Abstract
Acidic fibroblast growth factor (aFGF; also known as FGF-1) is a potent neurotrophic factor that affects neuronal survival in the injured spinal cord. However, the pathological changes that occur with spinal cord injury (SCI) and the attribution to aFGF of a neuroprotective effect during SCI are still elusive. In this study, we demonstrated that rat SCI, when treated with aFGF, showed significant functional recovery as indicated by the Basso, Beattie, and Bresnahan locomotor rating scale and the combined behavior score (p < 0.01–0.001). Furthermore proteomics and bioinformatics approaches were adapted to investigate changes in the global protein profile of the damaged spinal cord tissue when experimental rats were treated either with or without aFGF at 24 h after injury. We found that 51 protein spots, resolvable by two-dimensional PAGE, had significant differential expression. Using hierarchical clustering analysis, these proteins were categorized into five major expression patterns. Noticeably proteins involved in the process of secondary injury, such as astrocyte activation (glial fibrillary acidic protein), inflammation (S100B), and scar formation (keratan sulfate proteoglycan lumican), which lead to the blocking of injured spinal cord regeneration, were down-regulated in the contusive spinal cord after treatment with aFGF. We propose that aFGF might initiate a series of biological processes to prevent or attenuate secondary injury and that this, in turn, leads to an improvement in functional recovery. Moreover the quantitative expression level of these proteins was verified by quantitative real time PCR. Furthermore we identified various potential neuroprotective protein factors that are induced by aFGF and may be involved in the spinal cord repair processes of SCI rats. Thus, our results could have a remarkable impact on clinical developments in the area of spinal cord injury therapy.
Spinal cord injury (SCI)1 is a costly disease as well as the most frequent cause of mortality and morbidity in every medical care system around the world (1, 2). Traumatic axonal injury is a primary sequela of contusive SCI and is characterized by axonal swelling and disconnection of the ascending sensory and descending motor tracts (3). Contusive SCI seems to initiate a complicated cascade of pathophysiological changes, a process called secondary injury, including fiber deformation, increased vascular permeability, local ischemia, intraneuronal edema, and local demyelination. Moreover traumatic axonal injury typically involves a disruption of the axonal membrane, and this results in up-regulation of the entry of calcium, the influx of which initiates calpain activation (4, 5) and mitochondrial injury/swelling (6), resulting in cytochrome c release and caspase activation (7). Simultaneously the proliferation and hypertrophy of astrocytes around the injury site are characterized by increased immunoreactivity to glial fibrillary acidic proteins (GFAPs) (8).
Although many studies have focused on revealing the pathophysiology of SCI, the exact molecular mechanisms and the biochemical pathways that mediate secondary injury remain sketchy (9). Spinal cord injury often causes permanent neurological deficits mostly because the injured neurons lack regenerative ability, and the series of destructive processes that follow SCI results in a second wave of cell death and spreading tissue loss (10). Therefore, studies of the stimulation of axonal regeneration and of how to enhance neuroprotection to restore functional recovery after SCI have become imperative. Limited amounts of neurotrophic factors are produced in the adult CNS after injury, and delivery of these neurotrophic factors to the injured spinal cord has been shown to stimulate neuronal survival and regeneration (11–13). These results probably indicate that there is a deficiency in these neurotrophic factors that produces the absence of spontaneous regeneration in the spinal cord.
Acidic fibroblast growth factor (aFGF; also known as FGF-1), a normal constituent of the spinal cord, is a universal neurotrophic factor that promotes survival and sprouting in various neural systems (14, 15). Within synthesizing cells, the functioning of aFGF is limited by sequestration because of a lack of an amino-terminal signal peptide that would direct its secretion. It seems to be released only when cells that sequester it are lysed after injury (16). aFGF has been shown to prevent the die back phenomenon of the corticospinal tract and to promote sprouting and regeneration using Schwann cell guidance channels or peripheral nerve grafts across the injured site after complete thoracic transection in adult rats (17, 18). Previous studies showed that peripheral nerve graft and aFGF treatment are able to induce regrowth of catecholaminergic fibers and protection of cholinergic spinal cord neurons in spinal cord-transected rats (19). Furthermore aFGF is able to stimulate the functional recovery of chronic complete idiopathic transverse myelitis (20). Previous work in our laboratory (21) and by others (22) using recombinant aFGF was able to increase the survival of neurons and increase the intrinsic capacity of mature neurons for regrowth; as a result, paraplegic rats showed remarkable functional recovery. Although a previous study has illustrated that aFGF is able to protect PC12 cells from glutamate-induced toxicity via the phosphatidylinositol 3-kinase (PI3K)/Akt kinase signaling pathway in vitro (23), the molecular intracellular signaling pathways of aFGF-induced functional recovery in injured spinal cord in vivo are still not clearly or fully understood. To further advance this area of study, it is of great importance to reveal the underlying molecular mechanisms involved in this vital functional recovery process.
Previous studies have identified acute spinal cord injury-associated gene expression patterns by microarray. These genes have been functionally categorized into several different classes, namely cell death and apoptosis, inflammation-related genes, neuronal growth and differentiation, ion transport, cytoskeleton, signal transduction, and lipid metabolism (24–28). It should be noted that mRNA expression level does not always correlate with protein expression level. Moreover spinal cord injury differs from other neurodegenerative diseases, for example Alzheimer or Parkinson disease, in that there is no direct genetic defect involved (29). Therefore, changes in gene expression level may not be the first functional stage to occur in cells after traumatic nerve injury (30–32). Thus it is quite possible that proteomics analysis will be able to detect both protein expression changes not directly involving increased mRNA expression and those that do but also identify post-translational modifications (PTMs) that occur in response to spinal cord injury (33). Thus, using a proteomics approach to analyze the global protein profile of injured spinal cords may provide specific information that is not obtainable from microarray studies.
To date, there have been only two reports involving the study of protein profiles of spinal cord injury using a proteomics approach (34, 35). However, Ding et al. (34) did not dissect the injured spinal segments from the uninjured neighboring tissues, and therefore it is unknown whether the differentially expressed proteins detected in this study were localized in injured spinal region or elsewhere. Furthermore they only investigated protein expression changes using a rat transection-injured model. In fact, most traumatic spinal cord injuries in a clinical situation are typically not transection of the cord but are contusive or compressive. Thus far, there have been no previous reports using proteomics analysis that involve the study of contusive spinal cord injury mediated by a computerized New York University impactor. Furthermore no analysis has been conducted in which aFGF administration was carried out. For that reason, we aimed to investigate the epicenter of the contusive spinal cord at the acute phase of recovery by a proteomics approach. Moreover in the test subjects, aFGF was applied to the injured epicenter before the proteomics analysis to reveal any possible protective mechanisms that are induced by aFGF after spinal cord injury.
In the present study, we showed for the first time that aFGF is able to induce functional recovery of locomotion in contusive SCI rats. Quantitative high resolution two-dimensional (2D) PAGE followed by highly accurate LC-MS/MS analysis and a bioinformatics approach was used to functionally cluster and annotate the data collected by proteomics study, and we found that expression of GFAP, S100B, keratan sulfate proteoglycan lumican (KSPG) were up-regulated after SCI and then down-regulated following aFGF treatment. These proteins are known to be involved in the early and late pathological processes of secondary injury that results in the inhibition of regeneration of the injured spinal cord. Thus, we hypothesize that aFGF may be able to attenuate secondary injury and thus ultimately lead to improved functional recovery of hind limb locomotion of SCI rats. In conclusion, our results provide for the first time a better insight into the SCI regeneration mechanisms that occur under the influence of aFGF, and this should provide useful informative background data to help devise new therapeutic approaches to reduce the effects of secondary injury after SCI.
MATERIALS AND METHODS
Animals and Surgery—
The experimental animals were divided into three groups, namely sham (n = 6), SCI (n = 8), and SCI aFGF-treated groups (n = 6). The shams (control rats) only underwent a T9 laminectomy without weight-drop injury, and the animals remained with normal locomotion of the hind limbs after surgery. SCI was induced using the weight-drop device developed by New York University (36). The animal surgical procedure was performed according to a previous report (37). Briefly female adult Sprague-Dawley rats ranging from 240 to 280 g were anesthetized and laminectomized at the T9 vertebrae level. The dorsal surface of the spinal cords was compressed by dropping a 10-g rod from a height of 50 mm. In the case of the treated rats, a 5-μl Exmire microsyringe with a 33-gauge needle was then positioned at the midline of the spinal cord 2 mm rostral and caudal to the contusive center, and 2 μg of recombinant aFGF (recombinant human FGF, acidic, R&D Systems Inc.) was injected 0.8 mm into the dorsal column of the spinal cord. aFGF was dissolved in PBS solution (1 μg/μl); therefore vehicle injection with 2 μl of PBS containing no aFGF was also performed in the control SCI group. Antibiotics were given daily by subcutaneous injection into the animals for 7 days. Their bladders were evacuated manually twice a day. Animals were sacrificed at 24 h or 28 days following surgery. The spinal cords were immediately removed, and the spinal segments 0.5 cm rostral and caudal of the contusion epicenter were dissected and stored at −80 °C for 2D PAGE, Western blotting, and quantitative real time PCR analysis. Animal handling and experimental protocols were reviewed and approved by the animal studies subcommittee of Taipei Veterans General Hospital.
Behavior Analysis—
The Basso, Beattie, and Bresnahan (BBB) open field score and the combined behavior score (CBS) were used to evaluate locomotion in terms of hind limb functional improvement of rats with spinal cord contusion (36, 38). The BBB test was scored from 0 (no observable hind limb movement) to 21 points (normal hind movement), and the scores for the CBS test ranged from 0 (normal walking) to 100 (no movement of hind limb and no weight bearing). In this study, the behavior analysis was performed at 7, 14, 21, and 28 days after injury. The behavior test process was recorded by video camera, and two examiners were blind to each behavior evaluation group. The results are presented as mean ± S.E. The two tailed Mann-Whitney test and repeated measures analysis of variance were performed to evaluate the statistical significance of the results using SPSS 11.0 software. The p value obtained from these two methods was <0.05, indicating the statistical significance of the recovery rate between the SCI rats either treated with or without aFGF.
Sample Preparation for 2D PAGE—
Spinal segments (0.5 cm) were transferred into an Eppendorf tube containing MagNA Lyser Green beads (Roche Applied Science) on ice and homogenized in 1 ml of lysis buffer containing 50 mm Tris-HCl (pH 7.5), 250 mm NaCl, 5% Nonidet P-40, and protease inhibitor mixture tablets (Roche Applied Science) using the MagNA Lyser Instrument (Roche Applied Science). The homogenate was centrifuged at 12,000 × g for 30 min at 4 °C to remove insoluble material. The supernatant was transferred to a fresh autoclaved tube, and the proteins were then precipitated by cold acetone (stored at −20 °C) overnight followed by air drying of the pellet. The pellet was then resuspended in 100 μl of rehydration buffer containing 8 m urea, 4% CHAPS, and 0.5% IPG Buffer (pH 3–10 non-linear; Amersham Biosciences) for 2D PAGE analysis. The protein concentration of each sample was assessed using a Bio-Rad detergent compatible kit.
Analysis of the Protein Profiles Using 2D PAGE—
For the 2D PAGE, IEF and SDS-PAGE were performed according to the method described in the manufacturer's manual (Amersham Biosciences) with minor modifications. For the first-dimension IEF, pH 3–10 non-linear range IPG strips (18 cm) were rehydrated with 350 μl of solubilized sample (800-μg protein amount) for 12 h before the sample was separated by IEF at 150 V for 2 h, 500 V for 1 h, 1000 V for 1 h, 4000 V for 1 h, and finally 8000 V for 7 h. Prior to the second dimension SDS-PAGE, the IPG strips were equilibrated with 2 ml of equilibration buffer consisting of 50 mm Tris-HCl (pH 8.8), 6 m urea, 2% SDS, 30% glycerol, 50 mm DTT, and 0.01% bromphenol blue at 25 °C for 15 min followed by equilibration in 50 mm Tris-HCl (pH 8.8), 6 m urea, 2% SDS, 30% glycerol, 25% iodoacetamide, and 0.01% bromphenol blue at 25 °C for 15 min. The second dimensional SDS-PAGE used a 12.5% separating gel and was performed without a stacking gel. The equilibrated IPG gel strip was placed on top of the SDS-PAGE gel with appropriate pressure to ensure a firm contact between the strip and the gel slab. Electrophoresis was carried out at 25 mA/gel until the tracking dye reached the bottom of the gel. The 2D PAGE gel was then stained with Coomassie Blue.
Detection and Quantitative Analysis of the 2D PAGE Gels—
2D PAGE images were obtained and the amount of protein in each spot was estimated using ImageMaster 2D Elite software version 5.0 (Amersham Biosciences). The volume of a protein spot was defined as the sum of the intensities of the pixel units within the protein spot. To correct quantitative variations in the intensity of protein spots, spot volumes were normalized as a percentage of the total volume of all the spots present in a gel.
