
- 19.Minczuk, M., Piwowarski, J., Papworth, M. A., Awiszus, K., Schalinski, S., Dziembowski, A., Dmochowska, A., Bartnik, E., Tokatlidis, K., Stepien, P. P., and Borowski, P. (2002) Nucleic Acids Res. 30 5074-5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dziembowski, A., Malewicz, M., Minczuk, M., Golik, P., Dmochowska, A., and Stepien, P. P. (1998) Mol. Gen. Genet. 260 108-114 [DOI] [PubMed] [Google Scholar]
- 21.Dmochowska, A., Golik, P., and Stepien, P. P. (1995) Curr. Genet. 28 108-112 [DOI] [PubMed] [Google Scholar]
- 22.Margossian, S. P., Li, H., Zassenhaus, H. P., and Butow, R. A. (1996) Cell 84 199-209 [DOI] [PubMed] [Google Scholar]
- 23.Chen, H. W., Rainey, R. N., Balatoni, C. E., Dawson, D. W., Troke, J. J., Wasiak, S., Hong, J. S., McBride, H. M., Koehler, C. M., Teitell, M. A., and French, S. W. (2006) Mol. Cell. Biol. 26 8475-8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briani, F., Del Favero, M., Capizzuto, R., Consonni, C., Zangrossi, S., Greco, C., De Gioia, L., Tortora, P., and Deho, G. (2007) Biochimie (Paris) 89 145-157 [DOI] [PubMed] [Google Scholar]
- 25.Chan, H. M., Narita, M., Lowe, S. W., and Livingston, D. M. (2005) Genes Dev. 19 196-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu, Z., Vijayakumar, S., Chen, C. F., Chen, P. L., and Lee, W. H. (2004) Biochemistry 43 4781-4790 [DOI] [PubMed] [Google Scholar]
- 27.Szczesny, R. J., Obriot, H., Paczkowska, A., Jedrzejczak, R., Dmochowska, A., Bartnik, E., Formstecher, P., Polakowska, R., and Stepien, P. P. (2007) Biol. Cell 99 323-332 [DOI] [PubMed] [Google Scholar]
- 28.Bogenhagen, D. F., Rousseau, D., and Burke, S. (2007) J. Biol. Chem. [DOI] [PubMed]
- 29.Wang, Y., and Bogenhagen, D. F. (2006) J. Biol. Chem. 281 25791-25802 [DOI] [PubMed] [Google Scholar]
- 30.Bogenhagen, D. F., Wang, Y., Shen, E. L., and Kobayashi, R. (2003) Mol. Cell. Proteomics 2 1205-1216 [DOI] [PubMed] [Google Scholar]
- 31.Minczuk, M., Lilpop, J., Boros, J., and Stepien, P. P. (2005) Biochim. Biophys. Acta 1729 81-87 [DOI] [PubMed] [Google Scholar]
- 32.Pereira, M., Mason, P., Szczesny, R. J., Maddukuri, L., Dziwura, S., Jedrzejczak, R., Paul, E., Wojcik, A., Dybczynska, L., Tudek, B., Bartnik, E., Klysik, J., Bohr, V. A., and Stepien, P. P. (2007) Mech. Ageing Dev. 128 609-617 [DOI] [PubMed] [Google Scholar]
- 33.Anderson, S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H., Coulson, A. R., Drouin, J., Eperon, I. C., Nierlich, D. P., Roe, B. A., Sanger, F., Schreier, P. H., Smith, A. J., Staden, R., and Young, I. G. (1981) Nature 290 457-465 [DOI] [PubMed] [Google Scholar]
- 34.King, M. P., and Attardi, G. (1989) Science 246 500-503 [DOI] [PubMed] [Google Scholar]
- 35.Forabosco, A.., Zaffe, D., and Tosato, L. (1972) Boll. Soc. Ital. Biol. Sper. 48 33-36 [PubMed] [Google Scholar]
- 36.Wu, G., Jiang, X., Lee, W. H., and Chen, P. L. (2003) Cancer Res. 63 2589-2595 [PubMed] [Google Scholar]
- 37.Dimri, G. P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E. E., Linskens, M., Rubelj, I., and Pereira-Smith, O. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9363-9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rustin, P., Chretien, D., Bourgeron, T., Gerard, B., Rotig, A., Saudubray, J. M., and Munnich, A. (1994) Clin. Chim. Acta 228 35-51 [DOI] [PubMed] [Google Scholar]
- 39.Trounce, I. A., Kim, Y. L., Jun, A. S., and Wallace, D. C. (1996) Methods Enzymol. 264 484-509 [DOI] [PubMed] [Google Scholar]
