Abstract
Quantitation of protein abundance is a vital component in the proteomic analysis of biological systems, which can be achieved by differential stable isotopic labeling. To analyze tissue-derived samples, the isotopic labeling can be performed using chemical labeling of the peptides post-digestion. Standard chemical labeling procedures often require many manual sample handling steps, reducing the accuracy of measurements. Here, we describe a fully automated, online (in nanoLC columns), labeling procedure, which allows protein quantitation using differential isotopic dimethyl labeling of peptide N termini and lysine residues. We show that the method allows reliable quantitation over a wide dynamic range and can be used to quantify differential protein abundances in lysates and, more targeted, differences in composition between purified protein complexes. We apply the method to determine the differences in composition between bovine liver and spleen 20 S core proteasome complexes. We find that although all catalytically active immunoproteasome subunits were up-regulated in spleen (compared with liver), only one of the normal catalytic subunits was down-regulated, suggesting that the tissue-specific immunoproteasome assembly is more diverse than previously assumed.
Comparative quantitation of proteins between biological samples is an important endeavor in the identification of proteins that play specific roles in biological pathways or diseases. In the past, the most common way to analyze and compare whole proteomes was by two-dimensional polyacrylamide gel electrophoresis (PAGE), with quantitative information being gleaned from the intensity of the spots observed after staining the separated proteins in the gel. Perhaps the most common quantitation methods in proteomics in use today are based on the application of differential stable isotopic labeling of protein samples (using for example 2H, 13C, 15N, or 18O), combined with detection by mass spectrometry, allowing both identification and quantitation of the sample components (1, 2). There are two main distinguishable methods commonly used for stable isotopic labeling of samples. The first is the incorporation of stable isotopes in proteins by supplying these isotopes to the growth media consumed and metabolized by cells or smaller organisms, generally termed metabolic labeling. The isotope label is then either incorporated as the single carbon or nitrogen source (3) or incorporated via specific auxotrophic amino acids that contain isotopes, called SILAC (stable-isotope labeled amino acids in cell culture) (4). The second method involves isotope labeling of isolated proteins or peptides with chemically identical tags that are isotopomers. The latter method is particularly advantageous for human or animal tissue samples where metabolic-based incorporation cannot easily be achieved (1).
Many approaches have been described for the chemical labeling of proteins and peptides to allow quantitation (1). These include the labeling of free cysteines in proteins by ICAT (isotope-coded affinity tagging) (5) and the labeling of free amines in the peptides obtained after protein digestion using N-hydroxysuccinimide esters like those used in the iTRAQ (isobaric Tagging for Relative and Absolute Quantification) approach (6). One of the most efficient reactions for chemical labeling of peptides is the specific dimethylation of free amines (peptide N termini and Lysine residues) by reductive amination using formaldehyde and cyanoborohydride (7). In this reaction, which is performed in near neutral conditions (between pH 6 and pH 8.5), the primary amines react with formaldehyde to create a Schiff base, which is then reduced by the cyanoborohydride. The label causes a mass increase of 28 Da per primary amine for regular formaldehyde and a mass increase of 32 Da when deuterated formaldehyde is used. The reaction is fast, does not create spurious side products, and does not adversely affect the identification of the peptides from the MS/MS spectra.
As all chemical labeling procedures are sensitive to small deviations in the reaction conditions and to human error in the handling of samples prior to and during labeling events, accuracy and reproducibility of quantitative experiments can benefit from automated labeling procedures. Here, we present a fully automated, online, and on-column sequential stable isotope labeling procedure based on the dimethylation of primary amines. We show that on-column double labeling, which we use in combination with high resolution MS/MS analysis (8, 9), is as sensitive as offline labeling, whereas the overall on-column procedure is more efficient and open to automation. The method is validated using model samples of varying complexity and can be used both to analyze protein levels in whole lysates as well as in specific proteins or in protein complexes. Finally, we apply the method to determine quantitatively differences in composition between proteasomes purified from different tissues.
