Abstract
MUC1, a transmembrane mucin, is a key modulator of several signaling pathways that affect oncogenesis, motility, and cell morphology. The interaction of MUC1 cytoplasmic tail (MUC1CT) with signal transducers and its nuclear translocation and subsequent biological responses are believed to be regulated by phosphorylation status, but the precise mechanisms by which this occurs remain poorly defined. We detected a novel association between the Met receptor tyrosine kinase and the MUC1CT. Met catalyzed phosphorylation of tyrosine at YHPM in the MUC1CT. Stimulation of S2-013.MUC1F pancreatic cancer cells with hepatocyte growth factor facilitated nuclear localization of MUC1CT, as determined by real time confocal imaging analysis. MUC1 overexpression also facilitated faster turnover of Met. Phosphorylation of MUC1CT by Met enhanced its interaction with p53, which led to suppression of AP1 transcription factor activity through interactions at the MMP1 promoter, ultimately leading to reduced transcription of MMP1. This correlated with a decrease in hepatocyte growth factor-induced invasiveness when MUC1 was overexpressed. The results demonstrate that MUC1 modulates Met-mediated oncogenic signaling in cancer.
Met, a receptor tyrosine kinase of the semaphorin family that regulates cell growth, invasion, and cell survival, is a heterodimeric molecule consisting of a 50-kDa extracellular α subunit and a 145-kDa transmembrane β subunit (1). The cytoplasmic tail of Met contains a kinase domain and a C-terminal docking site, which recruits adaptor proteins (2). These interactions are modulated by HGF,2 a ligand for Met that stimulates phosphorylation at different residues of Met and on other substrates. Deregulated activation or overexpression of Met elicits tumorigenic and metastatic effects and has been reported in several cancers, including pancreatic adenocarcinoma (3–5). HGF stimulation also induces degradation of Met and terminates Met signaling, thus bringing cells back to homeostasis.
Human MUC1, a type I transmembrane protein normally expressed on the apical surface of ductal epithelia, is overexpressed and aberrantly glycosylated in several cancers, including pancreatic adenocarcinoma (6, 7). MUC1 is synthesized as a single polypeptide chain but exists on the cell surface as a heterodimer. Co-translational proteolytic cleavage of the full-length protein results in two associated fragments as follows: a large extracellular polypeptide containing the tandem repeat domain that can be released from the cell surface; and a polypeptide consisting of the short extracellular region, the transmembrane domain, and the cytoplasmic tail that exists as an integral membrane protein (8, 9).
The cytoplasmic tail of MUC1 (MUC1CT) contains multiple serine, threonine, and tyrosine residues, which are phosphorylated and involved in different signaling cascades in response to changing cell surface conditions, including spatial reorganization or stimulation with growth factors and cytokines (8). Changes in the phosphorylation status of MUC1CT are believed to modulate its affinity for mediators of signal transduction, including β-catenin, p53, and Grb2-SOS.
The processes that regulate phosphorylation of the MUC1CT and the specific kinases that catalyze these reactions are now being elucidated. The MUC1CT is a substrate for GSK3β, c-Src, EGFR, and platelet-derived growth factor receptor-β (10–13). Activated EGFR is reported to phosphorylate the MUC1CT on tyrosine at a YEKV motif that functions as a binding site for the c-Src Src homology 2 domain (12). EGFR-mediated phosphorylation of MUC1CT induces binding of MUC1CT to c-Src and enhances the binding affinity of MUC1CT to β-catenin. Interactions of MUC1CT with β-catenin are diminished by GSK3-β-mediated phosphorylation (12). MUC1CT is translocated to the nucleus in association with β-catenin, where it may influence the activity of the latter as a transcriptional co-activator (12, 16). Nuclear translocation of MUC1CT is mediated by its interaction with importin β and nucleoporin Nup62 (17).
The in vivo phosphorylation status of MUC1CT and the kinases that act on it in pancreatic cancer have not been well characterized. We present evidence that the Met receptor tyrosine kinase interacts with MUC1 in pancreatic cancer cells, and catalyzes phosphorylation of the MUC1CT in response to stimulation by HGF. Overexpression of MUC1 in pancreatic adenocarcinoma cells down-regulated conventional Met-mediated signaling and inhibited motility and invasion. HGF stimulation facilitated interaction of MUC1CT with p53, enabled p53-mediated suppression of AP1 transcriptional activity, and decreased MMP1 expression. We conclude that Met-mediated phosphorylation of MUC1 modulates signaling related to motility and invasion in pancreatic cancer.
EXPERIMENTAL PROCEDURES
Cell Culture—Panc-1 was obtained from the American Type Culture Collection (Manassas, VA). S2-013 is a cloned subline of a human pancreatic tumor cell line (SUIT-2) derived from a liver metastasis (18). HPAF2 pancreatic tumor cells have been described previously (19, 20). FLAG epitope-tagged MUC1 (MUC1F) transfectants of the S2-013 cell line (S2-013.MUC1F), Panc1 cell line (Panc1.MUC1F), and cytosolic tail-deleted MUC1 transfectants of the S2-013 cell line (S2-013.CT3), and Panc1 (Panc1.CT3) were cultured as described previously (21). MDA-MB-435 cells, previously described as breast cancer cells but recently reported to be of melanoma origin (22, 23), were stably transduced with pLNCX.1 retroviral vectors expressing wild type MUC1 or empty vector neo controls, as described previously (24), to produce MDA-MB-435.MUC1 and MDA-MB-435.neo, respectively. HPAF2 cells were maintained in Eagle's minimal essential medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, nonessential amino acids, sodium pyruvate, and penicillin/streptomycin under similar conditions.
Immunoblotting and Immunoprecipitation—Cell lysate proteins were resolved on 10 or 14% NOVEX Tris-glycine denaturing polyacrylamide gels (Invitrogen) in a 1× SDS-PAGE buffer (1 g/liter SDS, 3 g/liter Tris base, and 14.4 g/liter glycine). Immunoblotting and immunoprecipitations were performed as described previously (14).
Biotinylation Assay—Cells serum-starved for 24 h were surface-biotinylated with EZ-link Sulfo-NHS-SS-Biotin (Pierce) on ice followed by washing and quenching of free biotin, and then incubated at 37 °C in the presence or absence of 100 ng/ml HGF (100 ng/ml, PeproTech, Rocky Hill, NJ) for 2 h. Following the 2-h incubation, cell monolayers were glutathione-stripped as described by Graeve et al. (25). Cells were incubated in two 20-min washes of glutathione solution (60 mm glutathione, 0.083 m NaCl, with 0.083 m NaCl, and 1% bovine serum albumin added before use) at 0 °C to remove the cell surface biotin groups. The remaining biotinylated proteins, which were endocytosed prior to biotin stripping, were recovered from the RIPA cell lysates by incubation with neutravidin beads (Pierce).
