Abstract
Fructosamine oxidases (FAOX) catalyze the oxidative deglycation of low molecular weight fructosamines (Amadori products). These proteins are of interest in developing an enzyme to deglycate proteins implicated in diabetic complications. We report here the crystal structures of FAOX-II from the fungi Aspergillus fumigatus, in free form and in complex with the inhibitor fructosyl-thioacetate, at 1.75 and 1.6Å resolution, respectively. FAOX-II is a two domain FAD-enzyme with an overall topology that is most similar to that of monomeric sarcosine oxidase. Active site residues Tyr-60, Arg-112 and Lys-368 bind the carboxylic portion of the fructosamine, whereas Glu-280 and Arg-411 bind the fructosyl portion. From structure-guided sequence comparison, Glu-280 was identified as a signature residue for FAOX activity. Two flexible surface loops become ordered upon binding of the inhibitor in a catalytic site that is about 12Å deep, providing an explanation for the very low activity of FAOX enzymes toward protein-bound fructosamines, which would have difficulty accessing the active site. Structure-based mutagenesis showed that substitution of Glu-280 and Arg-411 eliminates enzyme activity. In contrast, modification of other active site residues or of amino acids in the flexible active site loops has little effect, highlighting these regions as potential targets in designing an enzyme that will accept larger substrates.
Fructosamines are formed by condensation of glucose with the amino group of amino acids or proteins. Fructosamines are formed spontaneously, i.e. non-enzymatically, at a rate that depends on temperature, sugar anomerization rate, concentration, and turnover rate of the target proteins. In medicine, protein-bound fructosamines (also named glycated proteins) have attracted much attention since their formation is increased in diabetes and taken to be in part responsible for diabetic complications. Fructosamines are relatively unstable compounds and are precursors for advanced glycation end products (AGEs),5 some of which cause proteins cross-linking, extracellular matrix stiffening, and activation of the receptor for AGEs (RAGEs) (1, 2). As an example, the fructosamine-derived lysine-arginine cross-link glucosepane is to date the single major cross-link known to accumulate in human collagen in diabetes and aging (3).
Several years ago our laboratory initiated a search for deglycating enzymes in soil organisms with the goal of finding enzymes that could deglycate proteins. In doing so we found enzymes with “amadoriase” activity toward low molecular weight substrates in soil samples, first in Pseudomonas sp. (4) and later in Aspergillus fumigatus (5–7). The latter turned out to have similar properties with the enzyme first published by Horiuchi et al. (8) under the name fructose amino acid oxidase (EC 1.5.3). Considerable work has since been published on these enzymes, which we are referring to in this work under the generic name fructosamine oxidases (FAOX).
In addition to FAOX enzymes, two different families of enzymes acting on fructosamines have been identified: fructosamine 3-kinases (found in mammals and birds) and fructoselysine-6-phosphate deglycase, which acts in functional association with fructoselysine-6-kinase (found in bacteria) (9, 10). All these enzymes ultimately catalyze the detachment of the sugar moiety from the amine of the fructosamine, leading to its “deglycation.” Those three families of enzymes use different catalytic mechanisms and have different physiological roles; fructosamine 3-kinase is an ATP-dependent protein-repair enzyme that clears fructosamines as they form on proteins in the cell, whereas the members of the last two families, fructoselysine-6-phosphate deglycase and FAOX, are microbial enzymes enabling the utilization of glycated amino acids as energy source.
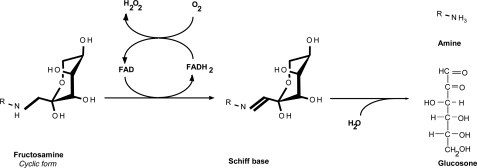
Fungal FAOXs are peroxisomal enzymes containing covalently bound FAD and catalyze the oxidation of the C-N bond between the nitrogen of the amino portion of the fructosamines and C1 of the fructosyl moiety. The resulting Schiff base is readily hydrolyzed to glucosone and an amino acid. After reduction, the reduced FAD is oxidized by molecular oxygen with concomitant release of hydrogen peroxide (Fig. 1).
FIGURE 1.
Reaction catalyzed by FAOX. FAOX catalyzes the oxidation of the C-N bond between the nitrogen of the amino portion of the fructosamines and C1 of the fructosyl moiety. The resulting Schiff base is readily hydrolyzed to glucosone and an amino acid. After the catalytic cycle, the reduced FAD is oxidized by molecular oxygen with concomitant release of hydrogen peroxide.
Many fungal species contain more than one FAOX in their genomes, suggesting complementary roles. A. fumigatus contains two FAOXs; FAOX-I, which acts best on fructosyl-ε-lysine and fructosyl aminocaproate, and FAOX-II, which has more affinity for fructose-α-glycine (5–7). All FAOXs act best on glycated amino acids, and their activity decreases as the size of the fructosamine increases and is almost undetectable on glycated proteins. The lack of enzymatic activity of FAOX enzymes toward glycated proteins frustratingly prevented them from being utilized as tools to revert protein glycation in in vitro or animal models to study the pathological consequences of protein glycation in aging and diabetes. To understand and possibly overcome by protein engineering, the lack of activity of FAOX toward glycated protein, we have solved the high resolution crystal structures of FAOX-II from A. fumigatus in free and inhibitor-bound forms.
MATERIALS AND METHODS
Expression and Purification of FAOX-II—An overnight 200-ml pre-culture (37 °C, LB containing 50 mg/liter ampicillin and 25 mg/liter chloramphenicol) of Escherichia coli BL21(DE3)pLysS carrying the expression plasmid of FAOX-II (6) was performed starting from a single freshly plated colony. 100 ml of this culture was used to inoculate 2 liters of LB medium containing 50 mg/liter ampicillin and 25 mg/liter chloramphenicol. Cells were grown at 37 °C under vigorous shaking until A600 reached 0.5, and isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.4 mm; the culture was grown for an additional 4 h at 37°C. Cells were pelleted by centrifugation at 6000 rpm for 10 min at 4 °C. The bacterial pellet was resuspended in 100 ml of 25 mm Tris, pH 8.0, 200 mm NaCl, 1 mg/ml hen egg lysozyme and submitted to three cycles of freezing and thawing. MgSO4 was added to a final concentration of 1 mm together with 20,000 units of DNase, and the preparation was incubated at 4 °C for 15 min before centrifugation at 16,000 rpm for 40 min at 4 °C. The supernatant was gently mixed with polyethylene glycol 6000 at a final concentration of 15% (w/v), and the preparation was centrifuged at 12,000 rpm for 20 min at 4 °C to give a pellet that contained >95% of FAOX-II. Resuspension in 25 mm Tris, pH 8.0, 200 mm NaCl was followed by dilution in buffer A (25 mm Tris, pH 8.0, 75 mm NaCl) to a final volume of 1100 ml before loading onto a 60-ml DEAE at 1.2 ml/min. The column was washed with 100 ml of buffer A, and the flow-through and washes, which contained FAOX-II, were collected. After adjusting the pH to 9.5 with 5 m NaOH, the volume was brought to 2000 ml by the addition of 800 ml of 25 mm Tris pH 9.5. The preparation was loaded on a 30-ml Q-Sepharose column at 5 ml/min, and the column was washed with 100 ml of buffer (25 mm Tris 8.0) before elution of proteins using a 300-ml gradient (0–100% B: 25 mm Tris, pH 8.0, 0.5 m NaCl) at 2 ml/min. Fractions were collected, and A450 nm was monitored to identify the fractions with the most intense yellow color (eluted at 15–30% B). The pooled fractions (∼16 ml) were concentrated to 40 mg/ml, and buffer was exchanged into 10 mm Tris, pH 8.0, and 10 mm NaCl, during which some protein precipitate formed and was removed by centrifugation. The final yield was 35 mg of pure protein with a specific activity of 3.3 units/mg using fructosyl-glycine as a substrate.
