Abstract
Transforming growth factor (TGF)-β1 induces fibroblast transdifferentiation to myofibroblasts, a process that requires the involvement of integrin-mediated signaling and focal adhesion kinase (FAK). FAK-related non-kinase (FRNK) is known for its role in inhibiting integrin-mediated cell migration; however, its role in myofibroblast differentiation has not been defined. Here, we report that FRNK abrogates TGF-β1-induced myofibroblast differentiation in vitro and in vivo. TGF-β1 can induce α-smooth muscle actin (α-SMA) expression in the presence or absence of FAK; however, TGF-β1-induced α-SMA expression is reduced (∼73%) in FAK-deficient fibroblasts. Although both ERK and p38 MAPK activation is required for maximal TGF-β1-induced α-SMA expression, ERK is the major signaling intermediate in cells that express FAK. In contrast, p38 MAPK is the dominant mediator of TGF-β1-induced α-SMA expression in FAK-deficient cells. FRNK overexpression blocks TGF-β1-induced ERK or p38 MAPK activation in the presence, and surprisingly, in the absence of FAK. The loss of FRNK was tested in vivo during experimentally induced pulmonary fibrosis in mice. FRNK knock-out mice have a greater increase in α-SMA-expressing cells in response to a pulmonary fibrotic stimulus in vivo, as compared with congenic wild type mice. This is the first time that FRNK loss has been shown to modify the pathobiology in any animal disease model. Together, the data demonstrate that FRNK negatively regulates myofibroblast differentiation in vitro and in vivo. These data further suggest that modulation FRNK expression may be a novel avenue for therapeutic intervention in tissue fibrosis.
Deranged tissue repair and remodeling or failed termination of the normal wound healing response may result in tissue fibrosis. After injury, fibroblasts migrate into the wound, proliferate, and differentiate to “activated” fibroblasts (1–4). The differentiated or activated fibroblasts are termed myofibroblasts and are present in abundance within fibrotic lesional areas in the lung, liver, and kidney (2, 4–6). Myofibroblasts secrete profibrotic cytokines and extracellular matrix proteins, such as collagen and fibronectin, and therefore have been presumed to act as effector cells of the fibrotic process (1, 2, 4–7).
Idiopathic pulmonary fibrosis is a devastating, untreatable, progressive fibrotic disease of the lung (8–11). In support of the importance of myofibroblasts in the pathogenesis of lung fibrosis, these cells localize to fibroblastic foci in pulmonary fibrotic lungs, and the extent of fibroblastic foci is a major prognostic factor for patients with idiopathic pulmonary fibrosis (8–11).
The phenotypic hallmarks of myofibroblasts are the expression of α-SMA2 and the incorporation of α-SMA into well organized cytoplasmic fibers (1, 4, 6). These fibers connect to robust, mature focal adhesions (1). α-SMA expression is associated with an enhanced contractile phenotype and can directly enhance the contractility of fibroblasts (12, 13). Inhibition of α-SMA integration into cytoplasmic fibers by α-SMA N-terminal peptides blocks cell contraction and collagen expression in rat lung fibroblasts (14). α-SMA-deficient mice have impaired vascular contractility and show increased renal fibrotic lesions in a unilateral ureteral obstruction model (15, 16). These data indicate that α-SMA plays a key role in regulation of myofibroblast functions in vitro and in vivo.
Although the myofibroblast is generally accepted as a key effector cell in the development of fibrosis, the molecular mechanisms and intracellular signaling events regulating myofibroblast differentiation are not completely understood. TGF-β is the best studied positive regulator of myofibroblast differentiation (17–19) and is widely accepted as a central mediator of the fibrotic responses in lung, liver, and kidney (5, 8, 20). Myofibroblast differentiation is dependent on the presence of active TGF-β, a rigid extracellular matrix, and the presence of the extra type III domain - A (EDA) fibronectin fragment (1, 21–23). Integrin receptors are responsible for transmitting these matrix environmental cues intracellularly (24, 25). In fact, integrins can participate in the activation of matrix-bound latent TGF-β through cystoskeletal-induced generation of tension (21, 26).
Integrin engagement with extracellular matrix proteins can activate focal adhesion kinase (FAK) and results in an increased phosphorylation of Tyr397 of FAK (25, 27–30). FAK has been demonstrated to have a critical role in integrin-mediated cell functions, such as cell migration and proliferation (25, 31–33). FAK has recently been shown to mediate the α-SMA up-regulation in response to TGF-β1 (34, 35), the α-SMA inhibition in response to fibroblast growth factor (36).
FRNK is an independently expressed cytoplasmic protein that is identical in sequence to the C-terminal region of FAK (25, 37, 38). It has been demonstrated that FRNK inhibits integrin-mediated FAK activation (Tyr397 phosphorylation) and blocks FAK-mediated cell migration and proliferation (25, 37, 39, 40). However, the role of FRNK in TGF-β1-induced myofibroblast differentiation has not been defined.
In this study, the effect of gain of FRNK and the FAK dependence of the effect of FRNK on TGF-β1-induced α-SMA expression were examined. Furthermore, because MAPKs are known downstream mediators of FAK signaling (41–43), we studied the FAK dependence of MAPK activation during myofibroblast differentiation. Lastly, we evaluated the effect of FRNK deficiency on myofibroblast differentiation in a murine model of experimentally induced pulmonary fibrosis.
EXPERIMENTAL PROCEDURES
Reagents—TGF-β1 was obtained from R & D Systems (Minneapolis, MN). The following purified polyclonal antibodies were purchased: anti-phospho-FAK (Tyr(P)397) (BIOSOURCE, Camarillo, CA), anti-FAK (recognizes both FAK C-terminal and FRNK; Upstate Biotechnology, Lake Placid, NY), anti-phospho-ERK (Cell Signaling Technology/New England Biolabs, Danvers, MA), anti-ERK IgG (Santa Cruz Biotechnology, Santa Cruz, CA), anti-p38 (Cell Signaling Technology), anti-phospho-p38 (Chemicon International, Temecula, CA), and anti-phospho-JNK (Cell Signaling). Purified Alexa Fluor 488 goat anti-mouse IgG was purchased from Molecular Probes (Carlsbad, CA). The following purified monoclonal antibodies were purchased: anti-α-SMA (American Research Products, Belmont, MA), Cy3-conjugated anti-α-SMA antibody (clone 1A4; Sigma-Aldrich), anti-hemagglutinin (HA) epitope tag (Santa Cruz Biotechnology), and anti-glyceraldehyde 3-phosphate dehydrogenase (G3PDH) (Research Diagnostics, Flanders, NJ). The selective inhibitors PD98059, U0126, U0124, and SB203580 were purchased from EMD Biosciences/Calbiochem and dissolved in Me2SO.
Animal Protocol—Animal interventions were approved by the Institutional Animal Care and Use Committee and the Animal Resources Facility at the University of Alabama at Birmingham and were performed under the guidelines for animal care recommended by the American Physiological Society. All of the animals were pathogen-free, housed in micro-isolator cages, and fed with autoclaved food and water. FRNK knock-out mice were generated as described (44) and were generously provided by Dr. J. Thomas Parsons. FRNK knock-out mice were C57BL/6 background (back-crossed with C57BL/6 mice for at least 12 generations before the current study). The genotype of FRNK knock-out (homozygous) mice and their wild type littermates was confirmed by PCR using genomic DNA extracted from tails as published (44), and the absence of FRNK expression in lung tissue from FRNK knock-out mice was confirmed by Western blot (data not shown). The administration of bleomycin and harvest of lung tissues were described previously (45, 46). Briefly, the animals (7–10 weeks) were anesthetized, and bleomycin (1 unit/kg of body weight in 50 μl of saline) or saline alone (50 μl) was slowly instilled through airway to lungs by using an intratracheal catheter. The skin wounds were closed, and the animals were allowed to recover. The animals were euthanized at day 18 after bleomycin instillation. The lungs were perfused with cold phosphate-buffered saline, inflated with tissue-preservation media Optimal Cutting Temperature compound (OCT), and sectioned for immunofluorescence analysis of α-SMA-positive cells. Whole lung homogenate supernatants were used for Western blot analysis. The harvested lungs were homogenized in 1% Nonidet P-40 lysis buffer (with the following inhibitors, 100 μm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 100 μm sodium vanadate, and 20 μg/ml TLCK) with a polytron (Brinkmann Instruments, Westbury, NY), and homogenate supernatants were extracted by centrifugation (14,000 × g for 20 min at 4 °C) (46).