Protein Identification by LC-MS/MS—
Selected Coomassie Blue-stained protein spots were manually excised from the 2D PAGE gel and cut into pieces; this was followed by destaining with 25 mm ammonium bicarbonate. The gel pieces were dehydrated with acetonitrile for 10 min, vacuum-dried, and then rehydrated with 55 mm DTT in 25 mm ammonium bicarbonate (pH 8.5) at 37 °C for 1 h. Subsequently the protein within the spot was alkylated with 100 mm iodoacetamide in 25 mm ammonium bicarbonate (pH 8.5) at room temperature for 1 h. The pieces were then washed twice with 50% acetonitrile in 25 mm ammonium bicarbonate (pH 8.5) for 15 min, dehydrated with acetonitrile for 10 min, dried, and finally rehydrated with a total of 25 ng of sequencing grade modified trypsin (Promega, Madison, WI) in 25 mm ammonium bicarbonate (pH 8.5) at 37 °C for 16 h. Following digestion, the tryptic peptides were extracted twice with 50% acetonitrile containing 5% formic acid for 15 min with moderate sonication. The extracted solutions were pooled and evaporated to dryness under vacuum. The dried pellet was then resuspended in 10 μl of 0.1% TFA.
Protein identification was performed on an integrated nano-LC-MS/MS system (Micromass) comprising a three-pump Micromass/Waters CapLC™ system with an autosampler, a stream select module configured with a precolumn, an analytical capillary column, and a Micromass Q-Tof Ultima™ atmospheric pressure ionization mass spectrometer fitted with a nano-LC sprayer operated under MassLynx™ 4.0 control.
MS/MS data acquisition was carried out by Micromass ProteinLynx™ Global Server (PGS) 2.0 data processing software in default mode and outputted as a single Mascot-searchable peak list (.pkl) file. The peak list files were used to query the Swiss-Prot version 50.8 database (release date, May 30, 2006; 234,112 sequences and 85,963,701 residues) using the Mascot program (release date, May 7, 2005; version 2.1, Matrix Science, London, UK) with the following parameters: taxonomy of Rodentia (rodents), peptide mass tolerance of 50 ppm, MS/MS ion mass tolerance of 0.25 Da, trypsin digestion with one missed cleavage, no fixed modification, and the variable modifications to be considered were methionine oxidation, cysteine carboxyamidomethylation, lysine acetylation, and phosphorylation of tyrosine, serine, and threonine. A total of 234,112 sequences and 85,963,701 residues in the database were actually searched. Only significant hits as defined by Mascot probability analysis were considered. Protein identifications were accepted with a statistically significant Mascot protein search score ≥40 that corresponded to an error probability of p < 0.05 in our data set. The protein identification with the highest score was selected to eliminate the redundancy of proteins that appeared in the database.
Cluster Analysis and Functional Classification of the Differentially Expressed Proteins—
A plot of the calibrated intensity for the expression value of each protein measured by ImageMaster 2D Elite software version 5.0 (Amersham Biosciences) among the different groups of samples (sham, SCI, and SCI + aFGF) was used in conjunction with an average linkage hierarchical clustering algorithm (Unweighted Pair Group Method with Arithmetic Mean (UPGMA)); this was done using Hierarchical Clustering Explorer 3.0 (39). The uncentered Pearson's correlation coefficient was determined as a measurement of the similarity metric, and the threshold value for the minimum similarity was 0.8. After clustering, each protein was allocated its place in a global temporal classification color heat map. We used the Bulk Gene Search System for Java (BGSSJ) (40) and Swiss-Prot protein knowledge database for the functional classification of the proteins.
Western Blotting—
Proteins extracts (50 μg) were separated by 12.5% SDS-PAGE and then transferred onto a PVDF membrane as described previously (41). The PVDF membrane was blocked with 1% nonfat milk in PBST (0.5% Tween 20 in PBS) for 1 h and probed with a series of primary antibodies (anti-heat shock protein 27 (Hsp27), 1:2000; anti-GAP43, 1:5000; and anti-stathmin, 1:5000) in 1% nonfat milk PBST. Finally after washing, the blots were treated with horseradish peroxidase-conjugated secondary antibody. Products were visualized by chemiluminescence according to the instructions provided by the manufacturer (Santa Cruz Biotechnology, Santa Cruz, CA). Images on x-ray films were scanned by a laser scanning densitometer (Amersham Biosciences), and band densities were quantified with the ImageQuant software version 5.2 (Amersham Biosciences). The results were expressed as mean ± S.D. A Student's t test was performed to evaluate statistical significances, and a p value <0.05 was regarded as statistically significant (n = 3 for each experiment).
Quantitative Real Time PCR (qPCR)—
For qPCR measurements, the spinal cord segments (0.5 cm) were collected at 24 h after surgery. Total RNA was extracted from each group (sham, SCI, and SCI + aFGF) using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and the RNA concentration was determined by measuring the absorbance at 260 nm on a spectrophotometer. The extracted total RNA was treated with RNase-free DNase I to eliminate potential contamination by genomic DNA. About 1 μg of total RNA from each sample was used for the first strand cDNA synthesis. The first strand cDNA synthesis was primed using oligo(dT) based on the SuperScript III First-Strand Synthesis kit (Invitrogen). The synthesized cDNA was used as a template for estimation of gene transcription in spinal cord tissue by qPCR. The selective SCI- and/or aFGF-responsive genes and their corresponding primers used in the qPCR are outlined in Table I. β-Actin served as an internal control for normalization.
Table I.
Primer sequences used for the quantitative real time PCR
| Gene | Sense primer (5′ to 3′) | Antisense primer (5′ to 3′) | Ref. and source |
|---|---|---|---|
| Calreticulin | ACGAGCCAAGATTGATGACC | TGGCCTCTACAGCTCATCCT | 102 |
| Synuclein | CAGCCAGGTCTCTTCCTCAC | TGAGCCTCTGTGGTTGACTG | 102 |
| Hsp27 | TGTCAGAGATCCGACAGACG | TACTGGGGATGGGTAGCAAG | 102 |
| GFAP | TGGCCACCAGTAACATGCAA | CAGTTGGCGGCGATAGTCAT | 103 |
| s100B | TGCCCTCATTGATGTCTTCCA | GAGAGAGCTCGTTGTTGATGAGCT | 103 |
| KSPG | GTTGAAAAGTGTGCCCATGGT | TTCATCAATATGGTCGATCTGGTT | 74 |
| UCHL1 | GAATGCCTTTCCCGGTGAA | AAGCGGACCTCTCCCTGCT | 104 |
| Actin | GGGAAATCGTGCGTGACATT | GCGGCAGTGGCCATCTC | RTPrimerDB ID: 255a |
The primer sequences for the actin quantitative real time PCR were adapted from the Real Time PCR Primer and Probe Database (RTPrimerDB). The ID number of the actin primers is 255.
Real time PCR amplification and analysis were carried out on a LightCycler 2.0 instrument (Roche Applied Science). The final volume of reaction was 20 μl, and LightCycler FastStart DNA Master SYBR Green I (Roche Applied Science) was used. The specificity of the SYBR Green PCR signal was confirmed by melting curve analysis and 2% agarose gel electrophoresis to demonstrate that only a single amplified product for each PCR was obtained. mRNA expression was quantified using the comparative cross-threshold (Ct; the PCR cycle numbers that were over the signal threshold that could be detected by the LightCycler 2.0) method. The Ct of the internal control gene β-actin was subtracted from the Ct of the target gene to obtain ΔCt. The normalized -fold changes of the mRNA expression of each target gene was expressed as 2−ΔΔDCt where ΔΔDCt is equal to the ΔCt of the SCI-treated sample with or without the aFGF ΔCt control.
RESULTS
Functional Recovery after aFGF Treatment in Rat Model with Contusive Spinal Cord Injury—
Initially we established the contusion model of spinal cord injury at the T9 segment of rats for this study. This incomplete SCI contusion model mimics a condition similar to that found in clinical patients with SCI. These SCI rats were made using a weight-drop device developed by New York University. The BBB (36) locomotor rating scale and the CBS (38) were used, and these showed that all of the rats in the contusion group (n = 8) and the contusion/aFGF-treated group (n = 6) suffered from complete paraplegic symptoms of the hind limbs (BBB score = 0; CBS = 100) at day 1 after SCI (Fig. 1). One week after surgery, the contusion group showed no recovery from the paraplegia. Overall this group only showed a very slight but not statistically significant improvement in hind limb locomotion throughout the 4 weeks after SCI. However, there was a significant improvement (p < 0.05) in the locomotor behavior analysis results for the aFGF-treated group when they were observed 1 week after surgery using both the BBB and the CBS analysis. Notably after monitoring the behavior analysis up to the 4th week postinjury, the improvement in locomotor function among the aFGF-treated rats was found to be statistically significant using both the BBB (Fig. 1A) and CBS methodologies (Fig. 1B); this was true for every time point compared with the SCI rats. This is probably the first report to demonstrate that aFGF is able to induce good functional recovery in contusive SCI rats.
Fig. 1.
The time course of hind limb functional recovery after contusive spinal cord injury. The time course of changes in the BBB score (A) and CBS (B) for the SCI rats that were untreated or treated with aFGF is shown. The recovery rate of the animals was assessed in a double blind manner. ○ and • denote the SCI rats either treated with (n = 6) or not treated with aFGF (n = 8), respectively. Statistical significance was evaluated using the Mann-Whitney test, which was performed using SPSS 11.0 statistical software. The results are reported as mean ± S.E. Error bars indicate standard error of the mean. *, p < 0.05; **, p < 0.01.
To further confirm the observed functional recovery of the SCI rats treated with aFGF, we investigated the expression of protein markers involved in neuronal regeneration using Western blotting. Growth-associated protein 43 (GAP-43) is a specific phosphoprotein of the nervous system and is formed at the leading edge of the elongation axon (43, 44). Importantly axonal transport and expression of GAP-43 occur only in neurons that are extending during development or are regenerating after injury (45). For these reasons, GAP-43 is regarded as a marker protein for spinal cord regeneration. Our Western blotting results showed that expression of GAP-43 in the contusion rats was much lower than among the sham rats (Fig. 2A). In contrast, expression of GAP-43 protein was found to recover to a level similar to that of the sham rats among the aFGF-treated SCI rats at 4 weeks after contusion. This result is consistent with a previous study (46). Stathmin is a potent microtubule-destabilizing factor and has been identified as a neuronal marker during the early stages of neural development (47). In addition, some studies have suggested that stathmin is able to regulate neurite outgrowth (48, 49). Our results provided evidence that aFGF is able to up-regulate stathmin expression in injured spinal cord tissue (Fig. 2B). Thus, based on our behavior testing (Fig. 1) and biochemical results (Fig. 2), these experiments clearly indicate that regeneration of injured axons and functional recovery of the hind limbs would seem to be induced in SCI rats that have been treated with aFGF.
Fig. 2.
Western blotting analysis of the protein extracts from spinal cords with antibodies raised against GAP-43 and stathmin. Recovery of the expression of neuronal regeneration markers GAP-43 (A) and stathmin (B) in the SCI rats treated with aFGF at 4 weeks after the contusion injury is shown. Actin was used as a protein loading control, and the sham animals were used as a control for GAP-43 and stathmin production. Representative blots are shown. The bar graphs show the quantitative analysis of the protein expression relative to actin. A Student's t test was performed to evaluate statistical significance. The results are presented as mean ± S.E. **, p < 0.01; *, p < 0.05.
2D PAGE Protein Profiles and Identification of Differentially Expressed Protein Spots from SCI Rats Untreated or Treated with aFGF—
To reveal the protective mechanism involved when SCI rats are treated with aFGF, we investigated and compared the protein profiles between sham and SCI rats either with or without aFGF treatment using an unbiased proteomics approach. Representative high resolution 2D PAGE was able to resolve hundreds of proteins extracted from the T9 spinal cord tissue as shown in Fig. 3. We found that these protein spots were largely distributed in an area with pI values ranging from 4.0 to 8.0, and the molecular mass of protein spots generally ranged from 6.5 to 220 kDa. The spinal cord proteome as visualized in this study contained about 970 detectable proteins on a single Coomassie Blue-stained 2D PAGE gel when analyzed by ImageMaster 5.0. Furthermore we used this software to identify the quantitatively differentially expressed proteins on the 2D PAGE gel when the sham and SCI rats either untreated or treated with aFGF treatment were compared. Among these detectable proteins spots, we were able to pinpoint a total of 51 that were either significantly increased or decreased in intensity in response to contusive spinal cord injury and/or SCI + aFGF treatment (-fold changes in protein expression, ≥1.3). These protein spots were then numerically labeled (Fig. 3).
