Abstract
The survival of Mycobacterium tuberculosis requires detoxification
of host ·NO. Oxygenated Mycobacterium tuberculosis truncated
hemoglobin N catalyzes the rapid oxidation of nitric oxide to innocuous
nitrate with a second-order rate constant
(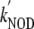 ≈ 745 ×
106 m-1·s-1), which is
∼15-fold faster than the reaction of horse heart myoglobin. We ask what
aspects of structure and/or dynamics give rise to this enhanced reactivity. A
first step is to expose what controls ligand/substrate binding to the heme. We
present evidence that the main barrier to ligand binding to deoxy-truncated
hemoglobin N (deoxy-trHbN) is the displacement of a distal cavity water
molecule, which is mainly stabilized by residue Tyr(B10) but not coordinated
to the heme iron. As observed in the Tyr(B10)/Gln(E11) apolar mutants, once
this kinetic barrier is lowered, CO and O2 binding is very rapid
with rates approaching 1-2 × 109
m-1·s-1. These large values almost
certainly represent the upper limit for ligand binding to a heme protein and
also indicate that the iron atom in trHbN is highly reactive. Kinetic
measurements on the photoproduct of the ·NO derivative of met-trHbN,
where both the ·NO and water can be directly followed, revealed that
water rebinding is quite fast (∼1.49 × 108
s-1) and is responsible for the low geminate yield in trHbN.
Molecular dynamics simulations, performed with trHbN and its distal mutants,
indicated that in the absence of a distal water molecule, ligand access to the
heme iron is not hindered. They also showed that a water molecule is
stabilized next to the heme iron through hydrogen-bonding with Tyr(B10) and
Gln(E11).
≈ 745 ×
106 m-1·s-1), which is
∼15-fold faster than the reaction of horse heart myoglobin. We ask what
aspects of structure and/or dynamics give rise to this enhanced reactivity. A
first step is to expose what controls ligand/substrate binding to the heme. We
present evidence that the main barrier to ligand binding to deoxy-truncated
hemoglobin N (deoxy-trHbN) is the displacement of a distal cavity water
molecule, which is mainly stabilized by residue Tyr(B10) but not coordinated
to the heme iron. As observed in the Tyr(B10)/Gln(E11) apolar mutants, once
this kinetic barrier is lowered, CO and O2 binding is very rapid
with rates approaching 1-2 × 109
m-1·s-1. These large values almost
certainly represent the upper limit for ligand binding to a heme protein and
also indicate that the iron atom in trHbN is highly reactive. Kinetic
measurements on the photoproduct of the ·NO derivative of met-trHbN,
where both the ·NO and water can be directly followed, revealed that
water rebinding is quite fast (∼1.49 × 108
s-1) and is responsible for the low geminate yield in trHbN.
Molecular dynamics simulations, performed with trHbN and its distal mutants,
indicated that in the absence of a distal water molecule, ligand access to the
heme iron is not hindered. They also showed that a water molecule is
stabilized next to the heme iron through hydrogen-bonding with Tyr(B10) and
Gln(E11).
·NO plays an important role in host defense against microbial pathogens by inhibiting or inactivating key enzymes such as the terminal respiratory oxidases (1-5) and the iron/sulfur protein aconitase (6, 7). ·NO also combines at near diffusion-limited rate with superoxide produced by respiring cells to form the highly oxidizing agent peroxynitrite (8, 9). ·NO-metabolizing reactions are, thus, required to defend microbial pathogens against ·NO poisoning.
In Mycobacterium tuberculosis the glbN gene encodes the
truncated hemoglobin N
(trHbN)4
(Fig. 1). Inactivation of
glbN in Mycobacterium bovis bacillus Calmette-Guérin
impairs the ability of stationary phase cells to protect aerobic respiration
from nitric oxide (·NO) inhibition, suggesting that trHbN may protect
M. tuberculosis from ·NO toxicity in vivo
(10). This functional
assessment is supported by the observation that trHbN catalyzes the rapid
oxidation of ·NO to nitrate
 ,
with a second-order rate constant
,
with a second-order rate constant
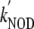 ≈ 745 ×
106 m-1·s-1 (23 °C)
(10). The nitric oxide
dioxygenase (NOD) reaction catalyzed by trHbN is at least 15-fold faster
(
≈ 745 ×
106 m-1·s-1 (23 °C)
(10). The nitric oxide
dioxygenase (NOD) reaction catalyzed by trHbN is at least 15-fold faster
(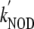 ≈ 45 ×
106 m-1·s-1 at 23 °C)
than the one recorded for horse heart myoglobin (Mb) and is almost as
efficient as the diffusion-controlled reaction of ·NO with free
≈ 45 ×
106 m-1·s-1 at 23 °C)
than the one recorded for horse heart myoglobin (Mb) and is almost as
efficient as the diffusion-controlled reaction of ·NO with free
 A critical issue in this
context is what aspects of structure and/or dynamics give rise to this
enhanced reactivity. A first step is to expose what controls ligand/substrate
binding to the heme.
A critical issue in this
context is what aspects of structure and/or dynamics give rise to this
enhanced reactivity. A first step is to expose what controls ligand/substrate
binding to the heme.
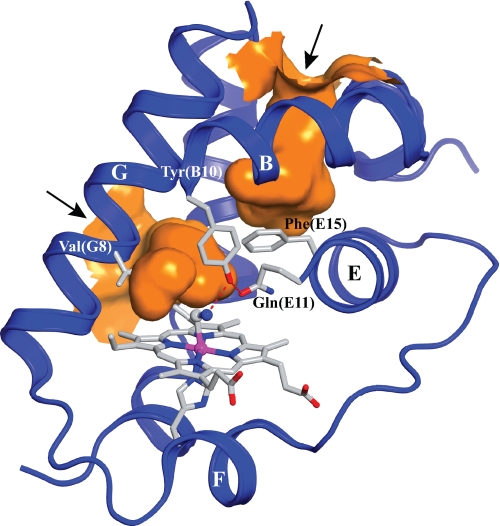
FIGURE 1.
View of the distal heme pocket and the tunnels of cyanomet-trHbN chain B under xenon pressure (PDB entry 1S56). Besides the protein backbone (blue ribbon, with labeled α-helices), the figure shows hydrogen-bonding (dashed red lines) between the distal residues Tyr(B10) and Gln(E11) and the heme-bound cyanide. The path of the two tunnels is shown in orange. The short tunnel (∼8 Å) connects the heme distal site to the outer solvent space at a location comprised between the central region of the G and H helices (left in the figure). The long tunnel (∼20 Å) extends from the heme distal cavity to a solvent access site located between the inter-helical loops AB and GH (upper part of the figure); note the gating role of Phe(E15) on the long tunnel. The arrows point to the tunnel entrance sites facing the solvent. The figure was produced using the PyMOL software (Delano Scientific).
Once the ligand/substrate accesses the distal heme pocket (DHP), the issue of reactivity focuses on local factors such as iron reactivity and steric effects originating within the DHP. Inspection of Mb and trHbN structures shows that in Mb the imidazole ring of the proximal His is in an eclipsed orientation with respect to the pyrrole nitrogen atoms. In contrast, that in trHbN is in a staggered geometry, suggesting reduced repulsive interactions between the imidazole ring and the pyrrole nitrogen atoms and a stronger heme-iron bond (higher iron reactivity). This assessment is supported by resonance Raman studies of deoxy-trHbN and Mb, also indicating a stronger Fe-His bond in trHbN (11). Based on the favorable proximal environment, one would anticipate faster combination rates and higher geminate yields for trHbN relative to Mb. Surprisingly both proteins bind O2 with relatively similar rates, and the geminate yields for CO are both comparably low in the few percent range at ambient conditions (12, 13). These observations reveal that significant distal factors dictate the binding properties of trHbN.
