Abstract
Transcription factors of the Sp/Klf (Krüppel-like factor) family regulate biological processes such as hematopoiesis, adipogenesis, and stem cell maintenance. Here we show that Bklf or Klf3 (Basic Krüppel-like factor) represses the Klf8 (Krüppel-like Factor 8) gene in vivo. Conversely, Eklf or Klf1 (Erythroid Krüppel-like factor) activates the Klf8 gene. Klf8 is driven by two promoters, both of which contain multiple CACCC sites. Klf3 can repress Klf1-mediated activation of both promoters. Chromatin immunoprecipitation experiments confirm that Klf3 occupies both Klf8 promoters in vivo. Interestingly, in Klf3 knock-out tissue Klf1 gains access, binds, and activates both Klf8 promoters. These results demonstrate direct competition between activating and repressing Klfs in vivo. Together with previous evidence that Klf1 directly activates the Klf3 gene, the results reveal an elaborate network of cross-talk within the Klf family. The recognition of cross-regulation and potential redundancy between Klf family members is critical to the interpretation of various Klf knock-out mice and the understanding of individual Klfs in particular contexts.
The Klf (Krüppel-like factor) family of sequence-specific DNA-binding proteins bind GC-rich regions and related CACCC sequences in DNA (1). Klf family members contain three conserved DNA-binding C2H2 zinc fingers that recognize similar consensus sites (2). The N-terminal domains of the various Klfs vary, and these different domains mediate different molecular functions. Some Klfs function predominantly as activators, such as Eklf/Klf1 (erythroid Klf) (3), some, such as Bklf/Klf3, are primarily transcriptional repressors (4), and others have been shown to have both activator and repressor roles, such as Klf4 and Klf5 (5). Many Klfs are broadly expressed, such as Klf3 and Klf8 (6, 7), but some are expressed only in specific tissues, such as the erythroid Klf, Klf1 (3). Members of the Klf family have been shown to play key roles in multiple biological processes including erythropoiesis (8–12), adipogenesis (13–15), and the self-renewal of embryonic stem cells (16–18).
Eklf (Klf1), the founding member of the Klf family (3), is a potent transcriptional activator known to bind a CACCC site in the adult β-globin promoter and to drive expression of the β-globin gene (19, 20). Accordingly, Klf1 knock-out mice die in utero at embryonic day 14.5 (E14.5)4 from severe anemia resulting from β-globin deficiency (10, 11). A number of additional Klf1 target genes have been recently identified such as Ahsp (α-hemoglobin stabilizing protein) (21) and dematin (22), a cytoskeletal protein required for erythrocyte membrane integrity. These were validated as direct Klf1 target genes through the use of a Klf1 knock-out erythroid cell line, B1.6, rescued by an inducible form of Klf1 (23). The B1.6 cells have also been used to identify another direct Klf1 target gene, Bklf/Klf3 (24).
Bklf (now known as Klf3) was originally identified through a low stringency cDNA screen using the zinc fingers of Klf1 as a probe (6). Klf3 is widely expressed but is particularly abundant in erythroid tissues (6). Klf3 has been demonstrated to act as a potent transcriptional repressor that recruits the co-repressor C-terminal binding protein (CtBP) to mediate gene repression (4). A critical PVDLT motif in the N-terminal repression domain of Klf3 directly contacts CtBP. Mutation of this motif dramatically reduces Klf3-mediated repression (4).
Here we have used microarrays to compare wild type and Klf3 knock-out fetal liver mRNA in order to identify Klf3 target genes. Klf8 was identified as a gene potentially repressed by Klf3. Klf8 is also a member of the Klf family. It is closely related to Klf3, with the zinc fingers of Klf3 and Klf8 showing 96% homology. Like Klf3, Klf8 binds strongly to CACCC boxes in vitro and is widely expressed (7). Klf8 also acts as a transcriptional repressor and interacts with the co-repressor CtBP through a PVDLS motif (7). There are two reports implicating Klf8 in γ-globin gene repression (25, 26). Klf8 has also been implicated in cell cycle progression and has been shown to be a downstream target of the cell cycle regulator focal adhesion kinase (27). It has been found to be up-regulated in a number of human cancer cells and primary tumor tissues, and induction of Klf8 in NIH3T3 cells results in accelerated cell growth (28). Furthermore, Klf8 can induce motility and invasiveness in normal epithelial cell lines through regulation of the E-cadherin gene (29). Klf8 has also been implicated in the repression of Klf4 (30), a gene that has a well documented role in cell cycle control (31) as well as being one of the four transcription factors that can reprogram mouse fibroblasts to embryonic stem cells (18).
Here we report that Klf8 is repressed by Klf3 in vivo and is up-regulated by Klf1. Transcript analysis indicates that Klf8 has two promoters, both containing multiple CACCC elements. Transactivation assays and chromatin immunoprecipitation experiments indicate that Klf3 represses Klf8 directly and that Klf1 activates Klf8 directly. Interestingly it appears that the activating and repressing Klfs, Klf1 and Klf3, respectively, compete for binding sites in the Klf8 promoters and that in the presence of Klf3, there is little Klf1 occupancy at either Klf8 promoter in vivo. Accordingly, Klf8 levels are low in normal erythroid tissues but high in erythroid cells from Klf3 knock-out mice.
EXPERIMENTAL PROCEDURES
Genotyping—Genotyping of the Klf3-null, heterozygous, and wild type mice was performed by PCR of both Klf3 (primers 5′-AAATGCACCTGGGAAGGCTGCAC-3′ (forward), 5′-CAGACTAGCATGTGGCGTTTCCTG-3′ (reverse)) and Neo (primers 5′-TGATGCAATGCGGCGGCTGCATAC-3′ (forward), 5′-CAGAAGAACTCGTCAAGAAGGCGA-3′ (reverse)).
RNA Extraction—Total RNA was extracted from freshly homogenized E14.5 fetal livers, CD71hi/TER-119hi sorted E15.5 fetal liver cells, Ctbp–/– and Ctbp+/– murine embryonic fibroblasts (MEFs), or B1.6 (23) cell lines as described previously (24).
Microarray Analysis—Total RNA from Klf3–/– and Klf3+/+ E14.5 fetal livers were hybridized to an Applied Biosystems (Foster City, CA) mouse genome survey microarray, and the arrays were scanned using the Applied Biosystems AB1700 Chemiluminescent Microarray analyzer at Human Genetic Signatures (Macquarie Park, Australia).
Real Time RT-PCR—Extracted RNA was used as a template for cDNA synthesis which was then used for quantitative real time RT-PCR as described previously (24).