Analysis of the Purity of FAOX-II—The final purified FAOX-II preparation showed a single band with an apparent mass of about 50 kDa by silver-stained SDS-PAGE. Dynamic light scattering analysis indicated that the sample was monodisperse and had an apparent mass of 47 kDa. Mass spectrometry analysis revealed one peak of mass 49,703 kDa, in excellent agreement with the calculated mass of a FAD-bound enzyme; no mass was observed at 48,931 kDa, expected for apoenzyme lacking FAD (the latter represented about 80% of FAOX-II in the bacterial crude extract and was removed during the purification). The A280/A450 ratio was 0.125, in good agreement with the calculated theoretical ratio of 0.124.
Mutagenesis—Mutagenesis was performed using a Stratagene QuikChange kit according to the manufacturer's instructions. The mutant proteins were expressed under the same conditions as the wild-type enzyme. The purification procedure was simplified and involved only the DEAE-Sepharose and the Q-Sepharose purifications steps. Purity of the mutant proteins was assayed by SDS-PAGE analysis using Coomassie Blue staining and was >70%.
Expression and Purification of Selenomethionine FAOX-II— An overnight 200-ml pre-culture (37 °C, LB ampi-chloramphenicol) of bacteria E. coli BL21(DE3)pLysS carrying the expression plasmid of FAOX-II was performed starting from a single freshly plated colony. The culture was centrifuged at 4000 rpm for 20 min at 4 °C, and the pellet was resuspended in 20 ml of a minimal medium (27 g/liter Na2HPO4, 13.5 g/liter KH2PO4, 4.5 g/liter NH4Cl, 2.25 g/liter NaCl, 4.5 mm MgSO4, 18 g/liter glucose, 2.25 mg/liter thiamine, 125 mg Fe(II)SO4·7H2O) and the antibiotics ampicillin (to 50 mg/liter) and chloramphenicol (25 mg/liter). Two liters of minimal media were inoculated with 12 ml of this preparation, and the culture was grown at 37 °C under vigorous shaking until A600 reached 0.5; at this stage, 200 mg of lysine, 200 mg of phenylalanine, 200 mg of threonine, 100 mg of isoleucine, 100 mg of leucine, 100 mg of valine, and 100 mg of selenomethionine (SeMet) were added to the culture. After 20 min isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.4 mm and CoCl2 and ZnCl2 to 0.1 mm, and the culture was pursued for another 6 h. The preparation of the bacterial extract and the purification procedure were performed identically as for wild-type FAOX-II with the exception that 1 mm dithiothreitol was added to all buffers to prevent selenomethionine oxidation. This procedure allowed the preparation of 10 mg of pure SeMet-FAOX-II. Mass spectrometric analysis indicated that all eight methionine residues were replaced by selenomethionine (data not shown).
Expression and Purification of FAOX-I—His-tagged FAOX-I from A. fumigatus was expressed and purified on a His-Trap column as described previously (7).
Crystallization and Cryoprotection—Crystals of wild-type and selenomethionine FAOX-II were obtained by the sitting drop vapor diffusion method at 20 °C by mixing 1 μl of the protein solution (15 mg/ml in 10 mm Tris buffer, pH 8.0) in the presence or absence of a 3 mm concentration of the inhibitor fructosyl thioacetate (FSA) with 1 μl of the reservoir containing 0.1 m Hepes, pH 7.4, 10% isopropanol, and 18% polyethylene glycol 4000. Crystals took 2 weeks to grow and are monoclinic, with space group P21, and contain 2 FAOX-II molecules in the asymmetric unit to give a calculated Matthews coefficient of 2.1 Å3/Da. SeMet crystals grew under the same conditions as the wild type and were isomorphous. Crystals with and without inhibitor grew under similar conditions but had different unit cell dimensions (Table 1); the smaller SeMet crystals had more regular morphology and were used for data collection. The crystals were transferred to a cryoprotectant solution containing 0.1 m Hepes, pH 7.4, 20% isopropanol, 18% PEG 4000 and dunked in liquid nitrogen for data collection.
TABLE 1.
Data collection, phasing, and refinement statistics Numbers in parentheses refer to the highest resolution shell. EPE, Hepes.
