Abstract
Investigating the proteome of intracellular pathogens is often hampered by inadequate methodologies to purify the pathogen free of host cell material. This has also precluded direct proteome analysis of the intracellular, amastigote form of Leishmania spp., protozoan parasites that cause a spectrum of diseases that affect some 12 million patients worldwide. Here a method is presented that combines classic, isopycnic density centrifugation with fluorescent particle sorting for purification by exploiting transgenic, fluorescent parasites to allow direct proteome analysis of the purified organisms. By this approach the proteome of intracellular Leishmania mexicana amastigotes was compared with that of extracellular promastigotes that are transmitted by insect vectors. In total, 509 different proteins were identified by mass spectrometry and database search. This number corresponds to ∼6% of gene products predicted from the reference genome of Leishmania major. Intracellular amastigotes synthesized significantly more proteins with basic pI and showed a greater abundance of enzymes of fatty acid catabolism, which may reflect their living in acidic habitats and metabolic adaptation to nutrient availability, respectively. Bioinformatics analyses of the genes corresponding to the protein data sets produced clear evidence for skewed codon usage and translational bias in these organisms. Moreover analysis of the subset of genes whose products were more abundant in amastigotes revealed characteristic sequence motifs in 3′-untranslated regions that have been linked to translational control elements. This suggests that proteome data sets may be used to identify regulatory elements in mRNAs. Last but not least, at 6% coverage the proteome identified all vaccine antigens tested to date. Thus, the present data set provides a valuable resource for selection of candidate vaccine antigens.
Leishmania spp. are unicellular parasites of the trypanosomatid family and the causative agents of a spectrum of diseases, the leishmaniases, that affect some 12 million people worldwide (1). They oscillate between free living, flagellated promastigotes transmitted by blood sucking insect vectors and intracellular, non-flagellated amastigotes. These dwell in vacuoles akin to late endosomes/early lysosomes primarily in phagocytic cells of vertebrate hosts (2). This life cycle brings about dramatic morphological and molecular changes provoked by, and evolved to cope with, the change in habitat (3). Comprehensive molecular analysis of these changes in amastigotes may reveal targets for drug development and vaccine design, but our current understanding remains very rudimentary.
The 32.8-Mbp genome sequence of Leishmania major was completed in 2005 and predicts ∼8300 ORFs organized in 133 polycistronic units of directional gene clusters spread over 36 chromosomes (4) (GeneDB). Other species are being sequenced, and data suggest a very high degree of conservation and synteny of polycistronic units throughout the genus (5) (GeneDB). This information allowed the adaptation of highly processive methods such as microarray analyses (6, 7) to compare life cycle forms also of unsequenced Leishmania spp. (8, 9). However, trypanosomatids transcribe almost their entire genome constitutively and regulate gene expression mostly post-transcriptionally and at the translational level. Because of the latter and because the relationship between mRNA and protein levels may not always be proportional, there is very limited analytical power in transcriptional profiles of the response to changes in habitat unlike e.g. in malaria parasites (10). For this reason, cataloguing molecular changes when Leishmania transforms from promastigotes to amastigotes has been pursued by proteomics (11–17).
A major handicap for proteome studies has been contamination of amastigotes with host cell material, which prevents direct proteome analysis. Amastigote-like forms can be grown host cell-free under conditions of low pH and higher temperature (22) that mimic the intracellular habitat, and these have been used as a substitute for the intracellular forms (13, 14, 17–19). However, these axenic amastigotes can only be grown from a few species, and although they display a number of biochemical markers of the intracellular stage (3), they fail to synthesize some major products of true amastigotes, e.g. the secreted amastigote-specific proteophosphoglycan in the case of Leishmania mexicana. Furthermore recent genome-wide mRNA profiling suggests that axenic amastigotes are more closely related to promastigotes than to ex vivo isolated amastigotes (8). Thus, current data sets on amastigote proteomes likely underestimate differences and could miss important changes. Overcoming the technical hurdles to intracellular parasite purification therefore seems crucial.
Here we report a novel purification protocol based on fluorescent parasites and fluorescent particle sorting to isolate amastigotes from their intracellular habitat in sufficient quantity and purity for direct proteome analysis. We applied this protocol to investigate the proteome of L. mexicana amastigotes and compared it with that of promastigotes. In total, 509 proteins (∼6% of predicted ORFs) were identified, and 34 were more abundant in amastigotes. Bioinformatics analysis of these data sets revealed a number of general characteristics of the parasite proteome and yielded novel insight into the biology of the parasite.
EXPERIMENTAL PROCEDURES
Growth and Differentiation of L. mexicana—
L. mexicana mexicana (MNYC/BZ/62/M379) expressing DsRed (20) were maintained in selective medium as described previously (21). Amastigotes were obtained and maintained in Schneider's Drosophila medium (22) supplemented with 20 μg/ml hygromycin B (Roche Applied Science). For proteome analyses, promastigotes were harvested in late logarithmic phase at a cell density of 6–7 × 107 parasites/ml.
Infection of Mice and Bone Marrow-derived Macrophages—
All animal experiments were approved by an ethics committee and licensed by the legal authority. BALB/c and C57BL/6 mice were purchased from Charles River, Sulzfeld, Germany and maintained in a conventional animal facility. Mice were infected with stationary phase promastigotes at the base of the tail where lesions developed. Mice with lesions were killed by cervical dislocation, and lesion tissue was excised for parasite isolation.
Macrophages were differentiated from bone marrow of 6–8-week-old female mice as described previously (23) and infected with single cell axenic amastigotes at a multiplicity of infection of 7–10. Infected cells were incubated for 24 h at 34 °C and 5% CO2 before harvesting.
Processing of Promastigotes and Isolation of Amastigotes from Infected Bone Marrow-derived Macrophages or Lesions—
Promastigotes in late logarithmic growth phase were harvested by centrifugation and processed according to Ref. 14. Briefly 2 × 108 parasites were washed once with PBS (Invitrogen, 14190-094) and three times with Tris/sucrose buffer (10 mm Tris-HCl, pH 7.4, 0.25 m sucrose). Parasites were resuspended in incomplete lysis buffer (2% N-decyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (SB3–10; Calbiochem), 2% CHAPS, 7 m urea, 2 m thiourea, 100 μg/ml leupeptin, 500 μg/ml Pefabloc (Roche Applied Science), 25 μl/ml Pefabloc protector (Roche Applied Science), 68.5 ng/ml pepstatin A, 174.2 μg/ml PMSF, 1 mm 1,10-phenanthroline, 1 mm each EDTA/EGTA, 25 μg/ml N-[N-(l-3-trans-carboxirane-2-carbonyl)-l-leucyl]-agmatine (E-64; Roche Applied Science); shock frozen in liquid nitrogen; and stored at −80 °C until 2-DE1 analysis.
Infected bone marrow-derived macrophages were abraded in homogenization buffer (20 mm HEPES-KOH, pH 7.3, 0.25 m sucrose supplemented with “Complete Mini” (Roche Applied Science)). Cells were lysed by shear force in 1-ml portions of homogenization buffer using a 1-ml syringe fitted with a 26-gauge needle. Nuclei were pelleted for 2 min at 100 × g, and supernatants were loaded onto discontinuous sucrose gradients of 3 ml each: 60, 40, and 20% (w/w) sucrose in HEPES saline (30 mm HEPES-KOH, pH 7.3, 0.1 m NaCl, 0.5 mm CaCl2, 0.5 mm MgCl2) (24). Gradients were centrifuged for 25 min at 700 × g. Parasites were harvested from the 40/60% interphase, diluted in PBS, and centrifuged for 10 min at 1200 × g.
The cells were resuspended in PBS supplemented with protease inhibitors (see above). 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) was added to the sample before sorting on a FACSDiVa (BD Biosciences) for DsRed+ and DAPI− events. Sorted parasites were collected in 50-ml tubes containing 5 ml of PBS with protease inhibitors and centrifuged at 1200 × g for 10 min. Supernatant was removed completely, and parasite pellets were lysed in incomplete lysis buffer as described above for promastigotes.
Excised lesion material was forced in PBS supplemented with Complete Mini through a 70-μm cell strainer (BD Biosciences). The material was centrifuged at 1200 × g at 4 °C. The resulting pellet was resuspended in an appropriate volume of PBS containing protease inhibitors and loaded onto the discontinuous sucrose gradient, and parasites were purified as above.