Real Time PCR Primers—Paired primers were designed as described previously (24). The sequences of forward and reverse primers used are as follows: 18S,5′-CACGGCCGGTACAGTGAAAC-3′ (forward) and 5′-AGAGGAGCGAGCGACCAA-3′ (reverse); Klf2, 5′-GCTGGCCGCGAAATGA-3′ and 5′-GAGAGGATGAAGTCCAACACGTT-3′; Klf3 exons 1a/2, 5′-CGGGCCTGGGTTTCTTG-3′ and 5′-GATCAAACATGAGCATCCTTTCAG-3′; Klf3 exons 1b/2, 5′-GGTGGAATTCTGTTCAGGTCAAC-3′ and 5′-CCACGCCTTCTAGGGTGTTCT-3′; Klf4, 5′-AGGCACACCTGCGAACTCA-3′ and 5′-TCGGAGCGGGCGAAT-3′; Klf5,5′-TCTCCCACCTGTCAGATACAACA-3′ and 5′-GACTTTGTATAAACTTTTGTGCAACCA-3′; Klf6,5′-CACATCAGCGCACTCACACA-3′ and 5′-CAAAACGCCACTCACAACCTT-3′; Klf7,5′-CACGACACCGGCTACTTCTCA-3′ and 5′-GGAGGTAGCGTTCCAACTCAAG-3′; Klf8,5′-CCAAAAGCTCTCACCTGAAAGC-3′ and 5′-AGCGAGCAAATTTCCAGGAA-3′ or 5′-TGGATGTCCGAATTAAATCAGAAA-3′ and 5′-GAAGGATCTCTGGTCGGAACAG-3′; Klf8 exon 1a, 5′-TTCGGAGGCGCAGAACC-3′; Klf8 exon 1c, 5′-CTGACACAGAAAGAGCTATGTACTCCAT-3′; Klf8 exon 3-exon 4 junction (reverse), 5′-GCCAGTGACCTGTTTGAATACCT-3′; Klf9,5′-GGCTGTGGGAAAGTCTATGGAA-3′ and 5′-AAGGGCCGTTCACCTGTATG-3′; Klf10, 5′-GCAGCCAACCATGCTCAAC-3′ and 5′-CCCCTCTCTGGGCTTTTCAG-3′; Klf12,5′-CGCCCTTGAGAACAGAATGC-3′ and 5′-GGGTAGTTGTGGACGTTTGGA-3′; Klf13, 5′-CGAGAAAGTTTACGGGAAATCTTC-3′ and 5′-CAGGCGAAAGGCCTCTCA-3′; Klf14,5′-TCCATGGACCGGTTCCAT-3′ and 5′-AGAGCCACAGACAGCGGTTAG-3′; Klf15,5′-AAGGCACCGGCGATCTC-3′ and 5′-CCGCGCGAATTTCTTCTC-3′; Klf16,5′-TCACACCTGCGGACTCACA-3′ and 5′-CAGAACGGGCGAACTTCTTG-3′; and β-actin,5′-GCTTCTTTGCAGCTCCTTCGT-3′ and 5′-CCAGCGCAGCGATATCG-3′.
Fluorescence-activated Cell Sorting—E15.5 fetal liver cells were stained with anti-CD71-PE and anti-TER-119 PE-Cy7 antibodies (BD Biosciences, Palo Alto, CA). CD71hi/TER-119hi cells were then purified by FACSAria sorting (BD Biosciences).
Antibody Generation—Anti-Klf1 and anti-Klf3 antibodies have been described previously (6). cDNA encoding full-length mKlf8 was amplified from 450 ng of heart cDNA by PCR using the primers 5′-ATTACTCGAGCAGTGGCTTGGAAAGTCAGATAC-3′ (forward) and 5′-ATTAGAATTCCAGCGTGCCTGCTGTG-3′ (reverse) and was cloned into EcoRI/XhoI pMT3 to form pMT3-mKlf8. cDNA encoding residues 1–260 of mKlf8 was amplified from pMT3-mKlf8 by PCR using the primers 5′-ATTAGGATCCATGGATAAATTCATAG-3′ (forward) and 5′-ATTAGAATTCCTCTTCTCCCTGCATGTGGAC-3′ (reverse), and the product was cloned in frame downstream of GST in the bacterial expression vector BamHI/EcoRI pGEX-2T (GE Healthcare Life Sciences). The GST-mKlf8 fusion protein was overexpressed in bacteria and purified on glutathione beads as described previously (32). Rabbits were inoculated with GST-mKlf8 protein, and serum was collected by the Institute of Medical and Veterinary Science (Adelaide, Australia).
Western Blotting—Western blot assays were performed to assess the relative levels of Klf8 in nuclear extracts from B1.6 cells and E14.5 fetal livers as has been described previously for other proteins (33). After analysis with anti-Klf8 antibody, the membrane was stripped in a solution of 62.5 mm Tris-HCl (pH 6.8), 2% SDS, 0.7% β-mercaptoethanol at 70 °C for 30 min. It was then incubated at room temperature for 1 h in several changes of 50 mm Tris-HCl (pH 7.4) containing 150 mm NaCl and 0.05% Tween 20 and was then probed with an anti-β-actin antibody (Sigma) as a loading control as described above.
EMSAs—Nuclear extracts were prepared, and EMSAs were carried out as described previously (6) When detecting Klf8, equal quantities of nuclear extracts were loaded in a total volume of 30 μl containing 50 μg/ml poly(dI-dC), 4.4 mm dithiothreitol, 100 μg/ml bovine serum albumin, 10 mm HEPES (pH 7.8), 50 mm KCl, 5 mm MgCl2, 1.07 mm EDTA, 6.67% glycerol, 1.33 mm MnCl2, 3.33 mm NaCl, 35 mm Tris-HCl (pH 7.5), 3.33 μm EGTA, 0.007% Brij35, 1 μl of preimmune serum or antibody (as appropriate) and radiolabeled probe. Oligonucleotides used in the synthesis of radiolabeled probes are as follows: mouse β-major globin CACCC probe, 5′-TAGAGCCACACCCTGGTAAG-3′ and 5′-CTTACCAGGGTGTGGCTCTA-3′; Klf8 probe1, 5′-AGGAGTGGGGGTGATGGTAC-3′ and 5′-GTACCATCACCCCCACTCCT-3′; Klf8 probe2, 5′-CATTATCTCCCCCTCTGCCA-3′ and 5′-TGGCAGAGGGGGAGATAATG-3′; Klf8 probe3, 5′-AGACCCGGGTGTGGTGTGGT-3′ and 5′-ACCACACCACACCCGGGTCT-3′; Klf8 probe4, 5′-GAGTTTGGGTGGGGTACAGT-3′ and 5′-ACTGTACCCCACCCAAACTG-3′; Klf8 probe5, 5′-GAGGTAGGGAGGGGAGGCCT-3′ and 5′-AGGCCTCCCCTCCCTACCTC-3′; Klf8 probe6, 5′-GCCGCTGGGTGGGGTCTGGG-3′ and 5′-CCCAGACCCCACCCAGCGGC-3′; Klf8 probe7, 5′-AACGCTGGGCGAGGGACGCT-3′ and 5′-AGCGTCCCTCGCCCAGCGTT-3′; Klf8 probe8, 5′-AATGAGGGGAGAGGAGGGTT-3′ and 5′-AACCCTCCTCTCCCCTCATT; Klf8 probe9, 5′-TACCACCACTCCCGCCACCG-3′ and 5′-CGGTGGCGGGAGTGGTGGTA-3′; Klf8 probe10, 5′-AGAACTGGGAGGGTTAAAAA-3′ and 5′-TTTTTAACCCTCCCAGTTCT-3′; Klf8 probe11, 5′-TACTGGCACTCCCCATTCTG-3′ and 5′-CAGAATGGGGAGTGCCAGTA-3′; Klf8 probe12, 5′-AATCACCACACCCTTCCCAA-3′ and 5′-TTGGGAAGGGTGTGGTGATT-3′; Klf8 probe13, 5′-GGGCCCGCCCCACCCCCTCCT-3′ and 5′-AGGAGGGGGTGGGGCGGGCCC-3′; Klf8 probe14, 5′-TTCTGCCCCACCCATCTCTA-3′ and 5′-TAGAGATGGGTGGGGCAGAA-3′; and Klf8 probe15, 5′-CTACTCCACTCCCCTCTAGC-3′ and 5′-GCTAGAGGGGAGTGGAGTAG-3′.