Cells and Cell Culture—Primary murine lung fibroblasts were derived from 7–10-week-old C57BL/6 mice and propagated as described previously (47, 48). Briefly, primary cultures of murine lung fibroblasts were established by mechanical disaggregation and enzymatic digestion (collagenase and DNase) of sterilely removed lungs and by cell culture. Fibroblasts were selected by differential adherence to tissue culture plastic and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 20 mm HEPES, and 100 units/ml penicillin/streptomycin/gentamycin. The cell identity of primary cultures was verified by the absence of tight junctions, by production of collagen types I and III, and by typical morphology on phase contrast as described previously (47, 48). Primary adult normal human lung fibroblasts (HL1–1608) were purchased from Cambrex (Walkersville, Maryland), and normal adult human lung fibroblasts (HL2–19LU) were purchased from American Type Culture Collection (Manassas, VA). Human lung fibroblasts were propagated and maintained in DMEM supplemented with 10% FBS, 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin. The experiments were performed on early passages (passage 2–9) of primary murine and human lung fibroblasts. The Tet-FAK fibroblast-like cells and their parental FAK-null embryonic fibroblasts were kindly provided by Dr. Steven K. Hanks. In the absence of doxycycline in cell culture medium, FAK expression is induced in Tet-FAK cells as described previously (49). In the presence of doxycycline (2 μg/ml), FAK expression in Tet-FAK cells is repressed (49). The Tet-FAK cells and their parental FAK-null cells were propagated and maintained in DMEM containing 10% FBS, 4.5 g/liter d-glucose, 0.6 g/liter glutamine, 1 mm sodium pyruvate, 0.1 mm nonessential amino acids, 100 units/ml penicillin/streptomycin, and 0.25 μg/ml amphotericin B as described previously (49).
Preparation, Amplification, and Purification of the Adenoviral FRNK (Ad-FRNK) Construct—The HA-tagged FRNK construct was reported previously (50). To efficiently deliver the FRNK cDNA into primary lung fibroblasts, the adenovirally mediated gene delivery system was used. The HA-tagged FRNK cDNA was engineered into a replication-deficient adenovirus vector by using the Adeno-X Expression System 2 according to the manufacturer's manual (Clontech, Mountain View, CA). The Ad-FRNK was rescued and amplified in 293 cells, purified by CsCl gradient centrifugation, and used to infect cells as described previously (46, 51). Adenoviral vectors encoding a green fluorescent protein (Ad-GFP) or a luciferase construct (Ad-Luc) were initially used to optimize the infection conditions on subconfluent (70–80%) cells cultured in serum-containing or serum-free conditions as described previously (46, 51). The cells in serum-free medium (DMEM with 1% BSA) were infected with Ad-FRNK or control vectors (Ad-Luc or Ad-GFP) 24 h prior to TGF-β1 treatment.
Signal Transduction Experiments without and with MAPK Inhibitors—Prior to TGF-β1 treatment, fibroblasts were quiescent by growing in DMEM with 0.2% FBS for 24 h, followed in serum-free media (DMEM with 1% BSA) for another 24 h. Quiescent cells were treated with either 10 ng/ml of TGF-β1 or vehicle (Me2SO) in serum-free media (DMEM with 1% BSA) for the time indicated in the text. The selective pharmacological inhibitors, PD98059 or U0126 (both are MEK inhibitors), or/and SB203580 (p38 MAPK inhibitor), or U0124 (inactive analog of U0126) were added to culture medium 1 h prior to TGF-β1 treatment.
Western Blotting—As described previously (33, 52). The cells were lysed in 1% Nonidet P-40 lysis buffer containing the following inhibitors, 100 μm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 100 μm sodium vanadate, and 20 μg/ml TLCK. Protein concentration of whole cell or whole lung lysate was determined by a BCA kit (Pierce). Equivalent micrograms (by BCA assay) of whole cell or lung detergent lysates were electrophoresed on a disulfide-reduced 7.5% SDS-PAGE, transferred to Immobilon-P membrane (Millipore Corp., Bedford, MA), probed, or stripped followed by reprobing with indicated antibodies, and developed with ECL system (GE Healthcare). FRNK protein level was determined by Western blot under non-disulfide-reduced conditions following immunoprecipitation of equivalent lysates as described previously (52). The expression of G3PDH protein was used as a loading control. Quantitative analysis (densitometry) of Western blots was performed by calculating the relative density (pixel density) of the immunoreactive bands after acquisition of the blot image (scanning) and analysis with Adobe Photoshop software. For analysis of band intensity, a specific band on the ECL-developed films was subjected to densitometric analysis (Adobe Photoshop). The densitometric readings were pooled and averaged from three independent experiments, and the results were normalized to the total protein level. The background of densitometric reading on the ECL-developed film was subtracted as described previously (33, 53).
Immunofluorescence Analysis—Cells cultured on glass coverslips were fixed in 4% buffered paraformaldehyde, permeabilized, and reacted sequentially with primary antibodies, followed by secondary antibodies. To detect HA-tagged FRNK expression, the cells were reacted with monoclonal anti-HA IgG (13 μg/ml), and followed by goat anti-mouse Alexa 488 IgG (10 μg/ml). To study the α-SMA-incorporated cytoskeletal fibers, the cells were reacted with Cy3-conjugated anti-α-SMA monoclonal antibodies (1:300). Digital fluorescence images were obtained by using a Leica microscope and Simple PCI software (Compix Inc.). The percentage of cells containing highly organized α-SMA-incorporated fibers were quantified. The results were pooled from eight randomly selected, non-overlapping fields (original magnification, 400×) from a total of three experiments (each performed in triplicate).
Frozen lung tissue sections (8–10 μm) were permeabilized, reacted with Cy3-conjugated monoclonal anti-α-SMA antibody (1:300 dilution), and counterstained with hematoxylin (Sigma-Aldrich). Digital fluorescence images (original magnification, 400×) of stained lung sections were obtained by using a Leica microscope and Simple PCI software (Compix Inc.). The areas containing α-SMA-positive cells versus total parenchymal area on lung sections was quantified by the Simple PCI software (Compix, Inc.). The results were pooled from a total of eight sections/animal (at least 50-μm apart). Lesions of comparable histology were used. Areas that represent airways and blood vessels were excluded. Five animals/group (bleomycin or saline) were analyzed.
Statistical Analysis—The data were analyzed using the t test analysis (Sigma Plot, SPSS Inc.) or one-way analysis of variance (SigmaStat, SPSS Inc.) as indicated and are expressed as the means ± S.D. p values <0.05 are considered statistically significant. All of the experiments were repeated at least three times.
RESULTS
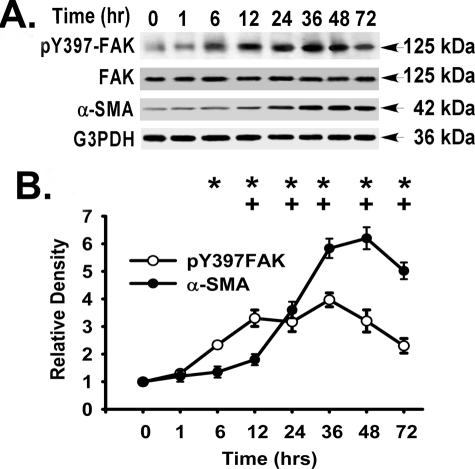
TGF-β1 Induces α-SMA Expression and the Activation of FAK, ERK, and p38 MAPK in Primary Murine and Human Lung Fibroblasts—To examine the role of FAK in TGF-β1-induced α-SMA expression (a hallmark of the myofibroblast phenotype) (1), we first compared the time courses of FAK activation (FAK tyrosine 397 phosphorylation) and α-SMA expression in primary murine lung fibroblasts in response to TGF-β1 stimulation (Fig. 1). FAK activation was persistently elevated from 6 to 72 h, and α-SMA expression was persistently elevated from 12 to 72 h (Fig. 1). This effect of TGF-β1 on α-SMA expression and FAK activation was confirmed in additional primary murine and human lung fibroblasts (at 36 h) (Fig. 2A).
FIGURE 1.
Time course of TGF-β1-induced FAK activation and α-SMA expression. Subconfluent (70–80%) primary murine lung fibroblasts were treated with TGF-β1 (10 ng/ml, 37 °C) in serum-free medium (DMEM, 1% BSA) for the indicated times. Equivalent amount of whole cell detergent lysates were Western blotted with the indicated antibodies as described under “Experimental Procedures.” A, representative time course of FAK activation (pY397) and α-SMA expression. B, densitometry of FAK activation (pY397-FAK normalized to total FAK protein) and α-SMA expression (normalized to G3PDH) from A. FAK activation (○); α-SMA expression (•). The data are presented as the means ± S.D. (n = 3). At the indicated times, + represents p < 0.05 for TGF-β1-induced α-SMA expression compared with control (without TGF-β1), and * represents p < 0.05 for TGF-β1-induced FAK activation compared with control (without TGF-β1) by analysis of variance.