There are several categories of distal effects that can modulate ligand binding. Steric effects from the side chains of distal residues can increase the barrier for binding through either static positioning or relaxation subsequent to ligand dissociation and diffusion. In our earlier work, which showed a dramatic increase in the geminate yield with increasing solvent viscosity, we postulated that viscosity-dependent relaxations of side chains were responsible for the large changes in the geminate yield (13). In the present study this hypothesis was reexamined along with consideration for another potential distal contribution arising from water occupying the DHP.
Ligand binding to ferrous (Fe2+) and ferric (Fe3+) Mb requires the displacement of a water molecule that is hydrogen-bonded to the distal His(E7) residue side chain (14-21). In ferric Mb, the distal water molecule coordinates as a weak ligand, whereas in the ferrous derivative it occupies a site in the DHP, blocking access to the heme iron, but at most it only transiently interacts with the ferrous heme iron (22). Kinetic data supports the concept that the distal water molecule increases the enthalpic contribution to the kinetic barrier by sterically hindering ligand access to the heme iron (14, 17-19, 23, 24). Static and dynamic steric contributions to such barrier have been previously shown for various DHP side chains including that of His(E7) in Mb (14, 15, 17-19, 23, 25-29). The His(E7)-stabilized DHP water molecule can be viewed as increasing the effective size of the sterically active His(E7) side chain. In agreement, photolysis experiments on Mb(Fe2+-CO) and Mb(Fe3+-NO) show that substitutions of His(E7) by different apolar residues resulted in enhanced ligand rebinding rates that are quantitatively related to the lack of occupancy of the distal water molecule (14-21, 30).
In the present work we examined the ligand binding properties of trHbN-bearing mutations at residues Tyr(B10) and Gln(E11). Our results indicate that both the main barrier to ligand binding to deoxy-trHbN and the origin of the low geminate yield are due to the presence of Tyr(B10)-stabilized water within the DHP at a site that blocks access to the heme iron. Such a proposal is further supported by the observation that in the Tyr(B10)/Gln(E11) double mutants, where side chain stabilization of the DHP water molecule is not possible, the combination rate becomes very rapid, with rates approaching those measured for diffusion-controlled reactions and the geminate yield increases by almost 2 orders of magnitude.
EXPERIMENTAL PROCEDURES
Mutagenesis, Expression, and Purification—Recombinant trHbN and mutants were expressed and purified as previously described (31).
Flash-photolysis Experiments—Laser flash-photolysis studies were carried out using the LKS.60 Spectrometer from Applied Photolysis (Leatherhead, UK) at 23 °C. Photolysis was initiated by a 5-ns pulse of light at 532 nm provided by a Brilliant B Nd:YAG laser (Quantel S.A., France). Absorbance changes were measured using the monochromator-filtered light from a 150-watt xenon arc lamp. Passing through the sample, the probe light beam was refocused on the slits (slits widths at 1 mm) of a second monochromator. Changes in transmitted probe light intensity were detected by a 1P28 PMT coupled with a HP 54830B DSO digital oscilloscope (Agilent Technologies Inc.) and transferred on a RISC platform PC (Acorn) for processing. An average of at least 10 kinetic traces from at least two separate experiments were averaged and analyzed with the instrument manufacturer software (Applied Photolysis) to obtain the rate constants. Plots of the pseudo first-order rate constants and plots showing absorbance changes after ·NO photolysis were obtained using the KaleidaGraph software (Synergy Software).
Protein samples for the flash-photolysis experiments were used at concentrations ranging from 1.5 to 10 μm and buffered in anaerobic 50 mm potassium phosphate, pH 7.5, containing 50 μm EDTA. The ferric and deoxy protein samples were prepared in a glove box as described previously (31) and put into a gastight quartz cuvette with a 5-mm path length. To obtain the desired complexes, the deoxy and ferric samples were equilibrated with different concentrations of either O2, CO, or ·NO provided by a series 4000 gas mixing system from Environics (Tolland, CT). Combination rates for CO and O2 were followed at wavelengths ascribed to maxima and minima in either the trHb(Fe2+-CO) or the trHb(Fe2+-O2) minus the trHb(Fe2+) differential spectra. To study the extent of water regulation on ligand binding to Mb and trHbN, ·NO recombination kinetics were followed over a broad timescale (ns-ms) and at specific wavelengths corresponding to isosbestic points between the (Fe3+-H2O), (Fe3+-NO), and (Fe3+) five-coordinate (5C) species (Fig. 2 and Ref. 21). Absorption spectra were recorded before and after time course measurements to ensure the integrity of the samples.
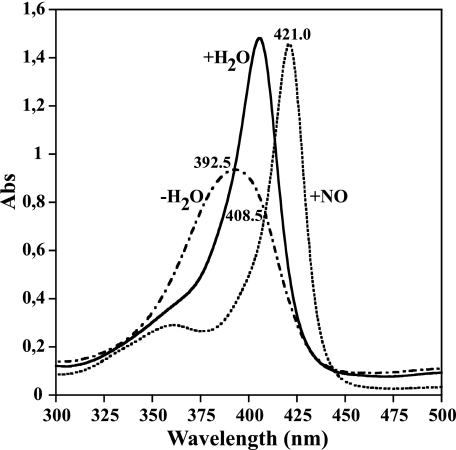
FIGURE 2.
Equilibrium absorption spectra of trHbN(Fe3+-H2O) (solid line), trHbN(Fe3+-NO) (dashed line), and trHbN Tyr(B10)Leu/Gln(E11)Val(Fe3+) mutant (dashed and dotted line) at pH 7.5. The protein concentrations were 10 μm. The Tyr(B10)Leu/Gln(E11)Val double mutant (31) is 5C in the ferric oxidation state and is analogous to the photoproduct generated when trHbN(Fe3+-NO) is dissociated. The isosbestic point at 408.5 nm was used to follow the water coordination subsequent to trHbN(Fe3+-NO) photolysis. At this wavelength we expect an increase in absorbance (Abs) when H2O binds to the heme iron and a decrease when ·NO replaces H2O. The experimental wavelength at 421 nm was employed to monitor the overall reaction trHbN(Fe3+-NO) → trHbN(Fe3+) → trHbN(Fe3+-H2O) → trHbN(Fe3+-NO).
Geminate and Solvent-phase Recombination Experiments—Geminate and solvent-phase recombination measurements were carried out using 8-ns 532-nm pulses at 1 Hz from a Nd:YAG laser (Minilite, Continuum, Santa Clara, CA) as a photodissociation source and a greatly attenuated continuous wave 442 nm probe beam from a He:Cd laser to monitor time-dependent changes in absorption. Details of the apparatus, data collection, and data display can be found in a previous publication and citations therein (13, 26, 32). The kinetic traces are displayed on a log-log plot of normalized absorbance (proportional to the survival probability of the photoproduct) versus time.
Kinetic measurements were typically carried out on solution samples (∼0.25-0.5 mm in heme) contained in standard 1-mm stoppered cuvettes placed in a custom-built dry N2-purged variable temperature cuvette holder (-15 to +65 °C). The one sol-gel encapsulated sample was prepared as a thin layer lining the bottom portion of a 10-mm diameter NMR tube as previously described (33) but with the protocol modified (no added glycerol) to minimize the increase in internal viscosity.
Molecular Dynamics Simulations—Simulations were performed using CHARMM (34) and the CHARMM22 all-atom potential energy parameter set (35) with ϕ, ψ cross-term map correction (CMAP) (36) and modified TIP3P waters (37). Electrostatic interactions were calculated via the Particle Mesh Ewald method (38) using a sixth-order spline interpolation for complementary function, with κ = 0.34 Å-1 and a fast-Fourier grid density of ≈ 1 Å-1. Cutoffs for the real space portion of the Particle Mesh Ewald calculation and the truncation of the Lennard-Jones interactions were 10 Å, with the latter smoothed via a shifting function over the range of 8 to 10 Å. The SHAKE algorithm (39) was used to constrain all covalent bonds involving hydrogen atoms. All simulations employed the leapfrog algorithm and an integration step of 1 fs. Coordinates were saved every ps. Non-bond and image lists were updated heuristically. All simulations were performed at constant pressure and temperature (NPT ensemble) of 1 atm and 298 K, respectively. The mass of the thermal piston was 20,000 kcal·mol-1·ps2, and the mass of the pressure piston equaled 1000 atomic mass unit. The net translation and rotation of the systems were removed every 10,000 steps.