Cell Culture—Murine erythroleukemia (MEL) cells, Drosophila melanogaster Schneider line 2 (SL-2) cells, COS, and B1.6 erythroblast cells (23) were cultured and induced as described previously (24). Ctbp1+/–Ctbp2+/– and Ctbp1–/–Ctbp2–/– MEFs (34) were cultured at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% (v/v) heat-inactivated fetal calf serum and 1% (v/v) penicillin, streptomycin, and glutamine solution (Invitrogen). Ctbp1–/–Ctbp2–/– MEFs were infected with pMSCV-puro-FLAGmCtbp2 and selected in puromycin (2.5 μg/ml) for 5 days. Resistant cells were pooled, and expression of FLAG-tagged mCtbp2 was assayed by Western blotting. For translation inhibition analyses, cycloheximide (made up in ethanol) was added as described previously (24). pcDNA3.Klf8 (provided by Stella Lee) was transfected into COS cells with FuGENE 6 (Roche Applied Science) following the manufacturer's instructions. Nuclear extracts were then prepared.
Chromatin Immunoprecipitation (ChIP)—ChIP was conducted with either anti-Klf3 or anti-Klf1 and preimmune serum using the cross-linker ethylene glycolbis[succinimidyl succinate] as described previously (35, 36) using a Biorupter™ (Diagenode sa, Liège, Belgium) for chromatin sonication with a 30 s on, 30 s off cycle for 20 min. Immunoprecipitated DNA was analyzed by real time PCR using the following Klf8 promoter primer sets: 4.5 kb up exon 1a, 5′-GGTTTCTGAGACCTAACACTTCACACT-3′ and 5′-CCATTTAGTCATCCAGCGAACAA-3′; 2.5 kb up exon 1a, 5′-TTTCAAGTGCTCAGCCATCATATATT-3′ and 5′-GGGCATTCATGAGCAGAAGTCT-3′; 0.6 kb up exon 1a, 5′-GGGCGGTGCCAAGCA-3′ and 5′-GCTCTTCCTAAACCAGCCAGAAC-3′; 0.3 kb up exon 1a, 5′-GGGAGGAAAAGCTAAGTTGTGTGA-3′ and 5′-TCCACCCCATCCACTGCTA-3′; 0.2 kb down exon 1a, 5′-CCAGCTCGTGCACACTGAA-3′ and 5′-GAAGCCTTAACATCAGGAGTGGAA-3′; 3 kb down exon 1a, 5′-AAAGTGGTTTCATGAGAGATAATTACAGA-3′ and 5′-GAGACAAACACAGACTGAGCAGAGA-3′; 33 kb down exon 1a, 5′-AACCTGGGTGCCTCCTTGTA-3′ and 5′-TCATGCCTTTGACTTTAGTGCTTT-3′; 0.5 kb up exon 1b, 5′-TTACTTCTGGGTTCATGGAAAGC-3′ and 5′-CACAGTGGGATCTCTTGTCCATT-3′; 4 kb up exon 1c, 5′-AACACTTTGGTTCCGTCCATTC-3′ and 5′-GAGGCTGTTGCCGTGACTATC-3′; 1.7 kb up exon 1c, 5′-AGAGTTTAGGCCACGTCAACAAA-3′ and 5′-TCTGATAAGACCAGGATTTTCTCAATT-3′; 0.2 kb up exon 1c, 5′-TCTGGGTGGATAGTTGACCAAGT-3′ and 5′CTTTTCGGTGCATCCTTAGTC-3′; and 1.8 kb down exon 1c, 5′-CCATGCACCGTGCTTTTCT-3′ and 5′-TGGAGCAGCCTGTATCATATATACCT-3′.
Vectors and Cloning of the Klf8 Promoters—Klf8 promoter fragments were amplified from FVBN murine genomic DNA by PCR using the forward primers 5′-CAGTGCTAGCTAGTGTACCGGCAT-3′, 5′-GGAAAAGCTAGCTTGTGTGATGCAG-3′, 5′-CTGAAGCTAGCCCAAGGTTAGGA-3′, 5′-CAGGTGCTAGCATCAACTGAGCTT-3′ and the reverse primer 5′-GGGACTTACTTTGGATATCTGCG-3′ for promoter 1a and the forward primers 5′-CCACTGCTAGCTTTTAAGGAACT-3′, 5′-CCTCTGGCTAGCCGCCTCTTCT-3′, 5′-CTAAGCTAGCACCGAAAAGGCT-3′, 5′-TCTCCCTAGCTAGCTCTTACTCC-3′ and the reverse primer 5′-GGGACTTACTTTGGATATCTGCG-3′ for promoter 1c.
The fragments were subcloned into EcoRI/NheI pGL4.10[luc2] (Promega Corporation, Madison, WI) to form pGL4.10[luc2]-Klf8prom1a(–83 + 38), pGL4.10[luc2]-Klf8prom1a(–350 + 38), pGL4.10[luc2]-Klf8prom1a(–700 + 38), pGL4.10[luc2]-Klf8prom1a(–990 + 38), pGL4.10[luc2]-Klf8prom1c(–60 + 52), pGL4.10[luc2]-Klf8prom1c(–127 + 52), pGL4.10[luc2]-Klf8prom1c(–241 + 52), or pGL4. 10[luc2]-Klf8prom1c(–353 + 52). pPac and pPac-Eklf were kindly provided by Menie Merika and Stuart Orkin (Harvard Medical School, Boston, MA), pPac-Bklf was kindly provided by José Perdomo (School of Molecular and Microbial Biosciences, Sydney, Australia).
Transactivation Assays—SL-2 cells were transfected in sixwell plates using the calcium phosphate method (37). Zero or 1000 ng of pPac-Eklf and 0, 10, 100, or 1000 ng of pPac-Bklf were transfected along with 1 μg of pGL4.10[luc2], pGL4.10[luc2]-Klf8prom1(–83 + 38), pGL4.10[luc2]-Klf8prom1a(–350 + 38), pGL4.10[luc2]-Klf8prom1a(–700 + 38), pGL4.10[luc2]-Klf8prom1a(–990 + 38), pGL4.10[luc2]-Klf8prom1c(–60 + 52), pGL4.10[luc2]-Klf8prom1c(–127 + 52), pGL4.10[luc2]-Klf8prom1c(–241 + 52), or pGL4. 10[luc2]-Klf8prom1c(–353 + 52). For all transfections, 1 μg of pGL4.74[hRluc/TK] (Promega) was included as a transfection control. Forty-eight hours post-transfection, the cells were lysed and assayed for luciferase activity by using a dual luciferase reporter assay system (Promega) per the manufacturer's instructions. In all cases, reporter activity was normalized with respect to Renilla luciferase levels.
RESULTS
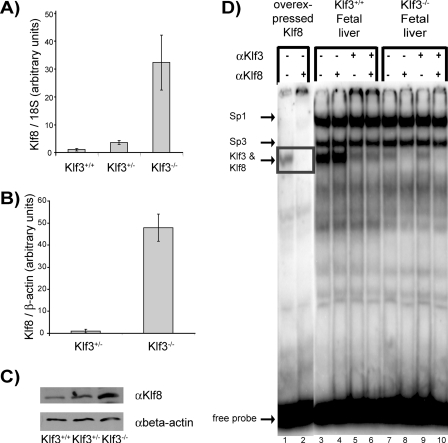
Klf8 Is De-repressed in the Absence of Klf3—To identify Klf3 target genes, microarray analysis comparing Klf3+/+ and Klf3–/– E14.5 fetal liver mRNA was undertaken. The fetal liver was chosen as a target tissue because Klf3 is highly abundant in this tissue (6). Klf8 mRNA was found to be significantly up-regulated (more than 10-fold) in the Klf3–/– samples. This result was confirmed by real time RT-PCR. Analysis of Klf8 mRNA in Klf3+/+, Klf3+/–, and Klf3–/– E14.5 fetal livers by real time RT-PCR showed that Klf8 is de-repressed in the absence of Klf3 (Fig. 1A). mRNA from Klf3+/– and Klf3–/– CD71hi/TER-119hi sorted fetal liver cells was also analyzed using real time RT-PCR (Fig. 1B). A highly significant de-repression of Klf8 was observed in the absence of Klf3 in these sorted erythroid cells (50-fold increase in expression in Klf3–/– erythroid cells compared with 10-fold in unsorted fetal liver cells).