In Vitro Kinase Assay—1 μg of peptides in 10 μl of reaction buffer (Met reaction buffer: 40 mm MOPS, pH 7.5, 1 mm EGTA) were incubated at 37 °C for 30 min along with 10 μCi of [γ-32P]ATP and 500 ng of active human recombinant Met (Upstate Cell Signaling Solutions, Lake Placid, NY). The reaction was stopped by boiling the samples in reducing SDS-sample buffer. Samples were resolved on 16% Tricine gel. The gel was dried and analyzed by PhosphorImager analysis.
Metabolic Labeling of Cells and Radioimmunoprecipitation Analysis—Metabolic labeling was performed as described previously (13). Briefly, HPAF2 cells were serum-starved for 24 h and incubated in phosphate-free minimum essential medium at 37 °C for 2 h prior to labeling with 2 mCi/ml [32P]orthophosphate (ICN, Irvine, CA) for 2 h in the presence or absence of 100 ng/ml HGF. Whole cell lysates were prepared with ice-cold radioimmunoprecipitation assay buffer. Radiolabeled MUC1CT was immunoprecipitated by incubation with mAb CT2 (mAb against MUC1CT) and resolved by SDS-PAGE on a 14% gel.
Tandem MS Analysis of MUC1CT and Its Association Partners—Immunoprecipitated MUC1CT from cell lysates or in vitro phosphorylated MUC1CT peptides were resolved on 14% NOVEX Tris-glycine denaturing polyacrylamide gels (Invitrogen), silver-stained, and then individual bands were excised and trypsin-digested. Eluted peptides were analyzed using a Q-TOF Ultima tandem mass spectrometer (Micromass/Waters) with electrospray ionization as described previously (13).
Cell Migration and Invasion Assays—In vitro invasive potential of cells with and without HGF stimulation were assayed using the Biocoat migration or Matrigel invasion chambers (BD Biosciences). S2-013.MUC1F, S2-013.CT3, and S2-013.Neo cells (5 × 104) were seeded onto the upper chamber, which also had a coating of Matrigel in the invasion chambers. The lower chamber contained Dulbecco's modified Eagle's medium with or without 100 ng/ml HGF. The chambers were cultured at 37 °C for 24 h in a CO2 incubator. Cells on the upper portion of the membrane were wiped off with a cotton-tipped swab, and cells on the bottom of this membrane were fixed and stained with Diff-Quick staining kit (Allegiance) and observed by light microscopy. The numbers of invading cells were counted in five randomly selected observation fields per membrane at ×200 magnification.
Plasmids, Transient Transfections, and Luciferase Assays—MMP-1 luciferase reporter, wild type or mutated at the first ETS site 388/385 (GGAT to AAAT), AP-1-binding site (370 to 368 from GTC to TGG), or the second ETS site 348/345 (GGAA to AAAA) were a kind gift from Dr. Ralf Janknecht (Mayo Clinic, Rochester, MN) (26). An AP1-responsive luciferase plasmid was obtained from Promega. PGL2 basic was used as a control plasmid for transfection in luciferase studies. A dual luciferase reporter assay (Promega) was used to detect promoter activity in this study. A synthetic Renilla luciferase reporter plasmid pRL-SV40 (Promega) was used as a control for transfection efficiency. 10,000 cells/well were seeded into a 48-well plate and grown to ∼60% confluence and then serum-starved for 24 h and transfected using Lipofectamine 2000 (Invitrogen), with and without 100 ng/ml HGF, and cultured for another 24 h. The cells were then washed twice with phosphate-buffered saline and harvested with 200 μl of Passive Lysis buffer (Promega) and assayed for the luciferase activities. Each experiment was performed three times in triplicate.
Chromatin Immunoprecipitation (ChIP) Assays—5.0 × 107 S2-013MUC1F cells were prepared for ChIP assays by cross-linking for 30 min with 50 mm (10-fold molar excess) dimethyl 3,3′-dithiobispropionimidate·2 HCl (Pierce), followed by 10 min of cross-linking with 1% formaldehyde. The cross-linkers were quenched with 125 mm glycine in phosphate-buffered saline. The cells were scraped and sonicated to an average of 500 bp to prepare the chromatin as per the Affymetrix ChIP protocol, which was subjected to immunoprecipitation with either CT2 or IgG control. The immunoprecipitants were washed and eluted as suggested by the protocol, and 3.0 μl of the eluted product was subjected to real time PCR using primer sets for pGUSB (Superarray Bioscience Corp., Frederick, MD), pMMP1 (forward, TGGCAGAGTGTGTCTCCTTCGC; reverse, TCGAAGGTAAGTGATGGCTTCC), or a control region 1.5 kb upstream of pMMP1 (forward, TGCCTAGCACCAAGGAGCGAAGA; reverse, CGGAGTATGAGATAACTCCCC) as described in the Superarray protocol.
Statististical Analysis—Results are expressed as mean ± S.E. of 3–5 independent experiments, each treatment performed in triplicate. Statistical analysis was performed using the SPSS or GraphPad Prism statistical software. Student's t test was used when appropriate. p < 0.05 was considered significant.
RESULTS
MUC1CT Interacts with Met—Proteomics techniques were used to identify proteins that co-immunoprecipitated with MUC1 from S2-013 pancreatic adenocarcinoma cells, a moderately differentiated human pancreatic tumor cell line known to express O-glycosylated mucin-like proteins, including low levels of endogenous MUC1 (18). A cDNA construct encoding a FLAG epitope-tagged form of MUC1 (7) was overexpressed in the these cells (S2-013.MUC1F). Immunoprecipitations with the CT2 antibody, which binds to MUC1CT, were resolved on Tris-glycine SDS-PAGE and silver-stained (supplemental Fig. 1). Bands representing specific proteins that co-immunoprecipitated with MUC1 and were absent from isotype control lanes were excised from the gel and subjected to in-gel trypsin digestion and MS/MS analysis for protein identification. A fragment corresponding to the cytoplasmic tail of Met, FTVKVADFGLAR, was identified by this approach (Fig. 1A).
FIGURE 1.
Interaction of MUC1 with Met. A, identification of Met as an interaction partner for MUC1 by mass spectrometry. MUC1 was immunoprecipitated from S2-013.MUC1F and Panc1.MUC1F cells by mAb CT2 and resolved on SDS-PAGE. The gel was silver-stained, and specific bands not observed in IgG control lanes were excised and subjected to trypsin digestion followed by liquid chromatography-electrospray ionization-mass spectrometric analysis. This analysis identified a tryptic fragment of Met indicated in the illustration. B, interaction of MUC1 and Met was confirmed by reciprocal co-immunoprecipitation. MUC1 was immunoprecipitated (IP) from Panc1.MUC1F, S2-013.MUC1F, and HPAF2 pancreatic adenocarcinoma cells and immunoblotted (IB) for Met by using a polyclonal Ab for the cytoplasmic tail of Met. A reciprocal immunoprecipitation of Met and immunoblotting with HMFG2 mAb raised against the tandem repeat region of MUC1 was also performed (right panel).