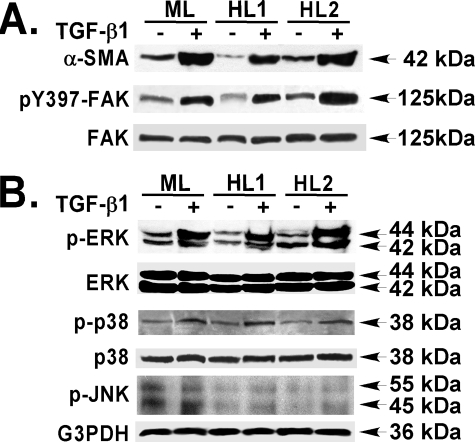
FIGURE 2.
TGF-β1 induces α-SMA expression, FAK activation, and MAPK activation in primary murine and human lung fibroblasts. Subconfluent primary murine (ML) and human (HL1 and HL2) lung fibroblasts were treated without or with TGF-β1 (10 ng/ml, 36 h) in serum-free medium (DMEM, 1% BSA) as in Fig. 1. Equivalent amounts of whole cell detergent lysates were Western blotted with the indicated antibodies. A, FAK activation (pY397) and α-SMA expression. B, ERK, p38, and JNK MAPK activation.
Because the MAPK pathway has been shown to exhibit cross-talk with that of TGF-β signaling pathway, we measured the activation of MAPK in response to TGF-β1. ERK activation was induced (∼4-fold), and p38 activation was induced (∼3-fold) at the time of peak of FAK activation and α-SMA expression (at 36 h) (Fig. 2B). In contrast, TGF-β1 had minimal effect on JNK activation (at 36 h) (Fig. 2B). The activation of both ERK and p38 MAPK in response to TGF-β1 were sustained (6–48 h) (data not shown), as shown in other cell types (42, 54, 55). Our data demonstrate that TGF-β1 can induce α-SMA expression, enhance FAK activation, and induce MAPK (ERK and p38) activation in primary murine and human lung fibroblasts.
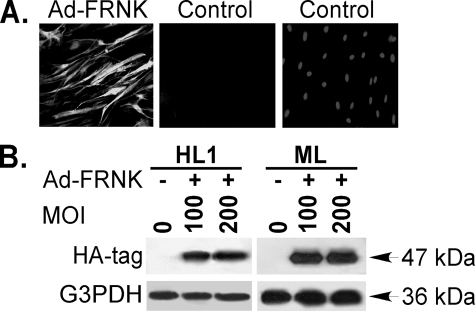
FRNK Abrogates TGF-β1-induced Incorporation of α-SMA into Cytoskeletal Fibers and α-SMA Expression—FRNK expression inhibits cell migration and proliferation in other cell types (25), but its effects on myofibroblast differentiation have not been examined. To determine the effect of FRNK on TGF-β1-induced α-SMA expression, we first validated that the adenoviral HA-tagged FRNK construct (Ad-FRNK) can achieve highly efficient FRNK expression in primary murine and human lung fibroblasts. The efficiency of Ad-FRNK infection was confirmed by immunofluorescent staining directed toward the FRNK HA-tag (Fig. 3A). At a multiplicity of infection (MOI) of 100, greater than 95% of the primary murine and human lung fibroblasts were infected by Ad-FRNK, with no increase in cell toxicity (data not shown). The capacity of Ad-FRNK to generate FRNK protein was also confirmed by Western blot by probing for the HA tag in whole cell detergent lysates from primary murine and human lung fibroblasts (Fig. 3B).
FIGURE 3.
Validation of adenovirus-mediated FRNK expression in lung fibroblasts. Subconfluent primary murine (ML) or human (HL1) lung fibroblasts were infected with the adenoviral HA-tagged FRNK vector (Ad-FRNK) at a MOI of 100 (A) or the indicated MOI (B) as described under “Experimental Procedures.” A, immunofluorescence analysis of HA-tag for FRNK expression in primary murine lung fibroblasts at 36 h without or with Ad-FRNK infection. Left panel, fibroblasts with Ad-FRNK infection and stained with anti-HA antibody. Middle panel, fibroblasts with vehicle only stained with anti-HA antibody or Right panel, counterstained with Hoechst (nuclear stain). The original magnification was 100×. B, fibroblasts were lysed at 36 h after Ad-FRNK infection. Equivalent amounts of whole cell detergent lysates were Western blotted with the indicated antibodies. FRNK expression in infected cells was determined by FRNK HA tag expression.
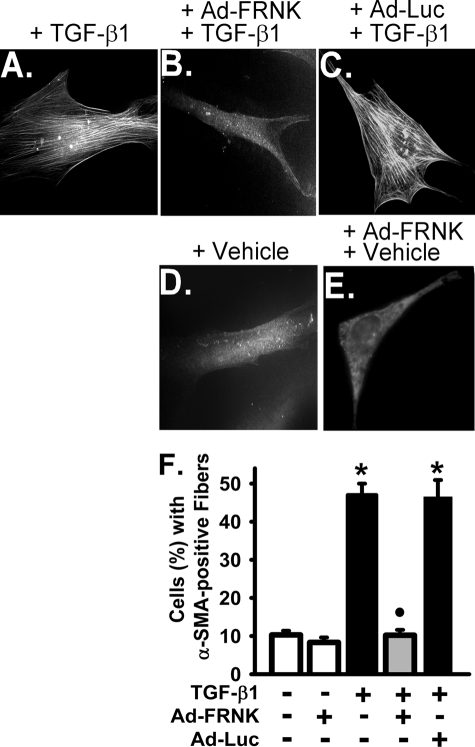
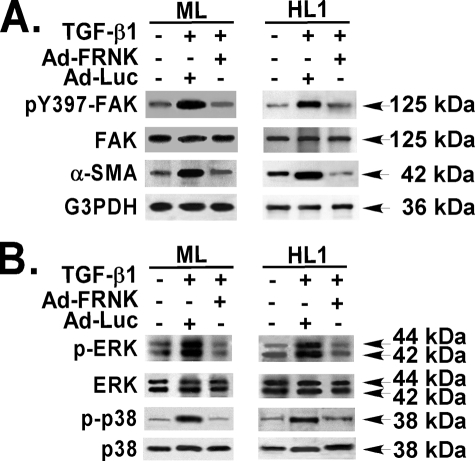
TGF-β1 induced incorporation of α-SMA into highly organized cytoskeletal fibers, a phenotypic hallmark of myofibroblast differentiation (1, 56), in primary murine lung fibroblasts (Fig. 4A). FRNK expression, as mediated by Ad-FRNK, completely blocked TGF-β1-induced incorporation of α-SMA into cytoskeletal fibers in these cells (Fig. 4B). Control transfections using Ad-Luc had no effect on TGF-β1-induced incorporation of α-SMA into cytoskeletal fibers (Fig. 4C), and FRNK expression mediated by Ad-FRNK had no effect on the basal (vehicle only) level of α-SMA incorporation into cytoskeletal fibers (Fig. 4E). FRNK expression also abrogated TGF-β1-induced α-SMA expression (by ∼85%) (Fig. 5A). Control transfection with Ad-Luc had no effect α-SMA protein expression (Fig. 5A). In summary, FRNK expression abrogates TGF-β1-induced myofibroblast differentiation by blocking the effects of TGF-β1 on α-SMA expression (Fig. 5) and on the formation of α-SMA-containing cytoskeletal fibers (Fig. 4).
FIGURE 4.
FRNK expression inhibits formation of α-SMA-containing cytoskeletal fibers in response to TGF-β1 in primary lung fibroblasts. Subconfluent primary murine lung fibroblasts were infected with Ad-FRNK or control Ad-Luc (adenoviral luciferase construct) at 100 MOI, treated with TGF-β1 (10 ng/ml, 36 h) in serum-free medium (DMEM, 1% BSA), and stained with Cy3-conjugated mouse anti-α-SMA IgG as described under”Experimental Procedures.” A, immunofluorescence analysis of α-SMA-containing fibers in primary murine lung fibroblasts with Cy3-conjugated mouse anti-α-SMA IgG (original magnification, 400×). B, quantification of the percentage of cells with highly organized α-SMA-containing fibers as described under “Experimental Procedures.” F, open bars represent vehicle-treated cells. Filled black bars represent TGF-β1-treated cells. The gray bar represents cells infected with Ad-FRNK and treated with TGF-β1. The data are presented as the means ± S.D. (n = 3, each performed in triplicate). * represents p < 0.01 for TGF-β1-treated cells compared with cells treated with vehicle only. • represents p < 0.01 for TGF-β1-treated cells compared with cells infected with Ad-FRNK and treated with TGF-β1.
FIGURE 5.