1-D and Two-dimensional Gel Electrophoresis—
DTT and Ampholytes were added to samples lysed in incomplete lysis buffer to a final concentration of 70 mm and 2%, respectively. Samples were mixed for 30 min at room temperature, and non-soluble material was cleared by centrifugation at room temperature (20,000 × g for 10 min). Supernatants were either used immediately or stored at −80 °C; the resulting pellet was kept and processed as described below.
To resolve promastigote and amastigote samples via 2-DE, maximum volumes for preparative gels (corresponding to 240–300 μg of protein as determined by Lowry assay were applied to the anodic side of an IEF gel (diameter of 1.5 and 2.5 mm for promastigotes and amastigotes, respectively). The first dimension gel was loaded onto a 23 × 30-cm 2-DE gel system with a resolution power of about 5000 protein species (25). Gels were stained by Coomassie Brilliant Blue G-250 (26). All visible protein spots were excised and processed for MALDI-TOF/TOF mass spectrometry as described previously (25).
The residual insoluble material was washed twice with 9 m urea, 70 mm DTT, 2% CHAPS. After washing, it was centrifuged for 30 min at 100,000 × g at 20 °C. The pellet was resolved in Laemmli buffer, agitated at 37 °C for 10 min, then boiled for 5 min, and centrifuged again for 30 min at 100,000 × g at 20 °C. The supernatant was loaded on a large 15% SDS-PAGE gel.
Sample lanes were cut in 96 slices, and every slice was divided in four pieces and transferred into a 96-well plate. Samples were washed three times with agitation for 30 min with 200 μl of destaining buffer and equilibrated in 200 μl of trypsin digestion buffer as described previously (25). The digested dried samples were resolved in 12 μl of 3% ACN, 0.1% formic acid. 6 μl were used for one LC-MS analysis. The remainder was stored at −80 °C.
Mass Spectrometry—
The peptides from excised spots from the 2-DE gels were analyzed by MALDI-TOF/TOF MS using a 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA) as described previously (25). After in-gel tryptic digestion peptides were dissolved in 1 μl of sample buffer (33% ACN, 0.1% TFA) of which 0.25 μl was mixed with 0.5 μl of matrix solution (α-cyano-4-hydroxycinnamic acid in 50% ACN, 0.3% TFA) and applied to a MALDI plate.
The genome of L. mexicana is not yet available, therefore protein identities were assigned by searching the L. major protein database (version GeneDB_protein_database_090704 containing 8217 sequences) and NCBInr (version 20070908 containing 5,454,477 sequences). Proteins were identified using Mascot version 2.1 (Matrix Science) MS/MS Ion Search with combined peptide mass fingerprint (PMF) and MS/MS information, allowing a peptide mass tolerance of 30 ppm and ±0.3 Da for the fragment mass tolerance. A maximum of one missed cleavage was allowed; oxidation of methionine, N-terminal acetylation of the protein, propionamide at cysteine residues, and N-terminal pyroglutamic acid formation were defined as variable modifications in these searches. For identification of a protein within a 2-DE spot the following identification criteria were used (23). A protein was considered identified by the MS analysis if at least 30% sequence coverage was obtained, or in case sequence coverage was between 15 and 30%, at least one MS/MS spectrum with Mascot identity combined with mass loss information and hypercleavage sites (27) was necessary to reach identification status. If sequence coverage was below 15%, two MS/MS spectra had to fit with the protein, or one MS/MS spectrum and additional information from 2-DE, e.g. from neighboring spots or pI or protein Mr, was necessary.
Processed slices of the 1-D gel were analyzed by nano-LC ESI-MS/MS. Peptides were separated on an Agilent 1100 series chromatography unit (Agilent Technologies, Palo Alto, CA). Samples were loaded onto a trap column (Zorbax 300SB, C18, 5 × 0.3 mm, Agilent Technologies) and washed for 5 min with a flow rate of 0.02 ml/min and buffer A (3% ACN, 0.1% formic acid). Peptides were eluted for 75 min using a gradient of 5 min of 3% buffer B (99.9% ACN, 0.1% formic acid) increasing to 15% buffer B within 3 min and over a 42-min period increasing to 45% buffer B increasing for 5 min to 90% buffer B onto a separation column (Zorbax 300SB, C18, 3.5 μm/150 × 0.075 mm, Agilent Technologies) with a flow rate of 0.3 μl/min. The column outlet was coupled on line to an LC/MSD Trap XCT mass spectrometer (Agilent Technologies). Spectra were recorded between 10 and 68 min of the run in standard/enhanced mode. Precursor masses with m/z ratio of 200–2200 were allowed. Database search was performed using Mascot (version 2.1) MS/MS Ion Search. A peptide mass tolerance of 0.4 Da and a fragment mass tolerance of ±0.4 Da were accepted. A maximum of one missed cleavage and the same variable modifications as defined above were allowed.
For ESI-MS/MS data analysis, the ion score cutoff was 26, and proteins were classified as identified either if at least two peptides with a Mascot score above the statistically relevant threshold (p < 0.05) were found or if only one peptide achieved the required Mascot score that at least four consecutive y- or b-ions with a significant signal to background ratio could be determined.
Electron Microscopy—
Parasite samples were fixed in 25% electron microscopy grade glutaraldehyde fixative and processed for transmission electron microscopy as described previously (28).
Bioinformatics Analyses—
3′-Untranslated mRNA regions (3′-UTR) analysis was performed using the oligo-analysis tool (29). The tool identifies oligomers ranging in length from 4 to 8 nt that are more frequent within the UTR sequence of a group of co-regulated genes compared with a set of non-regulated genes. In our case, 51 intergenic sequences from the L. major genome were chosen for analysis of regulated loci. This test set was compared against a random selection of intergenic sequences from annotated protein-coding genes on L. major chromosomes 4, 7, 12, 18, 21, 25, 28, 30, and 36, totaling 2677 sequences. Statistics and significance values were determined according to Ref. 30.
The 51 regulated test loci (see supplemental Table 3) represented the 3′-intergenic regions of 35 ORFs for which the respective proteins were solely identified in amastigote samples by the present proteome analysis and 16 additional 3′-intergenic regions of ORFs encoding proteins found to be more abundant in amastigotes by isotope-coded protein labeling (71). The cysteine proteinase B ORF array was excluded as synthesis of the homologous L. major ORFs appears not to be stage-specifically regulated (31). Leishmanolysin (gp63)-encoding loci were excluded for the same reason because the respective genes are not expressed in L. major amastigotes (32). For Leishmania infantum, 3′-UTRs of 45 corresponding orthologs were searched for the presence of the identified oligonucleotide motifs.
To test whether a motif lay within the boundaries of a presumptive 3′-UTR, we predicted putative splice sites of the downstream ORFs that define the 5′-UTR boundary. The longest predicted 5′-UTR was chosen, and the putative end of the 3′-UTR of the upstream gene was arbitrarily set 250 nt further upstream, effectively representing a conservative (short) estimate of 3′-UTR length. Splice sites of downstream ORFs were predicted with the algorithm developed by Benz et al. (33) in the form adapted by Gopal et al. (34) using the following parameters: a minimum length of polypyrimidine tract of 8 with up to two mismatches but at least six consecutive pyrimidines and a splice site predicted within the 400 nt upstream of the ATG (34).
Codon adaptation indices (CAIs) (35) were calculated as described by Carbone et al. (36). Subcellular localization was predicted according to Szafron et al. (37). Virtual 2-DE Gel images were generated with JVirGel (38).
RESULTS
Purification of Intracellular Amastigotes—
To date, amastigotes have been purified by filtration or by density gradient centrifugation relying on their physical properties. Here the latter was combined with fluorescence-activated cell sorting (FACS) exploiting transgenic L. mexicana stably expressing DsRed (20). The method uses lesion tissue or in vitro cultured, infected host cells (Fig. 1A). After homogenization or lysis and fractionation on a discontinuous sucrose gradient, fluorescent parasites that concentrate at the 40/60% interphase were collected. This fraction was further stained with DAPI. The stained suspension was sorted, selecting for red fluorescent parasites but excluding DAPI-stained events and DAPI-positive dead parasites. This effectively created a dump channel. DAPI-positive material was prominent, indicating that host cell nuclear material was a major contaminant. Sorted parasites were finally recovered from the collection buffer by centrifugation at 1200 × g, which separated them further from loose membrane. The pelleted parasites were directly resuspended in lysis buffer at a ratio of 100 μl of buffer/108 parasites. We routinely sorted >2 × 108 parasites in approximately 4 h at speeds of 20,000 events/s.