MATERIALS AND METHODS
NanoLC-MS/MS—
All analyses were performed on nanoLC-LTQ-Orbitrap at a resolution of 60,000 or nanoLC-LTQ-FTICR at a resolution of 100,000. (Thermo, San Jose, CA) mass spectrometers. For nanoLC, Agilent 1100 series LC systems were equipped with 20-mm Aqua C18 (Phenomenex, Torrance, CA) trapping columns (packed in-house, i.d.,1 100 μm; resin, 5 μm) and 250 mm ReproSil-Pur C18-AQ (Dr. Maisch GmbH, Ammerbuch) analytical columns (packed in-house, i.d., 50 μm; resin, 3 μm). Solvents used where 0.6% HAc (buffer A) and 0.6% HAc/80% acetonitrile (ACN) (buffer B). Trapping was performed at 5 μl/min for 10 min, and elution was achieved with a gradient of 0–45% B in 45 min, 45–100% B in 1 min, 100% B for 4 min The flow rate was passively split from 0.36 ml/min to 100 nL/min. Nanospray was achieved using a distally coated fused silica emitter (New Objective, Cambridge, MA) (outer diameter, 360 μm; i.d., 20 μm, tip i.d. 10 μm) biased to 1.8 kV. The mass spectrometer was operated in the data-dependent mode to automatically switch between MS and MS/MS. Survey full scan MS spectra were acquired from m/z 350 to m/z 1500, and the three most intense ions were fragmented in the linear ion trap using collisionally induced dissociation. The target ion setting was 5e5 for the Orbitrap, with a maximum fill-time of 250 ms and 1e6 for the FTICR, with a maximum fill-time of 250 ms. Fragment ion spectra were acquired in the LTQ with a target ion setting of 3e4 and a maximum fill-time of 500 ms. Dynamic exclusion for selected precursor ions was set at 30 s.
Online Labeling—
All online labeling steps were performed at a flow speed of 5 μl/min with a continuous flow of buffer A. All reagents were injected with the autosampler using short injection programs of 5–20 min, depending on the injected volume. After loading the trap column with the first sample in 5% formic acid (FA), light isotope labeling was performed by flushing the trap column with 40 μl of 0.04% formaldehyde and 6 mm cyanoborohydride in 50 mm sodium phosphate buffer, pH 7.5. After washing the trap column with 10 μl of 5% FA, the second sample was loaded. Heavy isotope labeling was then performed using 40 μl of 0.04% deuterated formaldehyde and 6 mm cyanoborohydride in 50 mm sodium phosphate buffer, pH 7.5. Finally, after washing with 40 μl of 5% FA, regular LC/MS analysis was performed as described above.
Proteasome Purification—
Proteasome samples were purified from bovine liver and spleen. The purification was monitored in each step using fluorescent probes for proteasome activity, followed by SDS-PAGE as has been described before (10). After homogenization of the tissues in phosphate buffered saline, the extracts were clarified by centrifugation (27000 × g, 4 °C, 50 min.). After precipitating unwanted proteins using 40% saturated ammonium sulfate and centrifugation, the proteasome-containing fraction was precipitated by increasing the ammonium sulfate concentration to 60% saturation. After centrifugation, precipitated proteins were dissolved in 10 mm Tris-HCl (pH 7.4) and dialyzed against the same buffer to remove all ammonium sulfate. The dialyzed solutions were further purified using a 10–40% sucrose gradient at 28,000 rpm at 4 °C for 16 h. Fractions containing liver and spleen proteasome activity were pooled for DEAE (diethylaminoethyl) separation (DEAE-Sephadex A25 and DEAE-Sephacel resin, respectively). After incubating the proteins with the DEAE resin for 30–60 min (80 mm potassium acetate buffer containing 20 mm Tris-HCl, pH 7.2, and 5 mm magnesium acetate for the Sephadex resin and 10 mm Tris-HCl, pH 8.0 containing 1 mm EDTA for the Sephacel resin), the resin was washed, and proteins were eluted using and increasing NaCl concentrations. All proteasome-containing fractions were pooled and concentrated by means of a nitrogen filter and protein concentrations determined using the Bradford assay (Bio-Rad). Proteasome preparation were stored at −80 °C. To check the purity of the proteasome preparations, samples were freeze-dried, boiled in reducing sample buffer, and analyzed by 12.5% SDS-PAGE using a PROTEAN II xi Cell system (Bio-Rad) followed by Coomassie Brilliant Blue staining. To visualize the active subunit composition of liver and spleen proteasome samples, 1 μg of proteasome was incubated with 500 nm of probe bodipyH (a close analog of the bodipyFL-probe described previously) for 1 h at 37 °C. Proteins were denatured by being boiled in reducing sample buffer and analyzed by 12% SDS-PAGE using the NuPAGE pre-cast gel system (Invitrogen). The gel was then scanned for fluorescence emission using a ProXPRESS two-dimensional Proteomic imaging system (Perkin Elmer).
Trypsin Digestion—
Whole tissue lysates were separated by 12% SDS-PAGE followed by staining with Coomassie Brilliant Blue. Gel lanes were cut into slices, which were washed with MilliQ and 0.6% HAc/80% acetonitrile (ACN). Prior to in-gel digestion, proteins were reduced with 1,4-dithiothreitol (6.5 mm) and alkylated with iodoacetamide reagent (54 mm). After thorough washing, pieces were rehydrated in trypsin solution (10 ng/μl) on ice. After addition of 30 ml of NH4HCO3 (50 mm, pH 8.5), samples were digested for 16 h at 37 °C. Supernatant of the digest was collected. The gel pieces were washed for 30 min in 5% formic acid at room temperature, after which the supernatant of this washing step was combined with the earlier fraction and stored at −30 C until the analysis. Purified proteins and proteins complexes were digested in-solution after reduction and alkylation. Digestion was performed using 50 mm NH4HCO3 at 37 °C for 16 h, with a protein/trypsin ratio of 1:100 by weight. Prior to injection, samples were diluted in MilliQ water containing 5% formic acid.