FRNK expression abrogates TGF-β1-induced α-SMA expression and the activation of FAK, ERK, and p38. Quiescent primary murine (ML) and human (HL1) lung fibroblasts were infected with Ad-FRNK or control vector (Ad-Luc) (both at 100 MOI), treated with TGF-β1 (10 ng/ml, 36 h) in serum-free medium (DMEM, 1% BSA) and detergent-lysed. A, equivalent amount of whole cell lysates were Western blotted with the indicated antibodies as described under “Experimental Procedures.” B, equivalent amounts of whole cell detergent lysates were also Western blotted for ERK and p38 with the indicated antibodies.
FRNK Abrogates TGF-β1-induced Activation of FAK, ERK, and p38 MAPK—Because FAK mediates myofibroblast differentiation, and MAPK can act as a signaling intermediate of both FAK and TGF-β1 signals, we tested the effect of Ad-FRNK on FAK and MAPK activation in response to TGF-β1. FRNK expression abrogated TGF-β1-induced FAK activation (by ∼95%), as measured by the proportion of the Tyr(P)397 form of FAK relative to total FAK (Fig. 5A). Control transfection with Ad-Luc had no effect on FAK activation (Fig. 5A) in these primary lung fibroblasts. Similarly, FRNK expression completely blocked TGF-β1-induced ERK activation (by >95%) and p38 activation (by ∼90%) (Fig. 5B). These data demonstrate that FRNK expression abrogates TGF-β1-induced FAK, ERK, and p38 activation.
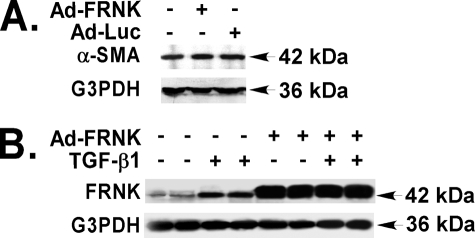
One consideration is the possibility that adenovirally mediated FRNK expression would result in massive overexpression of FRNK leading to the unmasking of nonphysiological actions. However, TGF-β1 itself induced FRNK protein expression by 3.7-fold (Fig. 6B), and adenovirally driven FRNK protein levels were only 5.1-fold greater than that detected after TGF-β1 stimulation (Fig. 6B).
FIGURE 6.
FRNK protein level is increased in primary lung fibroblasts in response to TGF-β1. A, quiescent primary murine lung fibroblasts infected with Ad-FRNK or control Ad-Luc (both at 100 MOI) were detergent-lysed. Equivalent amount of whole cell lysates was used to determine the α-SMA expression by Western blot as described under “Experimental Procedures.” B, quiescent primary murine lung fibroblasts infected with or without Ad-FRNK (at 100 MOI) were treated with TGF-β1 (10 ng/ml, 72 h) or vehicle in serum-free medium (DMEM, 1% BSA), and detergent-lysed. Equivalent amounts of whole cell detergent lysates were used to determine the expression of FRNK by Western blot.
FRNK Expression Abrogates TGF-β1-induced α-SMA Expression in the Presence or Absence of FAK—FRNK is thought to inhibit FAK-mediated signaling through its effects on recruitment of signaling proteins that bind to FAK. To examine whether the inhibitory effect of FRNK on TGF-β1-induced α-SMA expression is totally dependent on blocking FAK-mediated signaling, the effect of FRNK expression on α-SMA expression was examined in fibroblasts from the same parental line with regulatable FAK expression. This approach minimizes the effect of inherent differences in α-SMA expression seen among different cell types and lineages (57).
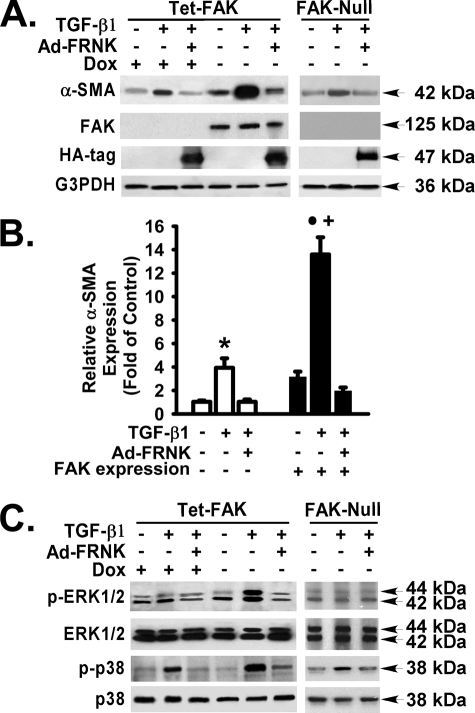
TGF-β1 was capable of inducing α-SMA expression in cells that express FAK, concordant with our results in primary cells (Fig. 7, A and B). Surprisingly, TGF-β1 was also capable of inducing α-SMA in FAK-deficient fibroblasts, albeit to a submaximal level (13.6 ± 2.5-fold versus 3.9 ± 1.4-fold in the FAK-deficient fibroblasts, p < 0.01) (Fig. 7B, fifth bar versus second bar). This partial FAK dependence was also noted under basal conditions, where FAK-deficient cells had a 3-fold lower level of α-SMA expression (Fig. 7B, fourth bar versus first bar), when compared with FAK-expressing cells. These data demonstrate that FAK has an independent impact on α-SMA induction by TGF-β1. Nonetheless, the FAK signaling pathway is required for maximal α-SMA expression by TGF-β1. Furthermore, these data demonstrate for the first time that FRNK expression can abrogate TGF-β1-induced α-SMA expression in the presence or absence of FAK.
FIGURE 7.
FRNK expression abrogates TGF-β1-induced α-SMA expression in the presence or absence of FAK. Tet-FAK fibroblasts were treated with doxycycline (Dox) to repress FAK expression or allowed to express FAK in the absence of doxycycline. Tet-FAK cells (with or without doxycycline) and their parental FAK-null fibroblasts were infected with or without Ad-FRNK and then treated with or without TGF-β1 (10 ng/ml, 36 h) in serum-free medium (DMEM, 1% BSA). A and C, equivalent amount of whole cell detergent lysates were Western blotted with the indicated antibodies. Representative results are shown. The FRNK HA tag is shown at 47 kDa. B, densitometry of α-SMA expression (normalized to G3PDH and relative to FAK-deficient cells without TGF-β1) as described under “Experimental Procedures.” α-SMA expression for FAK-deficient cells were pooled from results obtained in Tet-FAK cells (with doxycycline) and FAK-null fibroblasts. Open bars denote the results from FAK-deficient cells. Filled black bars denote the results from FAK-expressing cells. The data are presented as the means ± S.D. (n = 3). * represents p < 0.05 for TGF-β1-treated FAK-deficient cells compared with the same group of cells infected with Ad-FRNK or treated with vehicle only. • represents p < 0.01 for TGF-β1-treated FAK-expressing cells compared with the same group of cells infected with Ad-FRNK or treated with vehicle only. + represents p < 0.01 for TGF-β1-treated FAK-expressing cells compared with TGF-β1-treated FAK-deficient cells.
ERK and p38 MAPK Differentially Mediate TGF-β1-induced α-SMA Expression in a FAK-dependent Manner—Because we have shown that FRNK expression will block TGF-β-initiated MAPK activation and α-SMA expression, we sought to determine whether the effects of FRNK are a consequence of inhibiting FAK. Both ERK and p38 MAPK were activated in response to TGF-β1, in fibroblasts that express FAK (Fig. 7C; also see Fig. 2B). In contrast, only p38 was activated in response to TGF-β1, in FAK-deficient fibroblasts (Fig. 7C).
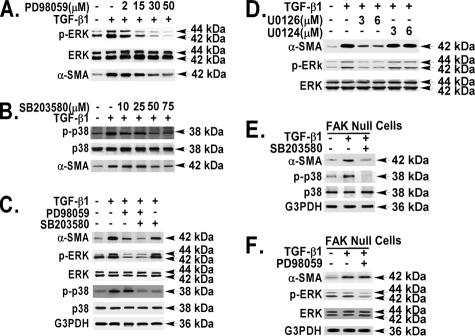
To delineate the importance of ERK and p38 MAPKs to the TGF-β1 response in α-SMA expression and its dependence on FAK, specific MAPK pharmacological inhibitors were used in cells that express FAK or lack FAK. As expected, the ERK inhibitor (PD98059) and the p38 inhibitor (SB203580) respectively, blocked TGF-β1-induced ERK and p38 MAPK activation in a selective and dose-dependent manner (Fig. 8, A and B). Inhibition of both ERK and p38 was required to completely abrogate the TGF-β1-induced α-SMA expression in fibroblasts that express FAK (> 95% by densitometry) (Fig. 8C, fourth lane versus second lane). ERK was the predominant MAPK (PD98059 blocked by ∼80%), whereas p38 played a minor role (SB203580 blocked by ∼15%) in the TGF-β1-induced α-SMA expression in FAK-expressing cells (Fig. 8C). These findings were confirmed by using a second ERK inhibitor (77% blockade using U0126) (Fig. 8D), whereas the inactive control analog, U0124, had no effect (Fig. 8D).