We performed reciprocal co-immunoprecipitations of MUC1 and Met from cell lysates of three human pancreatic adenocarcinoma cell lines as follows: Panc1.MUC1F (a poorly differentiated cell line overexpressing a FLAG-tagged MUC1 construct); S2-013.MUC1F; and HPAF2 (a highly differentiated cell line with high endogenous levels of MUC1). Immunoprecipitations utilizing anti-cytoplasmic tail antibodies for both molecules (Fig. 1B) were resolved on 10% SDS-PAGE to facilitate separation of pre-Met (unprocessed form) and Met and Western blotted, and the membranes were probed with HMFG-2 (a monoclonal antibody against the tandem repeat region of MUC1) or an antibody against the Met cytoplasmic tail. The immunoblotting results revealed consistent interactions between MUC1 and Met in S2-013.MUC1F and HPAF2 cells, and a weaker interaction was observed for Panc-1.MUC1F cells. Although the reason for the weaker interaction between MUC1 and Met in Panc1.MUC1F cells is not definitively known at this time, it is known that Panc1 is a poorly differentiated pancreatic cancer cell line (27) that attaches core 1 type short oligosaccharides (e.g. sialyl-Tn and sialyl-T) to MUC1 because of a lack of expression of core 2 GlcNAc transferase activity (28). In contrast, S2-013 and HPAF2 are moderately and well differentiated pancreatic cancer cell lines, respectively, that attach to MUC1 extended oligosaccharides such as sialyl Lewis A (7) and sialyl Lewis C. This raises the possibility that glycosylation of MUC1 influences its ability to interact with Met. Both pre-Met and Met were found to interact with MUC1.
MUC1 Facilitates Faster Turnover of Met—Because MUC1 and Met were in complex, we sought to determine whether overexpression of MUC1 modulates stability of Met by performing cell surface biotinylation experiments (25). S2-013.MUC1F and S2-013.Neo cells were serum-starved for 24 h and then surface-biotinylated with a cell-impermeable biotinylation reagent. The surface-biotinylated cells were stimulated with HGF (100 ng/ml) for 0, 15, 60, or 120 min. The cells were glutathione-stripped to remove biotin from the nonendocytosed cell surface proteins, thus specifically biotin-labeling proteins that were endocytosed. Ammonium chloride was used to block lysosomal degradation of endocytosed proteins. Proteins from whole cell lysates were immunoprecipitated by using Neutravidin beads and subsequently immunoblotted for Met. HGF stimulation caused a significant reduction in total levels of Met protein in a temporal manner in S2-013.MUC1F cells (Fig. 2A). However, steady state levels of Met were comparatively stable in S2-013.Neo cells (which do not overexpress MUC1), suggesting that MUC1 overexpression facilitated faster turnover of Met during stimulation with HGF. The increased turnover of Met in S2-013.MUC1F cells following HGF stimulation was also associated with reduced binding to Gab1, as demonstrated by immunoprecipitation followed by immunoblotting (Fig. 2B). However, the interactions of Met with Gab1 were higher in S2-013.MUC1F cells under serum-starved steady state conditions, as compared with the S2-013.Neo cells.
FIGURE 2.
MUC1 promotes endocytosis, lysosomal degradation, and reduced signaling of Met. A, effect of MUC1 overexpression on Met endocytosis was determined in S2-013.MUC1F or S2-013.neo cells. Serum-starved cells were surface-biotinylated on ice and then incubated at 37 °C for 2 h in the absence (serum-free or SF) or presence of HGF (H) to allow endocytosis. Surface-biotinylated proteins from RIPA lysates were recovered on neutravidin beads and analyzed by SDS-PAGE, followed by Western blotting with a polyclonal Ab against Met. 1st 3 lanes indicate the total levels of surface-biotinylated Met after 2 h of incubation at 0, 37, or 37 °C in the presence of HGF and ammonium chloride, and 4th to 6th lanes represent the levels of endocytosed Met (detected by glutathione stripping or GS) under serum-free conditions or HGF stimulation with and without ammonium chloride. B, effect of HGF stimulation on Met signaling. S2-013.MUC1F or S2-013.neo cells were stimulated with HGF for different times. Lysates were immunoprecipitated with Met cytoplasmic tail Ab or IgG control. The immunoprecipitants were resolved on SDS-PAGE and immunoblotted (IB) with Abs against Gab1 or Met.
In Vitro Phosphorylation of MUC1CT by Met—We investigated whether Met phosphorylated the MUC1CT by performing in vitro kinase assays with a 66-mer MUC1CT peptide (MUC1CT-p66, representing the C-terminal 66 residues of 72-residue-long MUC1CT) and recombinant active human Met kinase, in the presence of [γ-32P]ATP. Autoradiograms from 32P-labeled in vitro kinase reactions showed phosphorylation of MUC1CT-p66 in the presence of Met (Fig. 3A, 3rd lane). No phosphorylation was observed in the absence of peptide (Fig. 3A, 2nd lane), or kinase (1st lane). Parallel reactions were applied to SDS-PAGE followed by silver staining (Fig. 3A, bottom panel). Bands corresponding to the MUC1CT peptide were digested with trypsin and sequenced by tandem mass spectrometry. The mass spectrometric analysis of MUC1CT revealed phosphorylation at tyrosine at the YHPM motif (or Tyr-1203, NCBI accession number P15941; Fig. 3B). Furthermore, MUC1CT peptide synthetically pre-phosphorylated at tyrosine in the YHPM motif (MUC1CT-p66-pYHPM) was not phosphorylated by Met (Fig. 3A).
FIGURE 3.