FIGURE 1.
Klf8 protein and mRNA are de-repressed in Klf3 null murine fetal livers. A, real time RT-PCR analysis of E14.5 fetal livers. Littermates with two embryos/genotype were analyzed. Expression of Klf8 was normalized with 18 S rRNA levels and then normalized again with respect to the Klf3+/+ sample which was set at 1. The error bars show S.E. B, real time RT-PCR analysis of sorted E15.5 TER119hi/CD71hi E15.5 fetal liver cells. Samples were taken from littermates with three (Klf3+/–) or four (Klf3–/–) embryos/genotype. Expression of Klf8 was normalized with β-actin and then normalized again with respect to the Klf3+/– sample which was set at 1. The error bars show S.E. C, Western blot of Klf8 in E14.5 fetal livers. Nuclear extracts were prepared from fetal liver cells from wild type, heterozygous, and knock-out littermates and analyzed by Western blotting using a Klf8 antibody. The blot was stripped and reprobed with β-actin antibody as a loading control. D, EMSA experiment of equal quantities of nuclear extracts, prepared from E14.5 fetal liver cells from wild type and knock-out littermates. A double-stranded oligonucleotide probe containing the β-globin CACCC box was used. Klf8 overexpressed in COS cells was used as a positive control. Klf3 antibody (αKlf3) was used to shift Klf3 protein that would mask any signal from Klf8 protein.
We wished to determine whether up-regulation of Klf8 also occurred at the protein level in Klf3–/– animals. Western blotting revealed that Klf8 protein levels were elevated ∼5-fold in the absence of Klf3 in E14.5 fetal livers (Fig. 1C). Both Klf3 and Klf8 have been previously shown to bind to the critical CACCC box of the β-major globin promoter (6, 7). Klf8 DNA binding activity was assessed in both Klf3+/+ and Klf3–/– E14.5 fetal liver samples by band retardation experiments (Fig. 1D). Because Klf3 and Klf8 co-migrate in these experiments, Klf3 antibody was used to shift away the Klf3 protein to reveal the band produced by Klf8 protein. Upon inspection, a band corresponding to Klf8 can be detected in the Klf3–/– fetal liver samples (Fig. 1D, lane 7). This band is supershifted by the addition of Klf8 antibody (Fig. 1D, lane 8). No such supershift was observed in the Klf3+/+ samples, suggesting that there is no detectable Klf8 DNA binding activity in the Klf3+/+ samples (the residual bands observed are not Klf8 and are presumably generated by an as yet uncharacterized CACCC-binding protein). Taken together these data suggest that Klf3 represses Klf8 expression either directly or indirectly.
Klf8 Is De-repressed in the Absence of CtBP Proteins—CtBP proteins are transcriptional co-factors that bind transcription factors and repress gene expression. In mammals there are two CtBP proteins, CtBP1 and CtBP2. They bind to a number of transcription factors including Klf3, Klf8, and Klf12 through Pro-Xaa-Asp-Leu-Ser (PXDLS) and related motifs (4, 7, 38). Mutations in a PVDLT motif in Klf3 prevent it from binding to CtBP and impair Klf3-mediated repression in cellular assays (4). To investigate whether CtBPs were involved in Klf3-mediated repression of Klf8, we assessed Klf8 levels in Ctbp+/– (i.e. Ctbp1+/– Ctbp2+/–) and Ctbp–/– (i.e. Ctbp1–/– Ctbp2–/–) MEFs (34). We also assessed Ctbp–/– MEFs that had been rescued with murine Ctbp2. Klf8 mRNA was found to be significantly de-repressed in Ctbp–/– MEFs compared with Ctbp+/– MEFs by real time RT-PCR (Fig. 2). Moreover Ctbp–/– MEFs rescued with CtBP2 showed a down-regulation of Klf8 mRNA. Thus Klf8 expression is regulated by CtBP, and Klf3 and CtBP appear to act together to repress expression of Klf8.
FIGURE 2.
Ctbp contributes to repression of Klf8. Real time RT-PCR analysis of Klf8 mRNA levels in Ctbp+/–, Ctbp–/–, and Ctbp–/– rescued MEFs. Expression of Klf8 was normalized with 18 S rRNA levels and then normalized again with respect to the Ctbp+/+ sample, which was set at 1. The error bars show S.E.
Klf8 Is Up-regulated in the Presence of Klf1—The B1.6 line is an erythroid cell line derived from Klf1-null knock-out mice into which a transgene encoding a tamoxifen-inducible Klf1-ER protein has been introduced (23, 39). We used this cell line to investigate whether Klf8 and other members of the Klf family are activated by Klf1 (Fig. 3A). We have recently reported that the downstream Klf3 promoter, promoter 1b, is directly activated by Klf1. Klf10 is also modestly up-regulated upon induction of Klf1-ER in the B1.6 cell line (24).
FIGURE 3.
Klf8 protein and mRNA are up-regulated upon induction of Klf1-ER. A, real time RT-PCR analysis of Klf mRNA levels before and after induction of Klf1-ER by tamoxifen in B1.6 cells. Expression of each Klf was normalized with 18 S RNA levels and then normalized again with respect to the detectable uninduced samples, which were set at 1. The error bars show S.E. B, Western blot of Klf8 in nuclear extracts prepared both before and after induction of Klf1-ER by tamoxifen in B1.6 cells. Nuclear extracts were run on a 10% polyacrylamide gel and transferred to a nitrocellulose membrane via Western blotting. The membrane was probed with Klf8 antibody and then was stripped and reprobed with β-actin antibody as a loading control. C, EMSA experiment of Klf8 in uninduced (lane 1) and induced (lanes 2 and 3) B1.6 cells. Anti-Klf8 antibody was added to lane 3. Equal quantities of nuclear extracts were used in EMSA using a double-stranded oligonucleotide probe containing the β-globin CACCC box.
Several Klfs (Klf2, Klf4, Klf5, Klf12, Klf14, and Klf15) were not detectable in the B1.6 cell line either before or after Klf1-ER induction. Others (Klf6, Klf7, Klf9, Klf13, and Klf16) showed a negligible change in expression upon induction. Interestingly, in addition to Klf10 and Klf3, Klf8 mRNA was significantly up-regulated upon induction of Klf1-ER (Fig. 3A). Western blotting confirmed that Klf8 protein levels are also significantly increased upon induction (Fig. 3B). Klf8 DNA binding activity was also increased as shown by EMSA experiments, where induction of Klf1-ER resulted in the appearance of a band (Fig. 3C, lane 2) that could be supershifted by the addition of anti-Klf8 antibody (Fig. 3C, lane 3) and not by an anti-Klf3 antibody (data not shown). These data suggest that in addition to being a target of the transcriptional repressor Klf3, Klf8 may also be a target of the transcriptional activator, Klf1.
The Klf8 Locus Contains Two Promoters—In silico studies revealed two major Klf8 transcriptional start sites that generate mRNAs containing various alternative 5′ exons. The initiation AUG lies in a common exon (exon 2) downstream of these alternative leader exons. Accordingly, transcripts containing these alternative 5′ exons are expected to encode identical Klf8 proteins. We have termed the two 5′-most untranslated exons from the upstream transcript (RefSeq accession number NM_173780) exon 1a and 1b and the first three untranslated exons from the downstream transcript (RefSeq accession number BC070442) exons 1c, 1d, and 1e (Fig. 4A). Thus there appear to be two promoters of Klf8. The upstream promoter we call promoter 1a, and the downstream promoter we call promoter 1c. A similar arrangement has recently been reported for the closely related Klf3 gene, which has alternative first exons 1a and 1b and correspondingly two promoters (24).