Data Processing and Analysis—
Raw MS data were converted to peak lists using Bioworks Browser software, version 3.1.1. For protein identification, MS/MS data were compared with the International Protein Index murine (v3.36, 51424 entries searched) or bovine (v3.22, 32915 entries searched) data bases (depending on the sample analyzed) using Mascot Version 2.1 (Matrix Science) with trypsin at the enzyme, allowing 1 missed cleavage. Precursor and fragment mass tolerances were set at 6 ppm and 0.9 Da deviation, respectively. As fixed modifications, carbamidomethylation (Cys) and dimethylation (Lys, N-terminal) were set as well as the following variable modifications: dimethylation: 2H(4) (Lys, N-terminal) and Oxidation (Met). Proteasome proteins were identified with a minimal Mascot protein score of 48 using at least two identified peptides with a Mascot ion score of >35 and an expect value of <0.005. At these settings, the false discovery rate was 0.68% as determined using a decoy data base. The only exception was the protein PSME2, which was identified with only one peptide (Mascot peptide score 48) fulfilling those criteria. The Mascot MS/MS annotation of that peptide is supplied in Fig. S5. Relative peptide abundance was calculated from extracted ion chromatograms of the different isotopic variants using MS-Quant software (11). Very low intensity peptides (extracted ion chromatogram intensity < 3000) as well as peptides with a poor extracted ion chromatogram or MS/MS spectrum were excluded after manual inspection. The program StatQuant2 was used to normalize the quantitation and calculate standard deviations of the log 2 of the ratios of all quantified peptides per protein. Where appropriate, quotation experiments were repeated with the light and heavy labels switched. All quantitation results shown are listed in tables S1–S4. All identification results have been uploaded to PRIDE (under project title “Sequential Labeling for Protein Quantitation”) and identification details for the proteasome proteins are supplied in table S5.
RESULTS
Online Sequential Labeling—
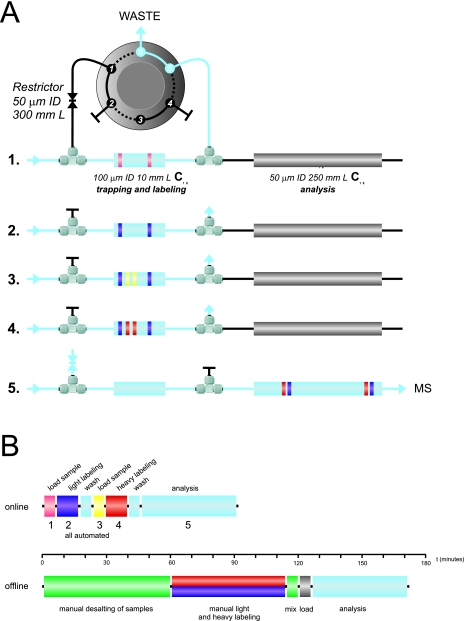
To test the feasibility of performing the dimethylation reaction on-column, we designed an online reaction protocol consisting of 5 main steps, all of which were performed on our regular nanoLC system, consisting of a six-port switching valve, a 100-μm i.d. trapping column, and a 50 μm i.d. analytical column (Fig. 1A) (13). During sample loading and online labeling (steps 1 to 4), the restrictor is closed and the flow speed is 5 μl/min. The first step is the loading of the first peptide sample onto the trapping column, which is then chemically labeled by flushing the trapping column with 40 μl of light labeling reagent (CH2O/NaBH3CN) in the second step. After a short wash with 5% formic acid, the second peptide sample is loaded onto the trapping column in step three followed by chemical labeling of that sample in step four when the column is flushed with the heavy labeling reagent (CD2O/NaBH3CN). Finally, again after a short wash with 5% FA, in the fifth step the valve switches, and the flow speed is increased to ∼400 μl/min. This results in a pressure of ∼150 bar and an effective flow of ∼100 nl/min for a regular LC-MS analytical gradient to analyze all labeled peptides. The minimal time for the protocol is around 90 min, compared with about 170 min for an offline labeling (including desalting of the sample) and LC-MS analysis (Fig. 1B). As the procedure is fully automated, no handling of the samples is required after digestion and loading the samples in the autosampler. In the offline method, several additional manual steps are required, including drying and mixing the samples, steps that are sensitive to sample loss and human error, respectively.
Fig. 1.