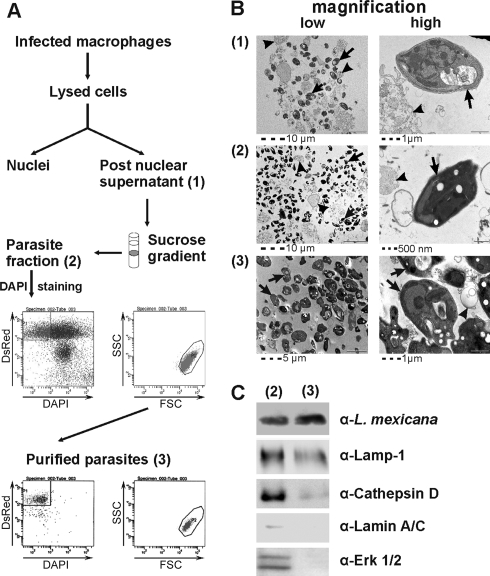
Fig. 1.
A, flow chart of amastigote purification for proteome analysis. Macrophages infected for 24 h with L. mexicana::DsRed were lysed, and nuclei were pelleted at low centrifugal force. The postnuclear supernatant (1) was overlaid onto a discontinuous sucrose gradient, and the parasite-containing fraction (2) was harvested and sorted by FACS. Gates were set on DsRed+/DAPI− and forward scatter (FSC)/side scatter (SSC) (gray population). Sorted parasites (3) were reanalyzed (bottom dot plots). B, analytical electron microscopy of parasite-containing fractions. Postnuclear supernatant (1) off the sucrose gradient (2) still contain host cell material (black arrowheads) that is absent in sorted parasites (3). Black arrows, free amastigotes; black arrowheads, loose membranes. C, Western blot analysis of parasite-containing fraction after the sucrose gradient (2) and post-FACS (3) monitoring the indicated host cell proteins and parasite antigen. Antibodies used were α-amastigotes (polyclonal rabbit anti-L. mexicana), α-Lamp-1 (clone 1D4B, BD Pharmingen), α-Cathepsin D (AF1029, R&D Systems), α-Lamin A/C (clone 14, BD Pharmingen), and α-ERK-1/2 (clone C-14, Santa Cruz Biotechnology).
This strategy allowed us to obtain highly pure parasites as demonstrated by cytofluorimetric reanalysis and analytical electron microscopy on aliquots taken at different steps during the purification (Fig. 1B). Of note, the phagosomal membrane was mostly intact after the density gradient but was non-continuous and broken after sorting. Western blot analysis (Fig. 1C) confirmed the depletion of cellular contaminants (e.g. Lamin) and enrichment of parasite material and retention of phagosomal proteins (LAMP-1 and Cathepsin) and thus corroborated the electron microscopy analysis.
Reproducibility was assessed by comparing purification in three individual biological experiments because yields of ∼2 × 108 parasites/sort may not suffice for all proteome applications. Bone marrow-derived macrophages were infected with axenic amastigotes for 24 h and lysed, and amastigotes were purified as described above. Parasite pellets were resuspended in lysis buffer, and lysates were subjected to 2-DE. Protein spot patterns were highly reproducible (not shown) indicating that material purified on individual sorts can be pooled for analysis if required.
Proteome Analysis—
Having established an efficient method to purify amastigotes from host cells, L. mexicana pro- and amastigotes were prepared for comparative proteome analysis. Promastigote culture conditions were optimized for reproducible growth, and cultures were harvested in late logarithmic phase as this population samples a range of cycling and non-cycling parasites akin to the likely population structure of lesion- or host cell-derived amastigotes. Amastigotes were purified by the described method from non-ulcerated lesions from BALB/c mice and in vitro infected bone marrow macrophages. Parasite lysates were processed for 2-DE, and in addition, material that remained insoluble in the 2-DE buffer was dissolved in SDS sample buffer and separated by one-dimensional SDS-PAGE (not shown) to be analyzed separately. All visible spots in the Coomassie-stained gels, i.e. 391 spots from promastigotes and 490 spots isolated from two gels from amastigotes (see supplemental and supportive information) prepared from infected macrophages were excised and processed for MALDI MS/MS analysis with 74 and 49% success in spot identification, respectively. Lanes from SDS-PAGE separating the urea-insoluble material were cut in 96 slices and processed for nano-LC ESI-MS/MS. Proteins were assigned by database searches based on PMFs or MS/MS ion search (for spectra, peptide assignments, and identification see supplemental and supportive data). The Leishmania genome contains many tandem arrays of identical or highly similar genes. In these cases protein assignment to an individual gene copy was not possible, and operationally PMFs and MS/MS spectra-derived sequences were therefore assigned to the respective gene product array. Although the L. mexicana genome is not yet sequenced, the validity of this strategy (14) is based on the very high degree of sequence conservation and synteny between Leishmania species (5). In total, 509 Leishmania proteins were assigned. Results from MALDI MS/MS- and ESI-MS/MS-analyzed samples were largely complementary because only 10% of the proteins were identified in both sets (Fig. 2). Compared with the genome sequence-predicted proteome, a conservative estimate of coverage counting only distinct proteins was ∼5.5% for promastigotes and 2.5% for amastigotes. Total coverage was ∼6%, and the list of all proteins identified is shown in supplemental Table 1. Only five spots in the amastigote sample corresponded to mouse proteins, indicating that contamination with host cell material was <2.3% (5 of 214; see supplemental Tables 1 and 2). The respective, identified peptides were assigned to vimentin and vacuolar ATPase A and B subunits, whereas another set matched HSP70 chaperones (most likely BiP), and one was assigned to an unnamed mouse protein.
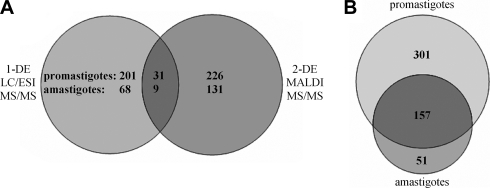
Fig. 2.
Complementary protein sets identified by MALDI MS/MS and nano-LC ESI-MS/MS. A, proteins identified by 2-DE MALDI MS/MS and/or SDS-PAGE with subsequent LC ESI-MS/MS in promastigotes and amastigotes, respectively. In total, 509 leishmanial proteins were identified (458 and 208 for promastigotes and amastigotes, respectively). B, number of identified proteins in different life cycle stages. 1-DE, one-dimensional gel electrophoresis.
General Characteristics of Pro- and Amastigote Proteomes—
To reveal general characteristics of the protein sets identified, bioinformatics analyses based on the L. major reference genome were pursued. These included calculating theoretical pI and molecular weight coordinates for the data sets, investigating bias in codon usage, and classifying inferred proteins based on likely function and predicted subcellular localization.
Visual inspection of 2-DE patterns of pro- and amastigote lysates suggested that relatively more basic proteins were present in amastigote samples (Fig. 3). We computed theoretical 2-DE plots for all predicted ORFs and the gene products identified in our electrophoretic analyses from pro- and amastigotes (Fig. 4). The genome-derived proteome showed a biphasic distribution reminiscent of the shape of a lung. When computing 2-DE coordinates of the pro- and amastigote-derived data sets, a shift in the pattern with more proteins with higher pI values was noted in the amastigote probe set; this is in perfect agreement with the experimental data sampling the abundant proteins from L. mexicana amastigotes (compare Figs. 3 and 4).
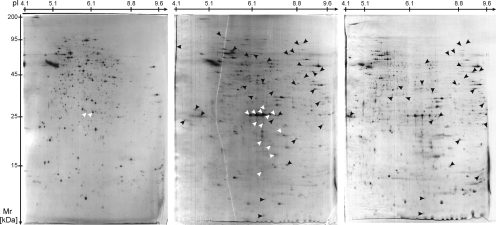
Fig. 3.
Comparison of 2-DE-separated proteome of L. mexicana::DsRed pro- and amastigotes. Left, promastigotes; middle, amastigotes isolated from bone marrow-derived macrophages; right, lesion amastigotes from infected BALB/c mice. White arrowheads point to spots identified as DsRed, and black arrowheads point to 31 spots only identified in amastigotes; the remaining 14 spots are in the second gel. Black arrowheads in the proteome of lesion amastigotes point to 26 spots assigned by PDQuest as being identical to marked spots in the 2-DE gel of amastigotes isolated from macrophages. For enlarged images and spot numbers see supplemental Figs. 1–3.
