Abstract
Adiponectin is an adipokine with potent anti-inflammatory properties. Treatment of macrophages with adiponectin results in a suppression of lipopolysaccharide (LPS)-stimulated cytokine production. Here we investigated the transcriptional and post-transcriptional mechanisms by which adiponectin suppresses LPS-stimulated tumor necrosis factor (TNF)-α production. Treatment of RAW 264.7 macrophages with LPS increased TNF-α promoter-driven luciferase activity (TNF-α promoter/Luc activity) by 20-fold over basal. After culture with 1 μg/ml globular adiponectin (gAcrp) for 18 h, TNF-α promoter/Luc activity was increased even in the absence of LPS; further challenge with LPS only increased TNF-α promoter/Luc activity by 1.4-fold. Treatment with gAcrp decreased LPS-stimulated ERK1/2 phosphorylation and IκB degradation and suppressed the ability of LPS to increase the DNA binding activity of Egr-1 and p65. gAcrp also suppressed LPS-mediated stabilization of TNF-α mRNA. In controls cells, the half-life of TNF-α mRNA was increased from ∼30 min at base line to ∼80 min in response to LPS. After treatment with gAcrp for 18 h, LPS failed to increase TNF-α mRNA stability. This gAcrp-mediated loss of stimulus-induced stabilization of TNF-α mRNA required the presence of the TNF-α 3′-untranslated region and was associated with an increase in expression and RNA binding activity of tristetraprolin, an mRNA-binding protein that destabilizes TNF-α mRNA. In summary, these data characterize the complex transcriptional and post-transcriptional effects of gAcrp on LPS-stimulated TNF-α expression in macrophages. gAcrp treatment profoundly suppressed the ability of LPS to increase TNF-α transcription and reduced the stimulus-induced stabilization of TNF-α mRNA in response to LPS.
Adiponectin (Acrp30) is an adipokine secreted by adipose tissue that regulates glucose and lipid metabolism in liver and muscle (1, 2). Adiponectin also influences the activity of the innate immune system, acting as a mediator between adipose tissue and inflammatory responses (3). Full-length adiponectin is a 30-kDa protein containing a collagen-like domain as well as a C1q-like globular head domain at the carboxyl terminus. The collagen-like domain can be cleaved from the full-length protein to generate globular adiponectin (gAcrp)3 (1). Adiponectin acts on target tissues via the activation of adiponectin receptors 1 and 2 (AdipoR1 and AdipoR2) (4). Although both AdipoR1 and AdipoR2 are expressed in monocytes and macrophages (5), accumulating evidence indicates that AdipoR1 is critical to the anti-inflammatory effects of adiponectin (6). The anti-inflammatory activity of adiponectin has been implicated as a potential mechanism for the antiatherogenic properties of adiponectin (7) as well as the potential therapeutic effects of adiponectin in alcoholic and non-alcoholic liver injury (8).
Adiponectin targets multiple sites to dampen inflammatory responses. Adiponectin inhibits the growth of myelomonocytic progenitor cells (9). Furthermore adiponectin decreases the ability of mature macrophages to respond to activation (9), suppressing phagocytic activity, as well as lipopolysaccharide (LPS)-stimulated cytokine production in macrophages (9–11). In response to long term treatment with either the globular or full-length forms of adiponectin, macrophages develop a decreased sensitivity to LPS-stimulated signaling (9). The phenotypic characteristics of adiponectin-treated macrophages are similar to those observed in response to repeated/prolonged exposure to LPS/endotoxin or what is often called “endotoxin tolerance.” Interestingly several recent reports demonstrate that short term treatment of macrophages with adiponectin first increases the expression of inflammatory cytokines, such as TNF-α and interleukin (IL)-6 (12, 13). Continued exposure to adiponectin then promotes the expression of anti-inflammatory mediators, such as IL-10 (12, 14), and the eventual development of tolerance to proinflammatory signals, including resistance to subsequent challenge with the toll-like receptor 4 ligand, LPS, and the toll-like receptor 3 ligand, poly(I·C) (12, 13).
TNF-α expression is regulated at transcriptional, post-transcriptional, and translational levels (15–17). Transcriptional regulation of TNF-α can involve the activation of distinct sets of transcription factors binding to at least two regions of the TNF-α promoter, which contains NFκB, Egr-1, cAMP-response element, and AP-1 binding sites (18, 19). The TNF-α mRNA, like other short lived mRNAs, contains A + U-rich elements (AREs) in its 3′-untranslated region (UTR) that function as post-transcriptional regulatory elements, contributing to control of TNF-α mRNA nuclear export (20), translational repression (21–23), and mRNA stability (24–27). The stability of the TNF-α mRNA is regulated by the binding of several RNA-binding proteins, including tristetraprolin (TTP), HuR, and AUF-1 to the 3′-UTR (17). Recent data have also suggested an important role for microribonucleoprotein-related proteins, including fragile X mental retardation-related protein 1 and argonaute 2, in the post-transcriptional regulation of TNF-α mRNA (28, 29). Micro-RNAs and microribonucleoprotein-related proteins appear to be involved in both translational repression and activation (29, 30) as well as mRNA stability (31).
To better understand the molecular mechanisms by which adiponectin desensitizes macrophages to LPS-stimulated TNF-α production, here we investigated the impact of 18-h treatment of RAW 264.7 macrophages with gAcrp on the transcriptional and post-transcriptional regulation of LPS-stimulated TNF-α expression. Long term treatment with gAcrp increased TNF-α promoter-driven luciferase activity. gAcrp disrupted the regulation of TNF-α transcription and caused a shift in the activity of key transcription factors from the initially important contributions of NFκB and Egr-1 to a predominant role of AP-1 in maintaining TNF-α promoter activity. We also found that gAcrp suppressed LPS-mediated stabilization of TNF-α mRNA; this effect of gAcrp was associated with a decreased ability of LPS to stimulate phosphorylation of p38 MAPK and an increase in the interaction of TTP with the 3′-UTR of the TNF-α mRNA.
EXPERIMENTAL PROCEDURES
Materials—LPS from Escherichia coli serotype 026:B6 (tissue culture-tested, L-2654) was purchased from Sigma; all experiments were carried out with a single lot of LPS (Lot number 064K4077). Recombinant human gAcrp expressed in E. coli was purchased from Peprotech, Inc. (Rocky Hill, NJ). gAcrp preparations contained less than 0.2 ng of LPS/μg of protein. Endotoxin contamination was routinely monitored in the laboratory using a kinetic chromogenic test based on the Limulus amebocyte lysate assay (Kinetic-QCL, BioWhittaker, Walkersville, MD).
All cell culture reagents were from Invitrogen. Antibodies were from the following sources: Egr-1, p50, p65, TTP, AP-1, Hsc70, and phospho-ERK (Santa Cruz Biotechnology, Santa Cruz, CA); IκB (Cell Signaling Technology Inc.); phospho-p38 (Promega); and total ERK1/2 (Upstate Biotechnology, Lake Placid, NY). Anti-rabbit and anti-mouse IgG-peroxidase were purchased from Chemicon (Indianapolis, IN). Endotoxin-free plasmid preparation kits were from Qiagen (Valencia, CA). Luciferase reporter constructs have been described previously. pTNF-α-LUC (containing –615 to +1 of the TNF-α promoter and mutated/truncated constructs in a luciferase reporter) (19) was from N. Mackman, University of California, San Diego, CA. pTNF-α-5′UTR-LUC (containing –993 to +110 of the TNF-α promoter and 5′-UTR in pGL2 vector) and pTNF-α-5′UTR-LUC-3′UTR (containing –993 to +100 of the TNF-α promoter and 5′-UTR as well as the 833-base TNF-α 3′-UTR in pGL2 vector) luciferase reporters were from Joyce and co-workers (32). SV40-LUC-3′UTR luciferase reporter (containing the TNF-α 3′-UTR in pGL2 vector) and the SV40-LUC-3′UTR-ARE deletion were from Kruys and co-workers (33). pAP1-Luc and pNFκB-Luc cis reporter plasmids were from Stratagene (La Jolla, CA).