A, schematic showing the LC-MS setup and the main steps involved in the sequential online stable isotope labeling of two samples and subsequent LC-MS/MS analysis. Initially, the six-port valve closes the restrictor, directing the flow via the trapping column to the waste. In step 1, the first peptide sample (pink) is loaded onto the trapping column. The peptides now retained by the trapping column are chemically labeled in step 2 (purple) by flushing the trapping column with labeling reagent. Following a short wash, the second peptide mixture (orange) is loaded on the trapping column in step 3. During step 4, the trapping column is flushed with the second labeling reagent, containing the heavy isotope, labeling all peptides from the second sample (purple). After this step, the valve switches (opening the restrictor) and the flow speed increases, thus allowing flow over the analytical column to the mass spectrometer to perform a regular LC-MS analysis of all peptides. B, the estimated time involved in all of the steps, both automated online (top) and manual offline (bottom) procedures, with color coding taken from A. In green are the steps that are not required in the online method. Indicated in the middle is a time-scale providing an indication of the time involved.
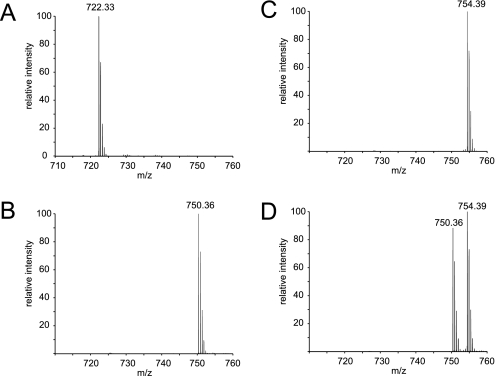
When performing a sequential labeling online it is of vital importance that the reaction goes to completion as otherwise the second label will also partially label the first sample, causing inaccurate quantitation. To test this, we first performed the online double labeling as described above, but although only injecting actual digested protein sample (bovine serum albumin (BSA), 100 fmol) in either the first (light) or the second (heavy) labeling step, though still performing the labeling in both steps. Fig. 2 shows the averaged mass spectrum of one of the trypic peptides of BSA, YICDNQDTISSK (amino acids 286–297), which is detected as a doubly charged peptide ion at an m/z value of 722.33 when cysteine residues are carbamidomethylated. These spectra are averaged over the complete retention time span of the unmethylated and the fully methylated version of the peptide, to be able to see any intermediates. In Fig. 2A, the unmodified peptide is seen from a regular LC-MS analysis. After dimethylation, the m/z of this peptide is expected to be 750 for the light label and 754 for the heavy label, as the peptide is dimethylated at both the N terminus as well as the C-terminal lysine residue. As can be seen for the light and heavy label in Fig. 2, B and C, respectively, the online reaction goes to completion because no input peptide or any intermediates are visible after labeling. Fig. 2D shows the same mass spectrum but now for a double labeling (two times 100 fmol) in which both the light and heavy labeled peptide are visible, in a ratio very close to 1:1. More examples of BSA peptides before and after double labeling can be found in supplementary Fig. S1. We also analyzed this sample with a Mascot search with all dimethylation events set as variable modifications to determine the extent of identification of partially dimethylated peptides. The result of that search can be found in Supplementary Fig. S2 and showed that at a Mascot ion score cutoff of 25, 98% of all identified spectra were dimethylated. Of the 157 identified spectra, four were not fully methylated and only one contained mixed light and heavy dimethylation. Because of the 30 s dynamic exclusion set in the mass spectrometer, these results strongly over-represent low intensity peptides, meaning the actual labeling percentage is much higher, as indicated in Fig. 2. To check for recovery of peptides after the extended washing steps, we compared an online double labeling run of two times 20 fmol tryptic BSA with a subsequent normal LC-MS run of 20 fmol tryptic BSA. The observed signal intensities were similar, showing a near 100% recovery of the peptides following isotopic labeling. This behavior was typical and observed for most tryptic peptides analyzed in this way.
Fig. 2.
Mass spectra of the doubly charged BSA peptide YICDNQDTISSK (amino acids 286–297) in its original form (panel A, calculated mono-isotopic mass 722.33) and after online labeling in either the light labeling step (panel B, calculated mono-isotopic mass 750.36), the heavy labeling step (panel C, calculated mono-isotopic mass 754.38), or both (panel D). All mass spectra are zoomed in on m/z range 710 to 760 and are averaged over the range in the chromatogram that includes the elution times of all three m/z values.