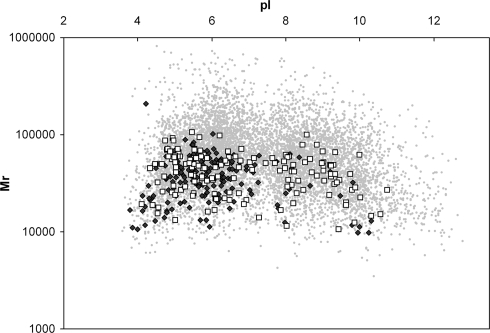
Fig. 4.
Amastigotes contain more proteins with basic pI. Shown is a virtual 2-DE gel image of L. major (Friedlin) reference genome ORFs (gray dots), ORFs corresponding to proteins identified in L. mexicana promastigotes (black dots), and ORFs corresponding to proteins identified in amastigotes (white squares). The genome shows a biphasic distribution with an average pI of 7.312 and a median of 6.9817, and it encodes for 50% acidic (pI < 7.0) proteins. The majority, 80% of protein homologues identified in L. mexicana promastigotes, were acidic in contrast to only 59% in amastigotes; this is significantly different p < 0.0001 (Fisher's exact test).
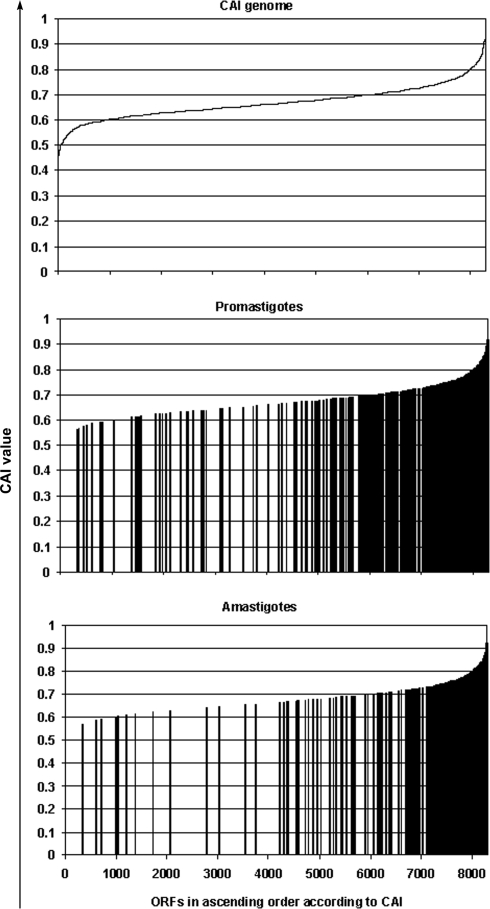
At ∼6% coverage only abundant proteins were detected. Codon usage was analyzed in this protein ensemble to seek evidence for translational bias. To this end, a CAI was calculated according to Carbone et al. (36). This iteratively computes a weight for each codon and an integrated measure for the whole of the respective ORF, its CAI. CAIs were calculated for all ORFs predicted from the L. major genome, and these were ranked on a scale of 0–8292 corresponding to the number of ORFs, excluding pseudogenes. Next ranks of genes encoding proteins detected in our pro- or amastigote-derived data sets were plotted (Fig. 5). The great majority of the respective genes exhibited high CAI values resulting in skewed median ranks of 7704 and 7958 (25–75th percentile ranks of 6732–8099 and 7304–8163, respectively) for pro- and amastigote data sets, indicating highly biased codon usage.
Fig. 5.
Codon bias in abundantly expressed ORFs. CAIs of all ORFs of the L. major (Friedlin) reference genome and of ORFs corresponding to identified proteins were plotted. x axis, 8292 ORFs of L. major in ascending order according to CAI value (top). CAI values of ORFs corresponding to proteins identified in the promastigote (middle) or amastigote (bottom) proteome are shown; each bar represents one ORF. The median CAI value of the genome was 0.66308865, for ORFs constituting the promastigote proteome the median was 0.762384, and for ORFs corresponding to amastigote proteome the median was 0.790777. The distribution of observed versus non-observed ranks was significantly different in promastigotes and amastigotes (Mann-Whitney test, p < 0.0001).
To analyze likely subcellular distribution of proteins in our probe sets in comparison with the whole genome, sequence-based predictive algorithms were used. This revealed that in the experimental sets, as expected, proteins predicted to be membrane-bound or secreted were underrepresented (Fig. 6). In consequence, soluble proteins with predicted cytoplasmic localization were overrepresented as were proteins with predicted mitochondrial localization. Of note, the major surface protein of promastigotes, the glycosylphosphatidylinositol-anchored gp63 was detected in several fractions but only by a single peptide in the 1-D SDS-PAGE-separated urea-insoluble material (not shown). In contrast, the non-glycosylphosphatidylinositol-anchored form, known to be produced in amastigotes where it is localized in the lumen of lysosomal structures (39), was detected solely in amastigote samples (Table I).
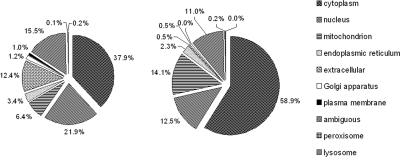
Fig. 6.
Pie chart of predicted subcellular localization of L. major reference genome ORFs (left) and ORFs corresponding to identified proteins from this study (right). Proteins were classified by Proteome Analyst 2.5. The program classifies proteins in nine classes (cytoplasm, nucleus, mitochondrion, endoplasmic reticulum, extracellular, Golgi apparatus, plasma membrane, peroxisome/glycosome, and lysosome). Proteins predicted to more than one location were classified as ambiguous. Proteins for which prediction was not possible (44% of the L. major ORFs) were omitted for clarity. Cytoplasmic and mitochondrial proteins were overrepresented (p < 0.0001). Proteins with predicted nuclear (p = 0.1411) or Golgi (p = 0.4739) localization were represented as expected. Proteins with extracellular (p < 0.0001) or plasma membrane (p < 0.05) localization were underrepresented.
Table I.
Proteins identified solely in amastigotes
| Sanger ID | NCBI accession no. | Spot no.a | Annotation |
|---|---|---|---|
| LmjF08.0320 | 245 | Mitochondria-associated ribonuclease | |
| LmjF09.0100 | 10 | Hypothetical protein, conserved | |
| LmjF18.0580 | 169 | Peroxisomal enoyl-CoA hydratase | |
| LmjF19.0710 | 213, SSP_8307 | Glycosomal malate dehydrogenase | |
| LmjF21.1710 | 296 | COX6 cytochrome c oxidase | |
| LmjF21.1760 | 112 | Centromere/microtubule-binding protein cbf5 | |
| LmjF22.1540 | SSP_4801 | Alanyl-tRNA synthetase | |
| LmjF23.0370 | 287, SSP_7006 | Hypothetical protein, conserved | |
| LmjF23.0760 | 164 | Mitochondrial RNA-binding protein | |
| LmjF24.1210 | SSP_9004 | Translation factor SUI1 | |
| LmjF25.2130b | 160, SSP_8408 | Succinyl-CoA synthetase | |
| LmjF25.2140b | 160, SSP_8408 | Succinyl-CoA synthetase | |
| LmjF26.1550 | 97 | Trifunctional enzyme α subunit | |
| LmjF27.1110 | 267, SSP_8207 | Mitochondrial RNA-binding protein 1 | |
| LmjF27.1220 | 122, SSP_6603 | Hypothetical protein, conserved | |
| LmjF27.1300 | 101, 102 | Hypothetical protein, conserved | |
| LmjF28.0490 | 107 | Propionyl-CoA carboxylase β chain | |
| LmjF29.0760 | 2 | Lipophosphoglycan biosynthetic protein | |
| LmjF31.1630b | 175 | Putative 3-ketoacyl-CoA thiolase-like | |
| LmjF31.1640b | 113, 175, SSP_8502 | Thiolase protein-like | |
| LmjF31.1900b | 289 | Ubiquitin fusion protein | |
| LmjF31.2030b | 289 | Ubiquitin fusion protein | |
| LmjF31.2250 | 70 | 3,2-trans-Enoyl-CoA isomerase | |
| LmjF32.0840 | 40 | Hypothetical protein, conserved | |
| LmjF33.0830 | 98 | 2,4-Dienoyl-CoA reductase | |
| LmjF33.1630 | 268 | Cyclophilin | |
| LmjF33.2390 | SSP_4808 | Heat shock protein | |
| LmjF34.3670 | 312 | Vacuolar ATP synthase | |
| LmjF35.1300 | SSP_4106 | Ubiquitin-conjugating enzyme E2 | |
| LmjF36.1160 | SSP_6414 | Hypothetical protein, conserved | |
| LmjF36.3100 | 242 | ATP synthase | |
| LmjF36.3780 | SSP_5302 | Hypothetical protein, conserved | |
| LmjF36.4360 | 151 | Proteasome regulatory ATPase subunit | |
| gi|68223993 | SSP_5402 | RNA helicase | |
| gi|14348750b | 223, 225, 226, SSP_0103 | CPB2 protein | |
| gi|1749812b | 226, SSP_0103 | CPB1 | |
| gi|886681b | 223, 225, 226, SSP_0103 | CPb19 | |
| gi|1730100b | 223, 225, 226, SSP_0103 | Cysteine proteinase B precursor | |
| gi|2780176b | SSP_0103 | Cysteine proteinase | |
| gi|461905b | SSP_0103 | Cysteine proteinase 2 precursor | |
| gi|9542b | SSP_0103 | Cysteine proteinase | |
| gi|51317309 | 41 | Leishmanolysin | |
| gi|146077037 | 90 | Stomatin-like protein |
45 protein spots identified in 2-DE analysis represented 34 proteins solely identified in amastigotes. Spot numbers refer to numbers indicated in supplemental figures representing gel images and tables detailing identification results. GeneDB Systematic IDs and NCBI accession numbers are indicated. In bold are proteins involved in carbohydrate, fatty acid, or energy metabolism.