Fig. 3.
Differential expression of proteins from contusive spinal cord untreated or treated with aFGF visualized by 2D PAGE. Protein extracted from the spinal cord tissue of the SCI rats was separated by 2D PAGE (first dimension, 18 cm, pH 3–10, non-linear gradient of IPG strips; second dimension, 12.5% SDS-PAGE) and visualized by Coomassie Blue staining. Representative examples of the 2D PAGE results from a sham T9 spinal cord segment (A), a 24-h postcontusion T9 spinal cord segment not treated with aFGF injection (B), and a 24 h postcontusion T9 spinal cord segment treated with aFGF injection (C) are shown. The numbered spots indicate corresponding differentially expressed protein spots between the sham and the contusive T9 spinal cord untreated or treated with aFGF. The proteins forming these spots were identified by LC-MS/MS mass spectrometry. The corresponding protein spot identities are shown in Table II. L1, landmark protein spot for gel matching. The orientation of the IEF strips is indicated at the top of gels. Molecular mass standards are shown on the right of the 2D PAGE gel.
To characterize these proteins, the protein spots from the 2D PAGE gel were excised from the Coomassie Blue-stained gels. These 51 differentially regulated protein spots were then identified using highly accurate LC-MS/MS mass spectrometry. The MS/MS spectra that were obtained by this state-of-the-art-technology were searched against the Mascot database on-line search engine (Matrix Science) to identify the proteins. The detailed results of the database search are shown in Table II. Mascot scores ≥40 are indicative of significant and extensive protein homology (p < 0.05).
Table II.
The list of proteins identified by LC-MS/MS
| Spot IDa | Protein Identity | Accession numberb | Abbreviation | Molecular weight/pIc | Scored | Match peptidee (n) | Sequence coveragef | SCI/shamg | aFGF/SCIh | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||||
| 1(188) | Contrapsin-like protease inhibitor 3 precursor | P05544 | CPI3 | 46,248/5.48 | 58 | 1 | 2 | ↑ | 3.25 | ↓ | 1.86 |
| 2(231) | Protein-disulfide isomerase A3 precursori | P11598 | PDIA3 | 56,588/5.88 | 271 | 6 | 16 | ↑ | 2.15 | ↓ | 2.64 |
| 3(253) | Calreticulin precursor | P18418 | CALR | 47,966/4.33 | 184 | 4 | 12 | ↑ | 2.08 | ↓ | 1.39 |
| 4(297) | Aldehyde dehydrogenase, mitochondrial | P11884 | ALDH2 | 56,453/6.63 | 86 | 2 | 5 | ↓ | 2.07 | ↓ | 1/∞ |
| 5(159) | Transferrini | P12346 | TRFE | 76,364/7.14 | 85 | 14 | 49 | ↑ | 8.37 | ↓ | 2.26 |
| 6(160) | Transferrini | P12346 | TRFE | 76,364/7.14 | 86 | 14 | 50 | ↑ | 3.30 | ↓ | 1.57 |
| 7(234) | Protein-disulfide isomerase A3 precursori | P11598 | PDIA3 | 56,588/5.88 | 311 | 8 | 16 | ↑ | 2.75 | ↓ | 1.65 |
| 8(152) | Transferrini | P12346 | TRFE | 76,364/7.14 | 86 | 12 | 51 | ↑ | 6.66 | ↓ | 5.25 |
| 9(624) | Rho GDP dissociation inhibitor 1 | Q99PT1 | GDIR | 23,262/5.12 | 96 | 4 | 12 | ↑ | 1.37 | ↓ | 1/∞ |
| 10(359) | β-Centractin | Q8R5C5 | ACTY | 42,255/5.98 | 128 | 2 | 8 | ↑ | 2.42 | ↓ | 2.28 |
| 11(398) | F-box only protein 2 | Q80UW2 | FBX2 | 33,655/4.21 | 50 | 1 | 5 | ↑ | ∞ | ↓ | 2.61 |
| 12(395) | Apolipoprotein A-IV precursor | P02651 | APOA4 | 44,429/5.12 | 506 | 15 | 45 | ↑ | ∞ | ↓ | 2.08 |
| 13(434) | Keratan sulfate proteoglycan lumican | P51885 | KSPG | 38,241/6.00 | 43 | 2 | 3 | ↑ | 2.33 | ↓ | 2.22 |
| 14(435) | G protein β2 subunit | P54313 | GBB2 | 37,176/5.60 | 76 | 3 | 13 | ↑ | 2.34 | ↓ | 1.93 |
| 15(424) | Tropomyosin β chain | P58775 | TPM2 | 32,817/4.66 | 463 | 8 | 22 | ↑ | 1.10 | ↓ | 1/∞ |
| 16(456) | Tropomyosin 1α chain | P04692 | TPM1 | 32,661/4.69 | 438 | 10 | 29 | ↑ | 2.06 | ↓ | 4.53 |
| 17(431) | Haptoglobin precursor | P06866 | HPT | 38,525/6.10 | 40 | 4 | 10 | ↑ | ∞ | ↓ | 2.68 |
| 18(486) | Apolipoprotein E precursor | P02650 | APOE | 35,731/5.23 | 561 | 16 | 36 | ↑ | 3.80 | ↓ | 2.32 |
| 19(467) | Malate dehydrogenase, cytoplasmic | O88989 | MDHC | 36,329/6.16 | 234 | 6 | 19 | ↓ | 1.39 | ↓ | 1.12 |
| 20(189) | Serum albumin precursori | P02770 | ALBU | 68,686/6.09 | 300 | 5 | 9 | ↑ | 5.83 | ↓ | 1.62 |
| 21(555) | Rho GDP dissociation inhibitor 1 | Q99PT1 | GDIR | 23,262/5.12 | 189 | 6 | 44 | ↑ | ∞ | ↓ | 1.65 |
| 22(1431) | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | Q00981 | UCHL1 | 24,822/5.14 | 342 | 8 | 40 | ↓ | 40.18 | ↓ | 1/∞ |
| 23(604) | Peroxiredoxin-2 | P35704 | PRDX2 | 21,639/5.34 | 97 | 2 | 14 | ↓ | 1.95 | ↓ | 1.26 |
| 24(183) | Serum albumin precursori | P02770 | ALBU | 68,686/6.09 | 224 | 5 | 9 | ↑ | 4.40 | ↓ | 1.56 |
| 25(601) | Apolipoprotein A-I precursor | P04639 | APOA1 | 30,069/5.52 | 265 | 7 | 20 | ↑ | 2.98 | ↓ | 2.86 |
| 26(629) | Phosphatidylethanolamine-binding protein 1 | P31044 | PEBP1 | 20,657/5.48 | 118 | 2 | 14 | ↑ | 2.29 | ↓ | 2.15 |
| 27(726) | Calmodulin (CaM) | P62161 | CALM | 16,696/4.09 | 265 | 6 | 39 | ↑ | 1.30 | ↓ | 1.65 |
| 28(879) | Protein S100-B | P04631 | S100B | 10,606/4.52 | 60 | 2 | 16 | ↑ | 2.07 | ↓ | 1.80 |
| 29(180) | Serum albumin precursori | P02770 | ALBU | 68,686/6.09 | 187 | 3 | 6 | ↑ | 3.36 | ↓ | 2.00 |
| 30(789) | Transthyretin precursor | P02767 | TTHY | 15,710/5.77 | 57 | 1 | 16 | ↑ | 3.99 | ↓ | 3.33 |
| 31(582) | Triose-phosphate isomerasei | P48500 | TPIS | 26,939/7.08 | 246 | 5 | 30 | ↓ | 1.45 | ↓ | 1.54 |
| 32(590) | Peroxiredoxin-6 | O35244 | PRDX6 | 24,672/5.65 | 181 | 4 | 25 | ↓ | 1.46 | ↑ | 1.02 |
| 33(736) | γ-Synuclein | Q9Z0F7 | SYUG | 13,152/4.68 | 241 | 6 | 32 | ↓ | 1.34 | ↓ | 1.17 |
| 34(794) | Galectin-1 | P11762 | LEG1 | 14,716/5.14 | 156 | 3 | 25 | ↑ | 1.50 | ↓ | 1.58 |
| 35(208) | Neurofilament triplet L protein | P19527 | NFL | 61,167/4.63 | 927 | 21 | 44 | ↓ | 1.75 | ↓ | 1.01 |
| 36(362) | Glutamine synthetase | P09606 | GLNA | 43,042/6.68 | 95 | 3 | 12 | ↓ | 1.27 | ↓ | 1.40 |
| 37(805) | Glial fibrillary acidic protein, astrocyte | P47819 | GFAP | 50,160/5.35 | 482 | 10 | 24 | ↑ | 2.60 | ↓ | 1.63 |
| 38(347) | Creatine kinase B-type | P07335 | KCRB | 42,685/5.33 | 430 | 9 | 29 | ↓ | 5.19 | ↑ | 3.52 |
| 39(490) | Lactoylglutathione lyase | Q6P7Q4 | LGUL | 20,675/5.12 | 98 | 3 | 22 | ↑ | 1.24 | ↓ | 4.16 |
| 40(438) | G protein β2 subunit | P54313 | GBB2 | 37,176/5.60 | 138 | 3 | 11 | ↑ | 3.71 | ↓ | 2.11 |
| 41(584) | Triose-phosphate isomerasei | P48500 | TPIS | 26,939/7.08 | 443 | 10 | 45 | ↑ | 2.59 | ↓ | 3.47 |
| 42(360) | Succinyl-CoA ligase | Q9Z2I9 | SUCB1 | 50,082/6.57 | 67 | 2 | 4 | ↑ | 1.42 | ↓ | 2.15 |
| 43(221) | α-Internexin | P23565 | AINX | 56,082/5.20 | 286 | 6 | 15 | ↓ | 3.41 | ↑ | 1.89 |
| 44(795) | Fatty acid-binding protein, brain | P55051 | FABPB | 14,723/5.47 | 235 | 5 | 46 | ↑ | 2.12 | ↓ | 1.97 |
| 45(413) | Isocitrate dehydrogenasei | Q99NA5 | IDH3A | 40,284/6.47 | 100 | 3 | 13 | ↑ | 2.36 | ↓ | 1.71 |
| 46(572) | Heat shock 27-kDa proteini | P42930 | HSP27 | 22,879/6.12 | 219 | 7 | 46 | ↑ | 1.59 | ↓ | 2.14 |
| 47(265) | Dihydropyrimidinase-related protein 2 | P47942 | DPYL2 | 62,239/5.95 | 79 | 2 | 5 | ↑ | 1.98 | ↓ | 2.87 |
| 48(402) | Isocitrate dehydrogenasei | Q99NA5 | IDH3A | 40,284/6.47 | 147 | 4 | 10 | ↑ | 2.31 | ↓ | 2.82 |
| 49(574) | Heat shock 27-kDa proteini | P42930 | HSP27 | 22,879/6.12 | 229 | 5 | 39 | ↑ | 1.58 | ↓ | 2.54 |
| 50(482) | Annexin A3 | P14669 | ANXA3 | 36,169/6.05 | 282 | 6 | 18 | ↓ | 1.37 | ↓ | 1/∞ |
| 51(271) | Rab GDP dissociation inhibitor | P50396 | GDI | 50,489/5.93 | 51 | 3 | 9 | ↑ | ∞ | ↓ | 1.50 |
The spot ID is the number of the significantly expressed protein spot shown in Fig. 3. Note that the numbers in parentheses indicate the numbering of the protein spots assigned by the ImageMaster 5.0 image analysis software.
The MS/MS spectra were searched against the Swiss-Prot protein knowledge database using Mascot software version 2.1 from Matrix Science to identify the proteins.
Theoretical pI and molecular weight are based on the Swiss-Prot protein knowledge database.
An MS/MS score ≥40 was regarded as significant with the p value being less than 0.05.
The total number of peptides assigned to the protein. The list of matched sequences and MS/MS spectra are shown in the supplemental data.
Sequence coverage (%) indicates the number of amino acids spanned by the assigned peptides divided by the sequence length.
The -fold change value of the protein expression level after SCI compared with the sham level. The symbols ↑ and ↓ denote up-regulation and down-regulation, respectively. ∞, these proteins were only expressed after SCI and were compared with sham, and therefore the ratio of SCI/sham is infinite.
The -fold change value of the protein expression level compared with SCI after aFGF treatment. The symbols ↑ and ↓ denote the up-regulation and down-regulation, respectively. 1/∞, these proteins disappeared after aFGF treatment and were compared with the SCI value, and therefore the ratio of aFGF/SCI = 0.
Some distinct proteins were detected as a chain of multiple spots, strongly suggesting that these were post-translationally modified forms of the protein and resulted from, for example, phosphorylation, which alters the pI of the protein.