| Crystal | FAOX-II (SeMet) | FAOX-II-FSA (SeMet) |
|---|---|---|
| Data collection | ||
| Cell dimensions | ||
| a (Å) | 74.3 | 87.1 |
| b (Å) | 54.3 | 53.4 |
| c (Å) | 106.3 | 103.3 |
| β (°) | 95.92 | 113.33 |
| Wavelength (Å) | 0.97929 | 1.08100 |
| Resolution (Å) | 20-1.75 (1.81-1.75) | 30-1.6 (1.66-1.6) |
| Rsym or Rmerge (%) | 6.9 (59.5) | 10.4 (24.0) |
| I/σI | 26.8 (2.9) | 14.6 (3.3) |
| Completeness (%) | 99.7 (98.4) | 91.3 (52.0) |
| Redundancy | 6.8 (6.2) | 6.8 (3.4) |
| Refinement | ||
| Resolution (Å) | 20-1.8 | 30-1.6 |
| No. of reflections | 74,880 | 105,596 |
| Rwork/Rfree (%) | 16.7/20.4 | 17.0/19.8 |
| No. of atoms | ||
| Protein | 6,678 | 7,018 |
| Ligand:FAD | 106 | 106 |
| Ligand:FSA | 32 | |
| Ligand:EPE | 15 | |
| Water | 842 | 925 |
| Average B-factors (Å2) | ||
| Protein | 17.5 | 16.6 |
| Ligands | 21.8 | 19.7 |
| Water | 29.3 | 30.0 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.013 | 0.010 |
| Bond angles (°) | 1.4 | 1.3 |
| Ramachandran | ||
| Most favored (%) | 88.4 | 88.7 |
| Additional allowed (%) | 11.6 | 11.3 |
| Disallowed (%) | 0 | 0 |
Data Collection and Structure Determination—Single wave-length SeMet anomalous dispersion data from a crystal containing the free form of FAOX-II were measured at beamline 19-ID at the Advanced Photon Source and processed using HKL2000 (11) (Table 1). The sites of 16 selenium atoms were determined and used for initial phasing to 1.8 Å resolution in SOLVE (12); phases were improved by solvent-flattening and non-crystallographic symmetry averaging in RESOLVE (13, 14) such that the mean figure of merit increased from 0.38 to 0.66. An initial FAOX-II model automatically built by RESOLVE provided the starting point for iterative cycles of interactive model building carried out in COOT (15) and refinement calculations in REFMAC (16). The quality of all models was assessed by PROCHECK (17) and MolProbity (18). The final refined model contained residues 3–57, 68–109, and 117–437 in molecule A, residues 3–57, 67–109, and 120–438 in molecule B, FAD cofactors in both molecules, and 842 waters. Diffraction data for FAOX-II bound to FSA were measured from a SeMet crystal. Data were measured at beamline X29 at the National Synchrotron Light Source and processed using HKL2000 (11) (Table 1). This structure was initially solved using the coordinates of the unbound FAOX-II. The final refined model of the FAOX-II includes residues 2–437 in molecule A, 2–438 in molecule B, FAD and FSA bound to both molecules, 926 waters, and a molecule of Hepes presumably from the crystallization mixture, bound to the surface of FAOX-II molecule B distant from the active site. Data and refinement statistics are provided in Table 1. Molecular figures were generated using PyMol (19).
Synthesis of Substrates and Inhibitor—Nα-(1-Deoxy-d-fructos-1-yl)-glycine, Nα-(1-deoxy-d-fructos-1-yl)-l-valine, Nα-(1-deoxy-d-fructos-1-yl)-l-glutamic acid, Nα-t-Boc-Nε-(1-deoxy-d-fructos-1-yl)-l-lysine (where t-Boc represents tert-butoxycarbonyl), Nε-(1-deoxy-d-fructos-1-yl)-l-lysine, Nε-(1-deoxy-d-fructos-1-yl)-amino caproic acid, Nα-t-Boc-Nε-(1-deoxy-d-ribulos-1-yl)-l-lysine, and Nε-(1-deoxy-d-ribulos-1-yl)-l-lysine were synthesized as previously described (20–23).
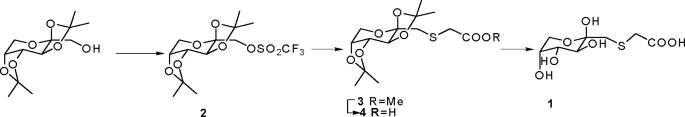
For synthesis of the inhibitor fructosyl-thioacetate (FSA), trifluoromethane sulfonic anhydride (300 mg, 1.1 mmol) was added into a solution of 2,3:4,5-di-O-isopropylidene-β-d-fructopyranose (260 mg, 1 mmol) in dichloromethane (10 ml). The resulting solution was stirred at room temperature for 1 h and evaporated under reduced pressure to give triflate 2 (see Scheme), which was dissolved in N,N-dimethylformamide (5 ml). Methyl-thioglycate (106 mg, 1 mmol) and cesium carbonate (325 mg, 1 mmol) were added, and the resulting mixture was stirred for 0.5 h and evaporated. The residue was purified by silica gel column chromatography (n-hexane/ethyl acetate 5/1) to give the ester 3 (260 mg, 74.7% yield), which was hydrolyzed by lithium hydroxide (9.5 mg, 4 mmol) in isopropanol/water (1:1, 5 ml) to give 4. The final product 1 was achieved by deprotection of 4 in TFA/water (4:1) at room temperature. Glycated bovine serum albumin and glycated histones were prepared as described previously (9).
SCHEME 1.
Enzymatic Assays—FAOX activity was measured as previously described by following the formation of hydrogen peroxide using the Amplex Red peroxidase assay (Molecular Probes) according to the manufacturer's instructions (7). Briefly, FAOX was incubated with the appropriate concentration of fructosamine in the presence of horseradish peroxidase (0.02 milliunits) and 10-acetyl-3,7-dihydroxyphenoxazine (50 μm) at 30 °C in a final volume of 200 μl, and fluorescence was detected at 590 nm using excitation at 540 nm in a microplate reader (Tecan Instrument). One unit of enzyme is defined as producing 1 mmol of hydrogen peroxide/min/mg of enzyme at 30 °C at a saturating concentration of the substrate.
RESULTS AND DISCUSSION
Previous attempts to obtain crystals from His-tagged FAOX-I (Amadoriase I) were fruitless, leading us to isolate and purify the non-tagged recombinant enzyme FAOX-II (Amadoriase II) instead. Crystals of FAOX-II in free form and bound to its FSA inhibitor grew under similar conditions, and their structures were determined to 1.75 and 1.6 Å resolution, respectively.
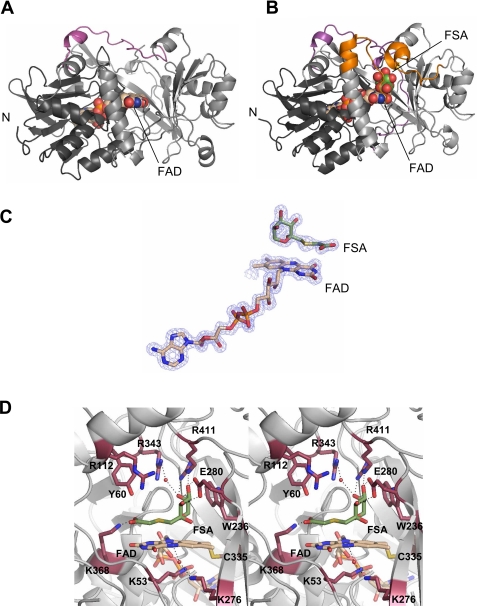
The free FAOX-II and inhibitor-bound FAOX-II-FSA structures each have two independent molecules in the asymmetric unit. Superposition of the Cα atoms of the two molecules in FAOX-II gives an r.m.s.d. of 0.3 Å; a similar superposition for the FAOX-II-FSA complex crystal gives a 0.2-Å r.m.s.d., indicating that the two molecules in each crystal are in nearly identical conformations. In the free FAOX-II structure, no continuous electron density was seen for residues 58–66 and 110–116, revealing that in the absence of ligand bound in the active site, these two surface regions are flexible. Aside from the two flexible segments near the active site, the conformation of the rest of the enzyme is very similar in the presence and absence of inhibitor, as evidenced by a low r.m.s.d. value of 0.5 for superposition of the Cα atoms of the free and complexed FAOX-II-FSA (Figs. 2, A and B). Unless otherwise indicated, structural details in the following sections are provided for molecule A in the FAOX-II-FSA crystal and are similar in both FAOX-II and FAOX-II-FSA structures.