Met mediates phosphorylation and nuclear localization of MUC1CT. A, recombinant active Met kinase phosphorylated MUC1CT-p66. MUC1CT-p66 or MUC1CTp66-pYHPM peptides were incubated with Met kinase in the presence of [γ-32P]ATP. The reaction mixtures were resolved on SDS-PAGE followed by autoradiography or the gel was stained with Coomassie Blue. B, MS/MS spectra of trypsinized phospho-MUC1CT-p66 from in vitro kinase reactions. MS/MS spectra of DTYHPMSEYPTYHTHGR peptide from Met-phosphorylated MUC1CT-p66 indicates phosphorylation of Y1. 32P labeling of HPAF2 cells was performed under HGF stimulation (C). HPAF2 cells were labeled with [32P]orthophosphate (250μCi/ml) in the presence (3rd lane) or absence (1st lane) of human recombinant HGF (100 ng/ml), lysed in RIPA buffer, immunoprecipitated (IP) with mAb CT2, and subjected to autoradiography or immunoblotting with mAb CT2. Immunoprecipitation with a nonspecific mAb was used as an IP control (2nd lane). Nuclear (N) and cytoplasmic (C) localization of MUC1CT from S2-013.MUC1F cells (D) and HPAF2 cells (E) phosphorylated at tyrosine at the YHPM site were determined by Western blotting with phosphorylation site-specific antibody. The polyvinylidene difluoride membrane was reprobed with antibodies against H2B and β-actin as controls for protein loading. F, left panel, S2-013.MUC1.CT-EYFP cells expressing MUC1 with C-terminally tagged EYFP were utilized for real time laser scanning confocal microscopy. The first two scans were taken under serum-starved conditions, and HGF (100 ng/ml) was added to the culture disk right after the second scan, and successive scans were taken for 15 min, every 10 s at 37 °C at ×100 magnification. After the first 15 min, the rest of the scans were performed every 2.5 min for a total period of 4 h. Representative real time confocal scans of a cluster of cells and a single cell are presented here. G, S2-013.MUC1F or S2-013.MUC1FHPM cells were serum-starved for 24 h, stimulated with HGF for 15 min, or left unstimulated, fixed, and permeabilized before incubation with mAb CT2 and the nuclear dye propidium iodide. The mAb CT2 (anti-MUC1CT) was visualized as blue (right panel) and detected by using a Cy5-conjugated secondary antibody, and nuclei were visualized in red. White arrows indicate localization of MUC1CT outside the nucleus.
Increased MUC1CT Phosphorylation by HGF Stimulation of Pancreatic Adenocarcinoma Cells—To investigate the in vivo phosphorylation of MUC1CT by Met in pancreatic adenocarcinoma cells, HPAF2 cells were serum-starved for 24 h followed by 2 h of incubation with 100 ng/ml recombinant human HGF, an activating ligand for Met, in the presence of [32P]orthophosphate. MUC1 was immunoprecipitated from cell lysates with CT2 mAb and separated by SDS-PAGE, and proteins were transferred onto a polyvinylidene difluoride membrane, which was then subjected to autoradiography. Cell lysates from unstimulated cells and lysates immunoprecipitated with an isotype identical Ab were used as controls. A duplicate gel was immunoblotted with CT2 antibody to identify the MUC1CT bands. Autoradiography revealed that in vivo labeling of HPAF2 cells with [32P]orthophosphate following stimulation with HGF induced a significant increase in phosphorylation of MUC1CT, as compared with unstimulated cells (Fig. 3C).
HGF Stimulation Causes Nuclear Localization of MUC1CT Phosphorylated at Tyrosine in the YHPM Motif—A rabbit polyclonal antibody specific for phosphorylated tyrosine at the YHPM site in MUC1CT (anti-pYHPM) was developed and utilized to evaluate the effect of HGF stimulation on MUC1CT phosphorylation. The antibody has been described previously (13). Nuclear and cytoplasmic extracts from HGF-stimulated S2-013.MUC1F and HPAF2 cells were resolved on SDS-PAGE followed by Western blotting with the anti-pYHPM antibody. The results revealed a significant increase in phosphorylation of tyrosine in the YHPM motif that was concordant with an increased duration of HGF stimulation, mostly observed in the nuclear fraction (Fig. 3, D and E).
Real time laser scanning confocal imaging was performed on S2-013 cells stably transfected with C-terminally tagged EYFP (S2-013.MUC1.CT-EYFP). The cells were serum-starved for 24 h and then stimulated with HGF (100 ng/ml) after two initial scans to establish steady state localization under unstimulated conditions. HGF stimulation significantly enhanced the nuclear localization of MUC1CT from cytoplasmic and cell surface locations within seconds to minutes of stimulation (Fig. 3F). By 15 min post-stimulation, there was significant turnover of MUC1CT.
The effects of HGF stimulation on nuclear localization of MUC1CT were also evaluated by immunofluorescence analysis with the CT2 mAb (raised against the MUC1CT) after paraformaldehyde fixation of the serum-starved or HGF-stimulated S2-013.MUC1 cells. HGF stimulation significantly enhanced the nuclear localization of MUC1CT (Fig. 3G). Furthermore, mutating a tyrosine in the YHPM motif to phenylalanine (S2-013.MUC1.FHPM) significantly abrogated the effect of HGF stimulation on nuclear localization of MUC1CT.
Responses to HGF Stimulation of Met Are Down-regulated by MUC1—We evaluated the effect of MUC1 overexpression on HGF stimulation of migration by in vitro motility and invasion assays performed in Boyden chambers with S2-013.MUC1F and S2-013.Neo. MUC1 expression has been shown to facilitate steady state in vitro motility and invasion in pancreatic adenocarcinoma cells (27, 28), and this was confirmed by the data presented in Fig. 4 (S2013.Neo cell migration is less than S2013.MUC1 under unstimulated conditions). Similar results were observed with the MUC1-transfected and control cancer cell lines MDA-MB-435.Neo and MDA-MB-435.MUC1 (data not shown). There was an ∼16-fold increase in the number of S2-013.Neo cells migrating to the other side of the filter in response to HGF-containing medium, as compared with nonstimulated controls. The magnitude of this increase in response to HGF stimulation was reduced by half (to 8-fold) for S2-013.MUC1F cells (Fig. 4A). The differences in HGF-stimulated motility of S2-013.MUC1F and S2-013.Neo cells were statistically significant (p < 0.0001). Similar results (although smaller in scale) were obtained when MDA-MB-435 cells were evaluated in the same manner. Stimulation of MDA-MB-435.Neo cells with HGF produced a 70% increase in motility, but there was only a 35% increase in motility in MDA-MB-435.MUC1 cells expressing MUC1 (data not shown, 4 independent experiments with 3 replicates each). Evidence of differences in motility between S2-013.MUC1F and S2-013.Neo cells was also reflected in the lamellipodial structures observed by Alexa488-Phalloidin staining, with and without HGF stimulation (Fig. 4A). S2-013.Neo cells showed significantly more lamellipodial structures during HGF stimulation, as compared with S2-013.MUC1F cells. Similarly, results of Matrigel invasion assays under HGF stimulation (Fig. 4B) revealed that S2-013.Neo cells exhibited greater than 8-fold increases in in vitro invasiveness as compared with S2-013.MUC1F cells, which showed modest increases of ∼2.5-fold (p < 0.0001). The cancer cell line MDA-MB-435.neo showed a 4.5-fold increase in invasiveness upon stimulation with HGF, which was decreased slightly to a 4.0-fold increase in MDA-MB-435.MUC1 cells expressing MUC1 (data not shown, 4 independent experiments with 3 replicates each). S2-013. MUC1.FHPM cells did not show significant abrogation of HGF-induced invasiveness (Fig. 4B).