Systems Setup—Coordinates for MD simulations were taken from the crystal structure of wild-type oxy-trHbN (PDB entry 1IDR). All ionizable residues were considered in their standard protonation state at pH 7, the histidines with the proton on ND1 position. Missing coordinates from the crystal structure were built using the internal coordinate definitions of CHARMM. For each subunit, the carboxyl terminal end was optimally positioned by performing 3-ns Langevin dynamics with a 1 fs time step and a friction coefficient FBETA of 5 ps-1 while keeping all coordinates from the crystal structure constrained. The converged structures were immersed in a rhombic dodecahedron unit cell containing pre-equilibrated TIP3P water molecules (8330 molecules). Six sodium ions were added to neutralize the charge of the systems. Water molecules within 2.8 Å of any protein atom were deleted yielding about 24,350 atoms for each system. Before the initiation of MD simulations, the energy of the solvated systems was minimized with 2 cycles of 500 steps of steepest descent followed by 500 steps of Adopted Basis Newton-Raphson minimizations. During energy minimization, the protein coordinates were kept constrained.
To increase sampling, two 20-ns trajectories were generated using the A and B crystal subunits in the absence of the coordinated dioxygen molecule. From these simulations, the last MD coordinates were taken to produce two 20-ns trajectories of deoxy-trHbN with and without a water molecule in the DHP, for a total of 4 20-ns trajectories. The water molecule was arbitrarily positioned in the DHP in a cavity located between the heme iron and the B10 residue, and the initial position was optimized with a short energy minimization, keeping all other coordinates constrained.
Because of their high structural similarities (31), mutant systems were built from equilibrated wild-type coordinates. Three trajectories were produced for each deoxy form of both Tyr(B10)Phe and Gln(E11)Val single mutants with a water molecule in the DHP, whereas five trajectories were produced for the Tyr(B10)Phe/Gln(E11)Val double mutant. The number of trajectories and their length were set to obtain a good sampling of the measured values. For each mutant, one 10-ns trajectory without a water molecule in the DHP was performed.
Analysis of the DHP Accessible Volume—The accessible volumes located in the DHP were studied using VOIDOO (40). The detected cavities were refined using a <0.2 Å grid spacing and a probe radius of 1.4 Å. One coordinate set every 10 ps was used for analysis.
RESULTS AND DISCUSSION
Kinetic Data Indicate That Tyr(B10) Mainly Contributes to the Kinetic Barrier to Ligand Binding to trHbN(Fe2+)
O2 and CO Binding to trHbN—Combination of
O2 and CO to most trHbN mutants was too rapid to be measured by
stopped-flow spectrophotometry. As a consequence, these reactions were studied
by laser flash-photolysis. Because the previously published binding rate
constants for O2 (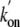 )
and CO (
)
and CO (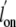 ) of trHbN have been
determined by stopped-flow spectrophotometry
(12), we reexamined the
reactions using flash-photolysis. The measured reactions for O2 and
CO corresponded to about 25 and 63%, respectively, that of the expected
changes in absorbance. The residuals of the fitted curves generated by
applying a single exponential mathematical term to the kinetic data were
nearly random, implying that under these conditions O2 and CO
combination processes are monophasic (Fig.
3). As shown in Table
1, the
) of trHbN have been
determined by stopped-flow spectrophotometry
(12), we reexamined the
reactions using flash-photolysis. The measured reactions for O2 and
CO corresponded to about 25 and 63%, respectively, that of the expected
changes in absorbance. The residuals of the fitted curves generated by
applying a single exponential mathematical term to the kinetic data were
nearly random, implying that under these conditions O2 and CO
combination processes are monophasic (Fig.
3). As shown in Table
1, the 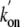 (55.8
× 106 m-1·s-1) value
is higher than that previously determined by stopped-flow spectrophotometry
(
(55.8
× 106 m-1·s-1) value
is higher than that previously determined by stopped-flow spectrophotometry
(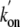 = 25 × 106
m-1·s-1), indicating that the starting
deoxy forms are different.
= 25 × 106
m-1·s-1), indicating that the starting
deoxy forms are different.
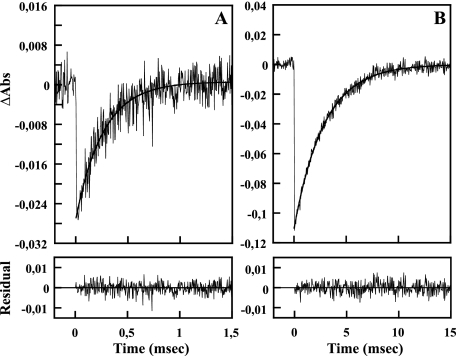
FIGURE 3.
The time courses of O2 (68. 7 μm) (panel A) and CO (50.7 μm) recombination to trHbN (5 μm in heme) (panel B) after photolysis in 50 mm potassium phosphate buffer, pH 7.5, 50 μm EDTA at 23 °C. The figure shows the single exponential fits and residuals to the kinetic traces measured at 411 and 420 nm for O2 and CO, respectively. Abs, absorbance units.
TABLE 1.
Kinetics constants for the reactions of trHbN and its mutants with O2 and CO
| Protein | k′on (O2) | l′on (CO) |
|---|---|---|
| 1 × 106m−1·s−1 | 1 × 106m−1·s−1 | |
| trHbN | 25a | 6.75a |
| 55.8 ± 4.1 (25%)b | 7.80 ± 0.15 (63%) | |
| Tyr(B10)Phe | 540.0 ± 14.3 (15%) | 77.2 ± 1.3 (74%) |
| Tyr(B10)Leu | 621.2 ± 7.0 (24%) | 92.56 ± 0.75 (83%) |
| Gln(E11)Val | 32.6 ± 1.0 (26%) | 6.81 ± 0.26 (56%) |
| Gln(E11)Ala | 37.5 ± 4.0 (29%) | 9.34 ± 0.27 (69%) |
| Tyr(B10)Phe/Gln(E11)Val | -c | 1119.2 ± 13.4 (33%) |
| Tyr(B10)Leu/Gln(E11)Val | 1811.8 ± 51.3 (8%) | 1148.4 ± 50.5 (15%) |
Rate determined by stopped-flow experiment (12).
Percentage of the expected amplitude measured for the reaction.
Binding of O2 may occur in the dead time of the apparatus.
O2 and CO Binding to trHbN Mutants—With the
exception of the double Tyr(B10)Phe/Gln(E11)Val mutant, all mutant proteins
bound O2 in a concentration-dependent manner requiring one
exponential term to fit the pseudo-first-order time courses. No reaction could
be measured for the Tyr(B10)Phe/Gln(E11)Val mutant, indicating either rapid
geminate rebinding after O2 dissociation or failure to
photodissociate the bound O2. As shown in
Table 1,
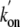 values of Tyr(B10)Phe (540
× 106 m-1·s-1) and
Tyr(B10)Leu (621.2 × 106
m-1·s-1) mutants were ∼10-fold
higher than that of trHbN, indicating that Tyr(B10) contributes significantly
to the energy barrier to O2 binding. In contrast,
values of Tyr(B10)Phe (540
× 106 m-1·s-1) and
Tyr(B10)Leu (621.2 × 106
m-1·s-1) mutants were ∼10-fold
higher than that of trHbN, indicating that Tyr(B10) contributes significantly
to the energy barrier to O2 binding. In contrast,
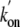 values for the oxygenation of
Gln(E11)Ala (37.5 × 106
m-1·s-1) and Gln(E11)Val (32.6 ×
106 m-1·s-1) mutants were
slightly lower than that of trHbN (55.8 × 106
m-1·s-1). Substituting both Tyr(B10)
and Gln(E11) for Leu and Val, respectively, creating the
Tyr(B10)Leu/Gln(E11)Val mutant, caused an additional increase of the
values for the oxygenation of
Gln(E11)Ala (37.5 × 106
m-1·s-1) and Gln(E11)Val (32.6 ×
106 m-1·s-1) mutants were
slightly lower than that of trHbN (55.8 × 106
m-1·s-1). Substituting both Tyr(B10)
and Gln(E11) for Leu and Val, respectively, creating the
Tyr(B10)Leu/Gln(E11)Val mutant, caused an additional increase of the
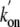 rate to 1811.8 ×
106 m-1·s-1.
rate to 1811.8 ×
106 m-1·s-1.