FIGURE 2.
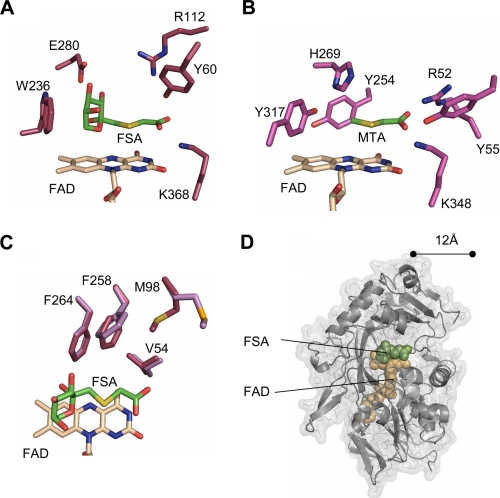
Crystal structure of FAOX-II. A, ribbon diagram of FAOX-II in free form. FAD-binding domain is in dark gray (left), the catalytic domain is in light gray (right), and FAD is in space-filling spheres with beige carbon atoms. The C-terminal portion absent in MSOX is in magenta. B, the FAOX-II-FSA complex. FSA is drawn as space-filling spheres (with green carbon atoms), and the flexible active site loops are in orange. C, 1.6 Å resolution Fo - Fc omit electron density contoured at 3.0σ for FAD and FSA. D, stereoview of FAOX-II active site. Side chains of residues surrounding FAD and FSA are shown; FAD is covalently linked to Cys-335.
FAOX-II is a two-domain protein belonging to the d-amino acid oxidase FAD-binding protein family. One domain contains a classic FAD binding motif that includes two β-sheets that are flanked by four α-helices on one side and consists of residues Ala-2—Gly-48, Ala-163—Ala-221, and Cys-335—Glu-382 (Figs. 2, A and B, in dark gray). The second, “catalytic” domain contains an 8-stranded mixed β-sheet that provides a curved wall of the active site substrate binding pocket, a long helix in a flap that covers the active site, and a second long helix at the periphery of the structure. An SSM structural homolog search (24) against the PDB data base identified monomeric sarcosine oxidase (MSOX, PDB code 1el9) from Bacillus subtilis, a protein sharing 18% sequence identity with FAOX-II (25), as the closest match, with an r.m.s.d. of 2.8 Å for 357 equivalent Cα atoms. The C-terminal 36 residues in FAOX-II form a loop over the substrate binding site which is missing in MSOX (Figs. 2, A and B, pink). l-Proline dehydrogenase (PDB code 1y56), glycine oxidase (PDB code 1ng3), the β chain of heterotetrameric sarcosine oxidase (PDB code 2gag), and N-methyl-l-tryptophan oxidase (PDB code 2apg) were also identified as structural homologs but share no significant sequence similarity with FAOX-II.
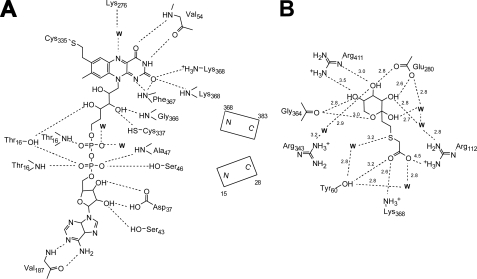
Flavin Binding—The FAD is bound to FAOX-II similarly in the presence and absence of the FSA inhibitor and in a manner that is also similar to that seen in MSOX (Fig. 2C). The cofactor is in an extended conformation and is nearly completely encased in the enzyme. In the two independent molecules in the free and inhibitor-bound structures, the isoaloxazine ring varied from planar to slightly bent (by up to 10°). The covalent, hydrogen-bonding and electrostatic interactions between FAD and FAOX-II are summarized in Fig. 3A. These include one covalent bond to Cys-335, 21 hydrogen bonds of which 3 are to solvent molecules, and 2 helix dipoles. The positive end of one of the helices (residues 367–383) is directed toward the FAD N1 and O2 atoms, helping to stabilize the electron-rich character of the flavin ring; the positive end of the other dipole points toward the negatively charged pyrophosphate group of FAD and serves to stabilize/neutralize its charge.
FIGURE 3.
Ligand binding to FAOX-II. A, interactions between FAD and FAOX-II. Hydrogen bonds are shown as dashed lines. B, FSA interactions with FAOX-II.
Arginine residues at positions 112, 343, and 411 and lysine residues at positions 53 and 368 contribute to a basic environment surrounding the FAD. Of particular interest is Lys-53, which approaches the si-side of the flavin aromatic ring; in MSOX, an Arg is found in the equivalent position. Lys-276 forms a water-mediated hydrogen bond with the flavin ring N5 atom. This interaction is also seen in MSOX and maize polyamine oxidase (PDB code 1b37), the latter of which has an unrelated protein fold. Glu-280 is the only acidic residue found in the flavin environment and is involved in hydrogen-bonding with the hydroxyl group of the fructosyl moiety of the inhibitor (see below).
FSA Inhibitor Binding—Studies on MSOX successfully took advantage of the replacement of the nitrogen of the C-N bond of the substrate by sulfur, tellurium, or selenium in the design of inhibitors (25, 26). We, therefore, synthesized an analog of fructosyl-glycine, the fructosamine for which FAOX-II displays the highest affinity, in which the α-nitrogen was replaced by a sulfur atom. This compound, FSA, was found to be a good competitive inhibitor with a Ki of 5 μm, i.e. ∼50-fold lower than the Km for fructosyl-glycine, without being a substrate of FAOX-II. Spectroscopic studies indicated that FSA formed a charge transfer species with FAOX, presumably due to the close proximity of the electron-rich sulfur of FSA and the flavin. The soaking of FAOX-II crystals in the presence of FSA caused their dissolution; co-crystallization was successfully used to obtain complex crystals. The FSA ligand could be located unambiguously on the re-side of the flavin ring (Figs. 2, B–D), with its fructosyl portion displaying a pyranose conformation, the most abundant form of fructosamines in solution (27). This finding confirms previous studies with stereospecific inhibitors adopting both furanose and pyranose conformations (28). The sulfur atom of the inhibitor, which would correspond to the nitrogen atom of a substrate, is 3.4 Å from C4α of FAD, whereas the adjacent carbon of the inhibitor, corresponding to the nascent imine carbon of a substrate, is 3.4 Å from N5 of FAD. Thus, the inhibitor appears to be bound in a position and orientation corresponding to a productive substrate binding mode. The interactions made by the ligand with the protein are summarized in Figs. 2B, 3B, and 4A and fall into three groups, those involving the inhibitor carboxylate, those involving the fructosyl group, and those forming a hydrophobic interface along one side of the inhibitor.