Visual Cluster Analysis and Functional Classification of Differentially Expressed Proteins—
To analyze these complex proteomics data, we used a bioinformatics tool to categorize the differentially expressed proteins from the sham spinal cords and the SCI spinal cords with or without aFGF treatment. We then adapted a data visualization and clustering program to monitor the genome-wide differences in gene expression with the aim of mining any meaningful patterns hidden within the proteomics data (50–52).
We uploaded the protein list from Table II with their accession numbers and expression levels, which had been quantified by ImageMaster 2D Elite software version 5.0 (Amersham Biosciences), into Hierarchical Clustering Explorer 3.0. Clustering allows us to extract groups of proteins that are tightly co-expressed over a range of different experimental conditions. Next the differentially expressed proteins were clustered using UPGMA algorithm into five categories with distinct expression profiles, and these were displayed using a color heat map (Fig. 4). With sham group as the control, it was found that expression of proteins from the SCI sample in cluster A was up-regulated, whereas the expression of the same proteins from the SCI samples treated with aFGF was down-regulated to a level more or less the same as that of the sham control (Fig. 4, cluster A). Expression of proteins from cluster B followed a pattern similar to that of cluster A, but the expression of proteins from aFGF-treated samples, although down-regulated when compared with SCI group, still had an overall protein expression level that was higher than that of the sham control (Fig. 4, cluster B). There was no significant difference between the sham and SCI samples in cluster C; however, the expression of these proteins was substantially down-regulation in samples treated with aFGF (Fig. 4, cluster C). In cluster D, down-regulation of protein expression was found in the SCI samples, and the expression was even further down-regulated in the samples treated with aFGF (Fig. 4, cluster D). The expression of proteins in cluster E from the SCI rats was drastically reduced, but expression of these proteins was recovered to a certain extent after the samples were treated with aFGF (Fig. 4, cluster E). In summary, the expression levels of proteins in clusters A, B, and E were recovered to a large extent to an expression level similar to that of the sham rats after the SCI rats were treated with aFGF. However, the expression of proteins in clusters C and D was further down-regulated in the SCI rats after treatment with aFGF. The biological relevance of the further down-regulation of proteins in response to aFGF in clusters C and D is not obvious. The protein expression levels of clusters A, B, C, D, and E were fully quantitatively analyzed using ImageMaster 2D Elite software version 5.0 (Amersham Biosciences). A selected protein spot for each cluster is shown in Fig. 5 using a zoomed location on the 2D PAGE.
Fig. 4.
Hierarchical clustering and functional classification of the changes in protein expression between the sham and SCI spinal cords untreated or treated with aFGF. The expression pattern for each protein was categorized by a UPGMA using Hierarchical Clustering Explorer 3.0. Using this, proteins with a similar expression were clustered into five different discrete groups (clusters A, B, C, D, and E) in a treelike organization. Each row in the color heat map indicates a single protein, and each column represents different groups of proteins from sham and SCI rats untreated or treated with aFGF. A high protein expression value for a specific protein spot is represented by a bright red color, and a low protein expression value is represented by a bright green color. A black color indicates that the protein spot was expressed at an average level; a gray color represents protein spots that were not detectable. Abbreviations (see Table II for definitions) for and the biological functions of the various proteins forming the spots are indicated. Further information about the differentially expressed protein spots identified is given in Table II. The functional classification of each protein was determined using BGSSJ and the Swiss-Prot protein knowledge base. These proteins were annotated into seven functional categories including structural molecular activity, metabolism, antioxidant/oxidoreductase activity, signaling transduction, regulation of apoptosis, enzyme regulator activity, and transporter acute phase response.
Fig. 5.
Representation of the temporal changes in protein expression for each discrete protein cluster. One typical protein spot was selected from each protein cluster for further quantitative analysis of its protein expression level using ImageMaster 2D platinum software version 5.0. The bar graphs show alteration in the protein expression levels of each selected spot for each of the five protein cluster groups (n = 3). The results are represented as mean ± S.E. The five selected protein spots are circled and specifically are KSPG, Hsp27, GFAP (astrocyte), Rho GDP dissociation inhibitor 1 (GDIR), UCHL1, and KCRB.
To reveal the biological relevance of the changes in protein expression in response to aFGF treatment, the identified proteins in each cluster were further annotated into seven functional categories according to the biological process in which they are involved using BGSSJ and the Swiss-Prot protein knowledge base. These categories included structural molecular activity, metabolism, antioxidant/oxidoreductase activity, signal transduction, regulation of apoptosis, enzyme regulator activity, and transporters/acute phase response (Fig. 4). The pie charts show the number of proteins and the proportion in each functional category found in each cluster (Fig. 6).
Fig. 6.
Functional categorization of the differentially expressed proteins identified from the spinal cord tissue of the SCI rats. Functional categorization of the protein was done by BGSSJ and the Swiss-Prot protein knowledge base. The pie charts represent each individual protein cluster from A to E as shown in Fig. 4. Proteins with the same or similar biological functions are grouped together in a sector of the circle in proportion to the number of proteins in the protein cluster.
It was found that proteins involving a function as transporter/acute phase response proteins only existed in clusters A and B with the ratios of 15% (n = 3) and 37% (n = 6), respectively. This result is consistent with a disruption of the blood-nerve barrier (53) after spinal cord injury. These proteins included transthyretin (TTHY), transferrin (TRFE), haptoglobin (HPT), albumin (ALBU), and protein S100B, and the appearance of these proteins has been implicated in the pathogenesis of brain contusion injury (54). Among these proteins, a strong up-regulation of TRFE (8.37-fold) and ALBU (5.83-fold) was shown after SCI, and HPT was only detectable in the injured spinal cord tissue (Table II) but not in sham-treated rat tissue. However, the elevated amount of HPT was substantially reduced in samples treated with aFGF. Furthermore the level of the inflammation protein S100B was elevated 2.07-fold after SCI. Thus, these results correspond well with the known pathology of traumatic central nervous system injury (brain and spinal cord), which involves inflammation coupled with a breakdown of the blood-brain barrier; this leads to extravasation of the plasma proteins into the central nervous system (54, 55). Most importantly, up-regulation of these proteins was attenuated in the injured spinal cord tissues after treatment with aFGF.
It is noticeable that expression of two proteins that are involved in structural molecular activity, KSPG and GFAP, were found in clusters A and B and were up-regulated 2.33- and 2.60-fold, respectively, in the SCI rat tissue (Table II and Fig. 5). It is well known that a larger number of activated astrocytes are produced after SCI and that this is followed by the secretion of chondroitin sulfate proteoglycans (CSPGs) in a large quantity (56). GFAP is a protein marker for astrocytes, and KSPG and CSPG are inhibitory extracellular matrix molecules. Furthermore KSPG is mainly expressed in macrophages and reactive microglia after spinal cord injury (57). Eventually the proteoglycans together with the astrocytes form scars around the damaged tissue that further inhibit neural regeneration of the SCI. Thus, it is plausible, as revealed by this work, that aFGF is able to initiate a transducing pathway attenuating the elevation of GFAP and KSPG after SCI (Fig. 5). This might help to generate favorable conditions for the process of neural regeneration.
Moreover there was also a group of proteins related to the regulation of apoptosis and signal transduction found in both clusters A and B, and these comprised Hsp27, phosphatidylethanolamine-binding protein 1 (PEBP1), and Rab GDP dissociation inhibitor (GDI). We found that the expression level of Hsp27 was increased 1.59-fold when detected in SCI tissue and that this expression level recovered to more or less the normal level when the SCI rats were treated with aFGF (Table II and Fig. 5). It is known that Hsp27 is able to directly interact with cytochrome c after its release from mitochondria and further suppresses the activation of caspase 3, which results in an inhibition of the initiation of apoptosis (42). PEBP1 is a Raf kinase inhibitor protein (RKIP), and the expression level of this protein was increased 2.29-fold (Table II) in the SCI rats. Such an increase may not favor the initiation of cell regeneration via the Raf/MEK/ERK pathway. Thus it is plausible that the application of aFGF is able to attenuate the elevation of PEBP1 in SCI tissue and help cell regeneration in the damaged tissue. Moreover Park et al. (58) reported that one function of Rab3A is to regulate the release of synaptic vesicles and neurotransmitters. Thus interaction between Rab3A and GDI may inhibit the functioning of Rab3A, and as a result of aFGF application, there might be a resumption of neurotransmitter release. Also most importantly, GDI has also been implicated in the inhibition of neurite outgrowth in vitro (59). Therefore, a decrease in the expression of RKIP and GDI after aFGF treatment may reflect attempted spinal cord regeneration.
Only two proteins, creatine kinase brain type (KCRB) and α-internexin (AINX), were found in cluster E, and these proteins were categorized into amino acid metabolism and structural molecule activity, respectively. The amount of KCRB was drastically reduced (−5.19-fold) in the SCI tissue, but there was a recovery of 3.52-fold when the SCI was treated with aFGF (Table II and Fig. 5). The major function of KCRB is to maintain a normal supply of ATP to the CNS tissues. Thus, a reduction in KCRB is indicative of an impairment of energy metabolism in the SCI tissue, and this result is consistent with a previous study (60). Most importantly, aFGF was able to attenuate this effect and potentially improve and restore neuronal function. On the other hand, AINX is a cytoskeleton protein of neuron cells, and a decrease in expression of this protein (−3.41-fold) was detected in the SCI tissue (Table II); this is indicative of a loss of neuron cells in the damaged tissues. Thus, the increased expression of AINX (1.89-fold) in the SCI tissue treated with aFGF (Table II) may reflect repair processes in the tissues treated with aFGF.
It is noteworthy that the antioxidant proteins peroxiredoxin-2 (PRDX2) and PRDX6 only appeared in cluster D. In SCI tissue, the expression of these two proteins was down-regulated 1.95- and 1.45-fold, respectively (Table II). In contrast, the expression of these two proteins recovered 1.26- and 1.02-fold, respectively, after the damaged tissue was treated with aFGF. These results clearly indicate that the ability of cells in the wounded area to clean up released free radicals induced by the SCI was disrupted by an insufficient amount of these two antioxidant proteins.
Confirmation of the Differential Expression of the Protein Genes Detected by Proteomics Analysis Using Quantitative Real Time PCR—
In this work, some of the highly differentially expressed proteins detected from the proteomics analysis were expected to be important for regeneration after SCI. Specifically these were GFAP (activation of astrocytes), S100B (regulation of inflammation), KSPG (scar formation), Hsp27 (regulation of apoptosis), and calreticulin (protein metabolism). All these proteins fell into clusters A and B (Fig. 4). To reveal whether the change in translational expression was coupled with a change in transcriptional expression for these protein genes, quantitative real time PCR was used. It was found that transcriptional expression of these proteins was elevated in the SCI samples (Table II and Fig. 5), and this overexpression was attenuated in the aFGF-treated SCI samples. It was interesting that the quantitative real time PCR results for the pattern of transcription expression of these protein genes was consistent with the pattern of protein expression derived from the proteomics analysis (Fig. 7). This result clearly indicates that regulation of the production of these proteins was indeed due to expression of their corresponding genes at the mRNA level, and this was triggered either directly or indirectly by the application of aFGF to the damage site. Synuclein and ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) were categorized as cluster D proteins (Fig. 4) among which expression was further down-regulated either without or with aFGF treatment of the SCI samples. When analyzed, transcription expression of the UCHL1 gene by real time PCR was consistent with translational expression results obtained from the proteomics data (Fig. 7A). The application of aFGF did not rescue this down-regulation of gene expression, and the impact of blockage of expression of this gene during the process of neural regeneration still remains to be resolved.
Fig. 7.
Independent evaluation by quantitative real time PCR of the differential transcriptional expression patterns of various selected protein genes that had been shown to be differentially expressed at the protein level. Various differentially expressed proteins as identified by 2D PAGE (Fig. 5), namely GFAP, S100B, KSPG, Hsp27, calreticulin, synuclein, and UCHL1, were chosen for further study of their transcription expression patterns using quantitative real time PCR. The protein expression pattern level (A) and mRNA expression pattern level (B) of these genes are compared. The bar graph displaying the mRNA expression level shows the -fold change after normalization against actin, a housekeeping gene, which was followed by normalization against the sham group. The results showed that the change in transcriptional expression pattern of these genes was consistent with the results derived from the proteomics analysis. The specificity of the quantitative real time PCR signal was confirmed by melting curve analysis and 2% agarose gel electrophoresis.