All mutants bound CO in a concentration-dependent manner requiring a single
exponential term to fit the kinetic traces. As observed for O2,
single substitutions at the Tyr(B10) position resulted in an ∼10-fold
increase of the 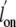 with values
attaining 77.2 × 106 and 92.56 × 106
m-1·s-1 for Tyr(B10)Phe and
Tyr(B10)Leu, respectively (Table
1). In contrast, the Gln(E11)Ala and Gln(E11)Val mutants showed
only small changes in the
with values
attaining 77.2 × 106 and 92.56 × 106
m-1·s-1 for Tyr(B10)Phe and
Tyr(B10)Leu, respectively (Table
1). In contrast, the Gln(E11)Ala and Gln(E11)Val mutants showed
only small changes in the 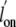 values. The Tyr(B10)Phe/Gln(E11)Val and Tyr(B10)Leu/Gln(E11)Val double mutants
combined with CO with similar rates (Table
1). These latter reactions are quite fast, approaching values for
diffusion-controlled reactions
(41,
42).
values. The Tyr(B10)Phe/Gln(E11)Val and Tyr(B10)Leu/Gln(E11)Val double mutants
combined with CO with similar rates (Table
1). These latter reactions are quite fast, approaching values for
diffusion-controlled reactions
(41,
42).
Table 1 shows that
replacement of Tyr(B10) with either Phe or Leu results in an over an order of
magnitude increase of both 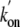 and
and 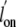 . That both ligands are
similarly affected implies a direct steric effect associated with the
Tyr(B10). Replacement of Gln(E11) with either Ala or Val has very little
influence on the binding rates; however, for the double mutant combining the
Tyr(B10) and Gln(E11) replacements, there is a synergistic effect that
substantially enhances both
. That both ligands are
similarly affected implies a direct steric effect associated with the
Tyr(B10). Replacement of Gln(E11) with either Ala or Val has very little
influence on the binding rates; however, for the double mutant combining the
Tyr(B10) and Gln(E11) replacements, there is a synergistic effect that
substantially enhances both 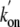 and
and 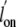 relative to the increase
due to the Tyr(B10) substitutions alone. The question remains as to what
structural and/or dynamical processes are responsible for these side
chain-specific effects on the combination rates.
relative to the increase
due to the Tyr(B10) substitutions alone. The question remains as to what
structural and/or dynamical processes are responsible for these side
chain-specific effects on the combination rates.
As a first step, we examined the nanosecond and slower recombination occurring subsequent to ligand photodissociation using an 8-ns laser pulse. In many instances geminate recombination, which occurs on the sub-microsecond time scale reflects the influence of the initial conformation before substantial relaxation. By monitoring the geminate recombination on these faster time scales, it is possible to establish whether the elements responsible for the very large differences in combination rates are operational from the onset, when the ligand is initially dissociated and localized within the local environment near the heme binding site.
Geminate and Solvent Phase Recombination
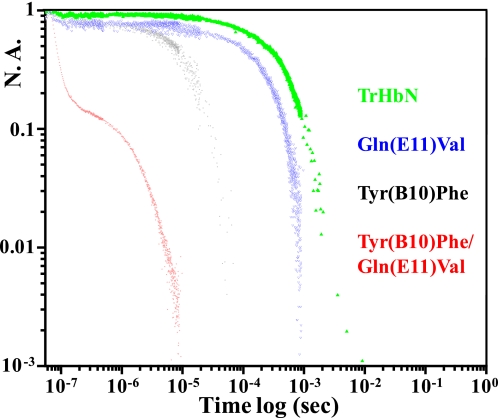
Fig. 4 compares kinetic traces of the geminate and solvent phase recombination of CO to trHbN and several distal mutants displayed on a log-log plot. The rebinding to trHbN (green trace) consists of a single exponential phase. This phase, which slows with decreasing concentrations of CO (not shown), is assigned to solvent phase recombination. The double mutant Tyr(B10)Phe/Gln(E11)Val (red trace) shows two very fast phases. Only the second phase slows in response to a decrease in CO concentration (data not shown), indicating that this kinetic phase is a very fast solvent phase recombination. The even faster recombination phase is consistent with the notion that it is a geminate recombination reaction based on both time scale and insensitivity to the external CO concentration. The recombination trace for the oxy derivative of this mutant showed two similar phases and geminate yields on the nanosecond time scale (data not shown). It was also concluded, based on the photolysis yield at 10-ns, that for the double B10/E11 mutant as well as the wild-type protein, there is a faster sub-nanosecond geminate phase for dioxygen that decreases the nanosecond quantum yield relative to the CO derivatives. In contrast to the CO derivative of the double B10/E11 mutant, which displays an exceptionally fast nanosecond geminate process with very large amplitude (>0.8) that is among the largest for any CO derivative of an Hb or Mb under ambient low viscosity conditions, there is almost no discernable geminate yield under these conditions for trHbN. The Tyr(B10)Leu/Gln(E11)Val double mutant exhibits very similar enhanced kinetics to those from the Tyr(B10)Phe/Gln(E11)Val double mutant both under low and high viscosity conditions (data not shown) The two single mutants Tyr(B10)Phe (black trace) and Gln(E11)Val (blue trace) manifest a measurable geminate process but with a geminate yield in the range of 0.2.
FIGURE 4.
Kinetic traces showing the recombination of CO subsequent to nanosecond photodissociation of the CO-saturated derivatives of wild-type trHbN and its distal mutants. The recombination traces are displayed on a log-log plot with the y axis corresponding to normalized absorbance and the x axis corresponding to time subsequent to photodissociation. The traces are color-coded as follows: wild-type trHbN in green, the Tyr(B10)Phe/Gln(E11)Val double mutant in red, the Tyr(B10)Phe single mutant in black, and the Gln(E11)Val single mutant in blue. All the samples except the double mutant are in solution phase at pH 7.5. The trace from the double B10/E11 mutant (red) is from a sample encapsulated in a thin porous sol-gel bathed in buffer. Essentially identical kinetics were obtained for the solution phase sample of the double B10/E11 mutant, but due to the low concentration of the sample that trace was of poor quality. N. A., normalized absorbance.
The geminate recombination data show that the factors that are responsible for the large differences in the combination rates, and the solvent phase kinetics are operative at early times subsequent to photodissociation. We now consider possible factors contributing to these differences. The enhancement of the binding rates and the solvent phase recombination in going from Tyr(B10) to Phe(B10) might be the result of a decrease in the effect of the tyrosine side chain relaxing to a position that blocks access to the heme iron. If this relaxation was fast enough, it could account for the very low geminate yield for trHbN. The further enhancement in combination rates seen for the double B10/E11 mutant could be attributable to a further reduction in steric factors due a change in the positioning of the Phe(B10) side chain due to the change in the E11 side chain.