FIGURE 4.
Comparison of FAOX-II and MSOX actives sites. A, FSA bound to FAOX-II. B, methyl-thioacetate (MTA) bound to MSOX. His-269 in MSOX and Glu-280 in FAOX-II bind to the fructosamine hydroxyl groups. The Arg-52–Tyr-55—Lys-348 triad in MSOX and the Tyr-60–Arg-112–Lys-368 triad in FAOX-II interact with the inhibitor carboxylate. C, FAOX-II hydrophobic pocket. Met-98 and Phe-258 undergo slight rearrangement upon FSA binding (light magenta in free FAOX-II, dark magenta in the ligand-bound structure). D, FAOX-II active site accessibility. The molecular surface of the inhibitor-bound FAOX-II structure is shown with the two flexible active site loops computationally removed. The FAD cofactor and FSA inhibitor are shown as beige and green space-filling spheres, respectively. With the flexible loops removed, both FAD and FSA are partially accessible to solvent and about 12 Å from the opening of the active site.
The FSA carboxylate oxygens are hydrogen-bound to the side chains of Tyr-60, Arg-112, and Lys-368 and to two water molecules (Fig. 4A). Interestingly, the MSOX active site contains Tyr-55, Arg-52, and Lys-348 in similar positions (Fig. 4B). To address their possible importance for enzyme function, these three residues were mutated to impair their interactions with the carboxylic portion of the substrate. Mutation of Tyr-60 to Phe, Arg-112 to Glu, and Lys-368 to Met resulted in decreases of Km for fructosyl-glycine by 3-, 10-, and 5-fold, respectively, without changing the Vmax significantly (Table 2).
TABLE 2.
Effect of amino acids substitution on kinetic properties of FAOX-II ND, not detected (detection limit is 0.1 nmol/min/mg). Fructosyl-glysine is used as a substrate.
| Km | Vmax | Structural role | |
|---|---|---|---|
| mm | mmol/min/mg | ||
| FAOX-II (wild type) | 0.24 | 3.30 | |
| E280L | ND | Fructosyl binding | |
| R411L | ND | ||
| Y60F | 0.86 ± 0.09 | 4.25 ± 0.23 | Carboxylate binding |
| R112E | 2.45 ± 0.18 | 3.97 ± 0.21 | |
| K368M | 1.25 ± 0.11 | 4.36 ± 0.16 | |
| E71S | 0.20 ± 0.07 | 2.59 ± 0.09 | Salt bridge between flexible regions |
| E71R | 0.21 ± 0.05 | 4.41 ± 0.31 | |
| R114E | 2.50 ± 0.14 | 2.94 ± 0.28 | |
| R114S | 0.18 ± 0.05 | 2.05 ± 0.13 | |
| E71R/R114E | 2.06 ± 0.18 | 3.92 ± 0.37 |
The fructosyl portion of the ligand is stabilized by a series of hydrogen bonds involving the side chains of Glu-280 and Arg-411, the main chain carbonyl group of residue Gly-364, and three water molecules which are in turn coordinated by Arg-112, Arg-364, and Glu-280 (Figs. 3B and 4A). Glu-280 was mutated to Leu to address the importance of the interactions between the enzyme and hydroxyl groups 3 and 4 of the fructosyl moiety of the inhibitor. The resulting E280L mutant protein was completely inactive (<0.1 milliunits), indicating that Glu-280 is important for binding the substrate or for orienting it properly in the catalytic site. Interestingly, Glu-280 is equivalent to His-269 in B. subtilis MSOX, a residue believed to be important in the orientation of the substrate (29). Mutation of Arg-411 to Ser also resulted in an inactive FAOX-II. In this case, however, the protein was unstable and precipitated after purification, suggesting that Arg-411 also plays a structural role, which is supported by the observation that it forms a salt bridge with Asp-339. It is, therefore, difficult to assess the importance of Arg-411 in substrate binding.
Conformational Change upon Inhibitor Binding—The side chains of four residues, Phe-264, Phe-258, Met-98, and Val-54, define a hydrophobic pocket that interacts with one face of the FSA (Fig. 4C). This feature suggests that fructosamines derived from amino acids with polar side chains would be poor substrates for FAOX-II compared with those derived from amino acids with aliphatic side chains. Accordingly, fructosyl-glutamic acid is a poor substrate for which the enzyme displays 40-fold lower affinity and 3-fold lower activity as compared with fructosyl-glycine, whereas fructosyl-valine was found to be as good a substrate as fructosyl-glycine (Table 3). Interestingly, the side chains of residues Phe-258 and Met-98 undergo slight conformational changes upon ligand binding (Fig. 4C).
TABLE 3.
Substrate specificity of FAOX-I and FAOX-II ND, not detected (detection limit is 0.1 nmol/min/mg). Boc, tert-butoxycarbonyl; BSA, bovine serum albumin.