FIGURE 4.
MUC1 abrogates HGF-stimulated motility and invasion. S2-013.Neo, S2-013.MUC1F, or S2-013.MUC1.FHPM cells were added to the upper compartment of migration or invasion chamber in serum-free culture media. HGF-containing media were added to the lower chamber. After 24 h of incubation, cells in the upper chamber were removed, and cells that had migrated or invaded onto the lower surface of the membrane were stained and counted in 10 different fields of light microscopy. Bars represent mean ± S.E. of migrating (A) or invading cell number (B) from three independent experiments performed in triplicate (*** indicates p < 0.001 and ** indicates p < 0.01). To study the actin cytoskeleton changes, cells were serum-starved for 24 h or stimulated with HGF for 2 h post-serum starvation. The actin cytoskeleton was visualized with Alexa-488-conjugated phalloidin (green), and the nuclei were stained with propidium iodide (red) and analyzed by laser scanning confocal microscopy (A). The horizontal size bars represent 20 μm.
MUC1 Abrogates HGF-stimulated Transcriptional Activation of MMP1—HGF stimulation of Met has been implicated in transcriptional activation of MMP1, leading to enhanced invasiveness in different cancer cell types (29, 30). We evaluated S2-013.MUC1F and S2-013.Neo cells for MMP1 mRNA expression by semiquantitative reverse transcription-PCR. MUC1 overexpressing cells showed no increase in MMP1 mRNA expression under HGF stimulation (Fig. 5A); however, MMP1 expression was enhanced in response to HGF in S2-013.Neo cells. As a control, stimulation with platelet-derived growth factor resulted in increased MMP1 expression in both cell types. To confirm these findings, we evaluated the activity of an MMP1 promoter luciferase reporter plasmid system. The results of these assays indicated significantly higher HGF-stimulated activation of the reporter system in S2-013.Neo and Panc1.Neo cells as compared with S2-013.MUC1F (Fig. 5B) and Panc1.MUC1F cells (Fig. 5C), respectively. However, expression of a truncated cytoplasmic tail construct of MUC1 (S2-013.CT3 and Panc1.CT3) did not abrogate the HGF-mediated increase in MMP1 expression. These results indicated that MUC1CT down-regulated HGF-mediated activation of MMP1 promoter activity.
FIGURE 5.
MUC1 abrogates HGF-stimulated activation of MMP1 expression. S2-013.MUC1F and S2-013.Neo cells were cultured to 60% confluence, serum-starved for 24 h, and then stimulated with platelet-derived growth factor (PDGF) (50 ng/ml) or HGF (100 ng/ml) for 24 h or left unstimulated. A, expression of MMP1 was determined by semiquantitative reverse transcription-PCR. The expression of β-actin was determined as a control. MMP1 promoter-luciferase reporter activity was assayed in S2-013.MUC1F, S2-013.CT3, and S2-013.Neo cells (B) as well as Panc1.MUC1F, Panc1.CT3, and Panc1.Neo cells (C) under HGF stimulation. Cells were cotransfected with –512hMMP1 luciferase reporter plasmid and Renilla luciferase plasmid (control for transfection efficiency). Cells were subsequently stimulated with HGF (100 ng/ml) or left unstimulated for 24 h. Cells were harvested, and proteins were assayed for dual luciferase activity. Three independent transfections were performed in triplicate, and the results are expressed as the mean of relative light units normalized with Renilla luciferase activity ± S.E. (*** indicates p < 0.001).
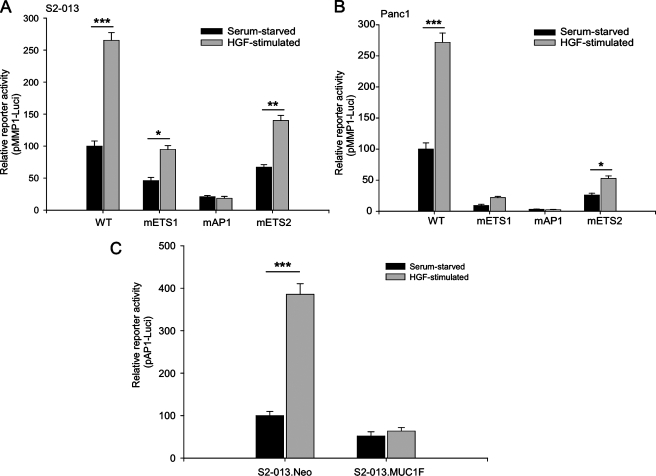
MUC1 Abrogates HGF Stimulation-mediated Activation of AP1 Transcription Factor Activity—We investigated the role of different DNA elements in the regulation of MMP1 expression upon HGF stimulation by utilizing MMP1 promoter-luciferase reporter constructs having mutations in the first ETS region (ETS1), the AP1-binding region, and the second ETS region (ETS2) (24). Assays with these promoter-luciferase reporter constructs showed that mutating the ETS regions significantly decreased luciferase reporter activity; however, the mutated ETS luciferase reporters still showed a 2-fold increase in reporter activity upon stimulation with HGF (Fig. 6, A and B). Constructs with mutations in the AP1-binding region showed greatest decrease in the reporter activity, and the effect of HGF stimulation was also totally abrogated. This supports the hypothesis that the AP1 transcription factor is involved in the HGF-stimulated regulation of MMP1 promoter activity. Because MUC1 also abrogated the effect of HGF stimulation, we evaluated the effect of MUC1 on AP1-dependent transcriptional activity under HGF stimulation by evaluating transcriptional activity with an AP1-responsive promoter-luciferase reporter construct. The S2-013.Neo cells showed a 4-fold increase in AP1-responsive luciferase reporter activity under HGF stimulation, as compared with serum-starved conditions (Fig. 6C). However, the S2-013.MUC1F cells showed no significant increase in AP1-responsive luciferase reporter activity under HGF stimulation.
FIGURE 6.
AP1 transcriptional activity is down-regulated by MUC1 and is crucial for HGF-mediated activation of MMP1 promoter activity. S2-013 (A) and Panc1 cells (B) were cotransfected with –512hMMP1 luciferase reporter plasmid or the mutant reporter plasmids and Renilla luciferase plasmid. The –512hMMP1 luciferase reporter plasmids were either wild type (WT) or contained mutations in the first ETS region (ETS1), the AP1-binding region (AP1), or the second ETS region (ETS2). Cells were subsequently stimulated with HGF (100 ng/ml) or left unstimulated for 24 h. Cells were harvested, and proteins were assayed for dual luciferase activity. S2-013.MUC1F and S2-013.Neo cells were co-transfected with pAP1-luciferase reporter plasmid and Renilla luciferase plasmid (C). Cells were subsequently stimulated with HGF (100 ng/ml) or left unstimulated for 24 h and analyzed for AP1-responsive promoter activity. Three independent transfections were performed in triplicate, and the results are expressed as the mean of relative light units normalized with Renilla luciferase activity ± S.E. (*** indicates p < 0.001, ** indicates p < 0.01, and * indicates p < 0.05).