FIGURE 8.
ERK and p38 are differentially involved in TGF-β1-induced α-SMA expression in the presence or absence of FAK. Pharmacologic inhibitor (PD98059, SB203580, or U0126) was added to quiescent primary murine lung fibroblasts (A–D) or FAK-null fibroblasts (E and F) 1 h before TGF-β1 treatment (10 ng/ml, 36 h). The cells were detergent-lysed. Equivalent amounts of whole cell lysates were Western blotted with the indicated antibodies. A and B, fibroblasts were incubated without or with the indicated concentrations of inhibitors (PD98059 or SB203580). C–F, fibroblasts were treated without or with TGF-β1 (10 ng/ml) in the absence or presence of PD98059 (30 μm), SB203580 (50 μm), or U0126 and U0124 at indicated concentrations.
In contrast to our finding in the FAK-expressing fibroblasts, p38 MAPK was solely responsible for the α-SMA expression in response to TGF-β1 in FAK-deficient cells. This is supported by our observation that the p38 inhibitor SB203580 blocked the α-SMA expression by ∼92% in the FAK-deficient cells (Fig. 8E), whereas the inhibition of ERK had no effect (Fig. 8F). These data demonstrate that the major MAPK utilized in the α-SMA expression pathway is ERK in FAK-expressing cells and is p38 in FAK-deficient cells (Fig. 8).
FRNK Inhibits TGF-β1-induced MAPK Activation and α-SMA Expression in the Absence of FAK—Because we show that FRNK expression inhibits both MAPK activation and α-SMA expression in the presence of FAK, we tested the effect of FRNK on these pathways in the absence of FAK. As expected, both ERK and p38 MAPK activation in response to TGF-β1 were blocked by FRNK, in fibroblasts that express FAK (Fig. 7C; also see Fig. 5B). Surprisingly, MAPK (p38) activation and α-SMA expression in response to TGF-β1 was also blocked by FRNK expression in FAK-deficient fibroblasts (Fig. 7, A and C). ERK was not activated in response to TGF-β1 in FAK-deficient cells (Fig. 7C). These data demonstrate that FRNK expression abrogates TGF-β1-induced α-SMA expression and MAPK activation in the presence or absence of FAK (Figs. 5 and 7).
We have demonstrated 1) MAPK kinases mediate α-SMA expression in response to TGF-β1, and 2) ERK is the major downstream signaling intermediate in the pathway from TGF-β1 to α-SMA in cells that express FAK, whereas 3) p38 is the major downstream signaling intermediate in the pathway from TGF-β1 to α-SMA in cells that lack FAK, and 4) FRNK expression will inhibit the α-SMA expression and MAPK activation in both FAK-expressing FAK-deficient fibroblasts. Therefore, we sought to determine whether FRNK inhibits myofibroblast differentiation in vivo during the pulmonary fibrotic process.
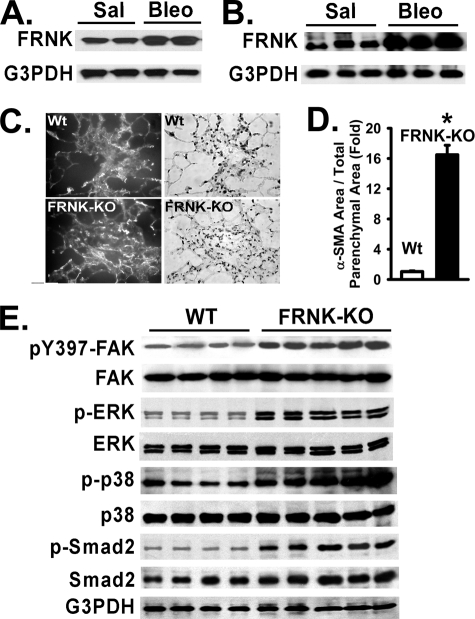
FRNK Mediates FAK and MAPK Activation and Myofibroblast Differentiation during Lung Fibrosis in Vivo—To determine the role of FRNK in myofibroblast differentiation in vivo during experimentally induced pulmonary fibrosis (bleomycin) (3, 5, 20, 45, 58–60), we first measured FRNK protein expression in normal and fibrotic lung tissue, and in primary fibroblast isolates from these lungs. FRNK protein levels were increased ∼3.2-fold in fibroblasts isolated from bleomycin-injured lungs, when compared with fibroblasts isolated from saline control lungs (Fig. 9A). Concordantly, FRNK protein was increased ∼4.7-fold in whole lung lysates from bleomycin-injured mice, as compared with that from saline controls (Fig. 9B). As a consequence of the loss of FRNK, FAK activation was increased 3-fold in bleomycin-treated FRNK knock-out mice, as compared with wild type (Fig. 9E). Unchallenged mice have similar levels of FAK activity (data not shown).
FIGURE 9.
FRNK expression is increased during lung injury, and myofibroblast mass is enhanced in lungs from bleomycin-challenged FRNK knock-out mice. Lungs were harvested from bleomycin-challenged mice (Bleo) or control saline-treated mice (Sal, 18 days after bleomycin challenge) as described under “Experimental Procedures.” A, primary lung fibroblasts were isolated from bleomycin-challenged (Bleo) or control saline-treated (Sal) mice and detergent-lysed. Equivalent amounts of whole cell lysate were Western blotted with the indicated antibodies. B, equivalent amount of whole lung lysate from bleomycin-challenged or control saline-treated mice were Western blotted with the indicated antibodies. C, α-SMA-positive cells on lung sections obtained from bleomycin-challenged FRNK knock-out (FRNK-KO) and bleomycin-challenged wild type (Wt) mice were analyzed by immunofluorescence staining for α-SMA as described under “Experimental Procedures.” Original magnification, 400×. Representative fluorescent images (Top left and bottom left) and bright field (Top right and bottom right) images from wild type mice (top panels) and FRNK-KO mice (bottom panels) are shown. D, the total area containing α-SMA-positive cells in lungs from FRNK-KO and wild type mice shown in C was quantitatively determined as described under “Experimental Procedures.” The data are presented as the means ± S.D. (n = 5/per group). * represents p < 0.001 for FRNK-KO mice compared with wild type controls. E, equivalent amounts of whole lung lysate from bleomycin-challenged FRNK-KO and bleomycin-challenged wild type (WT) mice were Western blotted with the indicated antibodies.
The activation of the signaling pathways involved in myofibroblast differentiation during bleomycin-induced pulmonary fibrosis was tested in wild type and FRNK knock-out mice. Both ERK and p38 MAPK activation was increased in whole lung lysates from bleomycin-injured FRNK knock-out mice, when compared with bleomycin-injured wild type controls (Fig. 9E). As evidence for TGF-β signaling, Smad2 phosphorylation was also examined and shown to be increased in whole lung lysates from bleomycin-injured FRNK knock-out mice, when compared with that in bleomycin-injured wild type controls (Fig. 9E).
Myofibroblast differentiation in wild type and FRNK knockout mice was measured by immunofluorescence staining of lung tissue with α-SMA and semi-quantitative morphometry. The myofibroblast cell area (α-SMA-positive cell area/total parenchymal area) was strikingly increased 16.5 ± 2.8-fold (p < 0.01) in bleomycin-treated lungs obtained from FRNK knockout mice, as compared with wild type mice (Fig. 9, C and D). If one focuses on the fibroproliferative lesions, the myofibroblast cell area was increased 3.2-fold in FRNK knock-out mice, as compared with wild type mice (data not shown). The myofibroblast cell area in unchallenged lung parenchyma of both FRNK knock-out and wild type murine controls was negligible and similar (data not shown). Taken together, these data suggest for the first time that FRNK acts as a brake for myofibroblast differentiation in vitro and in vivo.
DISCUSSION
Our studies show that FRNK expression mediated through adenovirus can abrogate TGF-β1-induced α-SMA expression and modulate MAPK activation in cells that either express FAK or lack FAK. Furthermore, endogenous FRNK expression is increased in fibroblasts in response to TGF-β stimulation in vitro and in response to the fibrotic agent bleomycin in vivo. The in vivo physiological relevance of FRNK is demonstrable through the consequences of the loss of FRNK in vivo. We show that FRNK-deficient mice have an enhancement of myofibroblast differentiation and MAPK activation in vivo in the TGF-β-dependent, fibrotic, bleomycin-injured animal model. Taken together, our data demonstrate that FRNK is a novel, negative regulator of myofibroblast differentiation and functions to limit myofibroblast generation in vivo, in response to tissue injury.