Post-translational Modification of the Proteins—
It is known that protein isoforms occur because of multiple PTM events, and such events can only be detected using a proteomics approach. This is one of the major pitfalls of microarray analysis, but proteome analysis is able to fill this niche. Several hundred different PTMs have been demonstrated including phosphorylation, acetylation, glycosylation, and myristoylation (61, 62). PTMs are important processes that are able to affect directly protein function, stability, conformation, binding interactions, and localization, all of which may be important to the events that also occur after SCI (32, 33). Our 2D PAGE results indicated that protein spots 46 and 49 were both Hsp27 (Fig. 3) and that although the molecular weight of the proteins in these spots is the same, they have pI values of 5.5 and 6.0, respectively. These different pI values suggest that these two proteins only differ in terms of their post-translation modification status, which may possibly be phosphorylation, acetylation, or another type of PTM. The protein expression profile of a Western blot using antibodies raised against this protein (Fig. 8) agreed with the protein expression profile detected by 2D PAGE. This suggests that Hsp27 has probably undergone PTM after SCI. It was noteworthy that expression levels of Hsp27 as detected by Western blotting were consistent with the level of transcriptional expression as detected by quantitative real time PCR (Fig. 7A). Similar results were obtained for other protein spots (Fig. 3) such as protein-disulfide isomerase A (spots 2 and 7), triose-phosphate isomerase (spots 31 and 41), IDH3A (spot 45 and 48), transferrin (spots 5, 6, and 8), and albumin (spots 20, 24, and 29). Further experiments are needed to verify the nature of these modifications and specific residues that are changed to understand their importance.
Fig. 8.
2D PAGE/Western blotting analysis for the identification of possible post-translation modification of Hsp27 from the spinal cord tissue of the SCI rat. Protein extracts from the spinal cords of sham (A) and SCI rats not treated (B) or treated with aFGF (C) were analyzed by 2D PAGE followed by transfer onto PVDF membrane. Western blotting was performed using antibody raised against Hsp27. The isoforms of Hsp27 protein have been circled, and the molecular masses and pI values of the various protein isoforms are shown. Note that the corresponding isoforms of Hsp27 protein identified in this experiment are identical to protein spots 46 and 49 in Fig. 3.
DISCUSSION
In this study, we demonstrated using animal behavior tests that a single bolus injection of aFGF is able to induce effective functional recovery after contusive spinal cord injury of rats in vivo (Fig. 1). Furthermore our Western blot results indicated that aFGF is able to up-regulate the protein expression of axonal regenerative protein markers such as GAP-43 and stathmin up to 4 weeks after SCI. This result is consistent with previous studies where both the transcriptional and translational expression levels of GAP-43 were found to be elevated in experimental animals treated with neutralizing antibody against CD95L or Nogo-A, which led to functional recovery of the hind limbs (27, 46). Noticeably previous studies have illustrated that there is an analogous relationship between rats and humans with respect to functional, electrophysiological, and morphological effects after contusive SCI (63). Hence in our experimental design, the contusive SCI rats mimicked the real situation of a clinical patient with a traumatic spinal cord injury, and this implies that aFGF may have potential as a promising therapeutic agent for the treatment of patients with spinal cord injury. Noticeably this is the first report to show that aFGF is able to induce an in vivo functional recovery effect in contusive spinal cord-injured rats.
Over the past few years, a large number of studies have been carried out to investigate the pathology and regenerative processes that take place in the injured spinal cord using both transcriptional and translational level strategies involving microarrays and proteomics (9, 34, 35), respectively. In addition to these previous reports, in this work, we used comparative proteomics techniques to screen for differences in protein expression levels between sham, contusion-injured, and contusion-injured aFGF-treated spinal cord tissue in rats. Moreover our results were further explored using a bioinformatics approach to annotate the protein function and protein expression patterns of our proteomics data. This approach allowed the systematic scrutinizing of the proteomics data and revealed some unexpected and interesting patterns that were hidden in the complex data set. These insights might help us to realize fully the pathological process during SCI and pinpoint the molecular basis of aFGF activity during the repair process.
It has been found previously that functionally related proteins are often coordinately expressed (51). The same type of results have been found in this work where some functionally related proteins from the aFGF-treated SCI rats could be annotated into different discrete clusters. Thus, five different expression profiles and seven major functional categories of proteins were determined that changed in response to aFGF treatment after SCI injury. Consequently the pathology of SCI and the neuroprotective effect of aFGF need to be discussed based on these functional categories, and this is done in the next few paragraphs.
Proteins Involved in Transporter Activity—
A previous study has shown that the blood-spinal cord barrier (BSCB) has an important role in regulating the fluid homeostasis of the spinal cord (55). According to our results, the strong up-regulation of transporter proteins such as albumin, transferrin, HPT, and TTHY in the injured spinal cord tissue would seem to be derived from the circulation and indicate a breakdown of the BSCB after contusive spinal cord injury. It is worth noting that from our 2D PAGE analysis we could only detect HPT in the contusive SCI group but not in the sham group. Moreover the up-regulation of albumin and transferrin was in agreement with a previous study using proteomics analysis of transection-injured spinal cord tissue of rats (34, 35). Most importantly, the up-regulation of these proteins was decreased in the aFGF-treated spinal cord tissue after SCI. Thus, based on this observation, our result probably implied that aFGF might create a favorable condition for a better recovery of the SCI rat that might benefit the maintenance of the functioning of BSCB. This phenomenon is consistent with a previous report that other growth factors such as brain-derived neurotrophic factor and insulin-like growth factor (IGF-1) are able to induce neuroprotection by attenuating BSCB permeability after SCI (64).
Proteins Involved in Inflammation, Astrocyte Activation, and Scar Formation—
Proteins involved in inflammation, astrocyte activation, and scar formation such as S100B, GFAP, and KSPG were only present in clusters A and B of our proteomics annotation. The up-regulation of these proteins that occurred after spinal cord injury was corrected to a level more or less the same as their normal expression level when the contusive spinal cord tissue was treated with aFGF (Fig. 4). S100B has a stimulating effect on astrocytes in vitro leading to glial proliferation (65). Furthermore S100B is primarily expressed in and might be secreted by astrocytes. Therefore S100B has been regarded as a protein marker of inflammation (66). On the other hand, a study of transection-injured spinal cords (34, 35) did not identify inflammation-related proteins among their proteomics data. These comparative results probably indicate that contusive injury to the spinal cord might induce a stronger inflammation effect than that with transection SCI. The inflammation response is normally regarded as a typical symptom during the early stages of SCI; thus the identification of inflammation-related proteins in the contusive injury of spinal cord probably suggests that the contusive animal model is a more suitable model when mimicking the clinical pathological changes of SCI. Taking this into account therefore, it would seem logical that the use of this contusive model system is able to reveal aFGF directly or indirectly inducing an anti-inflammation effect. Furthermore this might be beneficial to the repair process of the damage spinal cord.
Scar formation is regarded as a major mechanical obstacle to axonal regeneration after SCI, and these regions contain reactive astrocytes as a major component (67). A previous study has shown that spinal cord regeneration can be facilitated by neutralizing the scar using chondroitinase ABC (68). When the spinal cord is injured, many astrocytes in the injured region will switch to the reactive phenotype, a process called gliosis. Consequent to this, the reactive astrocytes are able to release inhibitory extracellular matrix molecules known as CSPG, which forms the major component of the glial scar after SCI. Moreover it has been found that mice that are deficient in the GFAP and vimentin genes show significantly improved axonal plasticity and functional recovery after spinal cord injury (69). Our results demonstrated that expression of GFAP at the transcriptional and translational levels was elevated after SCI. Most interestingly, our data indicated that aFGF is able to reduce the elevation in GFAP found in SCI rats, which should result in an attenuation of the gliosis effect. This probably contributes in part to the improved repair process in the treated injured spinal cords.
It is noteworthy that axonal regeneration after chondroitinase ABC treatment is still limited, suggesting that other inhibitory factors might exist that inhibit axonal regeneration after SCI (68). A previous study has shown that KSPG is another major extracellular matrix molecule, and it appears to establish boundaries for axonal growth after spinal cord injury (57). KSPG also plays an important role in glial scar formation after brain injury (70). Comparatively CSPG is mainly expressed by astrocytes, whereas the expression of KSPG is located in reactive microglia, macrophages, and oligodendrocyte progenitor cells. Our results show that the elevation in gene and protein expression levels of KSPG is attenuated by an application of aFGF, and this may help to produce an alleviation of the scar formation in the damaged tissue.
Taken together, our data demonstrate that administration of aFGF to SCI rats seems to be able to attenuate the major pathological process of SCI secondary injury including inflammation, astrocyte activation, and scar formation. This might lead to axonal regeneration as a result of attenuation of gene or protein expression of these three negative factors that affect axonal outgrowth after SCI.
Proteins Involved in Regulation of Apoptosis or Signal Transduction—
The pathology of SCI secondary injury is also characterized by a reduced expression of synaptic molecules such as Rab3A that are involved in vesicle fusion and neurotransmitter release (71). Recently microarray experiments have further demonstrated that Rab3A is decreased 6.8-fold after SCI (24). Furthermore we found that a substantial amount of Rab-GDI can be detected using 2D PAGE analysis in the SCI rats but not in the shams at 24 h after operation. Thus, it was plausible to find that administration of aFGF to the SCI rats was able to reduce the increase in Rab-GDI to about 1.5–1.6-fold (Table II). It is well documented that Rab-GDI is a key regulatory factor for the conversion of Rab3A-GDP to Rab3A-GTP (72). In addition, Mladinic and Wintzer (73) reported that expression of a larger amount of Rab-GDI in spinal cord-injured opossums exerts an adverse effect on axon regeneration of the damaged spinal cord. Similarly the use of antisense oligonucleotides to repress the expression of Rab-GDI can lead to a positive effect on PC12 cell neurite extension (59). Obviously the reduction of Rab-GDI in SCI rats treated with aFGF that was found in our animal model might indicate the importance of aFGF to the maintenance of normal neurotransmitter release, and this might benefit the repair process of the SCI rats.
It is known that Hsp27 is an intrinsic cellular survival chaperone, and expression of this protein is able to rescue neurons from neuron growth factor deprivation-derived apoptosis (75). In addition, Hsp27 is able to directly interact with cytochrome c to inhibit caspase activation, which will also result in the protection of neuronal cells from apoptosis (76). Furthermore Benn et al. (77) have discovered that Hsp27 is up-regulated and phosphorylated after nerve injury, and these process are required for neuronal survival. Our present results are consistent with previous reports (24) where it is stated that the amount of Hsp27 increases in the spinal cords of SCI rats (Fig. 3 and Table II). In addition, we also discovered two isoforms of Hsp27 in SCI tissue by 2D PAGE. Furthermore we used 2D Western blotting to confirm that there is a change in the Hsp27 isoform present during the regeneration process of the injured spinal cord (Fig. 8). Moreover 2D Western blotting indicated that the pI 5.5 protein spot disappears from the SCI tissue after treatment with calf intestinal alkaline phosphatase.2 This is consistent with a previous report indicating that the pI of phosphorylated Hsp27 is 5.5 (78). Thus protein spot 46 may be the phosphorylated form of Hsp27. All theses results imply that elevation of Hsp27 may be a crucial procedure in the survival of damaged neurons after SCI.
Although Hsp27 clearly increased in abundance in response to injury, which was attenuated by aFGF treatment, it is not clear whether the intensity of phosphorylation signal of the pI 5.5 and pI 6.0 proteins from Fig. 8 (or Table II) is significantly more than the amount of increase of the Hsp27 protein before and after treatment with aFGF. Therefore more experiments should be done to verify the possibility that Hsp27 undergoes PTM during the regeneration process.
In contrast, we also detected an increase in another survival-related protein, RKIP, in the SCI rats (Fig. 3 and Table II). RKIP is a member of the PEBP family, which have an important function as inhibitors of kinase signaling pathway. It is known that Raf-1 is able to phosphorylate and activate MEK, a kinase that activates ERK (79, 80). Yeung et al. (81, 82) reported that RKIP may be able to break the binding interaction between Raf-1 and MEK, which will result in an interruption in signaling through the Raf-1/MEK/ERK pathway. This kinase cascade is evolutionarily conserved and regulates many vital cellular function such as survival, proliferation, and differentiation (83). These results probably imply that the neurons damaged by SCI are not able to adapt to the survival process via the Raf-1/MEK/ERK signal transduction pathway. Instead under this situation, Hsp27 might play a temporary role helping to promote neuronal survival after SCI. Both Hsp27 and RKIP were reduced to their normal expression levels after the SCI rats were treated with aFGF (Fig. 4, cluster A), and therefore, it would seem, that the Raf-1/MEK/ERK pathway plays a crucial part in cell survival. Thus, the reduction in RKIP among the aFGF-treated SCI rats may help to reestablish the Raf-1/MEK/ERK pathway to some extent. This may aid the partial recovery among the SCI rats demonstrated in this work (Fig. 1). A previous study has shown that aFGF is able to protect PC12 cells from glutamate-induced apoptosis via activation of the PI3K/AKT/GSK-3β pathway. Furthermore the ERK, the Akt pathway, or both are crucial for neuron survival under cell death stress, and either ERK or Akt activation is enough to rescue PC12 cells from apoptosis induced by neuron growth factor withdraw (84, 85).