FIGURE 4.
Klf8 has two alternative promoters. A, schematic diagram of alternative Klf8 exons. Alternative start points of transcription at exons 1a and 1c and the start point of translation in exon 2 are shown. Intron lengths are given below. B and C, sequences of exon 1a (B) and exon 1c (C) and the region 1 kb upstream of the start of each exon. The start of each exon is given as +1. The exons are shown in bold type. Klf consensus sites (NCN CNC CCN) upstream of each exon are boxed.
Both Klf8 Promoters Are De-repressed in Klf3–/– CD71hi/TER-119hi Fetal Liver Cells—We conducted real time RT-PCR to determine whether transcripts from one or both Klf8 promoters were de-regulated in Klf3–/– compared with Klf3+/– CD71hi/TER-119hi sorted fetal liver cells. Forward primers specific for exons 1a and 1c were employed with a common reverse primer specific for the exon 2-exon 3 junction to detect transcripts arising from the upstream and downstream promoter respectively. Both transcripts displayed an ∼50-fold up-regulation in expression in Klf3–/– compared with Klf3+/– sorted fetal liver cells (data not shown). This result was consistent with the previous observation that total Klf8 transcript levels are de-repressed ∼50-fold in CD71hi/TER-119hi sorted fetal liver cells (Fig. 1B). This raises the possibility that Klf3 either directly or indirectly regulates the activity of both Klf8 promoters.
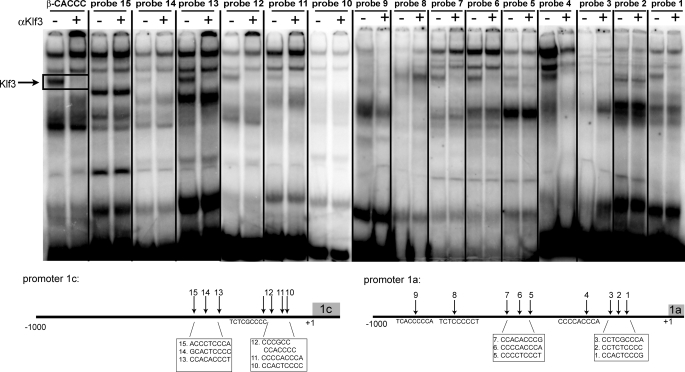
Klf3 Can Bind to CACCC Sequences Present in Both Klf8 Promoters in Vitro—Inspection of the two putative Klf8 promoters (upstream of exons 1a and 1c, respectively) reveals a number of sites that match the Klf binding consensus 5′-NCN CNC CCN-3′. Nine of the general Klf binding consensus sites were identified in Klf8 promoter 1a, three of which contain actual CACCC sequences (Fig. 4B). In Klf8 promoter 1c, seven general consensus sites were identified, and again three of these contain CACCC sequences (Fig. 4C). The sites that matched the Klf consensus in the Klf8 promoters were assessed for Klf3 binding using band retardation experiments (Fig. 5). The β-major globin CACCC probe, to which Klf3 had been previously shown to bind (6), was used as a positive control. This experiment confirms that Klf3 is able to bind to a number of Klf consensus sites in both Klf8 promoters in vitro, and as expected, Klf3 binds well to those sites containing actual CACCC sequences (probes 1, 4, 6, 7, 11, 12, 13).
FIGURE 5.
Klf3 binds to several CACCC sites in promoter 1a and 1c of Klf8. Nuclear extracts were prepared from E14.5 wild type fetal livers. These were used in EMSA using double-stranded oligonucleotide probes containing consensus sites found upstream of Klf8 exon 1a (probes 1–9), exon 1c (probes 10–15), and the β-globin CACCC box as a positive control. Schematic diagrams of consensus sequences in Klf8 promoter 1a and 1c used for EMSA are given below.
Klf1-ER Can Activate Both the Endogenous Klf8 Promoters—We previously reported that Klf3 promoters 1a and 1b were both dependent on Klf1 in erythroid cells, although the downstream promoter, promoter 1b, is more responsive to Klf1 in various assays (24). Because Klf1 also appears to regulate the expression of Klf8 (Fig. 3A), we wished to determine whether one or both Klf8 promoters were Klf1-responsive. Total RNA from induced and uninduced B1.6 cells was analyzed by real time RT-PCR. Prior to induction of Klf1-ER, transcripts containing exon 1a were undetected, whereas transcripts containing exon 1c were detected at very low levels. Upon Klf1-ER induction, comparable up-regulation of both transcripts was observed, similar to those seen in Fig. 3A (data not shown). This suggests that both Klf8 promoters are activated by Klf1-ER in B1.6 cells.
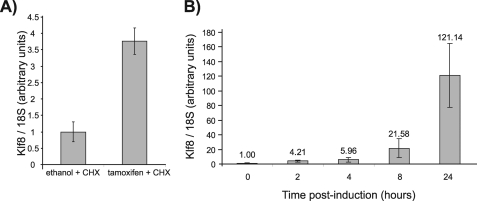
To ascertain whether Klf8 is a direct target gene of Klf1-ER, B1.6 cells were exposed to cycloheximide, a translation inhibitor, prior to and during induction with tamoxifen. The cells were harvested 8 h (rather than 48 h) after induction to minimize the cytotoxicity of cycloheximide. Klf8 mRNA was up-regulated by Klf1-ER in the presence of cycloheximide, suggesting that Klf8 is a direct target of Klf1 (Fig. 6A). A time course study of Klf8 mRNA levels following induction was also performed. Although indirect target genes would be expected to exhibit a lag phase before being expressed, Klf8 transcripts showed an up-regulation as early as 2 h post-induction (Fig. 6B). This short induction time is reminiscent of what was previously observed with the direct Klf1 target, Klf3 promoter 1b, in induced B1.6 cells (24). This provides further evidence that Klf8 is a direct target gene of Klf1.
FIGURE 6.
Klf8 is directly activated by Klf1-ER. B1.6 cells were exposed to cycloheximide (CHX) 30 min before induction with tamoxifen and were harvested for RNA 8 h thereafter (A) or harvested at numerous time points after tamoxifen induction in the absence of CHX (B). Klf8 mRNA levels were determined by real time RT-PCR. Expression of Klf8 was then normalized with 18 S RNA levels and then normalized again with respect to the ethanol control or 0-h time point which was set at 1. The error bars show S.E.
Klf3 Can Repress Klf1-mediated Activation at Both Klf8 Promoters in Transfection Assays—To further investigate the ability of Klf1 and Klf3 to control expression of Klf8 through both promoters, we performed transactivation experiments in Drosophila SL-2 cells. These cells are often used for Klf transactivation assays because they contain no detectable endogenous CACCC box-binding proteins that might interfere with analyses (40). Deletion series containing regions of the Klf8 promoters were cloned into the promoterless reporter construct pGL4.10[luc2]. The promoter-pGL4.10[luc2] constructs were transfected into SL-2 cells with 1 μg of a Klf1 expression vector, pPac-Eklf, and increasing concentrations of a Klf3 encoding vector, pPac-Bklf (10–1000 ng) (Fig. 7). When the shortest promoter 1a and 1c constructs that contained no CACCC box consensus sites (–83 to +38 for promoter 1a and –60 to +52 for promoter 1c) were examined, they showed no activation by Klf1 or subsequent repression by Klf3. However, Klf1-mediated activation of both promoter 1a and 1c was observed with longer constructs and increased with the number of CACCC boxes. Klf3 was able to repress this activation in all instances (Fig. 7). For example, Klf8 promoter 1c constructs containing four and five CACCC boxes were activated by Klf1 and subsequently repressed by Klf3 in a dose-dependent manner. The Klf8 promoter 1c construct containing only two CACCC boxes showed a similar pattern but with a lesser degree of activation (Fig. 7B). These results corroborate our previous results that suggest that both Klf1 and Klf3 regulate expression of Klf8 at both promoters, with Klf1 activating at both Klf8 promoters and Klf3 repressing. These results raise the possibility that Klf3 may directly compete with Klf1 for occupancy of CACCC box-binding sites at the Klf8 promoters.