Culture of RAW 264.7 Macrophages and Luciferase Assays—The murine RAW 264.7 macrophage-like cell line was routinely cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin at 37 °C and 5% CO2. For luciferase reporter assays, RAW 264.7 macrophages were grown in 6-well plates to 60–70% confluency and then transiently transfected with control and expression vectors using Superfect transfection reagent (Qiagen) according to the manufacturer's instructions. Cells were co-transfected with pTK-RL (Promega), an expression vector for Renilla luciferase under the control of the thymidine kinase promoter. Transfected cells were subcultured and seeded at 10.2 × 104/cm2 in 96-well plates. After 24 h, medium was removed, and cells were stimulated or not with gAcrp or LPS in Dulbecco's modified Eagle's medium/fetal bovine serum (times of treatment are indicated in the figure legends). Cells were treated for a minimum of 4 h with either gAcrp or LPS to ensure adequate expression and activity of luciferase. Cells were then extracted in lysis buffer, and luciferase activities were measured using the Dual-Luciferase assay system (Promega). Data were then expressed as a ratio of heterologous promoter-driven luciferase activity divided by the activity of the Renilla luciferase. Treatment with gAcrp for up to 18 h or LPS for up to 4 h had no effect on cell number, protein concentration, or Renilla luciferase activity (data not shown).
Western Blot Analysis—RAW 264.7 macrophages were treated with or without 1 μg/ml gAcrp for 18 h. Cells were stimulated or not with 100 ng/ml LPS for varying lengths of time as indicated in the figure legends. Phosphorylation of ERK1/2 and p38 MAPK and the quantity of IκB were measured by Western blot analysis as described previously (12). Immunoreactive TTP was also measured by Western blot and normalized to Hsc70 as a loading control.
Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assays—RAW 264.7 macrophages were treated with or without 1 μg/ml gAcrp for 0–18 h and then stimulated or not with 100 ng/ml LPS for 60 min. LPS stimulation of RAW 264.7 macrophages for 60 min results in a maximal activation of Egr-1 and p65 DNA binding activity under these conditions (34). Nuclei were isolated using the Nuclei EZ Prep kit from Sigma, and nuclear proteins were extracted (34). Nuclear extracts (5 μg of protein) were then used to assess DNA binding activity by electrophoretic mobility shift assay using an oligonucleotide probe for the sequence for the Egr-1 binding site in the TNF-α promoter (Integrated DNA Technologies, Inc., Coralville, IA) or for the consensus sequences for the NFκB, SP-1, or AP-1 binding site (Santa Cruz Biotechnology) as described previously (34). In some experiments, extracts were incubated with antibodies against Egr-1, p65, p50, or AP-1 or control IgG. Controls were also carried out in cells pretreated with polymyxin B, an antibiotic that binds to and inactivates LPS, prior to stimulation with gAcrp.
Real Time PCR and Northern Blot Analysis—Total RNA was isolated from RAW 264.7 macrophages using the RNeasy Micro kit (Qiagen) with on-column DNA digestion using the RNase-free DNase set (Qiagen) according to the manufacturer's instructions. For real time PCR, 200–300 ng of total RNA were reverse transcribed using the RETROscript kit (Ambion, Austin, TX) with random decamers as primers. Real time PCR amplification was performed in an MX3000P apparatus (Stratagene) using the Brilliant SYBR Green QPCR Master Mix (Stratagene). The relative amount of target mRNA was determined using the comparative threshold (Ct) method by normalizing target mRNA Ct values to those for β-actin (ΔCt). The primer sequences are as follows: TNF-α, F 5′-CCC TCA CAC TCA GAT CAT CTT CT-3′ and R 5′-GCT ACG ACG TGG GCT ACA G-3′; TTP, F 5′-TCTCTGCCATCTACGAGAGCC-3′ and R 5′-CCAGTCAGGCGAGAGGTGA-3′; and β-actin, F 5′-CTT TGC AGC TCC TTC GTT GC-3′ and R 5′-ACG ATG GAG GGG AAT ACA GC-3′. All primers used for real time PCR analysis were synthesized by Integrated DNA Technologies, Inc. For Northern blots, 10 μg of total RNA was electrophoresed through 1.1% agarose-formaldehyde gels, and the quantity of TNF-α mRNA was measured using four anti-sense oligonucleotides corresponding to different regions of the mature TNF-α mRNA as described previously (34). Quantity of 18 S RNA was measured as a loading control.
RNA Binding Assay—TTP binding to TNF-α mRNA was measured by immunoprecipitating TTP from RAW 264.7 macrophages cultured for 18 h with or without 1 μg/ml gAcrp. Cells were stimulated or not with 100 ng/ml LPS during the last 2 h of gAcrp exposure. Cells were removed from the cell culture dish, washed twice with cold phosphate-buffered saline, and fixed in 0.1% formaldehyde, and lysates were prepared as described previously (35, 36). TTP was immunoprecipitated from the cell lysates by overnight incubation with 10 μg/ml anti-TTP antibody followed by the addition of 50 μl of protein G-agarose beads. The beads were then washed, and RNA was purified (35, 36). The total RNA sample was reverse transcribed, and 5% of the total reverse transcribed product was used for PCR. The sequences of the primer pairs within the TNF-α 3′-UTR were as follows: F 5′-AGC CCC CAG TCT GTA TCC TT-3′ and R 5′-CTC CCT TTG CAG AAC TCA GG-3′. Products of the PCRs were then separated by agarose gel electrophoresis.
Statistical Analysis—Values reported are means ± S.E. Data were analyzed by the general linear models procedure followed by least square means analysis of differences between groups (SAS, Cary, NC). Statistical analysis of real time PCR data was performed using ΔCt values.