|
Fructosyl-glycine
|
Boc-fructosyl-ε-lysine
|
Fructosyl-ε-lysine
|
Fructosyl-amino caproate
|
Fructosyl-glutamate
|
Fructosyl-valine
|
Glycated BSAaactivity | Glycated histonesaactivity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | Km | Vmax | Km | Vmax | Km | Vmax | Km | Vmax | |||
| mm | mmol/min/mg | mm | mmol/min/mg | mm | mmol/min/mg | mm | mmol/min/mg | mm | mmol/min/mg | mm | mmol/min/mg | μmol/min/mg | μmol/min/mg | |
| FAOX-I | 5.8 ± 0.26 | 0.54 ± 0.03 | 0.11 ± 0.01 | 7.67 ± 0.34 | 0.08 ± 0.003 | 6.61 ± 0.12 | 0.35 ± 0.02 | 5.55 ± 0.21 | 11.3 ± 0.22 | 5.29 ± 0.31 | 5.20 ± 0.28 | 4.96 ± 0.31 | ND | 2.51 ± 0.21 |
| FAOX-II | 0.24 ± 0.04 | 3.30 ± 0.13 | 0.76 ± 0.06 | 5.79 ± 0.51 | 2.40 ± 0.31 | 1.51 ± 0.13 | 0.97 ± 0.08 | 4.03 ± 0.16 | 9.01 ± 1.10 | 0.92 ± 0.08 | 0.43 ± 0.05 | 3.16 ± 0.15 | ND | 0.70 ± 0.13 |
Tested at 10 mg/ml
The binding of FSA causes the reorganization and stabilization of two segments of the protein, corresponding to residues Gly-58 —Glu-66 and Gly-110 —Pro-119, for which only very weak and discontinuous electron density could be seen in the free FAOX-II crystal, suggesting that they are disordered (Figs. 2, A and B). Tyr-60 and Arg-112, involved in the binding of the substrate carboxylate group, are likely to mediate the stabilization of the two flexible regions by anchoring them to the ligand, thereby enclosing the substrate in the catalytic site, sheltered from the bulk solvent. FSA binding extends two helices and orders two loops which cover the active site; in the complex structure, a salt bridge between Glu-71 and Arg-114 is formed. The importance of this bridge appears to be negligible as indicated by mutagenesis; substitution of Glu-71 with Ser or Arg and of residue Arg-114 with Ser barely affected the Km or Vmax of the protein (Table 2). However, mutation of Arg-114 to Glu resulted in a 10-fold decreased affinity for the substrate, as previously observed (28). Because Arg-114 is near Arg-112, which interacts with the substrate, the effect of the charge reversal of the R114E mutation may be to alter local structure that extends to modulating the nearby Arg-112 conformation and substrate binding.
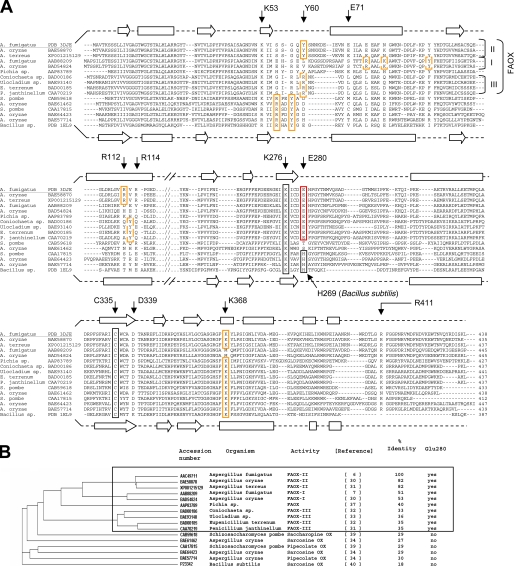
Identification of Glu-280 as a Signature Residue of FAOX Activity—The identification of Glu-280 as a residue involved in fructosyl binding and critical for FAOX-II activity suggests that this residue may be a signature for fructosamine oxidases, thus providing a guide for predicting the currently unknown functions of FAOX-II sequence homologs. A sequence data base search identified about 100 proteins sharing at least 27% sequence identity with A. fumigatus FAOX-II. Among these, eight are known to display FAOX activity, and three are known to oxidize sarcosine, one pipecolate, and a last saccharopine.
Using a FAOX-II and MSOX structural alignment as a partial guide, a sequence alignment and a phylogenic tree were constructed with the FAOX-II sequence homologs for which actual enzymatic activity has been reported (Figs. 5, A and B). Among the FAOX enzymes, three groups can be distinguished; FAOX-I enzymes, which act best on fructosyl-ε-lysine (7, 30), FAOX-IIs, which act best on fructosyl-glycine (6, 30, 31), and FAOX-IIIs, which also act on fructosamines but for which the substrate specificity is less well defined (31–33). It appears that residue Glu-280 is conserved in all FAOX enzymes but not in sarcosine oxidases (34, 40), pipecolate oxidases (34, 39), or saccharopine oxidase (39). In bacterial MSOX and most fungal sarcosine oxidases, a histidine (His-269 in the B. subtilis enzyme) is found in the position equivalent to Glu-280 in FAOX-II. This histidine plays an important role in substrate binding and positioning in the bacterial enzyme (29) and may, therefore, be considered as a signature residue for sarcosine oxidase activity. However, the Aspergillus oryzae enzyme does not have this histidine residue and has been reported to oxidize sarcosine (34). It is, however, possible that sarcosine is not the physiological substrate for that enzyme but, rather, a compound of related structure such as dimethylglycine.
FIGURE 5.
FAOX and related proteins. A, structure-guided sequence alignment. Residues involved in carboxylate binding (Tyr-60, Arg-112, and Lys-368) in FAOX-II, MSOX and related enzymes are boxed in orange. Putative carboxylate binding residues in FAOX-I and FAOX-III are in dashed orange boxes. Residue Glu-280, involved in the binding of the fructosyl moiety, is conserved in all FAOX enzymes but not in sarcosine, pipecolate, and saccharopine oxidases(red box); Glu-280 is, therefore, a signature residue for fructosamines oxidase activity. Arg-411 also binds the fructosyl moiety and forms a conserved salt bridge together with Asp-339, which appears to be required for enzyme stability. Residues Glu-71 and Arg-114 form a salt bridge connecting the two flexible regions upon ligand binding, which is conserved only in FAOX-II. Cys-335 is covalently attached to the 8α methyl group of the flavin (black box). Lys-276 makes a water-mediated hydrogen bond with N5 of the flavin (black box). Secondary structure elements for the FAOX-II and MSOX structures are shown as arrows (β-strands) and rectangles (α-helices). B, phylogenic tree of the sequence alignment. Sequence identity is related to FAOX-II from A. fumigatus.
FAOX-II Arg-411 hydrogen-bonds to the fructosyl hydroxyl groups 4 and 5 and is conserved among FAOX-I and FAOX-II sequences. Difficulties encountered in aligning the C-terminal part of FAOX-III with FAOX-II result in uncertainty whether the equivalent of Arg-411 is conserved in FAOX-III. Interestingly, FAOX-II Asp-339, which makes a salt bridge with Arg-411, is conserved among all FAOX, including FAOX-III. However, an aspartate is found in the equivalent position of several non-FAOX oxidases. Thus, neither Arg-411 nor Asp-339 qualifies as signatures for FAOX.