One hypothesis that our experimental data indicate is that SCI neuron cells might prefer a survival pathway mediated by Hsp27. Nonetheless the alternative Raf-1/MEK/ERK pathway might be turned on after the SCI neurons are treated with aFGF. The mechanism of this alternative change is still elusive. Recently it was found that brain-derived neurotrophic factor could activate ERK, which leads to inhibition of the heat shock transcription factor, and this finally causes Hsp27 down-regulation (86). It must be mentioned that, although Hsp27 overexpression is important for neuron survival activity, it is not sufficient to activate neurite elongation (87). Therefore, we proposed that there is a shift in the balance of the signaling pathways in response to aFGF treatment from the Hsp27 pathway to the Raf-1/MEK/ERK and/or PI3K/AKT/GSK-3β pathways in vivo and that this might help the partial recovery of the SCI rats demonstrated in this study.
Protein Function Involved in Antioxidant Activity—
Large amounts of reactive oxygen species (ROS), including hydrogen peroxide, hydroxyl radicals, and peroxynitrate, are produced within a few hours after spinal cord injury (88, 89). These neurotoxic factors contribute to the pathology of secondary damage of spinal cord injury and play an important role in the progression of spinal cord injury (90). The major function of PRDXs is as protective antioxidants; their peroxidase activity is able to reduce the toxicity incurred by the overproduction of ROS in cells after neuron damage (91, 92). Therefore, the balance between ROS production and degradation is crucial to the maintenance of cells in a healthy condition. The protective antioxidants PRDX2 and PRDX6 were only found in protein cluster D of our proteomics analysis. The amount of these proteins was substantially reduced in cells from SCI rats indicating that any excess amount of ROS produced in the damaged tissue will not be detoxified, which will create unfavorable conditions for survival and regeneration of the damaged neurons. Moreover we found that aFGF was unable to rescue this effect and allow normal production of these protective antioxidants. This is despite the fact that aFGF was shown to generate a favorable condition for the repair process with respect to transporter activity, apoptosis, or signaling transduction as mentioned above. Scott et al. (90) reported that an injection of uric acid, a natural scavenger of peroxynitrite, into the spinal cord of SCI rats was able to improve hind limb locomotor recovery. This suggests that antioxidants do play a part in the repair process of the damaged spinal cord. Moreover the application of aFGF was able to only partially induce locomotor recovery among the SCI rats as measured by our animal behavior analysis (Fig. 1). Thus, all these experimental results clearly indicated that a range of different stimulating factors is required for the full recovery of the damage spinal cord. Therefore we would like to suggest that a mixture therapeutic strategy should be designed using a combination of multiple factors such as aFGF, uric acid, and any other neurotrophic factors that may produce an ideal recovery of the damage spinal cord.
Protein Function Involved in Enzyme Regulator Activity—
A previous study has demonstrated that apoptosis of oligodendrocytes is a prominent feature of the secondary damage after SCI (93). It was known that a larger amount of neuropsin, a serine protease, was found to be located in oligodendrocytes, and neuropsin-deficient mice showed alleviation of the severity of apoptosis of the oligodendrocytes (94). In addition to neuropsin, other serine proteases such as tissue-type plasminogen activator and matrix metalloproteinases also play a detrimental role in the pathology after CNS injury (95, 96). In fact, serpins, known as the serine protease inhibitor protein family, play a key role in regulating the activity of serine proteases in vivo (97). Thus, alteration in the balance between serine proteases and serpins could result in a pathological condition. In this work, we identified a contrapsin-like protease inhibitor 3 (CPI3), which is known to have an enzyme regulator activity (Fig. 4, cluster B). We found that the amount of this protein was 3.25-fold (Table II, spot ID1) higher than that of the sham. Similar results have been found in brain injury detected by microarray (98) and in segmental nerve ligation injury detected by proteomics analysis (99). Interestingly it was found that CPI3 belongs to the subfamily of serpins (100), and its proposed function is to inactivate neuropsin and to control the level of neuropsin in adult mouse brain (101). Up-regulation of CPI3 in SCI rat might indicate the necessity of the damaged tissue to counteract the excess amount of proteases induced after SCI. We found that treatment of the SCI with aFGF might reduce nearly 50% of the amount of CPI3 (Table I, spot ID1). Nevertheless we do not expect that aFGF may play a direct role in regulating the amount of CPI3; instead the down-regulation of CPI3 may only reflect the condition becoming favorable for regeneration of the SCI after the treatment with aFGF. Therefore, revealing the function of these proteases and their cognate serpins could lead to the development of better therapeutic strategies for preventing cell death after traumatic CNS injury.
Potential Molecular Mechanism of the Regulatory Proteins Involved in the Regeneration of Spinal Cord Injury Induced by aFGF—
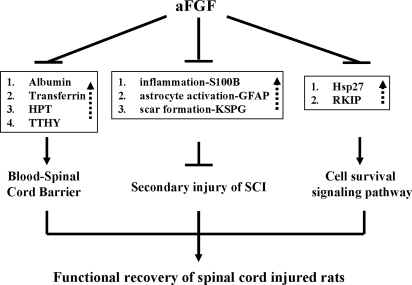
Taken together, our results revealed a possible protective pathway that is induced by aFGF (Fig. 9). Treatment with aFGF may attenuate SCI secondary injury by suppressing the overproduction of proteins involved in inflammation (S100B) and in scar formation (KSPG and GFAP) in the SCI tissue. Moreover aFGF may induce normal production of neurotransmitters (Rab-GDI) and alleviate the production of an inhibitor (RKIP) of the Raf/MEK/ERK pathway; this should create favorable conditions for the regeneration process and the repair of the SCI tissue.
Fig. 9.
A hypothetical protective pathway involving aFGF during the process of spinal cord repair in SCI rats. Basically aFGF may interact through three different discrete pathways to create favorable conditions for the functional recovery of a SCI rat. The first pathway is maintaining the blood-spinal cord barrier. Albumin, transferrin, HPT, and TTHY are proteins normally situated in the peripheral blood circulatory system. An increased level of these proteins was found in the damage spinal cord tissue, indicating a breakdown of the blood-spinal cord barrier. Administration of aFGF seemed to attenuate the increase in these proteins, showing that aFGF might be involved in maintaining the blood-spinal cord barrier across the damaged tissue. In turn, this may create favorable conditions for the functional recovery of the SCI rat. The second pathway is alleviation of the causes of secondary injury. The inhibitory effect of the S100B, GFAP, and KSPG proteins on the repair processes of the damage spinal cord might be alleviated by the suppression effect of aFGF, which reduced the elevation in these proteins in the damaged tissue. The third pathway is encouragement of the cell survival signal transduction pathway during functional recovery. The inhibitory effect of aFGF on the increase in RKIP after damage may encourage the initiation of the ERK signal transduction pathway for survival and regeneration of the damaged spinal cord. Note that solid arrows indicate activation, dotted arrows indicate that the amount of protein was increased in the damaged spinal cord tissue, and the barred lines indicate repression. Proteins with a similar function in spinal cord repair are boxed.
In conclusion, our results suggested that aFGF does improve functional recovery of contusive spinal cord-injured rats in an effective manner. We also found quite a few novel injury-associated and aFGF-responsive proteins in the adult spinal cord using an unbiased comparative proteomics technique. Thus, these results provide numerous insights into the degeneration/regeneration mechanisms that occur after SCI. These insights will help to provide data to aid the development of therapeutic approaches for the reduction of the effects of secondary injury after SCI. A study of the mechanisms behind the protective effect induced by the application of aFGF should be beneficial to patients with spinal cord injury in the future.
Supplementary Material
Acknowledgments
We show deepest gratitude to Dr. Shun-Fen Tzeng of National Cheng Kung University, Tainan, Taiwan for technical support with the animal surgery and help with the interpretation of the data. Protein identification was done in the Proteomic Research Center at National Yang Ming University and Core Facilities for Proteomic Research at the Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan.
Footnotes
Published, MCP Papers in Press, May 14, 2008, DOI 10.1074/mcp.M800076-MCP200
The abbreviations used are: SCI, spinal cord injury; aFGF, acidic fibroblast growth factor; ERK, extracellular signal-regulated kinase; GFAP, glial fibrillary acidic protein; CNS, central nervous system; PI3K, phosphatidylinositol 3-kinase; PTM, post-translational modification; 2D, two-dimensional; KSPG, keratan sulfate proteoglycan lumican; BBB, Basso, Beattie, and Bresnahan; CBS, combined behavior score; UPGMA, unweighted pair group method with arithmetic mean; BGSSJ, Bulk Gene Search System for Java; qPCR, quantitative real time PCR; Ct, cross-threshold; GAP-43, growth-associated protein 43; TTHY, transthyretin; TRFE, transferrin; HPT, haptoglobin; ALBU, albumin; CSPG, chondroitin sulfate proteoglycan; Hsp27, heat shock protein 27; PEBP1, phosphatidylethanolamine-binding protein 1; GDI, GDP dissociation inhibitor; RKIP, Raf kinase inhibitor protein; MEK, mitogen-activated protein kinase; KCRB, creatine kinase brain type; AINX, α-internexin; PRDX, peroxiredoxin; UCHL1, ubiquitin carboxyl-terminal hydrolase isozyme L1; BSCB, blood-spinal cord barrier; ROS, reactive oxygen species; CPI3, contrapsin-like protease inhibitor 3; ID, identification.
M.-C. Tsai, H. Cheng, and K.-F. Chak, unpublished data.