There are several observations that raise questions about the validity of this side chain-based steric explanation. The B10 side chain explanation can only account for the observed kinetics if one also invokes modulation of the positioning of the side chain by the E11 side chain. Arguments against that scenario come from the observation that the replacement of Gln(E11) with valine or alanine has a minimal effect on the on rates and on the geminate yield (data not shown for Gln(E11)Ala, which is essentially identical to that of the Gln(E11)Val mutant). Furthermore, the binding rates for Tyr(B10)Phe and Tyr(B10)Leu are very similar. Geminate recombination studies comparing these two mutants both at low and high viscosity show very little difference (data not shown), suggesting that if the B10 side chain contributes through a pure steric effect (due to the side chain alone), the leucine and phenylalanine side chains would have behaved similarly with respect to this proposed steric interaction. A similar steric effect by these two residues seems implausible given the difference in flexibility and volume of the two side chains. Finally, both the Tyr(B10)Phe and Tyr(B10)Leu double mutants with Gln(E11)Val show similar recombination kinetics. If the further substantial enhancement in combination rates, solvent phase recombination rates, geminate yield, and rates of geminate recombination were due to the further reduction in a steric effect due to a change in the B10-E11 interaction, it would seem very implausible that the two different B10 side chains would behave so similarly. Although these arguments are not definitive, they certainly weaken any explanation based solely on steric effects arising solely from the side chains of the B10 and E11 residues. Given these points together with the absence of large ligand binding induced conformational changes involving the proximal heme environment associated with trHbN (11), we consider yet additional factors that can contribute to the control of ligand binding kinetics.
Water in the DHP of Mb and Hb is known to significantly contribute to the kinetic barrier for ligand binding (14-21). In these two cases water occupies the DHP of the deoxy derivative and only populates the DHP of the liganded species subsequent to ligand dissociation and the onset of conformational fluctuations that open the so-called distal His(E7) gate. Thus, there is a delay between the moment of the ligand dissociation and the reentry of water back into the DHP. It has been claimed that this delayed water reentry process is essential for ensuring a high probability of escape by the dissociated ligand once the ligand has accessed the xenon cavities of Mb and Hb. Delayed occupancy of the DHP by water subsequent to ligand dissociation is also a significant factor contributing to the geminate yield for the slower geminate phase arising from the recombination after the ligand has access to the xenon cavities. If the dynamics of water-controlling occupancy of the DHP of trHbN is a factor contributing to the observed kinetic patterns, it would suggest that water participates very early in the recombination process and requires that the B10 and E11 side chains modulate its dynamics and/or occupancy factors. Thus, in contrast to Mb, where water is observed to enter the DHP of Mb only after 50 to 100 ns, in the case of trHbN the water would have to be present within a few nanoseconds if it is responsible for the low geminate yield in wild-type trHbN. The high yield and fast rates for the double B10/E11 mutant would be attributable to the very low occupancy of water within the DHP. To test this hypothesis we have conducted both kinetic measurements on the photoproduct of the ·NO derivative of met-trHbN, where both the ·NO and water can be directly followed, and MD simulations to establish the behavior of water within the DHP of trHbN as a function of B10 and E11 side chain substitutions.
Water Controls Ligand Binding to Ferric trHbN
Combined mutagenesis and spectroscopic studies indicated that Tyr(B10) and Gln(E11) residues stabilize the coordinated water molecule in ferric trHbN at 23 °C and pH 7.5 (31). Accordingly, the optical spectra of ferric double mutants bearing apolar residues at Tyr(B10) and Gln(E11) positions were found typical of ferric heme proteins with no water coordinated to the iron atom (31).
We used laser-flash photolysis of the trHbN(Fe3+-NO) complex to study the kinetics of water entry and binding to the heme iron at 23 °C and pH 7.5. Photodissociation of the horse heart Mb(Fe3+-NO) complex leaves the heme distal site in a ferric-dehydrated state 5C Mb(Fe3+) (21). After ·NO photolysis and escape, a water molecule enters the DHP and binds to the heme iron, forming the aquomet Mb state (Mb(Fe3+-H2O)). At longer times, ·NO displaces the bound water molecule to reestablish the equilibrium Mb(Fe3+-NO) complex.
For monitoring H2O kinetics in trHbN, the experimental wavelength is the isosbestic point between trHbN(Fe3+-NO) and trHbN 5C Tyr(B10)Phe/Gln(E11)Val(Fe3+) mutant, which is located near the Soret band of native ferric trHbN at 406 nm (Fig. 2). Fig. 5A shows the changes in absorbance at 408.5 nm. We interpret the experimental results in terms of three optical states: trHbN(Fe3+-NO), trHbN(Fe3+-H2O), and trHbN(Fe3+). Accordingly, after ·NO photolysis most of the ·NO directly rebinds without leaving trHbN. After the non-geminate fraction of ·NO escapes to solvent, forming a short-lived 5C ferric-dehydrated state, a water molecule rebinds very rapidly (1.49 × 108 s-1), forming the trHbN(Fe3+-H2O) state. Finally, under ·NO saturating conditions (∼1.8 mm), the bound H2O is rapidly displaced (2.0 × 105 s-1), leading to the decrease in absorbance seen in Fig. 5A. As expected, increasing the ·NO concentration shortens the duration of the trHbN(Fe3+-H2O) complex and has no effect on the rate of formation of trHbN(Fe3+-H2O) (Fig. 5B). As shown in Fig. 5A and Ref. 21, water binding to 5C ferric horse heart Mb after ·NO photolysis is significantly slower (5.7 × 106 s-1), suggesting a lower barrier for migration of water molecule in trHbN.
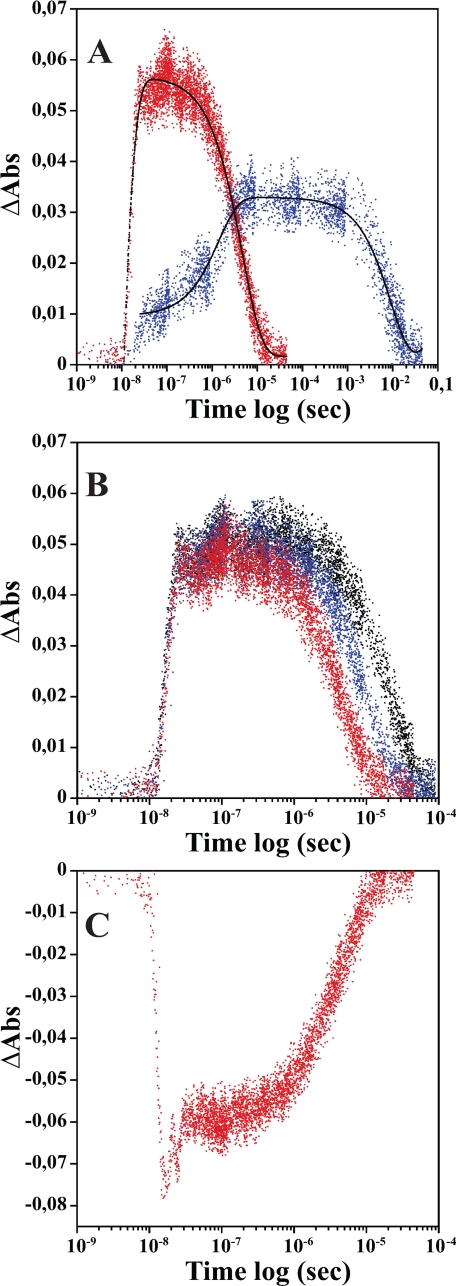
FIGURE 5.