HGF Stimulation Facilitates Interaction of MUC1CT with p53 and Activates p53 to Abrogate MMP1 Expression—Previous studies have reported interactions between MUC1 and p53 in the region of MUC1CT containing the Met phosphorylation site (33). Because it has also been reported that p53 can block the interaction of AP1 transcription factor with basal transcription machinery to abrogate MMP1 expression, we wanted to determine whether Met-phosphorylated MUC1 contributed to effects of p53 on MMP1 promoter activity (34–36). We investigated this possibility by performing co-immunoprecipitation of MUC1 and p53 from serum-starved and HGF-stimulated S2-013.MUC1F cells. Immunoprecipitations with the CT2 antibody against the MUC1CT were resolved on SDS-PAGE and subjected to immunoblotting with an anti-p53 pAb. HGF stimulation of S2-013.MUC1F cells significantly enhanced the interaction of MUC1CT with p53 (Fig. 7A). Furthermore, mutating a tyrosine in the YHPM motif to phenylalanine significantly abrogated this association, while mutating the same residue to glutamate (providing a phosphorylation-mimicking negative charge) significantly enhanced the interaction of MUC1 with p53 (Fig. 7B).
FIGURE 7.
Phosphorylation of MUC1CT by Met enhances interaction with p53 and activation of p53 further down-regulates MMP1 expression in the presence of MUC1. Serum-starved or HGF-stimulated S2-013.MUC1F cell extracts were co-immunoprecipitated (IP) with CT2 mAb or isotype control antibody and immunoblotted (IB) for p53 (A). S2-013.MUC1F, S2-013.MUC1.FHPM, and S2-013.MUC1.EHPM cells were also analyzed for the effect of phosphorylation abrogating or mimicking mutations at tyrosine in the YHPM motif for their effect on interaction of MUC1 with p53 by co-immunoprecipitations (B). Serum-starved or HGF-stimulated S2-013.MUC1F and S2-013.Neo cells were co-transfected with MMP1 promoter luciferase reporter constructs, and the promoter activity was assayed in the presence or absence of etoposide treatment (10 μm), to activate p53 (C) (* indicates p < 0.05). Promoter occupancy of MUC1CT at MMP1 promoter (pMMP1) under serum-starved and HGF-stimulated conditions was determined by performing ChIP assay on S2-013.MUC1 cells by utilizing CT2 mAb or control IgG followed by real time PCR with pMMP1 primers, primers to the 1.5-kb region upstream of pMMP1 or GUSB primers. The amount of pMMP1 immunoprecipitated with CT2 was normalized with input and plotted as relative amounts of pMMP1 immunoprecipitated with CT2 compared with the serum-starved conditions ± S.E. (D). The PCR products were also visualized on a gel to determine the specificity of amplification after 40 cycles (E). The experiment was performed in triplicate.
Having established the effect of HGF stimulation on interaction of MUC1CT with p53, we wanted to determine whether activation of p53 affected the MUC1-mediated abrogation of MMP1 promoter activation. For this purpose, we stimulated S2-013.MUC1F and S2-013.Neo cells with HGF in the presence or absence of 50 μm etoposide, an activator of p53 (37, 38). Etoposide treatment significantly decreased MMP1 expression in the presence or absence of HGF stimulation of S2-013.MUC1F cells (Fig. 7C). However, etoposide treatment enhanced MMP1 promoter activity in S2-013.Neo cells, suggesting that MUC1 overexpression augmented the inhibitory activity of p53 on the MMP1 promoter in pancreatic adenocarcinoma cells.
MUC1CT Physically Occupies the MMP1 Promoter and Promoter Occupancy Is Enhanced by HGF Stimulation—Given the evidence presented here that the MUC1CT was acting as a transcriptional co-regulator, and has been shown to physically occupy different promoter elements (33), we sought to determine whether MUC1CT was present in transcriptional complexes at the MMP1 promoter, and if promoter occupancy was regulated by HGF stimulation. A quantitative ChIP assay demonstrated greater than 3-fold increases in MMP1 promoter (pMMP1) occupancy by MUC1CT at the AP1-binding site following HGF stimulation as compared with serum-starved conditions at 24 h (Fig. 7, D and E). Control IgG ChIP assays showed no enrichment at the pMMP1 site. As an additional control, we tested for enrichment of other unrelated elements (GUSB promoter) when immunoprecipitated with CT2 from serum-starved and HGF-stimulated cells, and MMP1 promoter occupancy was normalized to GUSB promoter occupancy. Also, no enrichment was detected at sites 1.5 kb upstream of the MUC1-binding region in the MMP1 promoter.
DISCUSSION
In this study, we identified Met as a novel interaction partner and kinase for the MUC1CT. We demonstrated that MUC1 associates with Met in pancreatic tumor cells, and under conditions of HGF stimulation MUC1 promotes endocytosis and increased turnover of Met. Overexpression of MUC1 down-regulated HGF stimulation-induced interaction of Met with conventional downstream signal transducers and inhibited subsequent biological responses (motility and invasion). The results clearly demonstrate that MUC1 is a regulator of Met signaling in pancreatic adenocarcinoma cells.
Our results demonstrate that under serum-starved conditions, which might simulate the effects of autocrine growth factor stimulation, MUC1-overexpressing cells have higher Met signaling activity, as indicated by greater steady state interaction with Gab1. These findings suggest that MUC1 facilitates activation of Met in an environment that contains low levels of HGF. Hence, MUC1 expression may confer on tumors a greater propensity to metastasize when present in low HGF tissue environments. Conversely, MUC1 also down-regulated HGF-stimulated activation of motility and invasion under conditions of high HGF concentrations. Additional indirect evidence of this biological effect was provided by our observation of down-regulation of HGF-mediated transcriptional activation of MMP1 in MUC1-overexpressing cells, which is discussed further below. MMP1 is a well known target of Met activation and is a regulator of metastasis by virtue of its extracellular matrix degrading activity (32).
Previously, the C terminus of another transmembrane mucin expressed in the kidney, MUC20, was shown to associate with Met and impair downstream signaling through the Grb2-Ras-ERK1/2 pathway by blocking Grb2 recruitment to HGF-activated Met (39), without affecting signaling through Gab1/phosphatidylinositol 3-kinase pathways. That these effects were distinct from the pathways affected by interactions between MUC1 and Met suggests that interactions between cell surface mucins and Met are important and specific modulators of Met signaling activity. The tissue-specific patterns of expression of different transmembrane mucins may thus confer tissue-specific effects on Met signaling, both constitutive and HGF-stimulated.