This work was supported by the National Science Council of Taiwan under the National Research Project for Genomic Medicines (Grants NSC92-3112-B-010-002, NSC93-3112-B-010-024, and NSC94-3112-B-010-001), the Yen Tjing Ling Medical Foundation (Grant CI-95-10), the Veterans General Hospital-University System of Taiwan (Grant VGHUST97-P6-20), and a research grant from the Aim for the Top University Plan for Nation Yang Ming University.The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Ackery, A., Tator, C., and Krassioukov, A. ( 2004) A global perspective on spinal cord injury epidemiology. J. Neurotrauma 21, 1355–1370 [DOI] [PubMed] [Google Scholar]
- 2.Thuret, S., Moon, L. D., and Gage, F. H. ( 2006) Therapeutic interventions after spinal cord injury. Nat. Rev. Neurosci. 7, 628–643 [DOI] [PubMed] [Google Scholar]
- 3.Maxwell, W. L., Povlishock, J. T., and Graham, D. L. ( 1997) A mechanistic analysis of nondisruptive axonal injury: a review. J. Neurotrauma 14, 419–440 [DOI] [PubMed] [Google Scholar]
- 4.Buki, A., Koizumi, H., and Povlishock, J. T. ( 1999) Moderate posttraumatic hypothermia decreases early calpain-mediated proteolysis and concomitant cytoskeletal compromise in traumatic axonal injury. Exp. Neurol. 159, 319–328 [DOI] [PubMed] [Google Scholar]
- 5.Shields, D. C., Schaecher, K. E., Hogan, E. L., and Banik, N. L. ( 2000) Calpain activity and expression increased in activated glial and inflammatory cells in penumbra of spinal cord injury lesion. J. Neurosci. Res. 61, 146–150 [DOI] [PubMed] [Google Scholar]
- 6.Okonkwo, D. O., and Povlishock, J. T. ( 1999) An intrathecal bolus of cyclosporin A before injury preserves mitochondrial integrity and attenuates axonal disruption in traumatic brain injury. J. Cereb. Blood Flow Metab. 19, 443–451 [DOI] [PubMed] [Google Scholar]
- 7.Buki, A., Okonkwo, D. O., Wang, K. K., and Povlishock, J. T. ( 2000) Cytochrome c release and caspase activation in traumatic axonal injury. J. Neurosci. 20, 2825–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridet, J. L., Malhotra, S. K., Privat, A., and Gage, F. H. ( 1997) Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci. 20, 570–577 [DOI] [PubMed] [Google Scholar]
- 9.Bareyre, F. M., and Schwab, M. E. ( 2003) Inflammation, degeneration and regeneration in the injured spinal cord: insights from DNA microarrays. Trends Neurosci. 26, 555–563 [DOI] [PubMed] [Google Scholar]
- 10.Schwab, M. E., and Bartholdi, D. ( 1996) Degeneration and regeneration of axons in the lesioned spinal cord. Physiol. Rev. 76, 319–370 [DOI] [PubMed] [Google Scholar]
- 11.Grill, R., Murai, K., Blesch, A., Gage, F. H., and Tuszynski, M. H. ( 1997) Cellular delivery of neurotrophin-3 promotes corticospinal axonal growth and partial functional recovery after spinal cord injury. J. Neurosci. 17, 5560–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grill, R. J., Blesch, A., and Tuszynski, M. H. ( 1997) Robust growth of chronically injured spinal cord axons induced by grafts of genetically modified NGF-secreting cells. Exp. Neurol. 148, 444–452 [DOI] [PubMed] [Google Scholar]
- 13.Jin, Y., Fischer, I., Tessler, A., and Houle, J. D. ( 2002) Transplants of fibroblasts genetically modified to express BDNF promote axonal regeneration from supraspinal neurons following chronic spinal cord injury. Exp. Neurol. 177, 265–275 [DOI] [PubMed] [Google Scholar]
- 14.Elde, R., Cao, Y. H., Cintra, A., Brelje, T. C., Pelto-Huikko, M., Junttila, T., Fuxe, K., Pettersson, R. F., and Hokfelt, T. ( 1991) Prominent expression of acidic fibroblast growth factor in motor and sensory neurons. Neuron 7, 349–364 [DOI] [PubMed] [Google Scholar]
- 15.Koshinaga, M., Sanon, H. R., and Whittemore, S. R. ( 1993) Altered acidic and basic fibroblast growth factor expression following spinal cord injury. Exp. Neurol. 120, 32–48 [DOI] [PubMed] [Google Scholar]
- 16.Reuss, B., and von Bohlen und Halbach, O. ( 2003) Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res. 313, 139–157 [DOI] [PubMed] [Google Scholar]
- 17.Bregman, B. S., McAtee, M., Dai, H. N., and Kuhn, P. L. ( 1997) Neurotrophic factors increase axonal growth after spinal cord injury and transplantation in the adult rat. Exp. Neurol. 148, 475–494 [DOI] [PubMed] [Google Scholar]
- 18.Bregman, B. S., Coumans, J. V., Dai, H. N., Kuhn, P. L., Lynskey, J., McAtee, M., and Sandhu, F. ( 2002) Transplants and neurotrophic factors increase regeneration and recovery of function after spinal cord injury. Prog. Brain Res. 137, 257–273 [DOI] [PubMed] [Google Scholar]
- 19.Lee, Y. S., Lin, C. Y., Robertson, R. T., Yu, J., Deng, X., Hsiao, I., and Lin, V. W. ( 2006) Re-growth of catecholaminergic fibers and protection of cholinergic spinal cord neurons in spinal repaired rats. Eur. J. Neurosci. 23, 693–702 [DOI] [PubMed] [Google Scholar]
- 20.Lin, P. H., Chuang, T. Y., Liao, K. K., Cheng, H., and Shih, Y. S. ( 2006) Functional recovery of chronic complete idiopathic transverse myelitis after administration of neurotrophic factors. Spinal Cord 44, 254–257 [DOI] [PubMed] [Google Scholar]
- 21.Cheng, H., Cao, Y., and Olson, L. ( 1996) Spinal cord repair in adult paraplegic rats: partial restoration of hind limb function. Science 273, 510–513 [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. S., Hsiao, I., and Lin, V. W. ( 2002) Peripheral nerve grafts and aFGF restore partial hindlimb function in adult paraplegic rats. J. Neurotrauma 19, 1203–1216 [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto, M., Sagara, Y., Langford, D., Everall, I. P., Mallory, M., Everson, A., Digicaylioglu, M., and Masliah, E. ( 2002) Fibroblast growth factor 1 regulates signaling via the glycogen synthase kinase-3β pathway. Implications for neuroprotection. J. Biol. Chem. 277, 32985–32991 [DOI] [PubMed] [Google Scholar]
- 24.Carmel, J. B., Galante, A., Soteropoulos, P., Tolias, P., Recce, M., Young, W., and Hart, R. P. ( 2001) Gene expression profiling of acute spinal cord injury reveals spreading inflammatory signals and neuron loss. Physiol. Genomics 7, 201–213 [DOI] [PubMed] [Google Scholar]
- 25.Liu, C. L., Jin, A. M., and Tong, B. H. ( 2003) Detection of gene expression pattern in the early stage after spinal cord injury by gene chip. Chin. J. Traumatol. 6, 18–22 [PubMed] [Google Scholar]
- 26.Pan, J. Z., Jornsten, R., and Hart, R. P. ( 2004) Screening anti-inflammatory compounds in injured spinal cord with microarrays: a comparison of bioinformatics analysis approaches. Physiol. Genomics 17, 201–214 [DOI] [PubMed] [Google Scholar]
- 27.Bareyre, F. M., Haudenschild, B., and Schwab, M. E. ( 2002) Long-lasting sprouting and gene expression changes induced by the monoclonal antibody IN-1 in the adult spinal cord. J. Neurosci. 22, 7097–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesic, O., Svrakic, N. M., Xu, G. Y., McAdoo, D., Westlund, K. N., Hulsebosch, C. E., Ye, Z., Galante, A., Soteropoulos, P., Tolias, P., Young, W., Hart, R. P., and Perez-Polo, J. R. ( 2002) DNA microarray analysis of the contused spinal cord: effect of NMDA receptor inhibition. J. Neurosci. Res. 68, 406–423 [DOI] [PubMed] [Google Scholar]
- 29.Ottens, A. K., Kobeissy, F. H., Golden, E. C., Zhang, Z., Haskins, W. E., Chen, S. S., Hayes, R. L., Wang, K. K., and Denslow, N. D. ( 2006) Neuroproteomics in neurotrauma. Mass Spectrom. Rev. 25, 380–408 [DOI] [PubMed] [Google Scholar]
- 30.Denslow, N., Michel, M. E., Temple, M. D., Hsu, C. Y., Saatman, K., and Hayes, R. L. ( 2003) Application of proteomics technology to the field of neurotrauma. J. Neurotrauma 20, 401–407 [DOI] [PubMed] [Google Scholar]
- 31.Freeman, W. M., and Hemby, S. E. ( 2004) Proteomics for protein expression profiling in neuroscience. Neurochem. Res. 29, 1065–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, R. S., Kinoshita, Y., Johnson, M. D., Uo, T., Ho, J. T., McBee, J. K., Conrads, T. P., and Veenstra, T. D. ( 2002) Proteomic analysis in the neurosciences. Mol. Cell. Proteomics 1, 553–560 [DOI] [PubMed] [Google Scholar]
- 33.Husi, H., and Grant, S. G. ( 2001) Proteomics of the nervous system. Trends Neurosci. 24, 259–266 [DOI] [PubMed] [Google Scholar]
- 34.Ding, Q., Wu, Z., Guo, Y., Zhao, C., Jia, Y., Kong, F., Chen, B., Wang, H., Xiong, S., Que, H., Jing, S., and Liu, S. ( 2006) Proteome analysis of up-regulated proteins in the rat spinal cord induced by transection injury. Proteomics 6, 505–518 [DOI] [PubMed] [Google Scholar]
- 35.Kang, S. K., So, H. H., Moon, Y. S., and Kim, C. H. ( 2006) Proteomic analysis of injured spinal cord tissue proteins using 2-DE and MALDI-TOF MS. Proteomics 6, 2797–2812 [DOI] [PubMed] [Google Scholar]
- 36.Basso, D. M., Beattie, M. S., and Bresnahan, J. C. ( 1995) A sensitive and reliable locomotor rating scale for open field testing in rats. J. Neurotrauma 12, 1–21 [DOI] [PubMed] [Google Scholar]
- 37.Cheng, H., Wu, J. P., and Tzeng, S. F. ( 2002) Neuroprotection of glial cell line-derived neurotrophic factor in damaged spinal cords following contusive injury. J. Neurosci. Res. 69, 397–405 [DOI] [PubMed] [Google Scholar]
- 38.Gale, K., Kerasidis, H., and Wrathall, J. R. ( 1985) Spinal cord contusion in the rat: behavioral analysis of functional neurologic impairment. Exp. Neurol. 88, 123–134 [DOI] [PubMed] [Google Scholar]
- 39.Seo, J., Bakay, M., Chen, Y. W., Hilmer, S., Shneiderman, B., and Hoffman, E. P. ( 2004) Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays. Bioinformatics 20, 2534–2544 [DOI] [PubMed] [Google Scholar]
- 40.Juan, H. F., Wang, I. H., Huang, T. C., Li, J. J., Chen, S. T., and Huang, H. C. ( 2006) Proteomics analysis of a novel compound: cyclic RGD in breast carcinoma cell line MCF-7. Proteomics 6, 2991–3000 [DOI] [PubMed] [Google Scholar]
- 41.Pan, Y. H., Liao, C. C., Kuo, C. C., Duan, K. J., Liang, P. H., Yuan, H. S., Hu, S. T., and Chak, K. F. ( 2006) The critical roles of polyamines in regulating ColE7 production and restricting ColE7 uptake of the colicin-producing Escherichia coli. J. Biol. Chem. 281, 13083–13091 [DOI] [PubMed] [Google Scholar]
- 42.Pandey, P., Farber, R., Nakazawa, A., Kumar, S., Bharti, A., Nalin, C., Weichselbaum, R., Kufe, D., and Kharbanda, S. ( 2000) Hsp27 functions as a negative regulator of cytochrome c-dependent activation of procaspase-3. Oncogene 19, 1975–1981 [DOI] [PubMed] [Google Scholar]
- 43.Karns, L. R., Ng, S. C., Freeman, J. A., and Fishman, M. C. ( 1987) Cloning of complementary DNA for GAP-43, a neuronal growth-related protein. Science 236, 597–600 [DOI] [PubMed] [Google Scholar]
- 44.Basi, G. S., Jacobson, R. D., Virag, I., Schilling, J., and Skene, J. H. ( 1987) Primary structure and transcriptional regulation of GAP-43, a protein associated with nerve growth. Cell 49, 785–791 [DOI] [PubMed] [Google Scholar]
- 45.Skene, J. H. ( 1989) Axonal growth-associated proteins. Annu. Rev. Neurosci. 12, 127–156 [DOI] [PubMed] [Google Scholar]
- 46.Demjen, D., Klussmann, S., Kleber, S., Zuliani, C., Stieltjes, B., Metzger, C., Hirt, U. A., Walczak, H., Falk, W., Essig, M., Edler, L., Krammer, P. H., and Martin-Villalba, A. ( 2004) Neutralization of CD95 ligand promotes regeneration and functional recovery after spinal cord injury. Nat. Med. 10, 389–395 [DOI] [PubMed] [Google Scholar]
- 47.Stein, R., Mori, N., Matthews, K., Lo, L. C., and Anderson, D. J. ( 1988) The NGF-inducible SCG10 mRNA encodes a novel membrane-bound protein present in growth cones and abundant in developing neurons. Neuron 1, 463–476 [DOI] [PubMed] [Google Scholar]
- 48.Di, P. G., Lutjens, R., Osen-Sand, A., Sobel, A., Catsicas, S., and Grenningloh, G. ( 1997) Differential distribution of stathmin and SCG10 in developing neurons in culture. J. Neurosci. Res. 50, 1000–1009 [DOI] [PubMed] [Google Scholar]
- 49.Nakazawa, T., Nakano, I., Furuyama, T., Morii, H., Tamai, M., and Mori, N. ( 2000) The SCG10-related gene family in the developing rat retina: persistent expression of SCLIP and stathmin in mature ganglion cell layer. Brain Res. 861, 399–407 [DOI] [PubMed] [Google Scholar]
- 50.Tsai, J. M., Wang, H. C., Leu, J. H., Hsiao, H. H., Wang, A. H., Kou, G. H., and Lo, C. F. ( 2004) Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78, 11360–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, L. F., Hughes, T. R., Davierwala, A. P., Robinson, M. D., Stoughton, R., and Altschuler, S. J. ( 2002) Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 31, 255–265 [DOI] [PubMed] [Google Scholar]