Proteins that could not be assigned based on identified peptides to a single ORF but to tandem arrays of ORFs. In this case all corresponding IDs or accession numbers are indicated but were counted as one distinct identification only.
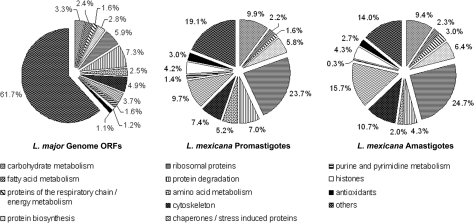
The distribution of proteins over functional groups was analyzed next to determine the representation of these classes in our data sets and to compare it with a theoretical frequency based on genome annotation (performed with Gene DB_Dataset version 5.1; Fig. 7). Predicted hypothetical proteins were underrepresented in both experimental data sets (11.5 and 17.1% versus 65.1% in the genome; p < 0.0001, χ2 analysis) suggesting that most gene products in this class are present in low copy numbers per parasite cell. If hypothetical proteins were excluded from the analysis, proteins with a role in protein degradation, amino acid metabolism, and purine/pyrimidine metabolism were underrepresented in the amastigote data set as were proteins with predicted but not classified functions (Fig. 7). In contrast, proteins with predicted functions in respiration, energy metabolism, and response to stress including chaperones were identified more frequently in the amastigote proteome.
Fig. 7.
Predicted functional classification of L. major reference genome ORFs (left) and ORFs corresponding to proteins identified in L. mexicana promastigotes (middle) and amastigotes (right). Functional classification was performed according to the Sanger database and “KEGG2.” Classifications are shown on the right.
Differentially Represented Proteins in Amastigotes—
A series of prominent spots in the 25-kDa range (pI 5.6 and higher) and numerous other spots (Fig. 3, e.g. arrowheads in middle and right panels) were more abundant or solely detectable in amastigote samples, and we next focused on the identity of these proteins. In total 56 such protein species were detected either in the 1-D gel electrophoresis or 2-DE approach (supplemental Tables 1–3). Proteins detected in the prominent spot series in the range of 25 kDa (Fig. 3) corresponded to DsRed, the fluorescent marker protein. This reflects properties of the expression system driving 7–10-fold increased synthesis in amastigotes (21). In 2-DE analyses, 45 protein spots corresponding to 34 proteins were only detectable in the amastigote probes (examples indicated in Fig. 3 by black arrowheads; see also Table I). Although undetected proteins may be present in promastigotes and escape detection because of low abundance, the above proteins define a differentially represented set and included cysteine proteinase B locus products, which are known to be more abundant in amastigotes (40, 41); proteins involved in fatty acid and energy metabolism; and proteins with predicted roles in RNA and protein processing or as chaperones. Furthermore we confirmed results of previous proteomics comparisons of promastigotes and axenic amastigotes of L. mexicana and Leishmania spp. causing visceral or mucocutaneous disease. These had identified several protein species of HSPs, β-tubulin, and the eEF-1α subunit indicative of proteolytic processing. Stage-specific protein species for eEF-1α (supplemental Fig. 2, spots 162 and 216) and HSP (supplemental Fig. 2, spot 84) in amastigotes were also observed here but were not identical to those reported for axenic amastigotes. Finally seven hypothetical proteins with unknown function were detected by 2-DE only in amastigotes (Table I).
Genome Distribution of ORFs Encoding Abundant Proteins—
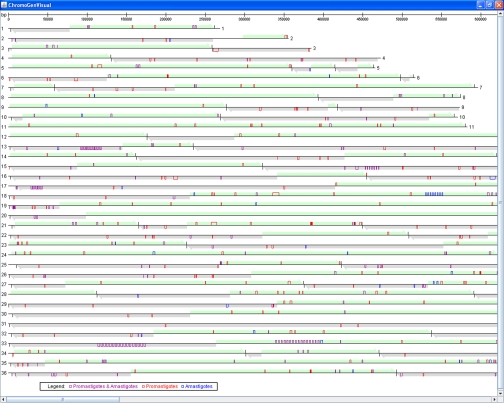
In trypanosomatids, genes are organized in polycistronic units that seem to be constitutively transcribed. Variation in expression levels of individual polycistronic units could contribute to differential protein representation, and abundant proteins may cluster to particular polycistronic units. To explore this hypothesis, we developed a tool to visualize chromosomal locations of ORFs representing proteins identified in our data sets and analyzed their distribution. As depicted (Fig. 8) no obvious clustering of genes encoding commonly or differentially detected proteins was observed. However, a number of polycistronic units of the reference genome of L. major did not contain any ORF encoding a protein identified in our analysis.
Fig. 8.
Screen shot of proteome-to-genome visualization tool. Chromosomes 1–36 are plotted with the predicted 133 polycistronic clusters indicated as arrows in light green and light gray. Black marks show ends of clusters. Boxes localize individual loci. Purple, ORFs corresponding to proteins identified in both life cycle stages; blue, ORFs corresponding to proteins identified only in amastigotes; red, ORFs corresponding to proteins identified only in promastigotes. The visualization tool is interactive: by clicking on a chromosome an enlarged image is displayed in a separate window. Clicking on a particular ORF redirects the user to the entry for this ORF in GeneDB on line.
Common Signatures in 3′-Untranslated mRNA Sequences of Differentially Expressed ORFs—
Regulatory sequences in 3′-UTRs are known to impact gene expression levels in trypanosomatids (see Ref. 42). Recently such elements were identified in genes differentially expressed in trypanosomes by adapting a nucleotide counting algorithm (43). Therefore, intergenic sequences 3′ of the genes coding for proteins differentially detected in amastigotes (Table I) were analyzed analogously. In addition, we included loci encoding proteins found to be more abundant in amastigotes by quantitative analysis using isotope-coded protein labeling (71). The analysis was performed on the respective set of 51 intergenic sequences from L. major (see supplemental Table 3 for the respective loci). A random selection of intergenic regions of 30% of all ORFs in the genome served as a control set. The oligonucleotide sequences AAGAGAA and/or TCTCCTTT were found within predicted 3′-UTRs in 21 (Table II) of the 51 test sequences; this was significantly different from the control set (p < 0.0001 and p = 0.0012 for TCTCCTTT and AAGAGAA, respectively). Furthermore a comparative analysis of the respective 3′-UTR of homologous genes in L. infantum indicated a high degree of conservation of the signatures between these species. Interestingly the pyrimidine-rich motif was also found in the 3′-UTR of the L. infantum amastin gene LinJ34.0840 that we included in the analysis as an experimentally analyzed stage-specific gene (44). In the amastin 3′-UTR, the motif TCTCCTTT starts 1587 bp downstream of the stop codon in a region known to regulate amastigote-specific expression at a translational level in response to the temperature shift when switching to mammalian hosts.
Table II.