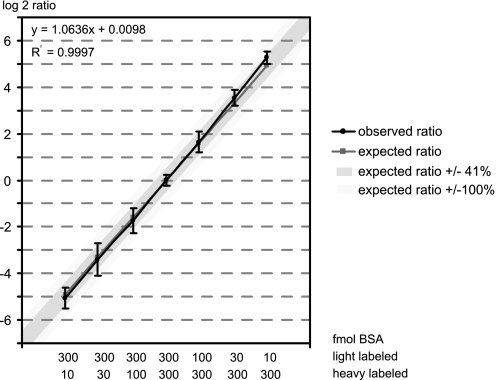
To test the dynamic range of the online labeling, different amounts of tryptic BSA peptides were loaded in the first and second sample loading step, varying between ratios of 30:1 to 1:30. Following the LC-MS analysis of the labeling, the ratio of BSA between the light and heavy label was determined from the average of the ten most intense peptide pairs using MS-Quant. As can be seen in Fig. 3, the method is linear in the base 2 log of the ratios and accurate over a wide dynamic range (the correlation between expected and observed ratios has an R2 of 0.9997). The average ratios for all combinations tested fell within a 0.5 deviation on a base 2 log scale, which equates to a variation of ±41%. The standard deviations calculated from the log 2 ratios of each set of 10 peptides tend to be larger when the second (heavy) sample contains less peptide compared with the first (light) sample. This shows that with this method ratios can be reliable calculated as low as for 1.5-fold changes, although usually a 2-fold change (100% variation) would be used as a cut-off in these types of experiments.
Fig. 3.
Linearity of the online double stable isotope labeling procedure. Plotted is the correlation between the expected (x axis) and determined (y axis) ratio for several online labeling experiments involving different amounts of BSA, as indicated below the graph. The ratios are calculated from the 10 most intense peptide peak pairs of each analysis. Plotted are the log 2 values of the actual ratios, with the pink line (squares) indicating the expected ratios and the blue line (circles) the observed ratios. The dark blue shading indicates the spread of 0.5 log units from the expected value and the light blue shading a spread of 1 log unit. Bars indicate the standard deviation between the ten quantified peptides for each sample.
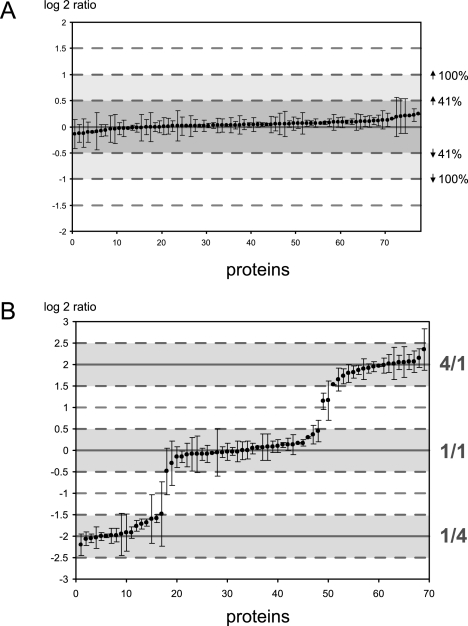
Of course, regular experiments would involve a large number of different proteins, with varying numbers of peptides that can be used for quantitation. To examine the behavior of the method under more complex conditions, a mouse lung tissue extract was run on a SDS-PAGE gel, cut into several bands, and the proteins present in the gel bands were subjected to in-gel tryptic digestion. These tryptic peptides were then first applied in equal amounts for heavy and light labeling, allowing us to determine the normal variation in the determined ratios introduced by the method. As can be seen in Fig. 4A, we quantified just under 80 proteins, from three gel bands, with at least two peptides per protein. Similar to the quantitation of tryptic BSA, the variation of the average ratios did not exceed ±41%, and the standard deviation of the ratios observed for different peptides of the same protein was small. A “rough check” for the efficiency of labeling was performed via a Mascot search with all dimethylation events set as variable modifications. In this experiment 98% of all identified peptides were fully labeled, 1.6% were partially labeled, and 0.4% had mixed light and heavy labeling at a Mascot ion score cutoff of 25. The result of the search can be found in supplementary Fig. S3 as well as in PRIDE (see “Materials and Methods” for details). Next, we mixed the gel bands in three different ratios (4:1, 1:1, and 1:4) and repeated the procedure. Fig. 4B shows that the quantified proteins nicely grouped into three ratios. With three exceptions, most likely caused by proteins being present in multiple slices of the gel (see “Discussion”), all proteins were again quantified within ±0.5 of the expected log 2 ratio (corresponding to ±41% of the actual ratios).
Fig. 4.
Quantified ratios from mouse lung tissue proteins. Indicated on the x axis are the number of proteins that were identified from several gel bands that were mixed either all in a 1:1 ratio (panel A) or with several gel bands mixed in different ratios (panel B). Ratios are calculated for all proteins from which at least two peptides were found that could be quantified and are plotted as the log 2 of the determined ratio. Shaded in blue, red, and green are the spread of 0.5 log units from the expected 1:1, 4:1, and 1:4 ratios, respectively. Bars indicate the standard deviation between the quantified peptides for each protein.