- 40.Coskun, P. E., Beal, M. F., and Wallace, D. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10726-10731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Santiago, B., Casademont, J., and Nunes, V. (2001) Eur. J. Hum. Genet. 9 279-285 [DOI] [PubMed] [Google Scholar]
- 42.Slomovic, S., Laufer, D., Geiger, D., and Schuster, G. (2005) Mol. Cell. Biol. 25 6427-6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shadel, G. S. (2004) Trends Genet. 20 513-519 [DOI] [PubMed] [Google Scholar]
- 44.Chrzanowska-Lightowlers, Z. M., Preiss, T., and Lightowlers, R. N. (1994) J. Biol. Chem. 269 27322-27328 [PubMed] [Google Scholar]
- 45.Piechota, J., Tomecki, R., Gewartowski, K., Szczesny, R., Dmochowska, A., Kudla, M., Dybczynska, L., Stepien, P. P., and Bartnik, E. (2006) Acta Biochim. Pol. 53 157-168 [PubMed] [Google Scholar]
- 46.Attardi, G., and Ching, E. (1979) Methods Enzymol. 56 66-79 [DOI] [PubMed] [Google Scholar]
- 47.Miranda, S., Foncea, R., Guerrero, J., and Leighton, F. (1999) Biochem. Biophys. Res. Commun. 258 44-49 [DOI] [PubMed] [Google Scholar]
- 48.Spiegelman, B. M. (2007) Novartis Found. Symp. 287 60-69 [PubMed] [Google Scholar]
- 49.Okamoto, K., and Shaw, J. M. (2005) Annu. Rev. Genet. 39 503-536 [DOI] [PubMed] [Google Scholar]
- 50.Wallace, D. C. (2005) Cold Spring Harbor Symp. Quant. Biol. 70 363-374 [DOI] [PubMed] [Google Scholar]
- 51.Eubel, H., Heinemeyer, J., Sunderhaus, S., and Braun, H. P. (2004) Plant Physiol. Biochem. 42 937-942 [DOI] [PubMed] [Google Scholar]
- 52.Eubel, H., Heinemeyer, J., and Braun, H. P. (2004) Plant Physiol. 134 1450-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stroh, A., Anderka, O., Pfeiffer, K., Yagi, T., Finel, M., Ludwig, B., and Schagger, H. (2004) J. Biol. Chem. 279 5000-5007 [DOI] [PubMed] [Google Scholar]
- 54.Genova, M. L., Bianchi, C., and Lenaz, G. (2003) Ital. J. Biochem. 52 58-61 [PubMed] [Google Scholar]
- 55.McKenzie, M., Lazarou, M., Thorburn, D. R., and Ryan, M. T. (2007) Anal. Biochem. 364 128-137 [DOI] [PubMed] [Google Scholar]
- 56.Stuart, R. (2002) Biochim. Biophys. Acta 1592 79-87 [DOI] [PubMed] [Google Scholar]
- 57.Kukat, A., Kukat, C., Brocher, J., Schafer, I., Krohne, G., Trounce, I. A., Villani, G., and Seibel, P. (2008) Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed]
- 58.Larsson, N. G., Wang, J., Wilhelmsson, H., Oldfors, A., Rustin, P., Lewandoski, M., Barsh, G. S., and Clayton, D. A. (1998) Nat. Genet. 18 231-236 [DOI] [PubMed] [Google Scholar]
- 59.Parisi, M. A., Xu, B., and Clayton, D. A. (1993) Mol. Cell. Biol. 13 1951-1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang, D., Kim, S. H., and Hamasaki, N. (2007) Mitochondrion 7 39-44 [DOI] [PubMed] [Google Scholar]
- 61.Morais, R., Desjardins, P., Turmel, C., and Zinkewich-Peotti, K. (1988) In Vitro Cell. Dev. Biol. 24 649-658 [DOI] [PubMed] [Google Scholar]
- 62.Itahana, K., Campisi, J., and Dimri, G. P. (2007) Methods Mol. Biol. 371 21-31 [DOI] [PubMed] [Google Scholar]
- 63.Li, C. H., Tzeng, S. L., Cheng, Y. W., and Kang, J. J. (2005) J. Biol. Chem. 280 26193-26199 [DOI] [PubMed] [Google Scholar]
- 64.Zhang, R., Poustovoitov, M. V., Ye, X., Santos, H. A., Chen, W., Daganzo, S. M., Erzberger, J. P., Serebriiskii, I. G., Canutescu, A. A., Dunbrack, R. L., Pehrson, J. R., Berger, J. M., Kaufman, P. D., and Adams, P. D. (2005) Dev. Cell 8 19-30 [DOI] [PubMed] [Google Scholar]
- 65.Funayama, R., Saito, M., Tanobe, H., and Ishikawa, F. (2006) J. Cell Biol. 175 869-880 [DOI] [PMC free article] [PubMed] [Google Scholar]