Sequence motifs in 3′-UTR characteristic of differentially expressed ORFs
Shown are Sanger IDs of L. major loci that encode gene products homologous to the 51 differentially expressed proteins detected in L. mexicana amastigotes and in which the predicted 3′-UTR contained one or both of two sequence motifs (denoted by x) that were significantly more frequently present when compared with a large control set of 30% of the genomic loci. For a list of all 51 loci in L. major and 45 homologous loci in L. infantum analyzed see supplemental Table 3. In 67 and 64% of cases, respectively, the motifs TCTCCTTT and AAGAGAA were at the same relative position within the predicted 3′-UTRs. In bold are proteins involved in carbohydrate, fatty acid, or energy metabolism.
|
L. major
|
L. infantum
|
|||||
|---|---|---|---|---|---|---|
| Sanger ID | Annotation | Motif
|
Sanger ID | Motif
|
||
| TCTCCTTTa | AAGAGAAb | TCTCCTTT | AAGAGAA | |||
| LmjF09.0100 | Hypothetical protein, conserved | x | LinJ09_V3.0120 | |||
| LmjF19.0710 | Glycosomal malate dehydrogenase | x | LinJ19_V3.0710 | x | ||
| LmjF23.0370 | Hypothetical protein, conserved | x | x | LinJ23_V3.0420 | x | x |
| LmjF23.0760 | Mitochondrial RNA-binding protein | x | LinJ23_V3.0930 | x | ||
| LmjF24.1210 | Translation factor SUI1 | x | LinJ24_V3.1240 | x | ||
| LmjF25.1420 | GTP-binding protein | x | x | LinJ25_V3.1460 | x | x |
| LmjF25.2130 | Succinyl-CoA synthetase α subunit | LinJ25_V3.2220 | x | |||
| LmjF26.1550 | Trifunctional enzyme α subunit | x | x | LinJ26_V3.1530 | x | x |
| LmjF27.1110 | Mitochondrial RNA-binding protein | x | LinJ27_V3.0980 | x | ||
| LmjF27.1220 | Hypothetical protein, conserved | x | x | LinJ27_V3.1100 | x | |
| LmjF27.1300 | Hypothetical protein, conserved | x | x | LinJ27_V3.1220 | x | x |
| LmjF28.2770 | Heat-shock protein hsp70 | x | x | LinJ28_V3.3060 | ||
| LmjF28.2780 | Heat-shock protein hsp70 | x | LinJ28_V3.2960 | |||
| LmjF32.0840 | Hypothetical protein, conserved | x | LinJ32_V3.0890 | x | x | |
| LmjF33.0830 | 2,4-Dienoyl-CoA reductase | x | LinJ33_V3.0870 | x | ||
| LmjF33.2390 | Heat-shock protein | x | LinJ33_V3.2520 | x | ||
| LmjF34.2580 | Hypothetical protein, conserved | x | x | LinJ34_V3.2410 | x | x |
| LmjF35.1300 | Ubiquitin-conjugating enzyme E2 | x | LinJ35_V3.1310 | x | ||
| LmjF36.1160 | Hypothetical protein, conserved | x | x | LinJ36_V3.1220 | x | |
| LmjF36.3100 | ATP synthase | LinJ36_V3.3250 | x | |||
| LmjF36.3780 | Hypothetical protein, conserved | x | LinJ36_V3.3970 | x | ||
| LmjF36.3990 | hs1vu complex proteolytic subunit-like | x | x | LinJ36_V3.4180 | x | |
| LmjF36.4360 | Proteasome-regulatory ATPase subunit | x | LinJ36_V3.4570 | x | x | |
p < 0.0001 by χ2 test for the indicated motif.
p = 0.0012 by χ2 test for the indicated motif.
DISCUSSION
Proteomics may be the method of choice for the analysis of developmental changes in trypanosomatids, but progress to characterize the proteome of Leishmania spp. parasites has been hampered by inadequate methods to separate the intracellular form from host cell material. By combining classic enrichment by isopycnic centrifugation with subsequent fluorescence-activated particle sorting we showed that intracellular parasites can be purified for direct analyses by proteomics. Purification required a “dump” fluorescence channel whereby the major host cell contaminants, i.e. nuclear material, labeled with a chromatin stain could be excluded. This improved sample purity critically but obviously at the expense of yield. Samples purified in this way showed 2–3% contamination with host proteins; this is far superior to what has been achieved without a dump channel strategy, e.g. with salmonella (45). These host proteins were likely components of the phagosome membrane delineating the habitat of the intracellular parasite. Inspection of the purified material by electron microscopy confirmed purity of the material and revealed the presence of phagosomal membranes that were co-purified. Thus, depletion of non-habitat-associated host proteins may have been even more efficient, and the approach may allow investigation of phagosomal components as well.
Exploiting this method, amastigotes were purified from model host cells, bone marrow-derived macrophages, to characterize their proteome and compare it with that of promastigotes. Products of a total of 509 distinguishable parasite ORFs were identified. This data set confirms the presence of the majority of proteins reported previously for L. infantum, Leishmania donovani, or Leishmania panamensis and together with a recent report on L. donovani (17) extended our knowledge significantly and brings coverage of the Leishmania genus proteome to ∼20% of predicted ORFs. This is still significantly lower than that for proteomics model organisms such as Bacillus subtilis where ∼30% of predicted proteins have been identified (46). Technical bias explains this in part as current data sets including this study show clear underrepresentation of secreted and membrane-bound proteins. Furthermore our approach to compare life cycle forms mainly relied on optical comparison of stained protein spots in 2-DE gels that allows only very limited quantitation (leading to underestimation of the number of differentially regulated proteins) and is prone to underrepresentation of membrane proteins but on the plus side resolves protein species. Optimizing sample preparation (47), improving analytical procedures (48), and isotopic labeling techniques (17, 18, 49, 50), however, will improve coverage and quantitation. This will be important to complement upcoming metabolomics studies (51) and fill gaps in models of parasite structure and metabolism.
Technical caveats aside, pro- and amastigote-derived data sets together are an informative representation of all abundant proteins and revealed interesting general characteristics and differences. Analysis of codon usage in these sets provided clear evidence for translational bias in Leishmania to increase efficacy of protein synthesis. Given the mainly post-transcriptional regulation of gene expression this may not be surprising but has not been tested on a similar scale before, seems not to be a general feature of parasites (52) or unicellular life, and is not a mere consequence of GC bias (53). Understanding codon usage bias and its link to translational efficiency will benefit the design of transgenes to steer expression levels in these organisms not least because Leishmania is marketed as a protein expression platform (54).
Amastigotes have to adapt to an acidic environment (2) where they actively maintain a neutral intracellular pH (55). In addition, amastigotes were shown to possess a comparatively low overall negative charge (56). Here experimental and theoretical analysis revealed a preponderance of basic proteins in amastigote probe sets that would be consistent with buffering protons and a change in total charge (although the latter may be largely due to reduction in polyanionic phosphoglycan synthesis). It is tempting to speculate that acidic environments constitute a selective force fixing mutations that lead to more basic amino acids in proteins produced by intracellular parasites. Of note, there is precedent for such a scenario. The proteome of Helicobacter pylori, a Gram-negative bacterium that colonizes the human stomach, arguably an acidic environment, displays a dominance of basic proteins (57) as does the predicted proteome of Coxiella burnetii (58), the Q-fever agent that like Leishmania spp. inhabits an acidic, late endosomal/early lysosomal organelle. The shift to higher average pI is in contrast to other bacteria thriving in neutral environments (58).
2-DE has the advantage that protein species can be separated and visualized. Thereby not only differentially expressed ORFs were identified but also a number of differentially processed protein variants, such as those of eEF-1α (14). It was proposed (14) that eEF-1α species may have roles in parasite host cell communication. In L. donovani infection eEF-1α reportedly activated host cell phosphatase SHP-1 whereby production of leishmanicidal NO may be suppressed (59). We were able to confirm several stage-specific processing events, but in our hands, the amastigote-specific eEF-1α species of ∼44 kDa (14) was also detected in promastigotes, whereas other species of this protein were not. Therefore, the relevance of these processed forms remains undefined but could be tested by studying processing-resistant mutant forms of these proteins in transgenic parasites.
Among the proteins that were more abundant in amastigotes were enzymes linked to respiration/energy metabolism and protein synthesis and proteins involved in stress responses. Enzymes are particularly informative with respect to metabolic adaptation to intracellular life, which, for Leishmania, we only begin to understand (3, 51). For example, hexose import seems critical because transporter-deficient L. mexicana mutants could not survive inside macrophages (60). However, hexoses may be limiting as amastigotes seem to require gluconeogenesis to thrive. Disruption of this pathway in L. major by deleting fructose-1,6-bisphosphatase severely attenuated the parasites (61). It has been proposed that gluconeogenesis depends on C2 bodies from the catabolism of amino acids as Leishmania lack genes encoding key enzymes for the glyoxylate pathway (61). Thus, hexoses and amino acids may be the primary carbon sources for amastigotes.