Despite the importance of FRNK in inhibiting integrin-mediated cell migration and proliferation through the inhibition of FAK, little progress has been made in the understanding of the mechanism by which FRNK expression is regulated. Nonetheless, FRNK expression is up-regulated in smooth muscle cells in vivo after balloon-induced vascular injury (37, 39). Furthermore, FRNK expression is increased in vascular smooth muscle cells plated on perlecan, with consequent impaired FAK activation and cell proliferation (61). Our study demonstrates for the first time that FRNK expression is up-regulated in response to a specific profibrotic cytokine, TGF-β1. Furthermore, FRNK expression is up-regulated in fibrotic lung tissue, and in primary isolates of fibroblasts from fibrotic lungs. Because TGF-β1 is considered a key mediator of both fibrosis and myofibroblast differentiation in the bleomycin model, we speculate that TGF-β1 is the physiological inducer of FRNK expression in vivo during the fibrotic response (1, 3, 4, 62).
It is well characterized that FRNK inhibits integrin-mediated cell migration through blocking FAK activation and FAK-mediated signaling (25, 37, 39, 40, 50). FRNK comprises the C-terminal region of FAK, including the proline-rich (SH3-binding domains) and focal adhesion targeting domains, but lacks the signaling kinase domain and integrin/cytokine-binding FERM domain. FRNK localizes to focal adhesions through its focal adhesion targeting domain and is thought to inhibit cell migration either through the competitive replacement of FAK in focal adhesions/contacts or by the competitive recruitment of critical signaling proteins away from FAK (25, 40, 63). Our demonstration of a role for FRNK in cell trans-differentiation is entirely novel. Our studies extend the described inhibitory effect of FRNK on integrin-mediated cell migration and proliferation to myofibroblast differentiation in vitro and in vivo. Furthermore, we show that the inhibitory effect of FRNK on TGF-β1-induced α-SMA expression is largely, but not solely, through inhibition of FAK-mediated signaling.
Other investigators have shown that expression of dominant negative FAK inhibits TGF-β1-induced α-SMA expression (34). Because complete deletion of FAK may have a different mechanism of action compared with the Tyr397 → Phe mutant FAK (34), we further tested the dependence of the α-SMA response to TGF-β, on the presence of FAK in both FAK-null and FAK-expressing cells. Surprisingly, we found that TGF-β1 can induce α-SMA expression in the absence of FAK, albeit to a much lower level. The presence of FAK facilitates basal α-SMA expression and significantly augments TGF-β1-induced α-SMA expression. This is supported by two lines of evidence. First, we found that the basal (no TGF-β) α-SMA expression level is 3-fold greater in cells with FAK compared with those without FAK. Second, we found that the maximal response of α-SMA to TGF-β1 is 3.6-fold greater in cells with FAK compared with cells without FAK (Fig. 7B, fifth bar versus second bar). These data demonstrate that the maximal level of α-SMA expression can be achieved through both FAK-independent and FAK-dependent signaling pathways (Fig. 10).
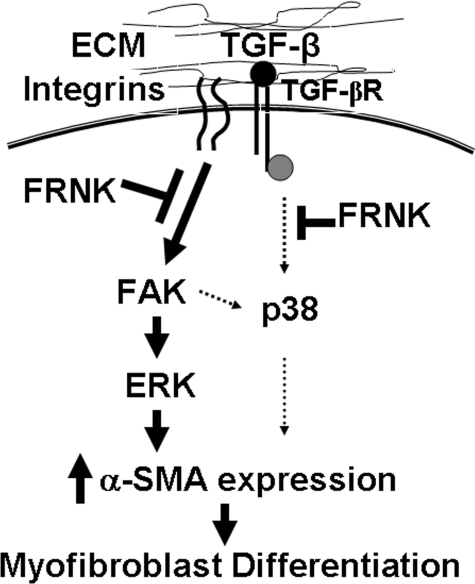
FIGURE 10.
Hypothetical mechanism whereby FRNK inhibits TGF-β-induced α-SMA expression in the presence and absence of FAK. TGF-β-induced α-SMA expression and myofibroblast differentiation requires the involvement of multiple signaling initiated through the integrins, extracellular matrix (ECM) environment, and TGF-β receptors (TGF-βR). Recent evidence demonstrates that certain integrins, in cooperation with the cytoskeleton, are capable of activating latent TGF-β through binding to latency-associated protein (21, 26, 68). TGF-β induces α-SMA expression through two pathways; the major pathway through ERK MAPK is FAK-dependent, and the minor pathway through p38 MAPK is FAK-independent. FRNK can inhibit TGF-β-induced α-SMA expression through inhibition of FAK activation and inhibition of the FAK/ERK-mediated pathway. FRNK also can inhibit the pathway of TGF-β-induced α-SMA expression mediated by p38, potentially through inhibition of critical signaling proteins upstream of p38 (i.e. Src kinase).
The enhanced α-SMA expression in the presence of FAK may be due to the role of FAK as an important mediator in integrin-mediated signaling, focal adhesion turnover, cytoskeleton remodeling, and mechanosensing processes (3, 4, 25, 32, 64–67). TGF-β1 alone is not sufficient to induce optimal α-SMA expression (1, 23). Integrin-mediated signaling and expression of EDA fibronectin (alternative splice form of fibronectin), as well as a rigid matrix environment, are required for TGF-β1-induced α-SMA expression and formation of mature focal adhesion/complexes (1, 23). The necessity of integrin-mediated signaling is further supported by the observation that TGF-β1 is not able to induce α-SMA expression in suspended human lung fibroblasts, although the response of Smad2 to TGF-β1 is intact in these cells (34). Recent evidence demonstrates that certain integrins, in cooperation with cytoskeletons, are capable of activating latent TGF-β through binding to latency-associated protein (21, 26, 68). Myofibroblast differentiation is associated with the formation of mature focal adhesion complexes and the incorporation of α-SMA into cytoskeletal fibers, and it also depends on the presence of a rigid matrix environment (1, 23). FAK regulates integrin-mediated focal adhesion and cytoskeletal remodeling and is required for mechanosensing of a rigid extracellular environment (25, 32, 67). Therefore, we would anticipate that expression of FRNK would inhibit myofibroblast differentiation, through inhibition of FAK activation. In fact, we show that TGF-β1-induced FAK activation and α-SMA expression are reduced in FRNK-expressing fibroblasts.
It is known that MAPK is required for TGF-β-induced expression of extracellular matrix proteins (41–43, 69). Although selective utilization of a MAPK has been reported for TGF-β-induced gene expression, for example, type I collagen expression (41–43, 54, 69–71), our observation that differential utilization of both ERK and p38 MAPK upon TGF-β1 stimulation in the same cell and in a FAK-dependent manner is novel. The differential role of ERK and p38 in TGF-β1-induced α-SMA expression was substantiated in this study by using complementary pharmacological inhibitors. We found that a complete abrogation of TGF-β1-induced α-SMA expression requires blockade of both ERK and p38 activation and that ERK is the major regulatory MAPK for TGF-β1-induced α-SMA expression in the presence of FAK. In contrast, we found that p38 is the major regulatory MAPK for TGF-β1-induced α-SMA expression in the absence of FAK but retains a minor regulatory role in FAK-expressing cells. Although the p38 MAPK pathway exists in wild type cells, the p38 pathway does not compensate for the loss of ERK activation in FAK-deficient cells, thereby resulting in a net reduction in the effect of TGF-β on α-SMA expression when ERK activation is blocked. Nonetheless, the presence of FAK enhances p38 activation in response to TGF-β1, as compared with that in FAK-deficient cells (Figs. 7C and 10).
We show that expression of FRNK, or deletion of FAK (as in FAK-null cells), completely abrogates ERK activation in response to TGF-β. This is consistent with previous findings of other investigators demonstrating that ERK activation induced by epidermal growth factor or serum is significantly impaired in FAK-deficient cells (72, 73). The impaired ERK activation they noted in suspended cells was rescued by expression of a constitutively active FAK, and expression of dominant negative FAK mutant blocked ERK activation, supporting the concept that FAK is an obligate upstream mediator of ERK activation (29, 33, 73). FAK has been shown to mediate ERK activation through multiple pathways. For example, binding of Grb2 to FAK (Y925) leads to subsequent activation of the Ras, Raf, MEK, and ERK signaling pathway (25). Alternatively, we have shown that the negative regulator of Ras activation, p120RasGAP, can be sequestered away through binding to active FAK, thereby augmenting Ras activity and downstream ERK activation (29, 33).
Our study also shows the novel finding that FRNK expression inhibits TGF-β1-induced p38 MAPK activation in the absence of FAK. This is the first demonstration of a FAK-independent physiological function for FRNK. In subsequent studies we will address the exact mechanism by which FRNK inhibits p38 activation. However, the FAK C-terminal domain (FRNK), contains two proline-rich regions, which have been shown to bind SH3 domain-containing proteins (i.e. p130Cas and GRAF) (25, 66). Src kinase contains an SH3 domain (74, 75), and Src has been shown to mediate TGF-β-induced p38 activation through phosphorylation of TGF-βRII independent of FAK and Smad (76–78). We therefore speculate that FRNK may bind to the SH3 domain of Src through its proline-rich region, thereby recruiting Src away from the TGF-βRII. This would have the effect of blunting Src-mediated phosphorylation of the TGF-βRII, and the subsequent downstream activation of p38 MAPK. Through this pathway, FRNK can inhibit p38 MAPK activation in a Smad- and FAK-independent manner.