Compositional Analysis of Protein Complexes—
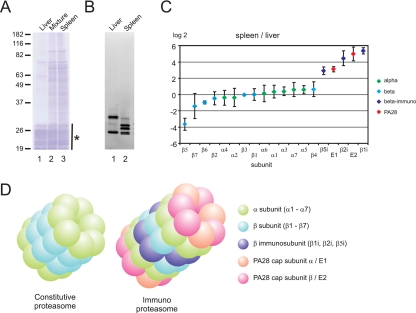
To test whether the method could be applied to analyze the differential composition of protein complexes, we purified the 20 S core of the proteasome from bovine liver and spleen samples, using ammonium sulfate precipitation, sucrose gradient centrifugation, and DEAE separation (see “Materials and Methods”). Following this procedure, relatively pure proteasome samples were obtained from both tissues, as evidenced by SDS-PAGE and Coomassie Brilliant Blue staining of the purification (Fig. 5A). To determine the presence and activity of catalytically active (β) subunits, the purified proteasomes were first incubated with a fluorescent probe followed by SDS-PAGE separation and fluorescence imaging. This revealed that both proteasome purifications contained a number of active (β) subunits, but also that there were significant differences in composition between these two proteasome samples (Fig. 5B). Most likely, these differences are caused by differences in the amount of constitutive proteasome and immunoproteasome between bovine liver and spleen samples, in agreement with previously reported data for mouse spleen and liver proteasomes (10). After in-solution tryptic digestion, we used the online dimethylation method to further characterize and quantify these differences, with the liver proteasome being light labeled and the spleen proteasome being heavy labeled. All Mascot identified proteasome components in the sample were quantified using MS-Quant, and the data set was normalized on the median of the average of all α proteasome subunits, as these were expected not to change between the two proteasome samples. In Fig. 5C, the base 2 logarithms of the measured ratios (intensity of the extracted ion chromatogram for the heavy labeled peptide divided by that of the light peptide) are plotted for all proteasome subunits. When using a cutoff value of 100% up- or down-regulation (which equals 1 unit on the log 2 scale), two proteins of the 20 S proteasome core were more abundant in the proteasome purification from liver (subunits β5 and β7). Three core proteins were more abundant in the spleen sample (β5i, β2I, and β1i), corresponding exactly to the three catalytically active immunoproteasome subunits. In addition, two other 20 S proteasome-associated proteins (PSME1 and PSME2) were found more abundantly in the spleen proteasome, which together can form an alternative cap of the proteasome that facilitates degradation of peptides for major histocompatibility complex (MHC) presentation (14), called the PA28 (proteasome activator) or REG complex. Fig. 5D displays a structural model of the differences between the regular constitutive proteasome and the immunoproteasome associated with the PA28 cap complex. To verify these results, the experiment was repeated with this time, the liver proteasome being heavy labeled and the spleen proteasome being light labeled. Although small differences were observed in the ratios calculated for the subunits having a log 2 ratio around 0, the same proteins were identified as being up- or down-regulated in the spleen sample, as compared with the liver proteasome (data not shown).
Fig. 5.
Quantitation of differences between purified bovine liver and spleen proteasome complexes. A, the purified proteasome samples are shown after SDS-PAGE, stained using Coomassie Brilliant Blue. Lane 1 shows the purified liver proteasome, lane 2 the mixture of both proteasome samples, and lane 3 the purified spleen proteasome. The location of the proteasome proteins on the gel is indicated with an asterisk. B, the same proteasome purifications of liver (lane 1) and spleen (lane 2), but now stained using a fluorescent probe for catalytically active subunits. C, the relative quantitation of all Mascot identified proteasome subunits, with α-subunits indicated in green, β-subunits indicated in light blue (normal subunits), or dark blue (immunosubunits) and PA28 subunits indicated in orange. Bars indicate the standard deviation between the quantified peptides for each protein. D, the structure of the constitutive proteasome core and of the immunoproteasome core with the PA28 cap complex attached.
DISCUSSION
Quantitative proteomics by using stable isotope labeling in combination with mass spectrometry is an extensively used technique for the comparison of protein abundance in complex biological samples. To analyze tissue-derived samples, chemical labeling after digestion is the most often used method to determine relative quantities of peptides and proteins in these samples. As the efficiency of chemical labeling of samples is often sensitive to small (manual) errors and differences in sample preparation and handling, automation of the labeling process can reduce these errors and allow more reproducible results. However, care should still be taken in sample preparation to avoid human error prior to isotopic labeling as is the case with any other derivatization method.
Online Labeling Method—
Here, we present a method for online, on-column, sequential derivatization of peptides, allowing relative quantitation of the peptides, by dimethylation using cyanoborohydride and either regular or deuterated formaldehyde (7). The online quantitation procedure provided reliable quantitation over a wide dynamic range and allowed us to quantify differences between proteasome complexes purified from different tissue samples.