High expression of HGF and Met has been reported in both early and late neoplastic ductal lesions of pancreatic cancer, suggesting that this may comprise an autocrine regulatory circuit (40). However, fully neoplastic and invasive cells lack the expression of Met, which has been further correlated with poor survival of cancer patients (40). It has been proposed for pancreatic cancer that up-regulation of Met receptor expression and HGF-Met interaction may have a crucial role in early stages of neoplasia, although subsequent loss may promote more aggressive behavior of tumor cells (40). MUC1-facilitated turnover of Met might reflect a similar biological phenomenon. Also, it is expected that activation of Met, which is well known to induce cell scattering, might be crucial during early phases of metastatic spread of cancer cells and also contribute to early stages of survival at different metastatic sites. However, once cells are established at different locations, they may turn off Met to facilitate growth of a compact tumor mass. MUC1-mediated endocytosis (which we demonstrate in this report) might down-regulate levels of Met at the cell surface under conditions of high HGF concentrations.
We demonstrated that Met is a kinase for MUC1CT, phosphorylating the latter at a tyrosine in the YHPM motif. What is the biological role of Met-mediated phosphorylation of MUC1CT? Previous studies have indicated that tyrosine phosphorylation of MUC1CT is crucial for its nuclear localization (8, 13). Real time confocal imaging of MUC1-GFP constructs as well as confocal imaging of fixed cells post-HGF stimulation were consistent with the hypothesis that there are pools of cell surface and intracellular (perhaps pre-endocytic or endocytic) MUC1CT under steady state conditions, and that stimulation with HGF induced nuclear localization of MUC1CT from these compartments within seconds to minutes of stimulation. Western blotting analysis of nuclear and cytoplasmic extracts of S2-013.MUC1F cells indicated that a majority of the YHPM phosphorylated fraction of MUC1CT is rapidly transported to and resident in the nucleus. It should be noted that although the in vitro studies prove that there is a direct kinase-substrate relationship between Met and MUC1, the phosphorylation of MUC1 detected upon HGF stimulation in cells could also be indirect and mediated through other kinases that are activated downstream of Met.
In this study, we also investigated the functional role of nuclear localization of MUC1CT post-HGF stimulation. We showed that HGF stimulation facilitated interaction of MUC1CT with p53, and phosphorylation of MUC1CT at tyrosine in the YHPM motif significantly enhanced its interaction with p53. The p53 protein has been reported previously to down-regulate MMP1 promoter activity by abrogating the interaction of AP1 transcription factor with basal transcriptional machinery (34–36). Our studies show that MUC1 blocks the HGF-mediated activation of AP1 transcriptional activity. Moreover, overexpression of MUC1 abrogates MMP1 promoter activity in a manner that is regulated by the AP1 transcription factor, which is further suppressed by etoposide-mediated activation of p53. However, control cells lacking MUC1 expression do not show p53-mediated inhibitory effects on MMP1 promoter activity, suggesting that interaction of p53 with MUC1CT facilitates the inhibitory effect of p53 on the MMP1 promoter.
The results presented here provide the first evidence that MUC1CT physically occupies the MMP1 promoter. The presence of MUC1CT in the transcriptional complex at the MMP1 promoter, which is further facilitated by HGF stimulation and occurs in the region of the promoter bound by p53 and AP1, significantly inhibited MMP1 promoter activity. Hence these studies suggest a transcriptional co-repressor role for MUC1CT.
Overall, this study provides insight into a mechanism by which MUC1, a cell surface associated mucin, engages in morphogenetic signal transduction, by directly influencing the steady state levels of protein expression of an associated receptor tyrosine kinase (Met), its downstream signaling, and by enacting a unique signal transduction pathway in which phosphorylated MUC1CT associates with p53 and modulates transcription of the MMP1 gene. Our findings demonstrate that signaling in response to HGF stimulation is influenced at multiple levels by interactions between Met and MUC1, including influences on conventional Met signaling, and the induction of a specific signaling pathway that involves the phosphorylated MUC1CT. Given that MUC1 and Met are overexpressed in pancreatic adenocarcinoma, further studies of MUC1 and Met signaling should provide additional insights into the molecular mechanisms that regulate invasion and metastasis of pancreatic cancer.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of Thomas Caffrey, Dr. Judith K. Christman, and Janice Taylor of the Confocal Laser Scanning Microscopy Core Facility, and Dr. Ralf Janknecht of Mayo Clinic, Rochester, for generously providing us with MMP1 promoter-luciferase reporter constructs.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA57362, U01 CA111294, CA09476 (training), P20 RR15635 from the COBRE Program of the NCRR, and P30 CA36727 from the NCI Cancer Center. This work was also supported by graduate studies assistantships (to P. K. S., J. P. E., and J. M. B.), the Nebraska Research Initiative, and the Danish National Research Council. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: HGF, hepatocyte growth factor; mAb, monoclonal antibody; MOPS, 4-morpholinepropanesulfonic acid; EYFP, enhanced yellow fluorescent protein; MS/MS, tandem mass spectrometry; ChIP, chromatin immunoprecipitation; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; EGFR, epidermal growth factor receptor.