RESULTS
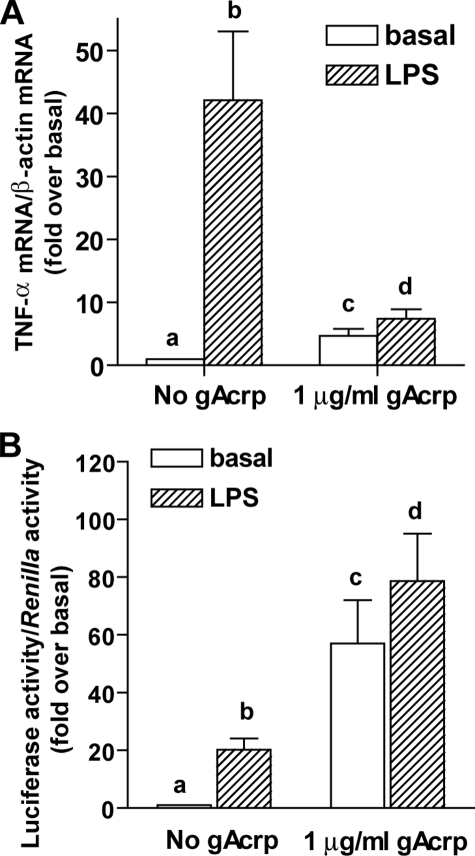
Adiponectin has potent anti-inflammatory effects on macrophages. We have reported previously that exposure of primary cultures of rat Kupffer cells to gAcrp or full-length adiponectin for 18 h suppresses subsequent LPS-stimulated TNF-α production (11). This anti-inflammatory effect of gAcrp can be modeled by treatment of RAW 264.7 macrophages with gAcrp for 8–18 h (12). Therefore, RAW 264.7 macrophages are a convenient cell model to study the molecular mechanisms for the anti-inflammatory effects of gAcrp. Here we investigated the transcriptional and post-transcriptional mechanisms by which gAcrp treatment desensitizes RAW 264.7 macrophages to LPS. Stimulation of RAW 264.7 macrophages with 100 ng/ml LPS for 2 h increased the accumulation of TNF-α mRNA by 40-fold (Fig. 1A). When cells were cultured overnight with 1 μg/ml gAcrp, basal (no LPS treatment) accumulation of TNF-α mRNA was increased 4–5-fold, but subsequent stimulation of TNF-α mRNA accumulation by LPS was greatly suppressed (Fig. 1A). To investigate the effects of adiponectin on the transcriptional regulation of TNF-α, RAW 264.7 macrophages were co-transfected with a luciferase reporter construct under the control of the TNF-α promoter and a Renilla luciferase reporter construct under the control of the thymidine kinase promoter. In this experimental design, luciferase activity, expressed relative to Renilla activity, provides an indication of rates of TNF-α transcription. (TNF-α promoter-driven luciferase activity is abbreviated here for convenience as “TNF-α promoter/Luc activity.”) In cells not treated with gAcrp, basal rates of TNF-α promoter/Luc activity were minimal; stimulation with LPS increased TNF-α promoter activity by 16-fold (Fig. 1B). In contrast, after overnight treatment with gAcrp, basal rates (no LPS treatment) of TNF-α promoter/Luc activity were increased more than 60-fold, but subsequent stimulation of TNF-α promoter/Luc activity with LPS was suppressed compared with controls (Fig. 1B). The differential effects of gAcrp on accumulation of TNF-α mRNA and TNF-α promoter/Luc activity suggest that gAcrp disrupts both transcriptional and post-transcriptional regulation of LPS-stimulated TNF-α expression.
FIGURE 1.
Differential effects of globular adiponectin on LPS-stimulated TNF-α mRNA accumulation and TNF-α promoter-driven luciferase activity. A, RAW 264.7 cells were cultured for 18 h in the presence or absence of 1 μg/ml gAcrp. Cells were then stimulated with 100 ng/ml LPS for 2 h, and accumulation of TNF-α mRNA was measured by real time PCR. Values represent TNF-α mRNA normalized to β-actin mRNA (means ± S.E., n = 8). Values with different superscript letters (a–d) are significantly different from each other (p < 0.05). B, RAW 264.7 macrophages were co-transfected with the TNF-α promoter-luciferase reporter and a Renilla luciferase control. Twenty-four hours after transfection, cells were then treated with or without 1 μg/ml gAcrp for 18 h and then stimulated or not with 100 ng/ml LPS for 4 h. Values represent relative luciferase activity (corrected for Renilla luciferase activity; means ± S.E., n = 10). Values with different superscript letters (a–d) are significantly different from each other (p < 0.05).
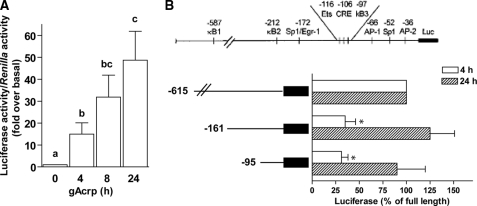
We first investigated the mechanisms by which gAcrp increased TNF-α promoter/Luc activity during overnight exposure to gAcrp. gAcrp stimulated TNF-α promoter/Luc activity within 4 h after treatment, and TNF-α promoter-driven luciferase activity continued to accumulate over time (Fig. 2A). Transcriptional activation of TNF-α in macrophages primarily involves the recruitment of Egr-1, NFκB, and AP-1 to the TNF-α promoter (18, 19). Using truncation mutants of the TNF-α promoter, we found that at early times of exposure to gAcrp multiple promoter elements contributed to gAcrp-stimulated TNF-α promoter/Luc activity. Truncations of the promoter to –161, deleting two κB sites as well as the Egr-1 site, decreased TNF-α promoter/Luc activity by 70% over the 4-h treatment with gAcrp. This is consistent with previous data demonstrating a key role for Egr-1 and NFκB in mediating increased TNF-α expression after 1–4-h exposure to gAcrp (12). However, after 24 h, TNF-α promoter/Luc activity was not affected by removal of these promoter elements (Fig. 2B). The –95 truncation, containing only two AP-1 sites and an SP-1 site, supported only 30% of gAcrp-stimulated TNF-α promoter/Luc activity after 4-h treatment with gAcrp. However, after 24 h, the –95 truncation was able to sustain full gAcrp-stimulated TNF-α promoter/Luc activity (Fig. 2B).
FIGURE 2.
Differential contributions of the Egr-1, NFκB, and AP-1 sites in the TNF-α promoter to gAcrp-stimulated TNF-α promoter-driven luciferase activity. A, RAW 264.7 macrophages were co-transfected with the TNF-α promoter-luciferase reporter and a Renilla luciferase control. Twenty-four hours after transfection, cells were then treated with 1 μg/ml gAcrp for 0–24 h, and TNF-α promoter-luciferase activity was measured. Values represent relative luciferase activity (corrected for Renilla luciferase activity; means ± S.E., n = 4–5). Values with different superscript letters (a–c) are significantly different from each other (p < 0.05). B, RAW 264.7 macrophages were transiently transfected with the full-length TNF-α promoter as well as a series of truncated promoters in which the κB1, κB2, and Egr-1 sites were removed (–161) or a further truncation removing the cAMP-response element (CRE) and κB3 sites (–95). RAW 264.7 macrophages were co-transfected with a Renilla luciferase reporter to control for transfection efficiency. Twenty-four hours after transfection, cells were then treated with 1μg/ml gAcrp for 24 h, and luciferase activity was measured. Values represent relative luciferase activity (corrected for Renilla luciferase activity) expressed as -fold over basal (basal luciferase activity for any of the constructs was not affected by the time of adiponectin treatment; means ± S.E., n = 4–5). *, p < 0.05 compared with full-length promoter.
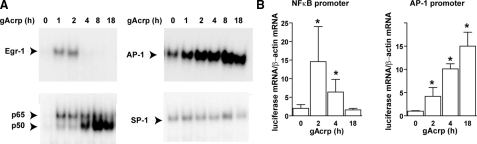
To further understand these differential effects of gAcrp on regulation of TNF-α promoter/Luc activity, we measured the effects of gAcrp on the DNA binding activity of Egr-1, NFκB, AP-1, and SP-1. DNA binding activities of Egr-1, AP-1, and NFκB increased in response to gAcrp, but there was no change in SP-1 DNA binding (Fig. 3A). Supershift assays were used to confirm the identify of each of the transcription factors (data not shown). Pretreatment with polymyxin B did not prevent the effects of gAcrp on the DNA binding activities of Egr-1, NFκB, or AP-1 (data not shown). Activation of Egr-1 was rapid and transient, whereas AP-1 binding continued to increase over time. NFκB DNA binding activity also increased rapidly with a shift from predominantly the p65 subunit to predominantly the p50 subunit over the 4-h exposure to gAcrp (Fig. 3A). To further define the changing activity of AP-1 and NFκB, we made use of luciferase cis reporter plasmids driven by promoters containing NFκB or AP-1 consensus binding sites. gAcrp transiently increased NFκB-dependent luciferase mRNA expression (Fig. 3B) with kinetics that paralleled the gAcrp-stimulated increase in p65 DNA binding activity. In contrast, the gAcrp-stimulated increase in AP-1-dependent luciferase mRNA accumulation continued to increase over time (Fig. 3B). Taken together, these data suggest that the regulation of TNF-α promoter activity by gAcrp at early time points is due primarily to Egr-1 and NFκB activation but that over time AP-1 becomes the critical element stimulating TNF-α promoter activity.