Online derivatization of peptides has been demonstrated before to enhance the fragmentation of the peptides in tandem mass spectrometry (15), but for online sequential labeling, it is very important that the labeling reaction goes to completion to prevent bleeding of the samples. In our method, no intermediates or unmodified peptides were present at detectable levels, as demonstrated in Fig. 2. Also, to prevent preferred binding of the first sample, the amount of peptide loaded on the trap column must be well below the capacity of the column. Therefore the method is especially suitable for analyzing differences between single proteins or purified protein complexes, where the total amount of protein is unlikely to exceed the capacity of the column. When analyzing a mixture of ratios generated from bands cut from a gel containing mouse lung extract, we noticed a few proteins with unexpected ratios. Because some proteins might be present in multiple isoforms, with varying molecular weights, they might be present in more than one of the bands that were used for this experiments, causing the ratio to shift away from the expected value. This will not be a significant problem, however, as in most applications either in-solution digests or similar gel bands, but from different samples, will be compared. Evidently, in-solution digests will not resolve isoforms of the proteins (16), but this would lead to inter-protein differences of quantified peptides rather than shifted ratios.
When automating the online labeling method, the stability of the reagent is another important issue. Using a colorimetric assay to determine cyanoborohydride concentrations (17) and by performing identical runs over a prolonged period of time, we found that the mixture of formaldehyde and cyanoborohydride was stable over at least 24 h when stored at 4 °C (supplemental Fig. S4), whereas many other reagents used for chemical labeling (like iTRAQ) often required the reagent to be mixed immediately prior to the reaction. This stability makes the dimethylation reaction very suitable for the online labeling procedure, as it allows automated labeling and analyses of the samples to take place overnight or even longer period of time. Although the throughput speed of our method is higher than offline labeling when two samples are compared (Fig. 1), offline labeling gets more favorable when larger numbers of samples have to be analyzed, as they can then all be labeled at the same time, rather than sequentially.
Compositional Analysis of Tissue-specific Proteasomes—
After comparing the 20 S core proteasomes purified from bovine liver and spleen we observed that proteasome subunit β5 was significantly less abundant in spleen and to a lesser extent also β7 (although the standard deviation for the latter was high). The immunoproteasome subunits β1i, β2I, and β5i were all more abundantly present in the spleen-derived proteasome preparation. These differences between the proteasome complexes purified from bovine liver and spleen can be partially explained by the high abundance of immunoproteasome complexes in spleen, compared with liver, in full agreement with previous observations (10). Still, it is remarkable that a single catalytic constitutive proteasome subunit is less abundant in the spleen-derived proteasome sample. This observation suggests that not only immunoproteasome complexes exist in which all constitutive active subunits are replaced by their immunoproteasome counterparts, but that “hybrid” proteasome complexes are present as well, in which only one of the β-subunits has been replaced by an immunosubunit. Such hybrid proteasome complexes have been suggested before, but their exact composition remains so far unclear (18). Also, because only β5 was relatively less abundant, and all three immunosubunits were more abundant, it cannot be excluded that β5 might be replaced by either β1i, β2I, or β5i in such a hybrid proteasome species.
Next to immunoproteasome subunits, we also found PSME1 and PSME2 to be almost uniquely present in the bovine spleen purified proteasomes. Together, these two highly homologous proteins form the alternative PA28 cap (also called 11 S regulator) of the 20 S proteasome, a heptameric ring containing most likely 4 subunits of PSME2 and 3 of PSME1 (14, 19). The PA28 cap can replace the 19 S regulator and is, like the catalytically active immunosubunits, up-regulated by interferon-γ (20). The fact that we found the 11 S complex in the proteasome purified from spleen, but did not find any subunits of the 19 S complex, might suggest that the 11 S regulator is more tightly bound to the proteasome core as the 19 S cap is known to dissociate under the isolation conditions.
CONCLUSION
Here, we presented an online, sequential labeling procedure using differential stable isotopic dimethlylation of N termini and lysine residues. The method allows automated sequential labeling and LC-MS/MS analysis of complex biological samples. We validated the method on model proteins and a whole cell lysate and identified the differences in composition between proteasome complexes purified from bovine liver and spleen. The method is efficient, performs similar to other chemical labeling methods, and can be generally applied. The method is especially suitable for the quantitative analysis of gel bands or purified protein complexes, and its full automation permits easier analysis of large sample numbers with a greatly reduced chance of inaccuracy due to human error.
Supplementary Material
Acknowledgments
We thank B. Rodenko and J. Neefjes (Netherlands Cancer Institute, Amsterdam, The Netherlands) and T. van de Goor and K. Kraiczek (Agilent Technologies, Waldbronn, Germany) for advice.
Footnotes
Published, MCP Papers in Press, June 4, 2008, DOI 10.1074/mcp.M800093-MCP200
The abbreviations used are: i.d., inner diameter; FA, formic acid; BSA, bovine serum albumin; PA, proteasome activator.
B. van Breukelen, H. van den Toorn, M. Drugan, and A. J. R. Heck, in preparation.
This work was supported by the Netherlands Proteomics Centre, Dutch Cancer Society (Koningin Wilhelmina Fonds) Grant 2005-3368 and the Agilent Technologies Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S5.