It is only through a complete understanding of the extensive cross-talk between integrin and TGF-β1 signaling pathways that mediate myofibroblast differentiation that we will be able to design efficient strategies to mitigate organ fibrosis and excess scar formation. Our study demonstrates that FAK mediates ERK activation necessary for optimal α-SMA expression in response to TGF-β1, and we show that a FAK-independent rescue pathway exists that requires p38 MAPK (Fig. 10). Interestingly, FRNK can block both pathways. This is physiologically relevant as endogenous FRNK is up-regulated in response to TGF-β1 in vitro and during the fibroproliferative process in vivo. Our observations in FRNK-deficient mice indicate that FRNK acts as a physiologic brake on the formation of myofibroblasts during the fibroproliferative process in vivo. Thus, to the extent that myofibroblasts mediate excess tissue fibrosis, FRNK may function to limit pathological tissue fibrosis in response to vascular and/or lung or other organ injury.
Acknowledgments
We thank Dr. J. T. Parsons for generously supplying FRNK knock-out mice and Dr. S. K. Hanks for generously providing Tet-FAK cells. We also thank members of our laboratory for technical help and stimulating discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants HL085324 (to Q. D.) and HL58655 (to M. A. O.). This work was also supported by a grant from the American Heart Association (to Q. D.) and a Veterans Affairs merit award (to M. A. O.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: α-SMA, α-smooth muscle actin; FAK, focal adhesion kinase; FRNK, FAK-related non-kinase; Ad-, adenoviral construct; TGF, transforming growth factor; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; HA, hemagglutinin; G3PDH, glyceraldehyde 3-phosphate dehydrogenase; TLCK, Nα-p-tosyl-l-lysine chloromethyl ketone; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; GFP, green fluorescent protein; Luc, luciferase; BSA, bovine serum albumin; MOI, multiplicity of infection; MEK, MAPK/ERK kinase.
References
- 1.Hinz, B., Phan, S. H., Thannickal, V. J., Galli, A., Bochaton-Piallat, M. L., and Gabbiani, G. (2007) Am. J. Pathol. 170 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olman, M. A. (2003) in Acute Respiratory Distress Syndrome (Matthay, M. A., and Lenfant, C., eds) pp. 313–354 Maracel Dekker, Inc., New York
- 3.Sheppard, D. (2006) Proc. Am. Thorac. Soc. 3 413–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan, S. H. (2002) Chest 122 286S–289S [DOI] [PubMed] [Google Scholar]
- 5.Desmouliere, A., Darby, I., and Gabbiani, G. (2003) Lab. Investig. 83 1689–1707 [DOI] [PubMed] [Google Scholar]
- 6.Desmouliere, A., Chaponnier, C., and Gabbiani, G. (2005) Wound Repair Regen. 13 7–12 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, K., Rekhter, M. D., Gordon, D., and Phan, S. H. (1994) Am. J. Pathol. 145 114–125 [PMC free article] [PubMed] [Google Scholar]
- 8.Crystal, R. G., Bitterman, P. B., Mossman, B., Schwarz, M. I., Sheppard, D., Almasy, L., Chapman, H. A., Friedman, S., King, T. E., Jr., Leinwand, L. A., Liotta, L., Martin, G. R., Schwartz, D. A., Wagner, C. R., and Musson, R. A. (2002) Am. J. Respir. Crit. Care Med. 166 236–246 [DOI] [PubMed] [Google Scholar]
- 9.King, T. E., Oostabel, U., Cordier, J., Dopico, G. A., Du Bois, R. M., Lynch, D. Lynch, J. P., Myers, J., Panos, R., Raghu, G., Schwartz, D., and Smith, C. M. American Thoracic Society and European Respiratory Society (2000) Am. J. Respir. Crit. Care Med. 161 646–66410673212 [Google Scholar]
- 10.Selman, M., King, T. E., and Pardo, A. (2001) Ann. Intern. Med. 134 136–151 [DOI] [PubMed] [Google Scholar]
- 11.King, T. E., Jr., Schwarz, M. I., Brown, K., Tooze, J. A., Colby, T. V., Waldron, J. A., Jr., Flint, A., Thurlbeck, W., and Chemiack, R. M. (2001) Am. J. Respir. Crit. Care Med. 164 1025–1032 [DOI] [PubMed] [Google Scholar]
- 12.Hinz, B., Celetta, G., Tomasek, J. J., Gabbiani, G., and Chaponnier, C. (2001) Mol. Biol. Cell 12 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinz, B., Dugina, V., Ballestrem, C., Wehrle-Haller, B., and Chaponnier, C. (2003) Mol. Biol. Cell 14 2508–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinz, B., Gabbiani, G., and Chaponnier, C. (2002) J. Cell Biochem. 157 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schildmeyer, L. A., Braun, R., Taffet, G., Debiasi, M., Burns, A. E., Bradley, A., and Schwartz, R. J. (2000) FASEB J. 14 2213–2220 [DOI] [PubMed] [Google Scholar]
- 16.Takeji, M., Moriyama, T., Oseto, S., Kawada, N., Hori, M., Imai, E., and Miwa, T. (2006) J. Biol. Chem. 281 40193–40200 [DOI] [PubMed] [Google Scholar]
- 17.Desmouliere, A. (1995) Cell Biol. Int. 19 471–476 [DOI] [PubMed] [Google Scholar]
- 18.Desmouliere, A., Geinoz, A., Gabbiani, F., and Gabbiani, G. (1993) J. Cell Biochem. 122 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodsick, J. E., Peters-Golden, M., Larios, J. M., Toews, G. B., Thannickal, V. J., and Moore, B. B. (2003) Am. J. Respir. Cell Mol. Biol. 29 537–544 [DOI] [PubMed] [Google Scholar]
- 20.Liu, X., Hu, H., and Yin, J. Q. (2006) Liver Int. 26 8–22 [DOI] [PubMed] [Google Scholar]
- 21.Wipff, P. J., Rifkin, D. B., Meister, J. J., and Hinz, B. (2007) J. Cell Biol. 179 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muro, A. F., Moretti, F. A., Moore, B. B., Yan, M., Atrasz, R. G., Wilke, C. A., Flaherty, K. R., Martinez, F. J., Tsui, J. L., Sheppard, D., Baralle, F. E., Toews, G. B., and White, E. S. (2007) Am. J. Respir. Crit. Care Med. 177 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagood, J. S., and Olman, M. A. (2007) Am. J. Respir. Cell Mol. Biol. 37 503–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giancotti, F. G., and Ruoslahti, E. (1999) Science 285 1028–1032 [DOI] [PubMed] [Google Scholar]
- 25.Parsons, J. T. (2003) J. Cell Sci. 116 1409–1416 [DOI] [PubMed] [Google Scholar]
- 26.Jenkins, R. G., Su, X., Su, G., Scotton, C. J., Camerer, E., Laurent, G. J., Davis, G. E., Chambers, R. C., Matthay, M. A., and Sheppard, D. (2006) J. Clin. Investig. 116 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calalb, M. B., Polte, T. R., and Hanks, S. K. (1995) Mol. Cell Biol. 15 954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlaepfer, D. D., and Hunter, T. (1997) J. Biol. Chem. 272 13189–13195 [DOI] [PubMed] [Google Scholar]
- 29.Hecker, T. P., Ding, Q., Rege, T. A., Hanks, S. K., and Gladson, C. L. (2004) Oncogene 23 3962–3971 [DOI] [PubMed] [Google Scholar]
- 30.Chen, H. C., Appeddu, P. A., Isoda, H., and Guan, J. L. (1996) J. Biol. Chem. 271 26329–26334 [DOI] [PubMed] [Google Scholar]
- 31.Schlaepfer, D. D., Mitra, S. K., and Ilic, D. (2004) Biochim. Biophys. Acta 1692 77–102 [DOI] [PubMed] [Google Scholar]
- 32.Hanks, S. K., Ryzhova, L., Shin, N. Y., and Brabek, J. (2003) Front. Biosci. 8 d982–d996 [DOI] [PubMed] [Google Scholar]