Convergent Evolution of the FAOX and MSOX Substrate Carboxylate Binding Site—As mentioned previously, FAOX-II residues Tyr-60, Arg-112, and Lys-368, which bind the substrate carboxylate group, are conserved as Tyr-56, Arg-53, and Lys-349 in bacterial MSOX. The bacterial MSOX carboxylate binding motif appears to be conserved not only among fungal sarcosine oxidases but also in pipecolate oxidases and saccharopine oxidase (Fig. 5A, orange boxes). The FAOX-II carboxylate binding residues are conserved in other FAOX-II enzymes, and an equivalent motif appears to be shared among FAOX-III; Arg-60, Arg-112, and Tyr-113 (in A. fumigatus FAOX-II numbering) in FAOX-III homology models are predicted to be able to hydrogen bond to the substrate carboxylate. This FAOX-III motif is similar to that of FAOX-II but with the swapping of the Arg/Lys and Tyr residues in sequence and space. In the case of FAOX-I, the carboxylate binding motif is not conserved, consistent with the higher specificity of FAOX-I for fructose-ε-lysine compared with fructosyl-glycine. In this substrate we expect the α-carboxylate in the larger fructosyl-ε-lysine to be shifted ∼4 Å relative to that in fructosyl-glycine. Structural modeling suggests that conserved FAOX-I residues Tyr-95, Arg-80, and Lys-84 (A. fumigatus FAOX-I numbering) may be properly positioned to interact with the fructosyl-ε-lysine α-carboxylate. In addition, FAOX-II Lys-368, which would potentially clash with the aliphatic side chain of fructosyl-ε-lysine, is replaced by a more compatible hydrophobic methionine residue in FAOX-I. It appears, therefore, that the triad Arg-Tyr-Lys has been selected independently for the binding of the α-carboxylic group of amino acid derivatives by FAOX-II, MSOX, and possibly FAOX-III and FAOX-I.
Implications for Active Site Accessibilities of Protein-bound Fructosamines—In FAOX-II, two flexible regions (residues Gly-58 —Glu-66 and Gly-110 —Pro-119) become ordered and cover the active site upon binding of the small FSA inhibitor (Figs. 2, A and B). It is unclear whether the closing of the active site is required for function; mutations designed to disrupt a salt bridge between the two loops do not compromise enzyme function, suggesting that the closure of the flexible loops is not critical for activity. The FSA inhibitor reveals that the substrate cleavage site is ∼12 Å from the opening of the active site cavity (Fig. 4D). This depth would be difficult for a fructosyl-lysyl residue covalently attached to a globular protein (such as bovine serum albumin) to access, consistent with the observation of a lack of activity of FAOX enzymes toward glycated bovine serum albumin (Table 3). The FAOX-II inhibitor complex structure indicates that suitable protein substrates would have the fructosyl-lysyl residue in a flexible loop or terminus to access the active site and that the enzyme two flexible loops would not be able to adopt a closed conformation (Fig. 4D). In this context it is interesting to note that glycated histones, which have a highly flexible and disordered C terminus domain containing a lysyl residue, are substrates for FAOX, although with activities ∼3 orders of magnitude lower than those observed with free fructosamines (Table 3).
Physiological Role of FAOX—In the initial studies that led to this work, our laboratory used the highly sterically inhibited and hydrophobic substrate fructosyl adamantane amine to isolate Amadoriase I and II (5, 6). However, even though these enzymes could metabolize this substrate and fructosyl ε-aminocaproate, kinetic considerations suggested that glycated amino acids might be the natural substrates for FAOX. Glycated amino acids can indeed be used as a sole energy source to sustain the growth of FAOX-containing fungi, as exemplified by the fact that the growth of Aspergillus nidulans on glycated amino acids could be prevented by knocking down the FAOX gene (35). The structure of FAOX-II provides additional arguments in favor of the hypothesis that glycated amino acids are the physiological substrates of FAOX. The catalytic site of these enzymes indeed appears to be well adapted to bind α-amino acid derivatives using a carboxylic binding motif that is shared by sarcosine oxidase.
Given the structural variety of amino acids, the presence of several FAOX in fungal genomes (e.g. up to 5 genes in Aspergillus niger) may help to cover a wide variety of fructosamine structures. FAOX-I for instance appears to be specialized in the utilization of fructosyl-ε-lysine, a fructosamine that is expected to be quantitatively important in nature since lysyl side chains are prime targets for protein glycation. However, for many fungi, even the presence of several FAOX may not be sufficient to allow efficient utilization of all glycated amino acids, as indicated for instance by the poor utilization of fructosyl glutamate by both FAOX-I and FAOX-II (Table 3).
This raises the question of the relative abundance of the various types of fructosamines in the natural environment. To the best of our knowledge, glycated amino acids have not yet been measured in fresh fruits or soils, which are among the natural locations where FAOX-containing fungi (e.g. Aspergillus) grow. Glycated amino acids have been reported on dried fruits, but they are likely to be formed during the drying process itself (36). Identification of glycated amino acids in the environment might be difficult since those relatively unstable compounds may be present in very small ecological niches and, possibly, during a well defined period of time. Finally, the presence of FAOX in the marine yeast Pichia sp. (37), a parasite of crabs and fishes, suggests that fructosamines can also be found in some marine ecosystems or in the animals that those marine yeasts parasitize.
Conclusions—This work was initiated to identify the structural basis of FAOX substrate recognition. The structure of FAOX-II-FSA identifies an active site pocket that is ∼12 Å deep, explaining why glycated lysyl residues bound to globular protein are poor substrates of the enzyme and only when they are located in very flexible regions of the protein. Two flexible loops are disordered in the free enzyme and close over the active site upon FSA binding. Mutations to disrupt the closed loop interactions do not significantly compromise enzyme activity, suggesting that increasing loop flexibility near the active site may be an approach in efforts to engineer a FAOX with increased activity on glycated protein substrates.
The characterization of the FAOX-II substrate-binding site in this work provides a basis for identifying likely substrates for related enzymes and a valuable structural scaffold for mutagenesis. Many groups have reported efforts, with the help of computerized models, to improve substrate selectivity of FAOX enzymes for fructosylvaline to develop enzymatic assays utilizing proteolytic digests of glycated hemoglobin (38). The FAOX-II structure will aid in this endeavor. Finally, the FAOX-II structure highlights the critical role of Glu-280 for fructosamine recognition. The presence of this residue allows one to discriminate between fructosamine oxidases and enzymes acting on other substrates in the FAOX/MSOX family of proteins.
Acknowledgments
We thank Drs. Bruce Palfey and Emile Van Schaftingen for helpful criticism of the manuscript. Data for this study were measured at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, and at beamline X29 of the National Synchrotron Light Source. The Argonne National Laboratory is operated by UChicago Argonne, LLC for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02–06CH11357. Financial support for National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and from the National Center for Research Resources of the National Institutes of Health.
The atomic coordinates and structure factors (codes 3DJD and 3DJE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants AG 18436 and EY07099 (to V. M. M.) and DK075897 (to V. C. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AGE, advanced glycation end product; FAOX, fructosamine oxidase; SeMet, selenomethionine; FSA, fructosyl thio-acetate; r.m.s.d., root mean square deviation; MSOX, monomeric sarcosine oxidase.