Kinetic traces illustrating the absorbance changes after photodissociation of trHbN(Fe3+-NO) and Mb(Fe3+-NO) at 23 °C. Panel A shows the absorbance (Abs) changes corresponding to the water coordination processes followed at 408.5 nm (red) for trHbN(Fe3+-NO) (8.31 μm in heme) and 410 nm (blue) for Mb(Fe3+-NO) (7.37 μm in heme) preequilibrated with 100% ·NO. The solid lines are the results of the exponential fits (black). Panel B shows the kinetic traces acquired at 410 nm for trHbN(Fe3+-NO) (9.6 μm in heme) preequilibrated with 25% (black), 50% (blue), and 100% (red) ·NO. Panel C shows kinetic traces obtained at 421 nm after photolysis of trHbN(Fe3+-NO) (8.31 μm in heme) preequilibrated with 100% ·NO.
Fig. 5C shows the kinetic trace obtained when the reaction is monitored at 421 nm. This wavelength corresponds to the maximum absorbance of the Soret band of the trHbN(Fe3+-NO) species (Fig. 2). At this wavelength we monitored the reaction trHbN(Fe3+-NO) → trHbN(Fe3+) → trHbN(Fe3+-H2O) → trHbN(Fe3+-NO). Initially a decrease in absorbance was observed (2.1 × 108 s-1), corresponding to the formation of 5C ferric-dehydrated state from trHbN(Fe3+-NO) followed by a small increase in absorbance (1.1 × 108 s-1) associated to (trHbN(Fe3+) → trHbN(Fe3+-H2O) and finally by a further increase in absorbance (2.3 × 105 s-1) corresponding to ·NO replacing the bound water molecule.
Thus, water appears to constitute the main barrier to ligand rebinding to trHbN. Unlike Mb and human HbA, where water does not appear to impact geminate recombination due to delayed reentry of water into the DHP subsequent to ligand dissociation, in the case of trHbN the water occupancy occurs on the time scale of the geminate recombination. This observation indicates that water has access to the reactive site over the heme iron on a time scale that is much faster than for Mb. This acceleration is consistent either with water being stabilized within the protein at a site near the heme iron or with water being able to enter the DHP from the solvent on a nanosecond time scale. Although the state of hydration of the DHP of deoxy-trHbN is not known, the present flash-photolysis experiments with trHbN(Fe3+-NO) strongly suggest that a non-coordinated water molecule, stabilized by Tyr(B10) and Gln(E11), may be close to the heme iron in deoxy-trHbN. To investigate the fate of a water molecule in the DHP of deoxy-trHbN and its distal mutants and to gain further insights into the role of Tyr(B10) and Gln(E11), MD simulations were performed.
Molecular Dynamics Simulations Suggest That Water May Constitute the Main Kinetic Barrier to Ligand Binding to trHbN(Fe2+)
All simulations showed stable trajectories with protein backbone root mean square deviations around 1 Å. The positioning of the different DHP residues B10, E11, CD1, and the free DHP water molecule was studied. Table 2 shows the average minimum interatomic distances between non-hydrogen atoms and the heme iron atom extracted from the different trajectories produced. To measure access to the heme iron atom (accessible volume), we used a probe of 1.4 Å radius, which approximates to the radius of a water molecule. The results are presented in Table 3 and are expressed as the fraction of MD snapshots showing an accessible volume over the iron.
TABLE 2.
Average minimum interatomic distances between non-hydrogen atoms and the heme iron
Other DHP residues (Leu(E7), Phe(B9), and Val(G8)) showed longer a distance from the iron, ranging from 5.5 to 7.5 Å.
|
Proteina
|
Iron-Residue distance Å
|
||||
|---|---|---|---|---|---|
| Water | B10 | E11 | F(CD1) | ||
| Å | |||||
| trHbN-w | 3.5 | 5.4 | 5.3 | 4.9 | |
| Tyr(B10)Phe-w | 4.9 | 7.2 | 4.4 | 4.7 | |
| Gln(E11)Val-w | 3.8 | 5.2 | 5.5 | 4.9 | |
| Tyr(B10)Phe/Gln(E11)Val-w | 6.8 | 5.5 | 4.9 | ||
| trHbN-d | 5.7 | 4.9 | 4.0 | ||
| Tyr(B10)Phe-d | 6.7 | 4.4 | 4.4 | ||
| Gln(E11)Val-d | 5.2 | 5.8 | 4.2 | ||
| Tyr(B10)Phe/Gln(E11)Val-db | 6.6 | 5.5 | 4.8 | ||
Protein with (w) and without (d) a water molecule in the DHP.
The water molecule escaped too fast the DHP to get a proper measurement.
TABLE 3.
Cavity formation frequency and volume over the iron atom
| Proteina | Frequency | Volume |
|---|---|---|
| % | Å3 | |
| trHbN-w | 0.7 | 50.9 ± 15.4 |
| Tyr(B10)Phe-w | 5.4 | 77.5 ± 38.7 |
| Gln(E11)Val-w | 0.6 | 74.0 ± 18.0 |
| Tyr(B10)Phe/Gln(E11)Val-w | 43.5 | 69.6 ± 27.4 |
| trHbN-d | 7.1 | 46.8 ± 18.5 |
| Tyr(B10)Phe-d | 5.0 | 62.2 ± 18.5 |
| Gln(E11)Val-d | 19.5 | 58.3 ± 23.4 |
| Tyr(B10)Phe/Gln(E11)Val-d | 54.7 | 81.8 ± 29.0 |
Protein with (w) and without (d) a water molecule in the DHP.
Wild-type trHbN(Fe2+) Trajectories—Two 20-ns trajectories were produced for the wild-type protein, and in both cases the water molecule was stabilized by strong hydrogen-bonds involving both the Gln(E11) and Tyr(B10) residues. As a result, the water molecule occupied a main position close to the iron atom at a mean H2O-iron distance of 3.5 Å. On some rare occasions (0.7% of MD frames) the water molecule left this main position to get closer to the Gln(E11) side chain, creating an accessible volume over the iron atom (Table 3).
In the absence of a water molecule, the Tyr(B10) hydroxyl group was hydrogen-bonded to the OE1 atom of Gln(E11). This H-bond pulled the Tyr(B10) side chain farther away from the heme iron atom at a mean minimum distance of 5.7 Å. This configuration prevailed in 89.3% of the time. In this configuration the B10 residue does not hamper ligand coordination, and the closest residue from the iron atom is Phe(CD1) at 4.0 Å. As a consequence, an accessible volume over the heme iron was found 10 times more often (7.1% of the MD frames) than in trHbN-hydrated trajectories.
Tyr(B10)Phe(Fe2+) Mutant Trajectories—In the Tyr(B10)Phe mutant the Gln(E11) residue pulled the water molecule away from the heme center at a mean distance of 4.9 Å. The water molecule no longer occupied a well defined position, being constantly in motion around the Gln(E11) side chain. Also, the Phe(B10) residue showed increased flexibility and explored two χ1 dihedral domains (χ1 in minus and trans), compared with only one for trHbN. Consequently, accessible volume over the heme iron atom was observed more frequently (∼8-fold) than in trHbN (Table 3). The average volume was also larger than in trHbN by ∼25 Å3. Consistent with MD simulations, kinetic data showed a similar increase in both O2 and CO combination rates.
In the absence of a water molecule, the access to the heme iron atom was
slightly reduced with respect to trHbN
(Table 3), predicting lower or
similar 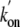 and
and
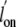 for the Tyr(B10)Phe mutant.
This is due to the Gln(E11) side chain which moves closer to the heme iron (by
0.9 Å), as also observed in the crystal structure of the cyanomet
derivative (31). MD data for
the Tyr(B10)Phe mutant are, thus, consistent with kinetic data if a water
molecule is present in the DHP.
for the Tyr(B10)Phe mutant.
This is due to the Gln(E11) side chain which moves closer to the heme iron (by
0.9 Å), as also observed in the crystal structure of the cyanomet
derivative (31). MD data for
the Tyr(B10)Phe mutant are, thus, consistent with kinetic data if a water
molecule is present in the DHP.