FIGURE 7.
Klf3 can repress activation mediated by Klf1 at Klf8 promoter 1a (A) and Klf8 promoter 1c (B) constructs. SL-2 cells were transfected with 1 μg of pGL4.10[luc2] containing regions of Klf8 promoter 1a or promoter 1c. Constructs cloned into the luciferase vector contained no CACCC sites to all CACCC sites. Empty pGL4.10[luc2] was used as a control. 1 μg of pPac.Eklf was used to activate promoter constructs. Increasing concentrations of pPac.Bklf (0.01, 0.1, or 1 μg) were used to repress promoter constructs. 1 μg of Renilla vector pGL4.74[HRluc/TK] was used as a transfection control. The average of four to six replicates/column is shown. The error bars show S.E. Dark closed ovals denote a CACCC site in the construct that Klf3 binds to in vitro, light closed ovals denote partial binding in vitro, and open ovals are CACCC sites that Klf3 does not bind in vitro.
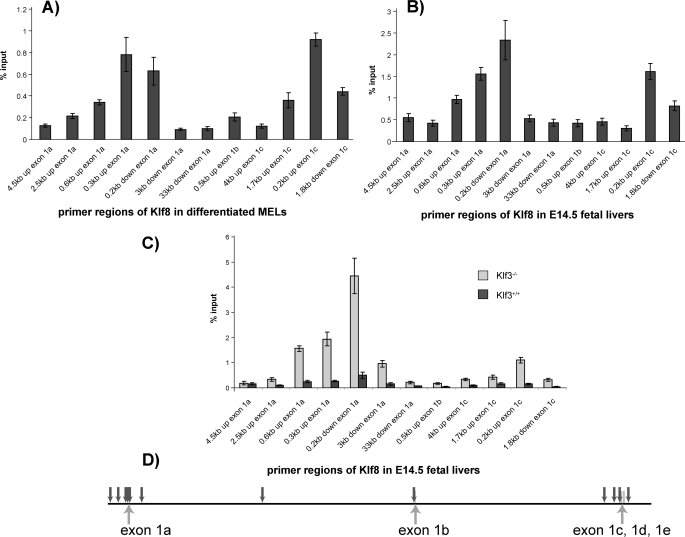
Klf1 and Klf3 Can Bind to Both Klf8 Promoters in Vivo—To confirm that Klf1 and Klf3 directly bind to the Klf8 promoters in vivo, we conducted ChIP assays to examine occupancy of Klf1 or Klf3 at the Klf8 promoters. First we tested for Klf3. Occupancy of Klf3 at the Klf8 promoters was analyzed in differentiated MEL cells and wild type E14.5 fetal livers (Fig. 8, A and B). Primers were designed to amplify ∼50 –100 bp of DNA lying ∼300 and 600 bp upstream of exon 1a and 200 bp upstream of exon 1c. These regions include CACCC box consensus sites. A number of primers were also designed to serve as negative controls. The relative locations compared with exons 1a, 1b, 1c, 1d, and 1e of all of these primers are shown (Fig. 8D). ChIPs from both differentiated MEL cells and wild type fetal livers showed an enrichment of Klf3 occupancy at and around both Klf8 promoter 1a and promoter 1c but not elsewhere in the locus.
FIGURE 8.
ChIP of promoter regions of Klf8. A and B, Klf3 at Klf8 promoter regions in differentiated MEL cells (A) and E14.5 fetal liver cells (B). C, Klf1 at Klf8 promoter regions in Klf3–/– and Klf3+/+ E14.5 fetal livers. Real time PCR analysis of genomic DNA immunoprecipitated by Klf3 or Klf1 antibody during ChIP. Primer regions refer to the area of the Klf8 locus that was amplified. The ChIP signal was normalized against input control and then normalized again with respect to the 4.5 kb upstream of exon 1a signal, which was set at 1. A shows an average of 8 ChIP experiments, B shows an average of four ChIPs for each of three different pooled litters, and C shows an average of 2 ChIPs for three or four embryos for each genotype. The error bars show S.E. D, schematic diagram showing the location of ChIP primers as downward arrows with respect to exons across the Klf8 locus with distances to scale.
These two ChIP experiments show that Klf3 is present at both of the Klf8 promoters in vivo in erythroid cells and tissue. Because our previous results had suggested that Klf3 and Klf1 may compete for binding to the Klf8 promoters, we tested occupancy of Klf1 at the Klf8 promoters in both the presence and absence of Klf3 (that is we compared samples from normal and Klf3 knock-out mice). Individual Klf3+/+ and Klf3–/– E14.5 fetal livers from littermates were subjected to ChIP experiments using Klf1 antiserum (Fig. 8C). Klf1 occupancy was observed at both promoter regions of Klf8 in the absence of Klf3 (i.e. in Klf3–/– fetal livers). However, in the presence of Klf3 (i.e. in wild type Klf3+/+ fetal livers), the signal is barely detectable above background.
DISCUSSION
Microarray analysis has previously been used to identify target genes for many transcription factors including members of the Klf family, such as Klf1 and Klf4 (19, 22, 41). We used this approach to identify Klf3 target genes. Only one gene, Klf8, was identified as being significantly de-repressed in our initial screen comparing Klf3+/+ and Klf3–/– E14.5 fetal livers. Klf8 mRNA and protein were found to be up-regulated in Klf3–/– E14.5 fetal liver. Klf8 mRNA was also up-regulated to a lesser extent (2–5-fold) in Klf3–/– adult tissues such as the heart, lung, spleen, and white adipose tissue (data not shown). Within the Klf family, Klf3 is most closely related to Klf8. They are 96% similar in their zinc finger domain (42) and have similar DNA binding preferences (6, 7). They are both broadly expressed at the mRNA level (6, 7, 42). Furthermore, they are both transcriptional repressors that operate through recruiting CtBP co-repressors (4, 7). It is possible that the up-regulation of Klf8 may in part compensate for the absence of Klf3 in Klf3 knock-out animals. The degree of compensation may vary in different tissues, because up-regulation of Klf8 mRNA varies between tissues (data not shown). Moreover, whereas the Klf3-null mice do not show a severe phenotype, they do show some differences to their wild type littermates such as a reduction in white adipose tissue, smaller size, and decreased viability (43). Thus Klf8 does not completely compensate for the absence of Klf3.
Functional redundancy is often seen within families of related proteins. One such example involves the basic helixloop-helix transcription factors MyoD, Myf5, and MRF4, which are involved in skeletal muscle development. Single knock-outs of either MyoD, Myf5 or MRF4 have no significant effect on muscle development (44–46), whereas a double knock-out of either MyoD and Myf5 or MyoD and MRF4 results in severe muscle deficiencies (45, 47). The cross-regulation seen among the myogenic transcription factors is consistent with the idea that genes with overlapping functions are coordinately cross-regulated so that the total amount of protein with equivalent function is controlled. We hypothesize that this cross-regulation occurs with Klf3 and Klf8 and serves to keep the total amount of Klf3 plus Klf8 constant.