FIGURE 3.
Time-dependent activation of Egr-1, NFκB, and AP-1 DNA binding and activity in response to globular adiponectin. A, RAW 264.7 macrophages were treated with 1 μg/ml gAcrp for 0–18 h. Nuclear extracts were then prepared and used to measure the binding of nuclear proteins to oligonucleotides specific for Egr-1, NFκB, AP-1, and SP-1 DNA binding sites. Images are representative of at least three independent experiments. B, RAW 264.7 macrophages were co-transected with either pAP1-Luc or NFκB-Luc cis reporter plasmids. Twenty-four hours after transfection, cells were then treated with 1 μg/ml gAcrp for 0–18 h, and luciferase mRNA were measured by quantitative real time PCR. Values are expressed relative to β-actin mRNA (means ± S.E., n = 4–5). *, p < 0.05 compared with cells not treated with gAcrp.
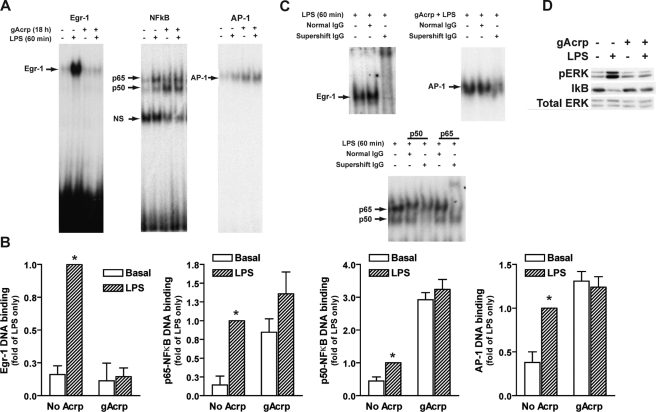
In addition to the impact of gAcrp on basal TNF-α promoter activity and mRNA accumulation, gAcrp also affected the ability of LPS to stimulate TNF-α promoter activity and mRNA accumulation (Fig. 1). Therefore, we investigated the effects of gAcrp on the ability of LPS to activate Egr-1, NFκB, and AP-1 DNA binding activity. In control cells, the DNA binding activity of these transcription factors was minimal but increased in response to LPS treatment (Fig. 4, A and B). However, after 18-h exposure to gAcrp, in addition to the changes in DNA binding activity in the absence of LPS already described in Fig. 3, the activation of these transcription factors in response to LPS was also affected by gAcrp. After 18-h treatment with gAcrp the following were observed: 1) Egr-1 DNA binding activity was undetectable in the absence of LPS and in response to LPS stimulation, 2) NFκB DNA binding activity was shifted to predominantly p50 in the absence of LPS; however, LPS-stimulated increases in p65 or p50 binding were suppressed after treatment with gAcrp, 3) AP-1 binding was increased in the absence of LPS but not further increased in response to LPS (Fig. 4, A and B). Supershift assays for each transcription factor are shown in Fig. 4C. Pretreatment of RAW 264.7 macrophages with polymyxin B, which binds to and inactivates LPS, prior to gAcrp stimulation did not affect DNA binding activity during overnight exposure to gAcrp (data not shown), indicating that the effects of gAcrp were not due to any contamination of the preparation with LPS.
FIGURE 4.
Overnight treatment with globular adiponectin disrupts Egr-1, NFκB, and AP-1 DNA binding activity. A, RAW 264.7 cells were cultured for 18 h in the presence or absence of 1 μg/ml gAcrp. Cells were then stimulated with 100 ng/ml LPS for 1 h. Nuclear extracts were then prepared and used to measure the binding of nuclear proteins to oligonucleotides specific for Egr-1, NFκB, and AP-1 DNA binding sites. NS, nonspecific band. B, densitometric analysis of electrophoretic mobility shift assays. Values represent means ± S.E. relative to cells stimulated with LPS (n = 3). *, p < 0.05 compared with cells not treated with LPS. C, supershift assays were carried out by incubating nuclear extracts with specific antibodies for each transcription factor or normal immunoglobulin control antibodies prior to the electrophoretic mobility shift assay. These controls are shown in the right-hand panels. Representative images are shown from three to five independent experiments. D, RAW 264.7 macrophages were cultured for 18 h in the presence or absence of 1 μg/ml gAcrp. Cells were then stimulated or not with 100 ng/ml LPS for 30 min. Lysates were prepared, and immunoreactive phosho-ERK1/2 (pERK) was assessed by Western blotting. Total IκB protein was also measured. ERK1/2 was measured as a loading control. Images are representative of four independent experiments.
We also investigated the impact of gAcrp on LPS-stimulated activation of cytosolic signaling pathways that contribute to activation of transcription factors that regulate TNF-α transcription. In control RAW 264.7 macrophages, LPS stimulated phosphorylation of ERK1/2, which is upstream of Egr-1 activity (37, 38), and decreased the quantity of IκB in the cytosol, a signaling event upstream of NFκB activation (39) (Fig. 4D). In contrast, overnight treatment with gAcrp greatly reduced the ability of LPS to increase phosphorylation of ERK1/2 and also prevented LPS-stimulated loss of IκB protein (Fig. 4D), consistent with the reduced ability of LPS to stimulate the DNA binding activity of Egr-1 and NFκB after exposure to gAcrp.
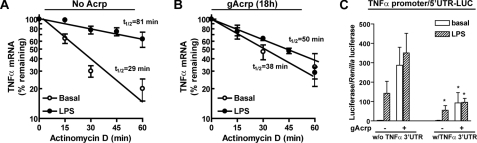
Because TNF-α mRNA accumulation was low in RAW 264.7 macrophages after overnight gAcrp despite high TNF-α promoter activity (see Fig. 1), we next tested the hypothesis that gAcrp also affected TNF-α mRNA stability. RAW 264.7 macrophages were cultured with or without gAcrp for 18 h and then stimulated or not with 100 ng/ml LPS for 2 h. Cells were then treated with actinomycin D for a further 0–60 min. The half-life of TNF-α mRNA was assessed by real time PCR (Fig. 5, A and B). The half-life of TNF-α mRNA at base line (no LPS or gAcrp treatments) was 29 min (Fig. 5A). Overnight treatment with gAcrp had no effect on TNF-α mRNA stability with a 38-min t½ (Fig. 5B). In cells not treated with gAcrp, LPS treatment stabilized TNF-α mRNA, increasing the half-life to 81 min (Fig. 5A). In contrast, after 18-h exposure to gAcrp, stimulation with LPS only increased the half-life of the TNF-α mRNA to 50 min (Fig. 5B).
FIGURE 5.