- 52.Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. ( 1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jimenez Hamann, M. C., Tator, C. H., and Shoichet, M. S. ( 2005) Injectable intrathecal delivery system for localized administration of EGF and FGF-2 to the injured rat spinal cord. Exp. Neurol. 194, 106–119 [DOI] [PubMed] [Google Scholar]
- 54.Liu, H. M., and Sturner, W. Q. ( 1988) Extravasation of plasma proteins in brain trauma. Forensic Sci. Int. 38, 285–295 [DOI] [PubMed] [Google Scholar]
- 55.Sharma, H. S. ( 2005) Pathophysiology of blood-spinal cord barrier in traumatic injury and repair. Curr. Pharm. Des. 11, 1353–1389 [DOI] [PubMed] [Google Scholar]
- 56.McKeon, R. J., Schreiber, R. C., Rudge, J. S., and Silver, J. ( 1991) Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J. Neurosci. 11, 3398–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones, L. L., and Tuszynski, M. H. ( 2002) Spinal cord injury elicits expression of keratan sulfate proteoglycans by macrophages, reactive microglia, and oligodendrocyte progenitors. J. Neurosci. 22, 4611–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park, J. B., Kim, J. S., Lee, J. Y., Kim, J., Seo, J. Y., and Kim, A. R. ( 2002) GTP binds to Rab3A in a complex with Ca2+/calmodulin. Biochem. J. 362, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leoni, C., Menegon, A., Benfenati, F., Toniolo, D., Pennuto, M., and Valtorta, F. ( 1999) Neurite extension occurs in the absence of regulated exocytosis in PC12 subclones. Mol. Biol. Cell 10, 2919–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aksenova, M., Butterfield, D. A., Zhang, S. X., Underwood, M., and Geddes, J. W. ( 2002) Increased protein oxidation and decreased creatine kinase BB expression and activity after spinal cord contusion injury. J. Neurotrauma 19, 491–502 [DOI] [PubMed] [Google Scholar]
- 61.Huber, L. A. ( 2003) Is proteomics heading in the wrong direction? Nat. Rev. Mol. Cell Biol. 4, 74–80 [DOI] [PubMed] [Google Scholar]
- 62.Garavelli, J. S. ( 2004) The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics 4, 1527–1533 [DOI] [PubMed] [Google Scholar]
- 63.Metz, G. A., Curt, A., van de Meent, H., Klusman, I., Schwab, M. E., and Dietz, V. ( 2000) Validation of the weight-drop contusion model in rats: a comparative study of human spinal cord injury. J. Neurotrauma 17, 1–17 [DOI] [PubMed] [Google Scholar]
- 64.Sharma, H. S. ( 2003) Neurotrophic factors attenuate microvascular permeability disturbances and axonal injury following trauma to the rat spinal cord. Acta Neurochir. Suppl. 86, 383–388 [DOI] [PubMed] [Google Scholar]
- 65.Selinfreund, R. H., Barger, S. W., Pledger, W. J., and Van Eldik, L. J. ( 1991) Neurotrophic protein S100 beta stimulates glial cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 88, 3554–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sen, J., and Belli, A. ( 2007) S100B in neuropathologic states: the CRP of the brain? J. Neurosci. Res. 85, 1373–1380 [DOI] [PubMed] [Google Scholar]
- 67.Davies, S. J., Fitch, M. T., Memberg, S. P., Hall, A. K., Raisman, G., and Silver, J. ( 1997) Regeneration of adult axons in white matter tracts of the central nervous system. Nature 390, 680–683 [DOI] [PubMed] [Google Scholar]
- 68.Bradbury, E. J., Moon, L. D., Popat, R. J., King, V. R., Bennett, G. S., Patel, P. N., Fawcett, J. W., and McMahon, S. B. ( 2002) Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640 [DOI] [PubMed] [Google Scholar]
- 69.Menet, V., Prieto, M., Privat, A., and Ribotta, M. ( 2003) Axonal plasticity and functional recovery after spinal cord injury in mice deficient in both glial fibrillary acidic protein and vimentin genes. Proc. Natl. Acad. Sci. U. S. A. 100, 8999–9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, H., Uchimura, K., and Kadomatsu, K. ( 2006) Brain keratan sulfate and glial scar formation. Ann. N. Y. Acad. Sci. 1086, 81–90 [DOI] [PubMed] [Google Scholar]
- 71.Grabs, D., Bergmann, M., Urban, M., Post, A., and Gratzl, M. ( 1996) Rab3 proteins and SNAP-25, essential components of the exocytosis machinery in conventional synapses, are absent from ribbon synapses of the mouse retina. Eur. J. Neurosci. 8, 162–168 [DOI] [PubMed] [Google Scholar]
- 72.Nuoffer, C., and Balch, W. E. ( 1994) GTPases: multifunctional molecular switches regulating vesicular traffic. Annu. Rev. Biochem. 63, 949–990 [DOI] [PubMed] [Google Scholar]
- 73.Mladinic, M., and Wintzer, M. ( 2002) Changes in mRNA content of developing opossum spinal cord at stages when regeneration can and cannot occur after injury. Brain Res. Brain Res. Rev. 40, 317–324 [DOI] [PubMed] [Google Scholar]
- 74.Tai, J. T., Brooks, E. E., Liang, S., Somogyi, R., Rosete, J. D., Lawn, R. M., and Shiffman, D. ( 2000) Determination of temporal expression patterns for multiple genes in the rat carotid artery injury model. Arterioscler. Thromb. Vasc. Biol. 20, 2184–2191 [DOI] [PubMed] [Google Scholar]
- 75.Lewis, S. E., Mannion, R. J., White, F. A., Coggeshall, R. E., Beggs, S., Costigan, M., Martin, J. L., Dillmann, W. H., and Woolf, C. J. ( 1999) A role for HSP27 in sensory neuron survival. J. Neurosci. 19, 8945–8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bruey, J. M., Ducasse, C., Bonniaud, P., Ravagnan, L., Susin, S. A., Diaz-Latoud, C., Gurbuxani, S., Arrigo, A. P., Kroemer, G., Solary, E., and Garrido, C. ( 2000) Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat. Cell Biol. 2, 645–652 [DOI] [PubMed] [Google Scholar]
- 77.Benn, S. C., Perrelet, D., Kato, A. C., Scholz, J., Decosterd, I., Mannion, R. J., Bakowska, J. C., and Woolf, C. J. ( 2002) Hsp27 upregulation and phosphorylation is required for injured sensory and motor neuron survival. Neuron 36, 45–56 [DOI] [PubMed] [Google Scholar]
- 78.Kumar, Y., Khachane, A., Belwal, M., Das, S., Somsundaram, K., and Tatu, U. ( 2004) ProteoMod: a new tool to quantitate protein post-translational modifications. Proteomics 4, 1672–1683 [DOI] [PubMed] [Google Scholar]
- 79.Morrison, D. K., and Cutler, R. E. ( 1997) The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9, 174–179 [DOI] [PubMed] [Google Scholar]
- 80.Morrison, D. K. ( 1995) Mechanisms regulating Raf-1 activity in signal transduction pathways. Mol. Reprod. Dev. 42, 507–514 [DOI] [PubMed] [Google Scholar]
- 81.Yeung, K., Janosch, P., McFerran, B., Rose, D. W., Mischak, H., Sedivy, J. M., and Kolch, W. ( 2000) Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the raf kinase inhibitor protein. Mol. Cell. Biol. 20, 3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yeung, K., Seitz, T., Li, S., Janosch, P., McFerran, B., Kaiser, C., Fee, F., Katsanakis, K. D., Rose, D. W., Mischak, H., Sedivy, J. M., and Kolch, W. ( 1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 401, 173–177 [DOI] [PubMed] [Google Scholar]
- 83.Wellbrock, C., Karasarides, M., and Marais, R. ( 2004) The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 84.Xia, Z., Dickens, M., Raingeaud, J., Davis, R. J., and Greenberg, M. E. ( 1995) Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 85.Namikawa, K., Honma, M., Abe, K., Takeda, M., Mansur, K., Obata, T., Miwa, A., Okado, H., and Kiyama, H. ( 2000) Akt/protein kinase B prevents injury-induced motoneuron death and accelerates axonal regeneration. J. Neurosci. 20, 2875–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krueger-Naug, A. M., Emsley, J. G., Myers, T. L., Currie, R. W., and Clarke, D. B. ( 2003) Administration of brain-derived neurotrophic factor suppresses the expression of heat shock protein 27 in rat retinal ganglion cells following axotomy. Neuroscience 116, 49–58 [DOI] [PubMed] [Google Scholar]
- 87.Nakagomi, S., Suzuki, Y., Namikawa, K., Kiryu-Seo, S., and Kiyama, H. ( 2003) Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J. Neurosci. 23, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee, Y. S., Sindhu, R. K., Lin, C. Y., Ehdaie, A., Lin, V. W., and Vaziri, N. D. ( 2004) Effects of nerve graft on nitric oxide synthase, NAD(P)H oxidase, and antioxidant enzymes in chronic spinal cord injury. Free Radic. Biol. Med. 36, 330–339 [DOI] [PubMed] [Google Scholar]
- 89.Olivas, A. D., and Noble-Haeusslein, L. J. ( 2006) Phospholipase A2 and spinal cord injury: a novel target for therapeutic intervention. Ann. Neurol. 59, 577–579 [DOI] [PubMed] [Google Scholar]
- 90.Scott, G. S., Cuzzocrea, S., Genovese, T., Koprowski, H., and Hooper, D. C. ( 2005) Uric acid protects against secondary damage after spinal cord injury. Proc. Natl. Acad. Sci. U. S. A. 102, 3483–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hofmann, B., Hecht, H. J., and Flohe, L. ( 2002) Peroxiredoxins. Biol. Chem. 383, 347–364 [DOI] [PubMed] [Google Scholar]
- 92.Peshenko, I. V., and Shichi, H. ( 2001) Oxidation of active center cysteine of bovine 1-Cys peroxiredoxin to the cysteine sulfenic acid form by peroxide and peroxynitrite. Free Radic. Biol. Med. 31, 292–303 [DOI] [PubMed] [Google Scholar]
- 93.Park, E., Velumian, A. A., and Fehlings, M. G. ( 2004) The role of excitotoxicity in secondary mechanisms of spinal cord injury: a review with an emphasis on the implications for white matter degeneration. J. Neurotrauma 21, 754–774 [DOI] [PubMed] [Google Scholar]
- 94.Terayama, R., Bando, Y., Murakami, K., Kato, K., Kishibe, M., and Yoshida, S. ( 2007) Neuropsin promotes oligodendrocyte death, demyelination and axonal degeneration after spinal cord injury. Neuroscience 148, 175–187 [DOI] [PubMed] [Google Scholar]
- 95.Abe, Y., Nakamura, H., Yoshino, O., Oya, T., and Kimura, T. ( 2003) Decreased neural damage after spinal cord injury in tPA-deficient mice. J. Neurotrauma 20, 43–57 [DOI] [PubMed] [Google Scholar]
- 96.Wells, J. E., Rice, T. K., Nuttall, R. K., Edwards, D. R., Zekki, H., Rivest, S., and Yong, V. W. ( 2003) An adverse role for matrix metalloproteinase 12 after spinal cord injury in mice. J. Neurosci. 23, 10107–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miranda, E., and Lomas, D. A. ( 2006) Neuroserpin: a serpin to think about. CMLS Cell. Mol. Life Sci. 63, 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matzilevich, D. A., Rall, J. M., Moore, A. N., Grill, R. J., and Dash, P. K. ( 2002) High-density microarray analysis of hippocampal gene expression following experimental brain injury. J. Neurosci. Res. 67, 646–663 [DOI] [PubMed] [Google Scholar]
- 99.Komori, N., Takemori, N., Kim, H. K., Singh, A., Hwang, S. H., Foreman, R. D., Chung, K., Chung, J. M., and Matsumoto, H. ( 2007) Proteomics study of neuropathic and nonneuropathic dorsal root ganglia: altered protein regulation following segmental spinal nerve ligation injury. Physiol. Genomics 29, 215–230 [DOI] [PubMed] [Google Scholar]
- 100.Ohkubo, K., Ogata, S., Misumi, Y., Takami, N., and Ikehara, Y. ( 1991) Molecular cloning and characterization of rat contrapsin-like protease inhibitor and related proteins. J. Biochem. 109, 243–250 [PubMed] [Google Scholar]
- 101.Kato, K., Kishi, T., Kamachi, T., Akisada, M., Oka, T., Midorikawa, R., Takio, K., Dohmae, N., Bird, P. I., Sun, J., Scott, F., Miyake, Y., Yamamoto, K., Machida, A., Tanaka, T., Matsumoto, K., Shibata, M., and Shiosaka, S. ( 2001) Serine proteinase inhibitor 3 and murinoglobulin I are potent inhibitors of neuropsin in adult mouse brain. J. Biol. Chem. 276, 14562–14571 [DOI] [PubMed] [Google Scholar]
- 102.Willis, D., Li, K. W., Zheng, J. Q., Chang, J. H., Smit, A., Kelly, T., Merianda, T. T., Sylvester, J., van Minnen, J., and Twiss, J. L. ( 2005) Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci. 25, 778–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raghavendra, V., Tanga, F. Y., and DeLeo, J. A. ( 2004) Complete Freunds adjuvant-induced peripheral inflammation evokes glial activation and proinflammatory cytokine expression in the CNS. Eur. J. Neurosci. 20, 467–473 [DOI] [PubMed] [Google Scholar]
- 104.Jimenez, C. R., Stam, F. J., Li, K. W., Gouwenberg, Y., Hornshaw, M. P., De Winter, F., Verhaagen, J., and Smit, A. B. ( 2005) Proteomics of the injured rat sciatic nerve reveals protein expression dynamics during regeneration. Mol. Cell. Proteomics 4, 120–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.