Klf3 is a potent transcriptional repressor and has been found to mediate repression through its interaction with CtBP corepressors (4). Mutations in Klf3 that prevent the binding of CtBP to Klf3 drastically reduce Klf3-mediated repression in reporter assays (4). Thus one would hypothesize that in the absence of CtBP, de-repression of Klf3 target genes, such as Klf8, would be observed. Consistent with this, Klf8 mRNA is elevated in Ctbp–/– MEFs. Moreover, when these cells are rescued with Ctbp2, the Klf8 levels are repressed. This is consistent with the hypothesis that the Klf3-CtBP complex represses Klf8 and indeed is the first in vivo demonstration beyond transient assays that Klf3 requires CtBP for repression.
As well as being repressed by Klf3, Klf8 expression was also found to be activated by Klf1. Induction of Klf1-ER in a Klf1-null cell line resulted in a significant increase in Klf8 mRNA, protein, and DNA binding activity. Like Klf3 (24), the Klf8 gene has two alternative promoters that we have termed promoter 1a and promoter 1c. In the case of Klf3, both promoters are dependent on Klf1 for activity in erythroid cells (24). Similarly, we find here that both Klf8 promoters are responsive to Klf1 in B1.6 (erythroid) cells.
Both Klf8 promoters contain a number of CACCC sites that Klf3 can bind in vitro and that Klf1 may also bind. Klf1 was shown to be able to activate both Klf8 promoters in transactivation assays. Klf3 was then able to repress this Klf1-mediated activation of both promoters. Deletion analysis suggested that at least one CACCC site was required to observe this activation and repression. Thus both Klf1 and Klf3 are able to act on both Klf8 promoters through the CACCC boxes, and as Klf3 levels were increased, Klf3 was able to out-compete Klf1 at both Klf8 promoters to repress expression of Klf8.
Klf3 was found to be present at both Klf8 promoters in vivo by ChIP in differentiated MEL cells and E14.5 fetal liver. Occupancy of Klf1 at the Klf8 promoters was also investigated. Although there was detectable occupancy of Klf1 at the Klf8 promoters in Klf3+/+ fetal livers, this was subtle, with signals barely above background. In contrast, in the Klf3–/– fetal livers there was a dramatic increase in Klf1 occupancy at both Klf8 promoters. This effect was more pronounced at the first promoter than the second. The large increase in Klf1 occupancy at the Klf8 promoters when Klf3 is not present supports the hypothesis that Klf3 and Klf1 compete for binding sites in the Klf8 promoters and that Klf3 out-competes Klf1 at these sites at least in wild type day E14.5 fetal liver and differentiated MEL cells. A schematic diagram illustrating this network of cross-regulation is given in Fig. 9.
FIGURE 9.
Schematic diagram of cross-regulation between Klf1, Klf3 and Klf8. Klf1 activates expression of the Klf3 and Klf8 gene promoters. The Klf8 promoters can also be repressed by Klf3. In tissues where both Klf1 and Klf3 are present, such as fetal liver, the proteins compete for binding, and this restricts Klf8 to low levels of expression. In the absence of Klf3, for example in the Klf3 null fetal liver, Klf1 gains access to the Klf8 promoters, resulting in de-repression of the gene and increased Klf8 protein.
Because of the high similarity in their DNA-binding zinc fingers, many Klfs bind very similar consensus sites, such as CACCC boxes (1). Klf1, Klf3, and Klf8 have all been shown to bind to similar CACCC boxes, such as the β-globin CACCC site, with comparable affinities in vitro (6, 7, 20). It has been thought that because of the similarity in their recognition sequences, different members of the Klf family may compete for binding to consensus sites in promoters of target genes in vivo to regulate gene expression. Several Sp/Klfs have been previously reported to compete for the same binding site in vitro, such as Sp1 with Klf13, Klf16, Klf4, and Klf9 (48–50). However, evidence for direct competition between the Klfs for the same promoters in vivo has not to our knowledge been previously demonstrated. Thus this competition between Klf1 and Klf3 at the Klf8 promoters may be the first description of competition between Klfs in vivo. This also illustrates a case where two Klfs that have previously been shown to bind to the same consensus sites in vitro (6, 20) have now been shown to compete for the same site in vivo. As has been proposed previously, it is likely that other Klf family members similarly compete for promoters in other tissues in vivo (5, 31, 42, 51).
Multiple Klfs are implicated in several processes, such as adipogenesis (13–15, 43, 52, 53) and stem cell maintenance (16, 17). Our results, together with recent results in stem cells and fibroblasts, suggest that feedback regulation and redundancy may occur within the Klf family, such that Klfs, 1, 2, 4, and 5, may (depending on which is present in which tissue) activate Klfs 3 and 8, and Klfs 3 and 8 may then feedback to temper the activation. Appreciation of this potential feedback mechanism and the overlapping functions of Klfs will be useful in interpreting experiments in multiple cell types.
Acknowledgments
We are grateful to Menie Merika and Stuart Orkin for providing pPac and pPac-Eklf, to Andrew Perkins for providing the B1.6 cells, and to Jeffery Hildebrand for providing the Ctbp–/– MEFs. We express our gratitude to Dale Hancock, Gareth Denyer, and Stella Lee for technical and scientific advice.
This work was supported, in whole or in part, by National Institutes of Health Grant HL073443. This work was also supported by grants from the Australian Research Council and National Health and Medical Research Council (to M. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: En, embryonic day n; CtBP, C-terminal binding protein; MEF, murine embryonic fibroblast; RT, reverse transcription; GST, glutathione S-transferase; EMSA, electrophoretic mobility shift assay; MEL, murine erythroleukemia; ChIP, chromatin immunoprecipitation; ER, estrogen receptor.