Overnight treatment with gAcrp suppresses the ability of LPS to stabilize TNF-α mRNA via the class II AU-rich element in the TNF-α 3′-UTR. A and B, RAW 264.7 macrophages were treated with (B) or without (A)1 μg/ml gAcrp for 18 h and then stimulated or not with 100 ng/ml LPS for 2 h prior to the addition of 5 μg/ml actinomycin D for up to 60 min. Accumulation of TNF-α mRNA was measured by real time PCR. Values represent TNF-α mRNA normalized to β-actin mRNA (means ± S.E., n = 3–4 independent experiments). C, RAW 264.7 macrophages were transiently transfected with luciferase reporters containing the full-length TNF-α promoter-5′-UTR with or without the TNF-α 3′-UTR inserted after the luciferase coding sequence. RAW 264.7 macrophages were co-transfected with a Renilla luciferase reporter. Twenty-four hours after transfection, cells were then treated with 1 μg/ml gAcrp for 18 h and then stimulated or not with 100 ng/ml LPS for 4 h, and luciferase activity was measured. Values represent relative luciferase activity (corrected for Renilla luciferase activity; means ± S.E., n = 5–6). *, p < 0.05 compared with constructs without (w/o) the 3′-UTR.
Sequences in the 5′-UTR and 3′-UTR of the TNF-α mRNA are important in the regulation of TNF-α mRNA stability. Therefore, we identified the cis-acting elements in the TNF-α mRNA required for gAcrp-induced destabilization of TNF-α mRNA. RAW 264.7 macrophages were transfected with constructs containing the TNF-α promoter and 5′-UTR with or without the TNF-α 3′-UTR after the luciferase coding sequence and then treated with gAcrp for 18 h. Treatment with gAcrp increased luciferase expression when the TNF-α 5′-UTR was included along with the promoter (Fig. 5C), similar to the response with only the TNF-α promoter present in the luciferase reporter (Fig. 1B). Inclusion of the TNF-α 3′-UTR in this reporter construct decreased both LPS- and gAcrp-stimulated luciferase expression, consistent with a role for the 3′-UTR in destabilization of TNF-α mRNA.
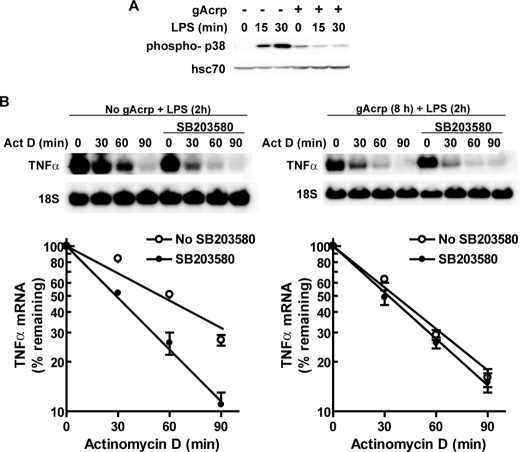
p38 MAPK activity has been shown to contribute to stabilization of TNF-α mRNA (40–42). Therefore, we investigated the effects of gAcrp on basal and LPS-stimulated p38 MAPK activation. Although LPS rapidly increased phospho-p38 in control cells, this activation was suppressed after long term treatment with gAcrp (Fig. 6A). If this loss of p38 activation contributed to the decrease in the ability of LPS to stabilize TNF-α mRNA after gAcrp, then pretreatment of cells with SB205380, a p38 MAPK inhibitor, should destabilize LPS-stimulated TNF-α mRNA in control cells but not in cells treated with gAcrp. We found that inhibition of p38 MAPK destabilized LPS-stimulated TNF-α mRNA in control cells but had no effect on TNF-α mRNA stability in cells treated with gAcrp (Fig. 6B).
FIGURE 6.
Destabilization of TNF-α mRNA by gAcrp is associated with a loss in LPS-stimulated phosphorylation of p38 MAPK. A, RAW 264.7 macrophages were cultured for 18 h in the presence or absence of 1 μg/ml gAcrp. Cells were then stimulated or not with 100 ng/ml LPS for 30 min. Lysates were prepared, and immunoreactive phosho-p38 was assessed by Western blotting. Hsc70 was measured as a loading control. Images are representative of four independent experiments. B, RAW 264.7 macrophages were treated with or without 1μg/ml gAcrp for 8 h and then preincubated with 10μm SB203580 for 30 min. Cells were then stimulated with 100 ng/ml LPS for 2 h followed by the addition of 5 μg/ml actinomycin D (Act D) for up to 90 min. Accumulation of TNF-α mRNA was measured by Northern blot and normalized to 18 S RNA. Values represent means ± S.E. (n = 4).
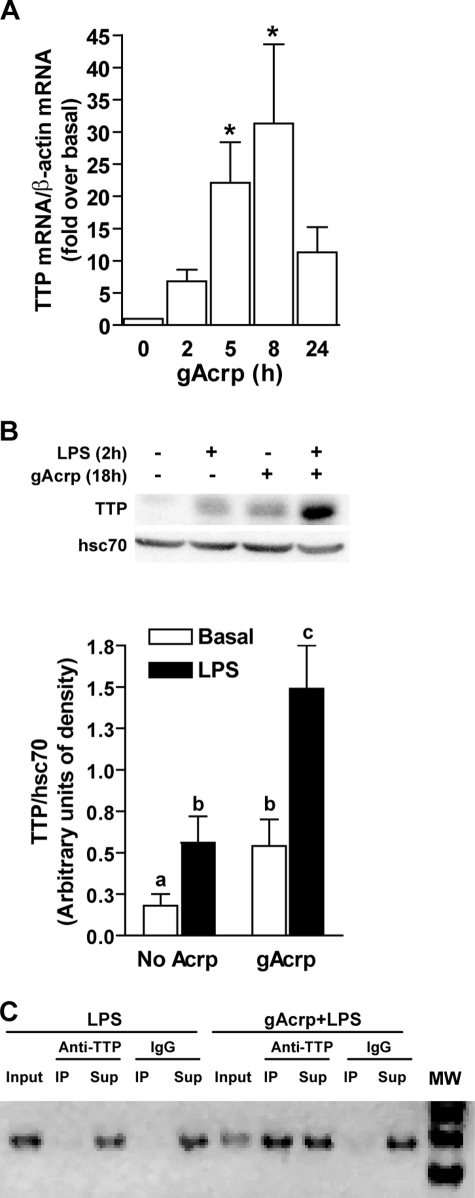
Stimulus-induced stabilization/destabilization of TNF-α mRNA is mediated by a number of mRNA-binding proteins, including the nuclear-cytoplasmic shuttling protein HuR, which acts to stabilize TNF-α mRNA, and TTP, which acts to destabilize TNF-α mRNA in a phosphorylation-dependent manner. TTP is expressed in resting macrophages (43) and can also be rapidly induced in response to LPS in macrophages via a p38 MAPK-dependent pathway (44). This induction results in the presence of multiple forms of differentially phosphorylated TTP thought to contribute to the regulation of TNF-α mRNA stability (44). Although TTP acts to destabilize TNF-α mRNA, p38 MAPK-mediated phosphorylation of TTP inactivates TTP activity in some cell types (45). Here we found that gAcrp increased the expression of TTP mRNA (Fig. 7A) and protein (Fig. 7B). In control cells, LPS increased TTP expression after 2 h (Fig. 7B); this LPS-induced increase was maintained even after overnight exposure to gAcrp (Fig. 7B).
FIGURE 7.
gAcrp increased the expression of TTP as well as its association with TNF-α 3′-UTR. A, RAW 264.7 macrophages were cultured with 1 μg/ml gAcrp for up to 24 h, and TTP mRNA was quantified by real time PCR. Values represent TTP mRNA normalized to β-actin mRNA (means ± S.E., n = 4). *, p < 0.05 compared with basal. B, RAW 264.7 macrophages were cultured with 1 μg/ml gAcrp for up to 18 h and then stimulated or not with 100 ng/ml LPS for 2 h. Cells were lysed, and immunoreactive TTP was measured by Western blot. Hsc70 was measured as a loading control. Values represent TTP normalized to Hsc70 (means ± S.E., n = 4). Values with different superscript letters (a–c) are significantly different from each other and compared with basal (p < 0.05). C, the association of TTP with TNF-α mRNA was assessed in an RNA immunoprecipitation assay. RAW 264.7 macrophages were cultured with or without gAcrp for 18 h and then stimulated with 100 ng/ml LPS for 2 h. Lysates were then incubated with antibody to TTP or control IgG. TNF-α 3′-UTR associated with the TTP was then measured as described under “Experimental Procedures.” Input and supernatants (Sup) used 20% of the volume before or after immunoprecipitation (IP), respectively. Similar results were observed in three separate experiments. MW, molecular mass markers.