Nutrients are also needed to fuel the energy metabolism. It is thought that this is mainly covered by fatty acid degradation as β-oxidation is highly up-regulated in amastigotes isolated from lesions (51, 62). Consistent with this observation, we found that several enzymes of fatty acid catabolism were more abundant in amastigotes, in particular enzymes such as dienoyl-CoA reductase and trans-enoyl-CoA isomerase involved in degrading unsaturated fatty acids. This was complemented by greater abundance of enzymes of the tricarboxylic acid cycle, e.g. succinyl-CoA synthetase, malate dehydrogenase, and finally cytochrome c oxidase as part of the respiratory chain. As these proteins are likely located in mitochondria, a higher contribution of the organelle to cell mass would be a trivial explanation for the observed increases, and a corresponding reduction in cytoplasm could also contribute to the noted overrepresentation of basic proteins (63). However, this seems unlikely as morphometric data suggest constant volumetric ratios of mitochondria and cell body throughout the Leishmania life cycle (64, 65). Also our findings regarding these metabolic adaptations are in good agreement with data recently published for L. donovani axenic amastigotes (17). Similar to the latter study, we have initiated isotope-coded protein labeling to quantitatively assess changes in protein amount between pro- and amastigotes, and preliminary data confirm the present 2-DE results (71)2 with a dynamic range much above 2-fold differences when normalized to cell number. However, interpreting quantitative proteomics data sets comparing pro- and amastigotes is not straightforward, and we would like to add a cautionary note. Protein yield per cell for the smaller amastigote life form is roughly half of what is obtained per promastigote cell. Thus, by normalizing to the total amount of protein, one compares in fact two amastigotes with one promastigote cell. Increases in histones and enzymes within a 2-fold range may merely reflect this ratio. The task, for example for histones, to wrap the genome remains constant, and therefore a 2-fold increase is expected. Similar caution is required for interpreting enzyme levels as the flux of metabolites per cell is likely to be the same if a 2-fold increase is observed in amastigotes. Thus, it will be important to define internal standards for normalization to interpret quantitative changes in particular proteins, and current conclusions may have to be reassessed.
Interestingly proteins with predicted RNA binding activities were among the newly identified proteins solely detected in amastigotes. RNA-binding proteins play an important role in regulating gene expression in trypanosomatids (42, 66, 67). Differences in RNA-binding proteins between life cycle stages are consistent with this view. Because we detected two predicted mitochondrial RNA-binding proteins differentially in amastigotes, this concept may extend to expression of mitochondrial genes also. RNA-binding proteins involved in regulating gene expression are thought to recognize sequence motifs in the 3′-UTR and are also implicated in modulating translation efficiency (42, 66, 67). Recently oligonucleotide counting was used to identify regulatory motifs in 3′-UTRs of Trypanosome genes (43). By analogy, we assumed that the set of proteins detected primarily or solely in amastigotes was in part regulated by RNA-binding proteins binding to sequence-specific motifs. We therefore adapted the oligonucleotide counting approach, and this identified specific short sequences within the 3′-UTR of several of the corresponding ORFs, and these motifs seemed conserved between the Leishmania species tested. Of note, these motifs were present in an experimentally defined regulatory region of L. infantum amastigote-specific amastin 3′-UTR (44). In this case, the regulatory region reportedly increased translation because abundance of mRNA encoding a reporter gene was not affected by the presence or absence of the region containing the motif, but protein levels were. The amastin regulatory region is thought to belong to a large family of degenerate retroposons (68). Intriguingly and consistent with the idea that such motifs may identify regions that affect translation, we found no correlation between ORFs encoding proteins present at higher levels in amastigotes and ORFs identified as up-regulated at the mRNA level by microarray analysis of L. mexicana lesion-derived amastigote mRNA (8). Thus, our proteome data sets are likely to prove useful in revealing regulatory properties of non-translated mRNA regions, and theoretically, the elements may provide a novel, additional feature to predict differentially translated mRNAs throughout the genome.
Although Leishmania gene expression is regulated mostly post-transcriptionally, microarray analyses showed that steady state transcript levels vary between different ORFs over a range of ∼2–3 orders of magnitude (8). Because genes are organized in polycistronic units in these organisms, we tested whether ORFs contributing to the abundant proteome may be clustered in particular polycistronic units that could indicate higher transcription rates for the respective units and possibly a novel feature of genome structure. We developed a visualization tool that projects proteome data onto the genome map, and this showed that ORFs encoding the abundant proteome as defined here are not clustered. For a number of polycistronic units of the L. major genome, no gene product was detected in the abundant proteome. We cannot distinguish at present whether this reflects blocks of genes absent in L. mexicana, low transcriptional activity, or other reasons as most units that were not represented in the proteome contained less than 44 ORFs. At a coverage of 6% we may simply have missed them for probabilistic reasons.
In summary, a novel colored approach to purify intracellular Leishmania parasites has been developed and used to compare the proteome of pro- and amastigotes of L. mexicana. More than 6% of predicted gene products were identified. Analysis of differentially represented proteins in the amastigote proteome confirmed current views on metabolic adaptations to intracellular life. Bioinformatics analyses revealed evidence for codon adaptation in Leishmania likely to increase translation efficiency of mRNAs and sequence signatures in 3′-UTRs that may be associated with translational control of gene expression. Importantly the presented proteome data sets comprised all L. mexicana homologues of vaccine antigens that have been tested to date with promising results in diverse models of experimental leishmaniases (see supplemental Table 1). Although secreted and surface membrane-bound proteins are underrepresented, the present ensemble provides a unique resource, e.g. for selecting novel vaccine antigens as protein abundance is an important parameter for protective vaccine antigen selection (69, 70).
Supplementary Material
Acknowledgments
We thank Harry P. de Koning and Daniel J. Bridges for the initial idea to analyze the CAI, Andrei Zinovyev for the help with the CAI calculation, and Katelyn Fenn for help in analyzing the 3′-UTR regions. We thank Keith Matthews for critical reading of the manuscript and many constructive comments and Thomas F. Meyer for generous support.
Footnotes
Published, MCP Papers in Press, May 12, 2008, DOI 10.1074/mcp.M700343-MCP200
The abbreviations used are: 2-DE, two-dimensional gel electrophoresis; 1-D, one-dimensional; CAI, codon adaptation index; DAPI, 4′,6-diamidino-2-phenylindole; PMF, peptide mass fingerprint; UTR, untranslated mRNA region; nt, nucleotides; FACS, fluorescence-activated cell sorting; HSP, heat-shock protein; NCBI, National Center for Biotechnology Information.