Gln(E11)Val(Fe2+) Mutant
Trajectories—In the Gln(E11)Val mutant, the DHP water molecule
occupied a position similar to that seen in trHbN
(Table 2) and was stabilized by
a strong H-bond to the Tyr(B10) hydroxyl group. Access to the heme iron was
slightly decreased with respect to trHbN, accounting for only 0.6% of the MD
frames analyzed. This is due to the absence of electrostatic attraction by the
E11 residue favoring water location near the Tyr(B10) hydroxyl group.
Consistent with these data, 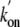 and
and 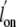 for the Gln(E11)Val
mutant were found similar to those of trHbN
(Table 1).
for the Gln(E11)Val
mutant were found similar to those of trHbN
(Table 1).
In contrast, in the absence of a DHP water molecule, cavity formation over the heme iron atom in the Gln(E11)Val mutant increased ∼3-fold compared with trHbN. In this case the Val(E11) side chain (5.8 Å) was unable to get as close to the heme iron atom as the Gln(E11) residue in the Tyr(B10)Phe mutant (4.4 Å). In trHbN, Tyr(B10) maintains the Gln(E11) side chain at a greater distance from the heme iron atom (4.9 Å) through H-bonding. As a consequence, the cavities detected over the iron atom in the Gln(E11)Val mutant were larger by about 10 Å3 than those formed in trHbN (Table 3). Thus, in contrast to kinetic data, MD simulations in absence of a distal water molecule would predict an increase in O2 and CO combination rates.
Tyr(B10)Phe/Gln(E11)Val(Fe2+) Mutant Trajectories—The B10/E11 double apolar substitution had a dramatic effect on the DHP water molecule stabilization. In all simulations, the water molecule rapidly escaped the protein matrix through the short tunnel. These results indicate that a water molecule is unlikely to reside within the DHP of the B10/E11 apolar mutants.
The analysis of the accessible volume revealed that apolar substitutions of
the B10/E11 pair increased the accessible volume by more than 15 Å
compared with trHbN (Table 3).
Additionally, in the absence of a water molecule, greater than 54.7% of the MD
data showed an accessible volume over the heme iron atom
(Table 3). Overall, the MD
results are in accord with the measured
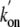 and
and
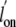 , which indicates that iron
coordination of small gaseous substrates in this case should only be limited
by their diffusion from the solvent to the tunnel and then to the active
site.
, which indicates that iron
coordination of small gaseous substrates in this case should only be limited
by their diffusion from the solvent to the tunnel and then to the active
site.
Conclusions—The present kinetic data and MD simulations
indicate that the main barrier to ligand binding from solvent and geminate
phase to deoxy-trHbN is the displacement of a non-coordinated distal site
water molecule, which is mainly stabilized by the Tyr(B10) residue. As
observed for trHbN Tyr(B10)/Gln(E11) double mutants, once this kinetic barrier
is eliminated, geminate yield is dramatically increased, and ligand binding is
very rapid, with rates approaching those measured for diffusion-controlled
reactions. Such a proposal is further supported by the observation that the
rates measured for ·NO binding to ferric heme-iron increases
dramatically in the Tyr(B10)Leu/Gln(E11)Val double mutant (1585 ×
106 m-1·s-1) compare with
wild-type trHbN (114.2 × 106
m-1·s-1), being as fast as CO and
O2 binding to the deoxy form of the double mutant (not shown).
These large combination rates almost certainly represent the upper limit for
ligand binding to a heme protein
(44,
45) and also indicate that the
heme iron in trHbN is highly reactive. Such rapid access to the active site is
attributed to the hydrophobic nature of the tunnels, which may favor rapid
docking and partitioning of the apolar gas into the polar distal heme cavity.
In turn, the rapid diffusion of apolar ligand to the active site may be
responsible for the efficient NOD reaction catalyzed by trHbN (745 ×
106 m-1·s-1). In addition
to ligand binding, water molecules in the DHP can participate actively in
other important processes including proton transfer reactions, catalysis,
folding, and redox processes. Recent quantum mechanics/molecular mechanics and
MD simulations with ferric trHbN suggest that formation of the
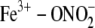 complex
triggers rapid hydration (a few nanoseconds) of the distal heme cavity, which
causes weakening of the Fe3+-O bond and rapid egress of the nitrate
ion from the active site (43).
Thus, water in the DHP facilitates the rapid release of NO3-, which
is necessary to guarantee an efficient NO detoxification and enhance survival
of the microorganism under stress conditions.
complex
triggers rapid hydration (a few nanoseconds) of the distal heme cavity, which
causes weakening of the Fe3+-O bond and rapid egress of the nitrate
ion from the active site (43).
Thus, water in the DHP facilitates the rapid release of NO3-, which
is necessary to guarantee an efficient NO detoxification and enhance survival
of the microorganism under stress conditions.
Photolysis experiments with trHbN(Fe3+-NO) indicates that water rebinds to the distal heme site at a rate of ∼1.49 × 108 s-1. Similar experiments with Mb(Fe3+-NO) and Mb(Fe2+-CO) estimated that water enters into the distal heme pocket at a rate of 5.7 × 106 and 9 × 106 s-1, respectively. The large difference in the rates of water rebinding emphasizes a lower barrier for water in trHbN, which can be attributed in part to the electrostatic interactions of water with the distal residues Tyr(B10) and Gln(E11). The difference in binding rates of water between Mb and trHbN may also be attributed to a much faster access to the DHP from the solvent due to the absence of a distal gate in trHbN (15, 19, 44, 45). The results of the geminate recombination studies are consistent with the water either being near the heme from the start or accessing the DHP on an unprecedented fast time scale. Whatever the situation, these results point to an important role for water in control of ligand reactivity in trHbN.
Acknowledgments
We are grateful to Dr. Beatrice A. Wittenberg and Dr. Jonathan B. Wittenberg from the Albert Einstein College of Medicine (New York) for insightful discussions. Dr. Martino Bolognesi is grateful to CIMAINA (Milano, Italy).
This work was supported, in whole or in part, by National Institutes of Health Grant 1-R01-AI052258 (to J. M. F.). This work was also supported by National Sciences and Engineering Research Council Grant 46306-01 (2005-2010) and Fonds Québécois de la Recherche sur la Nature et les Technologies Grant 104897 (M. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: trHbN, M. tuberculosis truncated hemoglobin N; ·NO, nitric oxide; NOD, nitric oxide dioxygenase; Hb, hemoglobin; Mb, myoglobin; MD, molecular dynamics; DHP, distal heme pocket; 5C, 5 coordinated.