Making use of an RNA immunoprecipitation assay to measure TTP-associated mRNA, we found that gAcrp increased the association of TNF-α mRNA to TTP (Fig. 7C). RAW 264.7 macrophages were cultured for 18 h with or without gAcrp and then stimulated with LPS for 2 h. Lysates were then incubated with antibodies to TTP or control IgG. TNF-α 3′-UTR was then amplified by PCR in the immunoprecipitates. In control cells stimulated with LPS, small but detectable levels of TNF-α 3′-UTR were associated with TTP. The quantity of the TNF-α 3′-UTR pulled down with antibody to TTP was greatly increased in cells treated with LPS after overnight treatment with gAcrp (Fig. 7C). These data suggest that the failure of LPS to stabilize TNF-α mRNA after treatment with gAcrp is due to an increased association with TTP.
DISCUSSION
Long term exposure to adiponectin desensitizes macrophages to subsequent stimulation of LPS-induced TNF-α production (10, 11). Here we identified adiponectin-mediated changes in the transcriptional and post-transcriptional regulation of TNF-α in RAW 264.7 macrophages. Treatment of RAW 264.7 macrophages with gAcrp profoundly decreased the responsivity to subsequent challenge with LPS. In particular, gAcrp decreased LPS-stimulated activation of key transcription factors controlling TNF-α transcription, including NFκB, Egr-1, and AP-1. Furthermore gAcrp also suppressed the ability of LPS to stabilize TNF-α mRNA. This loss of LPS-induced stabilization of TNF-α mRNA required the presence of the 3′-UTR of the TNF-α mRNA. Moreover loss of stimulus-induced stabilization of TNF-α mRNA was associated with an increase in the expression of TTP, an mRNA-binding protein that destabilizes TNF-α mRNA, as well as an increase in the association of the TNF-α mRNA with TTP.
Regulation of TNF-α production by macrophages is under complex regulatory controls; transcriptional regulation is both cell type- and stimulus-specific. For example, stimulation of TNF-α production by LPS in macrophages is primarily dependent on transcriptional activation via the κB3 site as well as contributions from binding of Egr-1 and c-Jun (18, 19). In contrast, activation of TNF-α production via the TNF-α receptor I involves phosphorylated ATF-2 binding to the cAMP-response element site in the TNF-α promoter (46). Further modulation of TNF-α mRNA stability is an additional important mechanism in the regulation of TNF-α biosynthesis in response to stimulation of macrophages by LPS as well as virus (25–27).
Adiponectin rapidly increases TNF-α promoter activity via a mechanism that involves both the activation of NFκB and Egr-1 (12). In contrast, after longer periods of exposure to adiponectin, here we report that the Egr-1 site no longer contributed to TNF-α expression and that the contribution from NFκB was greatly reduced. Deletion of either the Egr-1 or κB3 site in the TNF-α promoter had little effect on gAcrp-stimulated promoter activity after 24 h. Furthermore Egr-1 DNA binding activity returned to base line by 18 h, and the binding of nuclear proteins to an oligonucleotide containing the NFκB site shifted from p65-p50 heterodimers at early times of treatment with gAcrp to predominantly p50 homodimers. p50 homodimers act as transcriptional repressors in many systems (47). Moreover binding of p50 to the κB3 site is associated with the development of an LPS-tolerant phenotype (48), so it was unlikely that NFκB was critical to maintaining TNF-α promoter activity. Instead TNF-α promoter activity after 18-h exposure to gAcrp was primarily driven by the activity of AP-1 as evidenced by increased AP-1 DNA binding activity and AP-1 promoter-driven luciferase expression. Collectively these data indicate that adiponectin has a complex, time-dependent impact on the regulation of TNF-α expression in macrophages.
Despite the high TNF-α promoter activity, TNF-α mRNA accumulation was reduced after 16–18-h treatment with gAcrp. Modulation of TNF-α mRNA stability is an important component in the regulation of TNF-α biosynthesis in response to a number of activators (22). Stabilization of mRNAs contributes to the strong and rapid induction of genes in the inflammatory process. Here we show that gAcrp destabilized TNF-α mRNA in RAW 264.7 macrophages, suggesting that destabilization of inflammatory cytokine mRNAs is one of the anti-inflammatory mechanisms of adiponectin. The TNF-α mRNA, like other short lived mRNAs, contains AREs in its 3′-UTR that function as destabilizing elements as demonstrated in transgenic mice in which the TNF-α ARE is deleted (26) as well as in various cell culture systems (25, 27). In addition to the destabilizing activity of the TNF-α 3′-UTR, the AREs in the 3′-UTR allow for stabilization of the TNF-α mRNA in response to activation (24, 49). Loss of LPS-mediated stabilization of TNF-α mRNA by gAcrp involved the 3′-UTR of the TNF-α mRNA (Fig. 5).
Stability of the TNF-α mRNA is controlled by trans-acting factors that bind to the TNF-α mRNA. A large number of mRNA-binding proteins regulate both stabilization and destabilization (22). Of these, several proteins that bind to the TNF-α mRNA, specifically to its 3′-UTR, have been identified. One such mRNA-binding protein is TTP, a zinc finger protein induced by LPS in macrophages that acts to destabilize TNF-α mRNA (50, 51). TTP-deficient macrophages express elevated TNF-α mRNA because of an increase in the TNF-α mRNA half-life (51). Here we found that the loss of LPS-mediated stabilization of TNF-α mRNA after exposure to gAcrp was associated with an increase in the expression of TTP mRNA. TTP protein levels were also increased after 18 h of gAcrp treatment and then further increased after challenge with LPS (Fig. 7). Importantly gAcrp enhanced the association of TNF-α mRNA with TTP (Fig. 7C). TTP expression is under complex regulatory controls. Interestingly p38 MAPK stimulates both the expression of TTP mRNA and prevents TTP protein degradation (52). Recent data indicate that additional mechanisms also contribute to induction of TTP. For example, interferons increase TTP expression in macrophages (53). Because gAcrp treatment of RAW 264.7 macrophages suppressed LPS-stimulated p38 MAPK phosphorylation, it is likely that p38-independent mechanisms contribute to increased TTP expression after gAcrp treatment. Further studies are required to delineate the regulation of TTP expression in response to gAcrp in macrophages.
The mRNA destabilizing activity of TTP is regulated via p38 MAPK- and MAPK-activated protein kinase 2 (MK2)-dependent phosphorylation (40, 54). Serine phosphorylation of TTP reduces its affinity for the ARE in the TNF-α 3′-UTR, thus decreasing the destabilizing effects of TTP (40). Importantly treatment of RAW 264.7 macrophages with gAcrp also decreased LPS-stimulated p38 phosphorylation (Fig. 6). Although pretreatment of RAW 264.7 macrophages with SB203580 destabilized LPS-stimulated TNF-α mRNA in control cells, inhibition of p38 MAPK had no effect on TNF-α mRNA stability after exposure of cells to gAcrp for 8 h (Fig. 6). Taken together, these data suggest that not only did gAcrp increase the total quantity of TTP, but it also shifted the balance of TTP to its non-phosphorylated state as a result of decreased LPS-stimulated p38 MAPK activation, resulting in a loss of LPS-stimulated stabilization of TNF-α mRNA in the gAcrp-treated cells.