REFERENCES
- 1.Julka, S., and Regnier, F. E. ( 2005) Recent advancements in differential proteomics based on stable isotope coding. Brief. Funct. Genomic. Proteomic. 4, 158–177 [DOI] [PubMed] [Google Scholar]
- 2.Heck, A. J., and Krijgsveld, J. ( 2004) Mass spectrometry-based quantitative proteomics. Expert Rev. Proteomics 1, 317–326 [DOI] [PubMed] [Google Scholar]
- 3.Krijgsveld, J., Ketting, R. F., Mahmoudi, T., Johansen, J., Artal-Sanz, M., Verrijzer, C. P., Plasterk, R. H., and Heck, A. J. ( 2003) Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat. Biotechnol. 21, 927–931 [DOI] [PubMed] [Google Scholar]
- 4.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 5.Shiio, Y., and Aebersold, R. ( 2006) Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat. Protoc. 1, 139–145 [DOI] [PubMed] [Google Scholar]
- 6.Ross, P. L., Huang, Y. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., Khainovski, N., Pillai, S., Dey, S., Daniels, S., Purkayastha, S., Juhasz, P., Martin, S., Bartlet-Jones, M., He, F., Jacobson, A., and Pappin, D. J. ( 2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 7.Hsu, J. L., Huang, S. Y., Chow, N. H., and Chen, S. H. ( 2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal. Chem. 75, 6843–6852 [DOI] [PubMed] [Google Scholar]
- 8.Makarov, A., Denisov, E., Kholomeev, A., Balschun, W., Lange, O., Strupat, K., and Horning, S. ( 2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 9.Peterman, S. M., Dufresne, C. P., and Horning, S. ( 2005) The use of a hybrid linear trap/FT-ICR mass spectrometer for on-line high resolution/high mass accuracy bottom-up sequencing. J. Biomol. Tech. 16, 112–124 [PMC free article] [PubMed] [Google Scholar]
- 10.Berkers, C. R., van Leeuwen, F. W., Groothuis, T. A., Peperzak, V., van Tilburg, E. W., Borst, J., Neefjes, J. J., and Ovaa, H. ( 2007) Profiling proteasome activity in tissue with fluorescent probes. Mol. Pharm. 4, 739–748 [DOI] [PubMed] [Google Scholar]
- 11.Schulze, W. X., and Mann, M. ( 2004) A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 [DOI] [PubMed] [Google Scholar]
- 12.Deleted in proof
- 13.Meiring, H. D., van der Heeft, E., ten Hove, G. J., and de Jong, A. P. J. M. ( 2002) Nanoscale LC-MS(n): technical design and applications to peptide and protein analysis. J. Sep. Sci. 25, 557–568 [Google Scholar]
- 14.Zhang, Z., Krutchinsky, A., Endicott, S., Realini, C., Rechsteiner, M., and Standing, K. G. ( 1999) Proteasome activator 11S REG or PA28: recombinant REG alpha/REG beta hetero-oligomers are heptamers. Biochemistry 38, 5651–5658 [DOI] [PubMed] [Google Scholar]
- 15.Cardenas, M. S., van der Heeft, E., and de Jong, A. P. ( 1997) On-line derivatization of peptides for improved sequence analysis by micro-column liquid chromatography coupled with electrospray ionization-tandem mass spectrometry. Rapid Commun. Mass Spectrom. 11, 1271–1278 [DOI] [PubMed] [Google Scholar]
- 16.Kolkman, A., Dirksen, E. H., Slijper, M., and Heck, A. J. ( 2005) Double standards in quantitative proteomics: direct comparative assessment of difference in gel electrophoresis and metabolic stable isotope labeling. Mol. Cell. Proteomics 4, 255–266 [DOI] [PubMed] [Google Scholar]
- 17.Sorensen, K. ( 1994) Coomassie protein assay reagent used for quantitative determination of sodium cyanoborohydride (NaCNBH3). Anal. Biochem. 218, 231–233 [DOI] [PubMed] [Google Scholar]
- 18.Dahlmann, B., Ruppert, T., Kloetzel, P. M., and Kuehn, L. ( 2001) Subtypes of 20 S proteasomes from skeletal muscle. Biochimie 83, 295–299 [DOI] [PubMed] [Google Scholar]
- 19.Knowlton, J. R., Johnston, S. C., Whitby, F. G., Realini, C., Zhang, Z., Rechsteiner, M., and Hill, C. P. ( 1997) Structure of the proteasome activator REGalpha (PA28alpha). Nature 390, 639–643 [DOI] [PubMed] [Google Scholar]
- 20.Tanahashi, N., Yokota, K., Ahn, J. Y., Chung, C. H., Fujiwara, T., Takahashi, E., DeMartino, G. N., Slaughter, C. A., Toyonaga, T., Yamamura, K., Shimbara, N., and Tanaka, K. ( 1997) Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells 2, 195–211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.