References
- 1.Brunori, M. (2001) Trends Biochem. Sci. 26 21-23 [DOI] [PubMed] [Google Scholar]
- 2.Brunori, M., Giuffre, A., Sarti, P., Stubauer, G., and Wilson, M. T. (1999) Cell. Mol. Life Sci. 56 549-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevanin, T. M., Ioannidis, N., Mills, C. E., Kim, S. O., Hughes, M. N., and Poole, R. K. (2000) J. Biol. Chem. 275 35868-35875 [DOI] [PubMed] [Google Scholar]
- 4.Cleeter, M. W., Cooper, J. M., Darley-Usmar, V. M., Moncada, S., and Schapira, A. H. (1994) FEBS Lett. 345 50-54 [DOI] [PubMed] [Google Scholar]
- 5.Brown, G. C., and Cooper, C. E. (1994) FEBS Lett. 356 295-298 [DOI] [PubMed] [Google Scholar]
- 6.Gardner, P. R., Costantino, G., and Salzman, A. L. (1998) J. Biol. Chem. 273 26528-26533 [DOI] [PubMed] [Google Scholar]
- 7.Gardner, P. R., Costantino, G., Szabo, C., and Salzman, A. L. (1997) J. Biol. Chem. 272 25071-25076 [DOI] [PubMed] [Google Scholar]
- 8.Kissner, R., Nauser, T., Bugnon, P., Lye, P. G., and Koppenol, W. H. (1997) Chem. Res. Toxicol. 10 1285-1292 [DOI] [PubMed] [Google Scholar]
- 9.Pfeiffer, S., Gorren, A. C. F., Schmidt, K., Werner, E., Hansert, B., Bohle, D. S., and Mayer, B. (1997) J. Biol. Chem. 272 3465-3470 [DOI] [PubMed] [Google Scholar]
- 10.Ouellet, H., Ouellet, Y., Richard, C., Labarre, M., Wittenberg, B., Wittenberg, J., and Guertin, M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5902-5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuni, U., Ouellet, Y., Guertin, M., Friedman, J. M., and Yeh, S. R. (2004) J. Am. Chem. Soc. 126 2682-2683 [DOI] [PubMed] [Google Scholar]
- 12.Couture, M., Yeh, S. R., Wittenberg, B. A., Wittenberg, J. B., Ouellet, Y., Rousseau, D. L., and Guertin, M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11223-11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dantsker, D., Samuni, U., Ouellet, Y., Wittenberg, B. A., Wittenberg, J. B., Milani, M., Bolognesi, M., Guertin, M., and Friedman, J. M. (2004) J. Biol. Chem. 279 38844-38853 [DOI] [PubMed] [Google Scholar]
- 14.Springer, B. A., Sligar, S. G., Olson, J. S., and Phillips, G. N., Jr. (1994) Chem. Rev. 94 699-714 [Google Scholar]
- 15.Vojtechovsky, J., Chu, K., Berendzen, J., Sweet, R. M., and Schlichting, I. (1999) Biophys. J. 77 2153-2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohlfs, R. J., Mathews, A. J., Carver, T. E., Olson, J. S., Springer, B. A., Egeberg, K. D., and Sligar, S. G. (1990) J. Biol. Chem. 265 3168-3176 [PubMed] [Google Scholar]
- 17.Olson, J. S., and Phillips, G. N., Jr. (1996) J. Biol. Chem. 271 17593-17596 [DOI] [PubMed] [Google Scholar]
- 18.Olson, J. S., and Phillips, G. N., Jr. (1997) J. Biol. Inorg. Chem. 2 544-552 [Google Scholar]
- 19.Scott, E. E., Gibson, Q. H., and Olson, J. S. (2001) J. Biol. Chem. 276 5177-5188 [DOI] [PubMed] [Google Scholar]
- 20.Goldbeck, R. A., Bhaskaran, S., Ortega, C., Mendoza, J. L., Olson, J. S., Soman, J., Kliger, D. S., and Esquerra, R. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1254-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao, W., Christian, J. F., Champion, P. M., Rosca, F., and Sage, J. T. (2001) Biochemistry 40 5728-5737 [DOI] [PubMed] [Google Scholar]
- 22.Christian, J. F., Unno, M., Sage, J. T., Champion, P. M., Chien, E., and Sligar, S. G. (1997) Biochemistry 36 11198-11204 [DOI] [PubMed] [Google Scholar]
- 23.Quillin, M. L., Li, T., Olson, J. S., Phillips, G. N., Jr., Dou, Y., Ikeda-Saito, M., Regan, R., Carlson, M., Gibson, Q. H., Li, H., and et al. (1995) J. Mol. Biol. 245 416-436 [DOI] [PubMed] [Google Scholar]
- 24.Quillin, M. L., Arduini, R. M., Olson, J. S., and Phillips, G. N., Jr. (1993) J. Mol. Biol. 234 140-155 [DOI] [PubMed] [Google Scholar]
- 25.Dantsker, D., Samuni, U., Friedman, J. M., and Agmon, N. (2005) Biochim. Biophys. Acta 1749 234-251 [DOI] [PubMed] [Google Scholar]
- 26.Dantsker, D., Roche, C., Samuni, U., Blouin, G., Olson, J. S., and Friedman, J. M. (2005) J. Biol. Chem. 280 38740-38755 [DOI] [PubMed] [Google Scholar]
- 27.Samuni, U., Dantsker, D., Roche, C. J., and Friedman, J. M. (2007) Gene (Amst.) 398 234-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Draghi, F., Miele, A. E., Travaglini-Allocatelli, C., Vallone, B., Brunori, M., Gibson, Q. H., and Olson, J. S. (2002) J. Biol. Chem. 277 7509-7519 [DOI] [PubMed] [Google Scholar]
- 29.Samuni, U., Roche, C. J., Dantsker, D., and Friedman, J. M. (2007) J. Am Chem. Soc. 129 12756-12764 [DOI] [PubMed] [Google Scholar]
- 30.Eich, R. F., Li, T., Lemon, D. D., Doherty, D. H., Curry, S. R., Aitken, J. F., Mathews, A. J., Johnson, K. A., Smith, R. D., Phillips, G. N., Jr., and Olson, J. S. (1996) Biochemistry 35 6976-6983 [DOI] [PubMed] [Google Scholar]
- 31.Ouellet, Y., Milani, M., Couture, M., Bolognesi, M., and Guertin, M. (2006) Biochemistry 45 8770-8781 [DOI] [PubMed] [Google Scholar]
- 32.Samuni, U., Roche, C. J., Dantsker, D., Juszczak, L. J., and Friedman, J. M. (2006) Biochemistry 45 2820-2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuni, U., Dantsker, D., Khan, I., Friedman, A. J., Peterson, E., and Friedman, J. M. (2002) J. Biol. Chem. 277 25783-25790 [DOI] [PubMed] [Google Scholar]
- 34.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S., and Karplus, M. (1983) J. Comput. Chem. 4 187-217 [Google Scholar]
- 35.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., Joseph-McCarthy, D., Kuchnir, L., Kuczera, K., Lau, F. T. K., Mattos, C., Michnick, S., Ngo, T., Nguyen, D. T., Prodhom, B., Reiher, W. E., Roux, B., Schlenkrich, M., Smith, J. C., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz-Kuczera, J., Yin, D., and Karplus, M. (1998) J. Phys. Chem. B 102 3586-3616 [DOI] [PubMed] [Google Scholar]
- 36.Mackerell, A. D., Jr., Feig, M., and Brooks, C. L., III (2004) J. Comput. Chem. 25 1400-1415 [DOI] [PubMed] [Google Scholar]
- 37.Price, D. J., and Brooks, C. L., III (2004) J. Chem. Phys. 121 10096-10103 [DOI] [PubMed] [Google Scholar]
- 38.Feller, S. E., Pastor, R. W., Rojnuckarin, A., Bogusz, S., and Brooks, B. R. (1996) J. Phys. Chem. 100 17011-17020 [Google Scholar]
- 39.Ryckaert, J.-P., Ciccotti, G., and Berendsen, H. J. C. (1977) J. Comput. Phys. 23 327-341 [Google Scholar]
- 40.Kleywegt, G. J., and Jones, T. A. (1994) Acta Crystallogr. Sect. D 50 178-185 [DOI] [PubMed] [Google Scholar]
- 41.Huie, R. E., and Padmaja, S. (1993) Free Radic. Res. Commun. 18 195-199 [DOI] [PubMed] [Google Scholar]
- 42.Blough, N. V., and Zafiriou, O. C. (1985) Inorg. Chem. 24 3502-3504 [Google Scholar]
- 43.Marti, M. A., Bidon-Chanal, A., Crespo, A., Yeh, S. R., Guallar, V., Luque, F. J., and Estrin, D. A. (2008) J. Am. Chem. Soc. 130 1688-1693 [DOI] [PubMed] [Google Scholar]
- 44.Milani, M., Pesce, A., Ouellet, Y., Ascenzi, P., Guertin, M., and Bolognesi, M. (2001) EMBO J. 20 3902-3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolognesi, M., Cannillo, E., Ascenzi, P., Giacometti, G. M., Merli, A., and Brunori, M. (1982) J. Mol. Biol. 158 305-315 [DOI] [PubMed] [Google Scholar]