Although the mechanisms by which gAcrp acts to suppress LPS-mediated stabilization of TNF-α mRNA in macrophages is not yet understood, data in the literature are consistent with the hypothesis that IL-10 may be important in mediating the effects of gAcrp on stimulus-induced TNF-α mRNA stability. gAcrp rapidly increases the expression of IL-10 (12, 55). IL-10 is a potent anti-inflammatory cytokine, which mediates its effects via inhibition of cytokine transcription as well as the destabilization of specific cytokine and chemokine mRNA (56). IL-10 destabilizes the mRNA for TNF-α (42) as well as the CXC ligand 1 (57). Although the mechanisms by which IL-10 acts to destabilizes cytokine/chemokine mRNA are not completely understood, IL-10 has been reported to inhibit p38 MAPK activation and decreases the expression of HuR (21, 42).
Regulation of TNF-α expression, both at the level of TNF-α transcription and mRNA stability, has been implicated in the pathophysiology of a number of chronic inflammatory diseases, such as chronic ethanol-induced liver injury (58, 59), atherosclerosis (42), and chronic intestinal inflammation (21). In parallel, adiponectin-based therapies can decrease atherosclerosis in apolipoprotein-deficient mice (7) and ethanol-induced liver injury in mice (60). These protective effects of adiponectin are associated with normalization of TNF-α expression (58). Taken together, these data are consistent with the hypothesis that adiponectin “tone” during chronic inflammation is an important modulator of TNF-α production by macrophages because of changes in both transcriptional and post-transcriptional regulatory mechanisms.
This work was supported, in whole or in part, by National Institutes of Health Grants AA011975 and AA013868 (to L. E. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: gAcrp, globular adiponectin; AdipoR, adiponectin receptor; ARE, A + U-rich element; Egr-1, early growth response protein-1; ERK, extracellular signal-regulated kinase; IL, interleukin; LPS, lipopolysaccharide; Luc, luciferase; TNF-α, tumor necrosis factor α; TTP, tristetraprolin; UTR, untranslated region; MAPK, mitogen-activated protein kinase; Ct, comparative threshold; F, forward; R, reverse; TNF-α promoter/Luc activity, TNF-α promoter-driven luciferase activity.
References
- 1.Scherer, P. E., Williams, S., Fogliano, M., Baldini, G., and Lodish, H. F. (1995) J. Biol. Chem. 270 26746–26749 [DOI] [PubMed] [Google Scholar]
- 2.Berg, A. H., Combs, T. P., and Scherer, P. E. (2002) Trends Endocrinol. Metab. 13 84–89 [DOI] [PubMed] [Google Scholar]
- 3.Weisberg, S. P., McCann, D., Desai, M., Rosenbaum, M., Leibel, R. L., and Ferrante, A. W., Jr. (2003) J. Clin. Investig. 112 1796–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamauchi, T., Kamon, J., Ito, Y., Tsuchida, A., Yokomizo, T., Kita, S., Sugiyama, T., Miyagishi, M., Hara, K., Tsunoda, M., Murakami, K., Ohteki, T., Uchida, S., Takekawa, S., Waki, H., Tsuno, N. H., Shibata, Y., Terauchi, Y., Froguel, P., Tobe, K., Koyasu, S., Taira, K., Kitamura, T., Shimizu, T., Nagai, R., and Kadowaki, T. (2003) Nature 423 762–769 [DOI] [PubMed] [Google Scholar]
- 5.Chinetti, G., Zawadski, C., Fruchart, J. C., and Staels, B. (2004) Biochem. Biophys. Res. Commun. 314 151–158 [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi, N., Argueta, J. G., Masuhiro, Y., Kagishita, M., Nonaka, K., Saito, T., Hanazawa, S., and Yamashita, Y. (2005) FEBS Lett. 579 6821–6826 [DOI] [PubMed] [Google Scholar]
- 7.Okamoto, Y., Kihara, S., Ouchi, N., Nishida, M., Arita, Y., Kumada, M., Ohashi, K., Sakai, N., Shimomura, I., Kobayashi, H., Terasaka, N., Inaba, T., Funahashi, T., and Matsuzawa, Y. (2002) Circulation 106 2767–2770 [DOI] [PubMed] [Google Scholar]
- 8.Wang, Y., Lu, G., Wong, W. P., Vliegenthart, J. F., Gerwig, G. J., Lam, K. S., Cooper, G. J., and Xu, A. (2004) Proteomics 4 3933–3942 [DOI] [PubMed] [Google Scholar]
- 9.Yokota, T., Oritani, K., Takahashi, I., Ishikawa, J., Matsuyama, A., Ouchi, N., Kihara, S., Funahashi, T., Tenner, A. J., Tomiyama, Y., and Matsuzawa, Y. (2000) Blood 96 1723–1732 [PubMed] [Google Scholar]
- 10.Wulster-Radcliffe, M. C., Ajuwon, K. M., Wang, J., Christian, J. A., and Spurlock, M. E. (2004) Biochem. Biophys. Res. Commun. 316 924–929 [DOI] [PubMed] [Google Scholar]
- 11.Thakur, V., Pritchard, M. T., McMullen, M. R., and Nagy, L. E. (2006) Am. J. Physiol. 290 G998–G1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, P., McMullen, M. R., Huang, H., Thakur, V., and Nagy, L. E. (2007) J. Biol. Chem. 282 21695–21703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsatsanis, C., Zacharioudaki, V., Androulidaki, A., Dermitzaki, E., Charalampopoulos, I., Minas, V., Gravanis, A., and Margioris, A. N. (2005) Biochem. Biophys. Res. Commun. 335 1254–1263 [DOI] [PubMed] [Google Scholar]
- 14.Park, P., Huang, H., McMullen, M. R., Bryan, K., and Nagy, L. E. (2008) J. Leukoc. Biol. 83 1258–1266 [DOI] [PubMed] [Google Scholar]
- 15.Beutler, B. (1995) J. Investig. Med. 43 227–235 [PubMed] [Google Scholar]
- 16.Jacob, C. O. (1992) Immunol. Today 13 122–125 [DOI] [PubMed] [Google Scholar]
- 17.Jacob, C. O., Lee, S. K., and Strassmann, G. (1996) J. Immunol. 156 3043–3050 [PubMed] [Google Scholar]
- 18.Tsai, E. Y., Falvo, J. V., Tsytsykova, A. V., Barczak, A. K., Reimold, A. M., Glimcher, L. H., Fenton, M. J., Gordon, D. C., Dunn, I. F., and Goldfeld, A. E. (2000) Mol. Cell. Biol. 20 6084–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao, J., Mackman, N., Edgington, T. S., and Fan, S. T. (1997) J. Biol. Chem. 272 17795–17801 [DOI] [PubMed] [Google Scholar]
- 20.Dumitru, C. D., Ceci, J. D., Tsatsanis, C., Kontoyiannis, D., Stamatakis, K., Lin, J. H., Patriotis, C., Jenkins, N. A., Copeland, N. G., Kollias, G., and Tsichlis, P. N. (2000) Cell 103 1071–1083 [DOI] [PubMed] [Google Scholar]