References
- 1.Kaczynski, J., Cook, T., and Urrutia, R. (2003) Genome Biol. 4 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson, R., Fleetwood, J., Eaton, S., Crossley, M., and Bao, S. (2008) Int. J. Biochem. Cell Biol. 40 1996–2001 [DOI] [PubMed] [Google Scholar]
- 3.Miller, I. J., and Bieker, J. J. (1993) Mol. Cell. Biol, 13 2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner, J., and Crossley, M. (1998) EMBO J. 17 5129–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang, D. T., Pevsner, J., and Yang, V. W. (2000) Int. J. Biochem. Cell Biol. 32 1103–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crossley, M., Whitelaw, E., Perkins, A., Williams, G., Fujiwara, Y., and Orkin, S. H. (1996) Mol. Cell. Biol. 16 1695–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Vliet, J., Turner, J., and Crossley, M. (2000) Nucleic Acids Res. 28 1955–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu, P., Morris, P. E., Haar, J. L., Wani, M. A., Lingrel, J. B., Gaensler, K. M. L., and Lloyd, J. A. (2005) Blood 106 2566–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto, N., Kubo, A., Liu, H., Akita, K., Laub, F., Ramirez, F., Keller, G., and Friedman, S. L. (2006) Blood 107 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuez, B., Michalovich, D., Bygrave, A., Ploemacher, R., and Grosveld, F. (1995) Nature 375 316–318 [DOI] [PubMed] [Google Scholar]
- 11.Perkins, A. C., Sharpe, A. H., and Orkin, S. H. (1995) Nature 375 318–322 [DOI] [PubMed] [Google Scholar]
- 12.Wani, M. A., Means, R. T., Jr., and Lingrel, J. B. (1998) Transgenic Res. 7 229–238 [DOI] [PubMed] [Google Scholar]
- 13.Banerjee, S. S., Feinberg, M. W., Watanabe, M., Gray, S., Haspel, R. L., Denkinger, D. J., Kawahara, R., Hauner, H., and Jain, M. K. (2003) J. Biol. Chem. 278 2581–2584 [DOI] [PubMed] [Google Scholar]
- 14.Mori, T., Sakaue, H., Iguchi, H., Gomi, H., Okada, Y., Takashima, Y., Nakamura, K., Nakamura, T., Yamauchi, T., Kubota, N., Kadowaki, T., Matsuki, Y., Ogawa, W., Hiramatsu, R., and Kasuga, M. (2005) J. Biol. Chem. 280 12867–12875 [DOI] [PubMed] [Google Scholar]
- 15.Oishi, Y., Manabe, I., Tobe, K., Tsushima, K., Shindo, T., Fujiu, K., Nishimura, G., Maemura, K., Yamauchi, T., Kubota, N., Suzuki, R., Kitamura, T., Akira, S., Kadowaki, T., and Nagai, R. (2005) Cell Metab. 1 27–39 [DOI] [PubMed] [Google Scholar]
- 16.Jiang, J., Chan, Y. S., Loh, Y. H., Cai, J., Tong, G. Q., Lim, C. A., Robson, P., Zhong, S., and Ng, H. H. (2008) Nat. Cell Biol. 10 353–360 [DOI] [PubMed] [Google Scholar]
- 17.Kim, J., Chu, J., Shen, X., Wang, J., and Orkin, S. H. (2008) Cell 132 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi, K., and Yamanaka, S. (2006) Cell 126 663–676 [DOI] [PubMed] [Google Scholar]
- 19.Drissen, R., von Lindern, M., Kolbus, A., Driegen, S., Steinlein, P., Beug, H., Grosveld, F., and Philipsen, S. (2005) Mol. Cell Biol. 25 5205–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, W. C., Southwood, C. M., and Bieker, J. J. (1994) J. Biol. Chem. 269 1493–1500 [PubMed] [Google Scholar]
- 21.Keys, J. R., Tallack, M. R., Hodge, D. J., Cridland, S. O., David, R., and Perkins, A. C. (2007) Br. J. Haematol. 136 150–157 [DOI] [PubMed] [Google Scholar]
- 22.Hodge, D., Coghill, E., Keys, J., Maguire, T., Hartmann, B., McDowall, A., Weiss, M., Grimmond, S., and Perkins, A. (2006) Blood 107 3359–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coghill, E., Eccleston, S., Fox, V., Cerruti, L., Brown, C., Cunningham, J., Jane, S., and Perkins, A. (2001) Blood 97 1861–1868 [DOI] [PubMed] [Google Scholar]
- 24.Funnell, A. P., Maloney, C. A., Thompson, L. J., Keys, J., Tallack, M., Perkins, A. C., and Crossley, M. (2007) Mol. Cell Biol. 27 2777–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, P., Basu, P., Redmond, L. C., Morris, P. E., Rupon, J. W., Ginder, G. D., and Lloyd, J. A. (2005) Blood Cells Mol. Dis. 35 227–235 [DOI] [PubMed] [Google Scholar]
- 26.Hu, J. H., Navas, P., Cao, H., Stamatoyannopoulos, G., and Song, C. Z. (2007) J. Mol. Biol. 366 1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao, J., Bian, Z. C., Yee, K., Chen, B. P. C., Chien, S., and Guan, J.-L. (2003) Mol. Cell 11 1503–1515 [DOI] [PubMed] [Google Scholar]
- 28.Wang, X., and Zhao, J. (2007) Oncogene 26 456–461 [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., Zheng, M., Liu, G., Xia, W., McKeown-Longo, P. J., Hung, M. C., and Zhao, J. (2007) Cancer Res. 67 7184–7193 [DOI] [PubMed] [Google Scholar]
- 30.Wei, H., Wang, X., Gan, B., Urvalek, A. M., Melkoumian, Z. K., Guan, J.-L., and Zhao, J. (2006) J. Biol. Chem. 281 16664–16671 [DOI] [PubMed] [Google Scholar]
- 31.Ghaleb, A. M., Nandan, M. O., Chanchevalap, S., Dalton, W. B., Hisamuddin, I. M., and Yang, V. W. (2005) Cell Res. 15 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, D. B., and Johnson, K. S. (1988) Gene 67 31–40 [DOI] [PubMed] [Google Scholar]
- 33.Perdomo, J., Verger, A., Turner, J., and Crossley, M. (2005) Mol. Cell. Biol. 25 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildebrand, J. D., and Soriano, P. (2002) Mol. Cell Biol. 22 5296–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsberg, E. C., Downs, K. M., and Bresnick, E. H. (2000) Blood 96 334–339 [PubMed] [Google Scholar]
- 36.Zeng, P. Y., Vakoc, C. R., Chen, Z. C., Blobel, G. A., and Berger, S. L. (2006) BioTechniques 694,41, 696 698. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbour, NY
- 38.Schuierer, M., Hilger-Eversheim, K., Dobner, T., Bosserhoff, A. K., Moser, M., Turner, J., Crossley, M., and Buettner, R. (2001) J. Biol. Chem. 276 27944–27949 [DOI] [PubMed] [Google Scholar]
- 39.Littlewood, T. D., Hancock, D. C., Danielian, P. S., Parker, M. G., and Evan, G. I. (1995) Nucleic Acids Res. 23 1686–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courey, A. J., and Tjian, R. (1988) Cell 55 887–898 [DOI] [PubMed] [Google Scholar]
- 41.Patel, S., Xi, Z. F., Seo, E. Y., McGaughey, D., and Segre, J. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18668–18673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomberk, G., and Urrutia, R. (2005) Biochem. J. 392 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sue, N., Jack, B. H., Eaton, S. A., Pearson, R. C., Funnell, A. P., Turner, J., Czolij, R., Denyer, G., Bao, S., Molero-Navajas, J. C., Perkins, A., Fujiwara, Y., Orkin, S. H., Bell-Anderson, K., and Crossley, M. (2008) Mol. Cell Biol. 28 3967–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun, T., Rudnicki, M. A., Arnold, H. H., and Jaenisch, R. (1992) Cell 71 369–382 [DOI] [PubMed] [Google Scholar]
- 45.Rawls, A., Valdez, M. R., Zhang, W., Richardson, J., Klein, W. H., and Olson, E. N. (1998) Development 125 2349–2358 [DOI] [PubMed] [Google Scholar]
- 46.Rudnicki, M. A., Braun, T., Hinuma, S., and Jaenisch, R. (1992) Cell 71 383–390 [DOI] [PubMed] [Google Scholar]
- 47.Rudnicki, M. A., Schnegelsberg, P. N., Stead, R. H., Braun, T., Arnold, H. H., and Jaenisch, R. (1993) Cell 75 1351–1359 [DOI] [PubMed] [Google Scholar]
- 48.Kaczynski, J. A., Conley, A. A., Fernandez Zapico, M., Delgado, S. M., Zhang, J. S., and Urrutia, R. (2002) Biochem. J. 366 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sogawa, K., Kikuchi, Y., Imataka, H., and Fujii-Kuriyama, Y. (1993) J. Biochem. (Tokyo) 114 605–609 [DOI] [PubMed] [Google Scholar]
- 50.Zhang, W., Shields, J. M., Sogawa, K., Fujii-Kuriyama, Y., and Yang, V. W. (1998) J. Biol. Chem. 273 17917–17925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouwman, P., and Philipsen, S. (2002) Mol. Cell. Endocrinol. 195 27–38 [DOI] [PubMed] [Google Scholar]
- 52.Birsoy, K., Chen, Z., and Friedman, J. (2008) Cell Metab. 7 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamura, Y., Tanaka, Y., Kawamori, R., and Maeda, S. (2006) Mol. Endocrinol. 20 844–856 [DOI] [PubMed] [Google Scholar]