References
- 1.Monnier, V. M., Mustata, G. T., Biemel, K. L., Reihl, O., Lederer, M. O., Zhenyu, D., and Sell, D. R. (2005) Ann. N. Y. Acad. Sci. 1043 533-544 [DOI] [PubMed] [Google Scholar]
- 2.Wendt, T., Tanji, N., Guo, J., Hudson, B. I., Bierhaus, A., Ramasamy, R., Arnold, B., Nawroth, P. P., Yan, S. F., D'Agati, V., and Schmidt, A. M. (2003) J. Am. Soc. Nephrol. 14 1383-1395 [DOI] [PubMed] [Google Scholar]
- 3.Sell, D. R., Biemel, K. M., Reihl, O., Lederer, M. O., Strauch, C. M., and Monnier, V. M. (2005) J. Biol. Chem. 280 12310-12315 [DOI] [PubMed] [Google Scholar]
- 4.Saxena, A. K., Saxena, P., and Monnier, V. M. (1996) J. Biol. Chem. 271 32803-32809 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi, M., Pischetsrieder, M., and Monnier, V. M. (1997) J. Biol. Chem. 272 3437-3443 [DOI] [PubMed] [Google Scholar]
- 6.Takahashi, M., Pischetsrieder, M., and Monnier, V. M. (1997b) J. Biol. Chem. 272 12505-125057 [DOI] [PubMed] [Google Scholar]
- 7.Wu, X., Takahashi, M., Chen, S. G., and Monnier, V. M. (2000) Biochemistry 39 1515-1521 [DOI] [PubMed] [Google Scholar]
- 8.Horiuchi, T., Kurokawa, T., and Sati, N. (1989) Agric. Biol. Chem. 53 103-110 [Google Scholar]
- 9.Delpierre, G., Rider, M. H., Collard, F., Stroobant, V., Vanstapel, F., Santos, H., and Van Schaftingen, E. (2000) Diabetes 49 1627-1634 [DOI] [PubMed] [Google Scholar]
- 10.Wiame, E., Delpierre, G., Collard, F., and Van Schaftingen, E. (2002) J. Biol. Chem. 277 42523-42529 [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-325 [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terwilliger, T. C. (2000) Acta Crystallogr. Sect. D 56 965-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D 59 34-44 [Google Scholar]
- 15.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 16.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 17.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 18.Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B., Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007) Nucleic Acids Res. 34 375-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLano, W. L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA
- 20.Röper, H., Röper, S., and Heyns, K. (1983) Carbohydr. Res. 116 183-195 [Google Scholar]
- 21.Beksan, E., Schieberle, P., Robert, F., Blank, I., Fay, L. B., Schlichtherle-Cerny, H., and Hofmann, T. (2003) J. Agric. Food Chem. 51 5428-5436 [DOI] [PubMed] [Google Scholar]
- 22.Thornalley, P. J., Langborg, A., and Minhas, H. S. (1999) Biochem. J. 344 109-116 [PMC free article] [PubMed] [Google Scholar]
- 23.Grandhee, S. K., and Monnier, V. M. (1991) J. Biol. Chem. 266 11649-11653 [PubMed] [Google Scholar]
- 24.Krissinel, E., and Henrick, K. (2004) Acta Crystallogr. Sect. D 60 2256-2268 [DOI] [PubMed] [Google Scholar]
- 25.Trickey, P., Wagner, M. A., Jorns, M. S., and Mathews, F. S. (1999) Structure 7 331-345 [DOI] [PubMed] [Google Scholar]
- 26.Wagner, M. A., Trickey, P., Chen, Z. W., Mathews, F. S., and Jorns, M. S. (2000) Biochemistry 39 8813-8824 [DOI] [PubMed] [Google Scholar]
- 27.Tagami, U., Akashi, S., Mizukoshi, T., Suzuki, E., and Hirayama, K. (2000) J. Mass Spectrom. 35 131-138 [DOI] [PubMed] [Google Scholar]
- 28.Wu, X., Chen, S. G., Petrash, J. M., and Monnier, V. M. (2002) Biochemistry 41 4453-4458 [DOI] [PubMed] [Google Scholar]
- 29.Zhao, G., Song, H., Chen, Z. W., Mathews, F. S., and Jorns, M. S. (2002) Biochemistry 41 9751-9764 [DOI] [PubMed] [Google Scholar]
- 30.Akazawa, S., Karino, T., Yoshida, N., Katsuragi, T., and Tani, Y. (2004) Appl. Environ. Microbiol. 70 5882-5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, N., Sakai, Y., Isogai, A., Fukuya, H., Yagi, M., Tani, Y., and Kato, N. (1996) Eur. J. Biochem. 242 499-505 [DOI] [PubMed] [Google Scholar]
- 32.Hirokawa, K., Gomi, K., and Kajiyama, N. (2003) Biochem. Biophys. Res. Commun. 311 104-111 [DOI] [PubMed] [Google Scholar]
- 33.Fujiwara, M., Sumitani, J., Koga, S., Yoshioka, I., Kouzuma, T., Imamura, S., Kawaguchi, T., and Arai, M. (2006) J. Biosci. Bioeng. 102 241-243 [DOI] [PubMed] [Google Scholar]
- 34.Yoshida, N., Akazawa, S., Karino, T., Ishida, H., Hata, Y., Katsuragi, T., Tani, Y., and Takagi, H. (2007) J. Biosci. Bioeng. 104 424-427 [DOI] [PubMed] [Google Scholar]
- 35.Yoshida, N., Akazawa, S., Kuwahara, A., Katsuragi, T., and Tani, Y. (2005) FEMS Microbiol. Lett. 248 141-145 [DOI] [PubMed] [Google Scholar]
- 36.Davidek, T., Kraehenbuehl, K., Devaud, S., Robert, F., and Blank, I. (2005) Anal. Chem. 77 140-147 [DOI] [PubMed] [Google Scholar]
- 37.Ferri, S., Miura, S., Sakaguchi, A., Ishimura, F., Tsugawa, W., and Sode, K. (2004) Mar. Biotechnol. (NY) 6 625-632 [DOI] [PubMed] [Google Scholar]
- 38.Miura, S., Ferri, S., Tsugawa, W., Kim, S., and Sode, K. (2008) Protein Eng. Des. Sel., 4 233-239 [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, N., Akazawa, S., Katsuragi, T., and Tani, Y. (2004) J. Biosci. Bioeng. 97 278-280 [DOI] [PubMed] [Google Scholar]
- 40.Koyama, Y., Yamamoto-Otake, H., Suzuki, M., and Nakano, E. (1991) Agric. Biol. Chem. 55 1259-1263 [PubMed] [Google Scholar]