- 21.Kontoyiannis, D., Kotlyarov, A., Carballo, E., Alexopoulou, L., Blackshear, P. J., Gaestel, M., Davis, R., Flavell, R., and Kollias, G. (2001) EMBO J. 20 3760–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan, C. M., and Steitz, J. A. (2001) CMLS Cell. Mol. Life Sci. 58 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neininger, A., Kontoyiannis, D., Kotlyarov, A., Winzen, R., Eckert, R., Volk, H. D., Holtmann, H., Kollias, G., and Gaestel, M. (2002) J. Biol. Chem. 277 3065–3068 [DOI] [PubMed] [Google Scholar]
- 24.Dean, J. L., Wait, R., Mahtani, K. R., Sully, G., Clark, A. R., and Saklatvala, J. (2001) Mol. Cell. Biol. 21 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hel, Z., Skamene, E., and Radzioch, D. (1996) Mol. Cell. Biol. 16 5579–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kontoyiannis, D., Pasparakis, M., Pizarro, T. T., Cominelli, F., and Kollias, G. (1999) Immunity 10 387–398 [DOI] [PubMed] [Google Scholar]
- 27.Lagnado, C. A., Brown, C. Y., and Goodall, G. J. (1994) Mol. Cell. Biol. 14 7984–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garnon, J., Lachance, C., Di Marco, S., Hel, Z., Marion, D., Ruiz, M. C., Newkirk, M. M., Khandjian, E. W., and Radzioch, D. (2005) J. Biol. Chem. 280 5750–5763 [DOI] [PubMed] [Google Scholar]
- 29.Vasudevan, S., and Steitz, J. A. (2007) Cell 128 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han, J., and Ulevitch, R. J. (2005) Nat. Immunol. 6 1198–1205 [DOI] [PubMed] [Google Scholar]
- 31.Wang, F., Gao, F., and Jing, L. (2005) Med. Hypotheses 65 1082–1087 [DOI] [PubMed] [Google Scholar]
- 32.Steer, J. H., Kroeger, K. M., Abraham, L. J., and Joyce, D. A. (2000) J. Biol. Chem. 275 18432–18440 [DOI] [PubMed] [Google Scholar]
- 33.Marchant, A., Gueydan, C., Houzet, L., Amraoui, Z., Sels, A., Huez, G., Goldman, M., and Kruys, V. (1996) J. Inflamm. 46 114–123 [PubMed] [Google Scholar]
- 34.Shi, L., Kishore, R., McMullen, M. R., and Nagy, L. E. (2002) J. Biol. Chem. 277 14777–14785 [DOI] [PubMed] [Google Scholar]
- 35.Datta, S., Biswas, R., Novotny, M., Pavicic, P. G., Jr., Herjan, T., Mandal, P., and Hamilton, T. A. (2008) J. Immunol. 180 2545–2552 [DOI] [PubMed] [Google Scholar]
- 36.Niranjanakumari, S., Lasda, E., Brazas, R., and Garcia-Blanco, M. A. (2002) Methods 26 182–190 [DOI] [PubMed] [Google Scholar]
- 37.Shi, L., Kishore, R., McMullen, M., and Nagy, L. E. (2002) Am. J. Physiol. 282 C1205–C1211 [DOI] [PubMed] [Google Scholar]
- 38.Guha, M., O'Connell, M. A., Pawlinski, R., Hollis, A., McGovern, P., Yan, S. F., Stern, D., and Mackman, N. (2001) Blood 98 1429–1439 [DOI] [PubMed] [Google Scholar]
- 39.Bonizzi, G., and Karin, M. (2004) Trends Immunol. 25 280–288 [DOI] [PubMed] [Google Scholar]
- 40.Hitti, E., Iakovleva, T., Brook, M., Deppenmeier, S., Gruber, A. D., Radzioch, D., Clark, A. R., Blackshear, P. J., Kotlyarov, A., and Gaestel, M. (2006) Mol. Cell. Biol. 26 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deleault, K. M., Skinner, S. J., and Brooks, S. A. (2008) Mol. Immunol. 45 13–24 [DOI] [PubMed] [Google Scholar]
- 42.Rajasingh, J., Bord, E., Luedemann, C., Asai, J., Hamada, H., Thorne, T., Qin, G., Goukassian, D., Zhu, Y., Losordo, D. W., and Kishore, R. (2006) FASEB J. 20 2112–2114 [DOI] [PubMed] [Google Scholar]
- 43.Taylor, G. A., Thompson, M. J., Lai, W. S., and Blackshear, P. J. (1996) Mol. Endocrinol. 10 140–146 [DOI] [PubMed] [Google Scholar]
- 44.Mahtani, K. R., Brook, M., Dean, J. L., Sully, G., Saklatvala, J., and Clark, A. R. (2001) Mol. Cell. Biol. 21 6461–6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, W., Brauchle, M. A., Di Padova, F., Gram, H., New, L., Ono, K., Downey, J. S., and Han, J. (2001) Am. J. Physiol. 281 L499–L508 [DOI] [PubMed] [Google Scholar]
- 46.Brinkman, B. M., Telliez, J. B., Schievella, A. R., Lin, L. L., and Goldfeld, A. E. (1999) J. Biol. Chem. 274 30882–30886 [DOI] [PubMed] [Google Scholar]
- 47.Ghosh, S., May, M. J., and Kopp, E. B. (1998) Annu. Rev. Immunol. 16 225–260 [DOI] [PubMed] [Google Scholar]
- 48.Bohuslav, J., Kravchenko, V. V., Parry, G. C., Erlich, J. H., Gerondakis, S., Mackman, N., and Ulevitch, R. J. (1998) J. Clin. Investig. 102 1645–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brook, M., Sully, G., Clark, A. R., and Saklatvala, J. (2000) FEBS Lett. 483 57–61 [DOI] [PubMed] [Google Scholar]
- 50.Lai, W. S., Carballo, E., Thorn, J. M., Kennington, E. A., and Blackshear, P. J. (2000) J. Biol. Chem. 275 17827–17837 [DOI] [PubMed] [Google Scholar]
- 51.Carballo, E., Lai, W. S., and Blackshear, P. J. (1998) Science 281 1001–1005 [DOI] [PubMed] [Google Scholar]
- 52.Brook, M., Tchen, C. R., Santalucia, T., McIlrath, J., Arthur, J. S., Saklatvala, J., and Clark, A. R. (2006) Mol. Cell. Biol. 26 2408–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer, I., Schaljo, B., Vogl, C., Gattermeier, I., Kolbe, T., Muller, M., Blackshear, P. J., and Kovarik, P. (2006) Blood 107 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carballo, E., Cao, H., Lai, W. S., Kennington, E. A., Campbell, D., and Blackshear, P. J. (2001) J. Biol. Chem. 276 42580–42587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolf, A. M., Wolf, D., Rumpold, H., Enrich, B., and Tilg, H. (2004) Biochem. Biophys. Res. Commun. 323 630–635 [DOI] [PubMed] [Google Scholar]
- 56.Hamilton, T. A., Ohmori, Y., and Tebo, J. (2002) Immunol. Res. 25 229–245 [DOI] [PubMed] [Google Scholar]
- 57.Biswas, R., Datta, S., Gupta, J. D., Novotny, M., Tebo, J., and Hamilton, T. A. (2003) J. Immunol. 170 6202–6208 [DOI] [PubMed] [Google Scholar]
- 58.Kishore, R., McMullen, M. R., and Nagy, L. E. (2001) J. Biol. Chem. 276 41930–41937 [DOI] [PubMed] [Google Scholar]
- 59.McMullen, M. R., Cocuzzi, E., Hatzoglou, M., and Nagy, L. E. (2003) J. Biol. Chem. 278 38333–38341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu, A., Wang, Y., Keshaw, H., Xu, L. Y., Lam, K. S., and Cooper, G. J. (2003) J. Clin. Investig. 112 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]