References
- 1.Galimi, F., Brizzi, M. F., and Comoglio, P. M. (1993) Stem Cells 11 Suppl. 2, 22–30 [DOI] [PubMed] [Google Scholar]
- 2.Bolanos-Garcia, V. M. (2005) Mol. Cell. Biochem. 276 149–157 [DOI] [PubMed] [Google Scholar]
- 3.Gao, C. F., and Vande Woude, G. F. (2005) Cell Res. 15 49–51 [DOI] [PubMed] [Google Scholar]
- 4.Di Renzo, M. F., Poulsom, R., Olivero, M., Comoglio, P. M., and Lemoine, N. R. (1995) Cancer Res. 55 1129–1138 [PubMed] [Google Scholar]
- 5.Corso, S., Comoglio, P. M., and Giordano, S. (2005) Trends Mol. Med. 11 284–292 [DOI] [PubMed] [Google Scholar]
- 6.Hinoda, Y., Ikematsu, Y., Horinochi, M., Sato, S., Yamamoto, K., Nakano, T., Fukui, M., Suehiro, Y., Hamanaka, Y., Nishikawa, Y., Kida, H., Waki, S., Oka, M., Imai, K., and Yonezawa, S. (2003) J. Gastroenterol. 38 1162–1166 [DOI] [PubMed] [Google Scholar]
- 7.Burdick, M. D., Harris, A., Reid, C. J., Iwamura, T., and Hollingsworth, M. A. (1997) J. Biol. Chem. 272 24198–24202 [DOI] [PubMed] [Google Scholar]
- 8.Singh, P. K., and Hollingsworth, M. A. (2006) Trends Cell Biol. 16 467–476 [DOI] [PubMed] [Google Scholar]
- 9.Parry, S., Silverman, H. S., McDermott, K., Willis, A., Hollingsworth, M. A., and Harris, A. (2001) Biochem. Biophys. Res. Commun. 283 715–720 [DOI] [PubMed] [Google Scholar]
- 10.Li, Y., Bharti, A., Chen, D., Gong, J., and Kufe, D. (1998) Mol. Cell. Biol. 18 7216–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Y., Kuwahara, H., Ren, J., Wen, G., and Kufe, D. (2001) J. Biol. Chem. 276 6061–6064 [DOI] [PubMed] [Google Scholar]
- 12.Li, Y., Ren, J., Yu, W., Li, Q., Kuwahara, H., Yin, L., Carraway, K. L., III, and Kufe, D. (2001) J. Biol. Chem. 276 35239–35242 [DOI] [PubMed] [Google Scholar]
- 13.Singh, P. K., Wen, Y., Swanson, B. J., Shanmugam, K., Kazlauskas, A., Cerny, R. L., Gendler, S. J., and Hollingsworth, M. A. (2007) Cancer Res. 67 5201–5210 [DOI] [PubMed] [Google Scholar]
- 14.Deleted in proof
- 15.Deleted in proof
- 16.Wen, Y., Caffrey, T. C., Wheelock, M. J., Johnson, K. R., and Hollingsworth, M. A. (2003) J. Biol. Chem. 278 38029–38039 [DOI] [PubMed] [Google Scholar]
- 17.Leng, Y., Cao, C., Ren, J., Huang, L., Chen, D., Ito, M., and Kufe, D. (2007) J. Biol. Chem. 282 19321–19330 [DOI] [PubMed] [Google Scholar]
- 18.Iwamura, T., Taniguchi, S., Kitamura, N., Yamanari, H., Kojima, A., Hidaka, K., Setoguchi, T., and Katsuki, T. (1992) J. Gastroenterol. Hepatol. 7 512–519 [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y. W., Kern, H. F., Mullins, T. D., Koriwchak, M. J., and Metzgar, R. S. (1989) Pancreas 4 353–362 [DOI] [PubMed] [Google Scholar]
- 20.Metzgar, R. S., Gaillard, M. T., Levine, S. J., Tuck, F. L., Bossen, E. H., and Borowitz, M. J. (1982) Cancer Res. 42 601–608 [PubMed] [Google Scholar]
- 21.McDermott, K. M., Crocker, P. R., Harris, A., Burdick, M. D., Hinoda, Y., Hayashi, T., Imai, K., and Hollingsworth, M. A. (2001) Int. J. Cancer 94 783–791 [DOI] [PubMed] [Google Scholar]
- 22.Rae, J. M., Creighton, C. J., Meck, J. M., Haddad, B. R., and Johnson, M. D. (2007) Breast Cancer Res. Treat. 104 13–19 [DOI] [PubMed] [Google Scholar]
- 23.Lacroix, M. (2008) Cancer Chemother. Pharmacol., in press
- 24.Thompson, E. J., Shanmugam, K., Hattrup, C. L., Kotlarczyk, K. L., Gutierrez, A., Bradley, J. M., Mukherjee, P., and Gendler, S. J. (2006) Mol. Cancer Res. 4 489–497 [DOI] [PubMed] [Google Scholar]
- 25.Graeve, L., Drickamer, K., and Rodriguez-Boulan, E. (1989) J. Cell Biol. 109 2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs, B., Inwards, C. Y., and Janknecht, R. (2003) FEBS Lett. 553 104–108 [DOI] [PubMed] [Google Scholar]
- 27.Batra, S. K., Kern, H. F., Worlock, A. J., Metzgar, R. S., and Hollingsworth, M. A. (1991) J. Cell Sci. 100 841–849 [DOI] [PubMed] [Google Scholar]
- 28.Beum, P. V., Singh, J., Burdick, M., Hollingsworth, M. A., and Cheng, P. W. (1999) J. Biol. Chem. 274 24641–24648 [DOI] [PubMed] [Google Scholar]
- 29.Kohlgraf, K. G., Gawron, A. J., Higashi, M., Meza, J. L., Burdick, M. D., Kitajima, S., Kelly, D. L., Caffrey, T. C., and Hollingsworth, M. A. (2003) Cancer Res. 63 5011–5020 [PubMed] [Google Scholar]
- 30.Satoh, S., Hinoda, Y., Hayashi, T., Burdick, M. D., Imai, K., and Hollingsworth, M. A. (2000) Int. J. Cancer 88 507–518 [DOI] [PubMed] [Google Scholar]
- 31.Jinnin, M., Ihn, H., Mimura, Y., Asano, Y., Yamane, K., and Tamaki, K. (2005) Nucleic Acids Res. 33 3540–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jinnin, M., Ihn, H., Mimura, Y., Asano, Y., Yamane, K., and Tamaki, K. (2005) J. Investig. Dermatol. 124 324–330 [DOI] [PubMed] [Google Scholar]
- 33.Wei, X., Xu, H., and Kufe, D. (2005) Cancer Cell 7 167–178 [DOI] [PubMed] [Google Scholar]
- 34.Sun, Y., Sun, Y., Wenger, L., Rutter, J. L., Brinckerhoff, C. E., and Cheung, H. S. (1999) Ann. N. Y. Acad. Sci. 878 638–641 [DOI] [PubMed] [Google Scholar]
- 35.Sun, Y., Sun, Y., Wenger, L., Rutter, J. L., Brinckerhoff, C. E., and Cheung, H. S. (1999) J. Biol. Chem. 274 11535–11540 [DOI] [PubMed] [Google Scholar]
- 36.Sun, Y., Zeng, X. R., Wenger, L., Firestein, G. S., and Cheung, H. S. (2004) J. Cell. Biochem. 92 258–269 [DOI] [PubMed] [Google Scholar]
- 37.Bian, J., and Sun, Y. (1997) Mol. Cell. Biol. 17 6330–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tishler, R. B., Calderwood, S. K., Coleman, C. N., and Price, B. D. (1993) Cancer Res. 53 S2212–S2216 [PubMed] [Google Scholar]
- 39.Higuchi, T., Orita, T., Katsuya, K., Yamasaki, Y., Akiyama, K., Li, H., Yamamoto, T., Saito, Y., and Nakamura, M. (2004) Mol. Cell. Biol. 24 7456–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furukawa, T., Duguid, W. P., Kobari, M., Matsuno, S., and Tsao, M. S. (1995) Am. J. Pathol. 147 889–895 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.