- 33.Ding, Q., Grammer, J. R., Nelson, M. A., Guan, J. L., Stewart, J. E., Jr., and Gladson, C. L. (2005) J. Biol. Chem. 280 6802–6805 [DOI] [PubMed] [Google Scholar]
- 34.Thannickal, V. J., Lee, D. Y., White, E. S., Cui, Z., Larios, J. M., Chacon, R., Horowitz, J. C., Day, R. M., and Thomas, P. E. (2003) J. Biol. Chem. 278 12384–12389 [DOI] [PubMed] [Google Scholar]
- 35.Thomas, P. E., Peters-Golden, M., White, E. S., Thannickal, V. J., and Moore, B. B. (2007) Am. J. Physiol. Lung Cell Mol. Physiol. 293 L417–L428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenberg, R. S., Bernstein, A. M., Benezra, M., Gelman, I. H., Taliana, L., and Masur, S. K. (2006) FASEB J. 20 1006–1008 [DOI] [PubMed] [Google Scholar]
- 37.Hayasaka, H., Simon, K., Hershey, E. D., Masumoto, K. H., and Parsons, J. T. (2005) J. Cell Biochem. 95 1248–1263 [DOI] [PubMed] [Google Scholar]
- 38.Schaller, M. D., Borgman, C. A., and Parsons, J. T. (1993) Mol. Cell Biol. 13 785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, J. M., Mack, C. P., Nolan, K., Regan, C. P., Owens, G. K., and Parsons, J. T. (2001) Mol. Cell Biol. 21 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauck, C. R., Hsia, D. A., Puente, X. S., Cheresh, D. A., and Schlaepfer, D. D. (2002) EMBO J. 21 6289–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulder, K. M. (2000) Cytokine Growth Factor Rev. 11 23–35 [DOI] [PubMed] [Google Scholar]
- 42.Yu, L., Hebert, M. C., and Zhang, Y. (2002) EMBO J. 21 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue, J., and Mulder, K. M. (2000) J. Biol. Chem. 275 30765–30773 [DOI] [PubMed] [Google Scholar]
- 44.Hayasaka, H., Martin, K. H., Hershey, E. D., and Parsons, J. T. (2007) J. Cell Biochem. 102 947–954 [DOI] [PubMed] [Google Scholar]
- 45.Olman, M. A., Mackman, N., Gladson, C., Moser, K., and Loskutoff, D. (1995) J. Clin. Investig. 96 1621–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmons, W. L., White, K. E., Curiel, D. T., Williams, W. F., and Olman, M. A. (1998) Am. J. Respir. Cell Mol. Biol. 18 307–314 [DOI] [PubMed] [Google Scholar]
- 47.Olman, M. A., Hagood, J. S., Simmons, W. L., Fuller, G. M., Vinson, C., and White, K. E. (1999) Blood 94 2029–2038 [PubMed] [Google Scholar]
- 48.Hagood, J. S., Olman, M. A., Zeballos, J. G., White, K. E., and Fuller, G. M. (1996) Blood 87 3749. [PubMed] [Google Scholar]
- 49.Owen, J. D., Ruest, P. J., Fry, D. W., and Hanks, S. K. (1999) Mol. Cell Biol. 19 4806–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haskell, H., Natarajan, M., Hecker, T. P., Ding, Q., Stewart, J. E., Jr., Grammer, J. R., and Gladson, C. L. (2003) Clin. Cancer Res. 9 2157–2165 [PubMed] [Google Scholar]
- 51.Wu, H., Dmitriev, I., Kashentseva, E., Seki, T., Wang, M., and Curiel, D. T. (2002) J. Virol. 76 12775–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding, Q., Stewart, J. E., Jr., Olman, M. A., Klobe, M. R., and Gladson, C. L. (2003) J. Biol. Chem. 278 39882–39891 [DOI] [PubMed] [Google Scholar]
- 53.Ding, Q., Stewart, J. E., Jr., Prince, C. W., Chang, P. L., Trikha, M., Han, X., Grammer, J. R., and Gladson, C. L. (2002) Cancer Res. 62 533–643 [DOI] [PubMed] [Google Scholar]
- 54.Meyer-Ter-Vehn, T., Gebhardt, S., Sebald, W., Buttmann, M., Grehn, F., Schlunck, G., and Knaus, P. (2006) Investig. Ophthalmol. Vis. Sci. 47 1500–1509 [DOI] [PubMed] [Google Scholar]
- 55.Finlay, G. A., Thannickal, V. J., Fanburg, B. L., and Paulson, K. E. (2000) J. Biol. Chem. 275 27650–27656 [DOI] [PubMed] [Google Scholar]
- 56.Serini, G., Bochaton-Piallat, M., Ropraz, P., Geinoz, A., Borsi, L., Zardi, L., and Gabbiani, G. (1998) J. Cell Biol. 142 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dugina, V., Alexandrova, A., Chaponnier, C., Vasiliev, J., and Gabbiani, G. (1998) Exp. Cell Res. 238 481–490 [DOI] [PubMed] [Google Scholar]
- 58.Lazo, J. S., Hoyt, D. G., Sebti, S. M., and Pitt, B. R. (1990) Pharmacol. Ther. 47 347–358 [DOI] [PubMed] [Google Scholar]
- 59.Olman, M. A., Simmons, W. L., Pollman, D. J., Loftis, A. Y., Bini, A., Miller, E. J., Fuller, G. M., and Rivera, K. E. (1996) Am. J. Physiol. 271 L519–L526 [DOI] [PubMed] [Google Scholar]
- 60.Tager, A. M., Kradin, R. L., LaCamera, P., Bercury, S. D., Campanella, G. S., Leary, C. P., Polosukhin, V., Zhao, L. H., Sakamoto, H., Blackwell, T. S., and Luster, A. D. (2004) Am. J. Respir. Cell Mol. Biol. 31 395–404 [DOI] [PubMed] [Google Scholar]
- 61.Walker, H. A., Whitelock, J. M., Garl, P. J., and Nemenoff, R. A. (2003) Mol. Cell Biol. 14 1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gabbiani, G. (2003) J. Pathol. 200 500–503 [DOI] [PubMed] [Google Scholar]
- 63.Sieg, D. J., Hauck, C. R., and Schlaepfer, D. D. (1999) J. Cell Sci. 112 2677–2691 [DOI] [PubMed] [Google Scholar]
- 64.Schaller, M. D. (2004) J. Cell Biol. 166 157–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, X., Suetsugu, S., Cooper, L. A., Takenawa, T., and Guan, J. L. (2004) J. Biol. Chem. 279 9565–9576 [DOI] [PubMed] [Google Scholar]
- 66.Mitra, S. K., Hanson, D. A., and Schlaepfer, D. D. (2005) Nat. Rev. Mol. Cell Biol. 6 56–68 [DOI] [PubMed] [Google Scholar]
- 67.Wang, H. B., Dembo, M., Hanks, S. K., and Wang, Y. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11295–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munger, J. S., Huang, X., Kawakatsu, H., Griffiths, M. J., Dalton, S. L., Wu, J., Pittet, J. F., Kaminski, N., Garat, C., Matthay, M. A., Rifkin, D. B., and Sheppard, D. (1999) Cell 96 319–328 [DOI] [PubMed] [Google Scholar]
- 69.Kutz, S. M., Hordines, J., McKeown-Longo, P. J., and Higgins, P. J. (2001) J. Cell Sci. 114 3905–3914 [DOI] [PubMed] [Google Scholar]
- 70.Yang, M., Huang, H., Li, J., Li, D., and Wang, H. (2004) FASEB J. 18 1920–1921 [DOI] [PubMed] [Google Scholar]
- 71.Underwood, D. C., Osborn, R. R., Bochnowicz, S., Webb, E. F., Rieman, D. J., Lee, J. C., Romanic, A. M., Adams, J. L., Hay, D. W., and Griswold, D. E. (2001) Am. J. Physiol. 279 L895–L902 [DOI] [PubMed] [Google Scholar]
- 72.Hauck, C. R., Sieg, D. J., Hsia, D. A., loftus, J. C., Gaarde, W. A., Monia, B. P., and Schlaepfer, D. D. (2001) Cancer Res. 61 7079–7090 [PubMed] [Google Scholar]
- 73.Renshaw, M. W., Price, L. S., and Schwartz, M. A. (1999) J. Cell Biol. 147 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okutani, D., Lodyga, M., Han, B., and Liu, M. (2006) Am. J. Physiol. 291 L129–L141 [DOI] [PubMed] [Google Scholar]
- 75.Williams, J. C., Wierenga, R. K., and Saraste, M. (1998) Trends Biochem. Sci. 23 179–184 [DOI] [PubMed] [Google Scholar]
- 76.Galliher, A. J., and Schiemann, W. P. (2006) Breast Cancer Res. 8 R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galliher, A. J., and Schiemann, W. P. (2007) Cancer Res. 67 3752–3758 [DOI] [PubMed] [Google Scholar]
- 78.Galliher-Beckley, A. J., and Schiemann, W. P. (2008) Carcinogenesis 29 244–251 [DOI] [PMC free article] [PubMed] [Google Scholar]