A. Freiwald, D. Paape, M. Schmid, T. Aebischer, and P. R. Jungblut, unpublished data.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant Ae16/2-1 and European Commission Marie Curie Excellence Grant EXT-25435 (to T. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Herwaldt, B. L. ( 1999) Leishmaniasis. Lancet 354, 1191–1199 [DOI] [PubMed] [Google Scholar]
- 2.Antoine, J. C., Prina, E., Lang, T., and Courret, N. ( 1998) The biogenesis and properties of the parasitophorous vacuoles that harbour Leishmania in murine macrophages. Trends Microbiol. 6, 392–401 [DOI] [PubMed] [Google Scholar]
- 3.Burchmore, R. J., and Barrett, M. P. ( 2001) Life in vacuoles—nutrient acquisition by Leishmania amastigotes. Int. J. Parasitol. 31, 1311–1320 [DOI] [PubMed] [Google Scholar]
- 4.Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., Sisk, E., Rajandream, M. A., Adlem, E., Aert, R., Anupama, A., Apostolou, Z., Attipoe, P., Bason, N., Bauser, C., Beck, A., Beverley, S. M., Bianchettin, G., Borzym, K., Bothe, G., Bruschi, C. V., Collins, M., Cadag, E., Ciarloni, L., Clayton, C., Coulson, R. M., Cronin, A., Cruz, A. K., Davies, R. M., De Gaudenzi, J., Dobson, D. E., Duesterhoeft, A., Fazelina, G., Fosker, N., Frasch, A. C., Fraser, A., Fuchs, M., Gabel, C., Goble, A., Goffeau, A., Harris, D., Hertz-Fowler, C., Hilbert, H., Horn, D., Huang, Y., Klages, S., Knights, A., Kube, M., Larke, N., Litvin, L., Lord, A., Louie, T., Marra, M., Masuy, D., Matthews, K., Michaeli, S., Mottram, J. C., Muller-Auer, S., Munden, H., Nelson, S., Norbertczak, H., Oliver, K., O'neil, S., Pentony, M., Pohl, T. M., Price, C., Purnelle, B., Quail, M. A., Rabbinowitsch, E., Reinhardt, R., Rieger, M., Rinta, J., Robben, J., Robertson, L., Ruiz, J. C., Rutter, S., Saunders, D., Schafer, M., Schein, J., Schwartz, D. C., Seeger, K., Seyler, A., Sharp, S., Shin, H., Sivam, D., Squares, R., Squares, S., Tosato, V., Vogt, C., Volckaert, G., Wambutt, R., Warren, T., Wedler, H., Woodward, J., Zhou, S., Zimmermann, W., Smith, D. F., Blackwell, J. M., Stuart, K. D., Barrell, B., and Myler, P. J. ( 2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peacock, C. S., Seeger, K., Harris, D., Murphy, L., Ruiz, J. C., Quail, M. A., Peters, N., Adlem, E., Tivey, A., Aslett, M., Kerhornou, A., Ivens, A., Fraser, A., Rajandream, M. A., Carver, T., Norbertczak, H., Chillingworth, T., Hance, Z., Jagels, K., Moule, S., Ormond, D., Rutter, S., Squares, R., Whitehead, S., Rabbinowitsch, E., Arrowsmith, C., White, B., Thurston, S., Bringaud, F., Baldauf, S. L., Faulconbridge, A., Jeffares, D., Depledge, D. P., Oyola, S. O., Hilley, J. D., Brito, L. O., Tosi, L. R., Barrell, B., Cruz, A. K., Mottram, J. C., Smith, D. F., and Berriman, M. ( 2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 39, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan, R. C., Salotra, P., Goyal, N., Akopyants, N. S., Beverley, S. M., and Nakhasi, H. L. ( 2004) The application of gene expression microarray technology to kinetoplastid research. Curr. Mol. Med. 4, 611–621 [DOI] [PubMed] [Google Scholar]
- 7.Akopyants, N. S., Matlib, R. S., Bukanova, E. N., Smeds, M. R., Brownstein, B. H., Stormo, G. D., and Beverley, S. M. ( 2004) Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol. Biochem. Parasitol. 136, 71–86 [DOI] [PubMed] [Google Scholar]
- 8.Holzer, T. R., McMaster, W. R., and Forney, J. D. ( 2006) Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol. Biochem. Parasitol. 146, 198–218 [DOI] [PubMed] [Google Scholar]
- 9.Saxena, A., Lahav, T., Holland, N., Aggarwal, G., Anupama, A., Huang, Y., Volpin, H., Myler, P. J., and Zilberstein, D. ( 2007) Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol. Biochem. Parasitol. 152, 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall, N., Karras, M., Raine, J. D., Carlton, J. M., Kooij, T. W., Berriman, M., Florens, L., Janssen, C. S., Pain, A., Christophides, G. K., James, K., Rutherford, K., Harris, B., Harris, D., Churcher, C., Quail, M. A., Ormond, D., Doggett, J., Trueman, H. E., Mendoza, J., Bidwell, S. L., Rajandream, M. A., Carucci, D. J., Yates, J. R., III, Kafatos, F. C., Janse, C. J., Barrell, B., Turner, C. M., Waters, A. P., and Sinden, R. E. ( 2005) A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 [DOI] [PubMed] [Google Scholar]
- 11.Walker, J., Vasquez, J. J., Gomez, M. A., Drummelsmith, J., Burchmore, R., Girard, I., and Ouellette, M. ( 2006) Identification of developmentally-regulated proteins in Leishmania panamensis by proteome profiling of promastigotes and axenic amastigotes. Mol. Biochem. Parasitol. 147, 64–73 [DOI] [PubMed] [Google Scholar]
- 12.Dea-Ayuela, M. A., Rama-Iniguez, S., and Bolas-Fernandez, F. ( 2006) Proteomic analysis of antigens from Leishmania infantum promastigotes. Proteomics 6, 4187–4194 [DOI] [PubMed] [Google Scholar]
- 13.McNicoll, F., Drummelsmith, J., Muller, M., Madore, E., Boilard, N., Ouellette, M., and Papadopoulou, B. ( 2006) A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics 6, 3567–3581 [DOI] [PubMed] [Google Scholar]
- 14.Nugent, P. G., Karsani, S. A., Wait, R., Tempero, J., and Smith, D. F. ( 2004) Proteomic analysis of Leishmania mexicana differentiation. Mol. Biochem. Parasitol. 136, 51–62 [DOI] [PubMed] [Google Scholar]
- 15.Gongora, R., Acestor, N., Quadroni, M., Fasel, N., Saravia, N. G., and Walker, J. ( 2003) Mapping the proteome of Leishmania Viannia parasites using two-dimensional polyacrylamide gel electrophoresis and associated technologies. Biomedica 23, 153–160 [PubMed] [Google Scholar]
- 16.Gupta, S. K., Sisodia, B. S., Sinha, S., Hajela, K., Naik, S., Shasany, A. K., and Dube, A. ( 2007) Proteomic approach for identification and characterization of novel immunostimulatory proteins from soluble antigens of Leishmania donovani promastigotes. Proteomics 7, 816–823 [DOI] [PubMed] [Google Scholar]
- 17.Rosenzweig, D., Smith, D., Opperdoes, F., Stern, S., Olafson, R. W., and Zilberstein, D. ( 2008) Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J. 22, 590–602 [DOI] [PubMed] [Google Scholar]
- 18.Leifso, K., Cohen-Freue, G., Dogra, N., Murray, A., and McMaster, W. R. ( 2007) Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol. Biochem. Parasitol. 152, 35–46 [DOI] [PubMed] [Google Scholar]
- 19.Walker, J., Acestor, N., Gongora, R., Quadroni, M., Segura, I., Fasel, N., and Saravia, N. G. ( 2006) Comparative protein profiling identifies elongation factor-1β and tryparedoxin peroxidase as factors associated with metastasis in Leishmania guyanensis. Mol. Biochem. Parasitol. 145, 254–264 [DOI] [PubMed] [Google Scholar]
- 20.Sorensen, M., Lippuner, C., Kaiser, T., Misslitz, A., Aebischer, T., and Bumann, D. ( 2003) Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 552, 110–114 [DOI] [PubMed] [Google Scholar]
- 21.Misslitz, A., Mottram, J. C., Overath, P., and Aebischer, T. ( 2000) Targeted integration into a rRNA locus results in uniform and high level expression of transgenes in Leishmania amastigotes. Mol. Biochem. Parasitol. 107, 251–261 [DOI] [PubMed] [Google Scholar]
- 22.Bates, P. A. ( 1994) Complete developmental cycle of Leishmania mexicana in axenic culture. Parasitology 108, 1–9 [DOI] [PubMed] [Google Scholar]
- 23.Weinheber, N., Wolfram, M., Harbecke, D., and Aebischer, T. ( 1998) Phagocytosis of Leishmania mexicana amastigotes by macrophages leads to a sustained suppression of IL-12 production. Eur. J. Immunol. 28, 2467–2477 [DOI] [PubMed] [Google Scholar]
- 24.Chakraborty, P., Sturgill-Koszycki, S., and Russell, D. G. ( 1994) Isolation and characterization of pathogen-containing phagosomes. Methods Cell Biol. 45, 261–276 [DOI] [PubMed] [Google Scholar]
- 25.Zimny-Arndt, U., Schmid, M., Ackermann, R., and Jungblut, P. R. (2008) Classical proteomics: two-dimensional electrophoresis/MALDI mass spectrometry, in Methods in Molecular Biology: Mass Spectrometry of Proteins and Peptides (Lipton, M., and Pasa-Tolic, L., eds), Humana Press Inc., Totowa, NJ, in press
- 26.Doherty, N. S., Littman, B. H., Reilly, K., Swindell, A. C., Buss, J. M., and Anderson, N. L. ( 1998) Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis 19, 355–363 [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, F., Krah, A., Schmid, M., Jungblut, P. R., and Thiede, B. ( 2006) Distinctive mass losses of tryptic peptides generated by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight. Rapid Commun. Mass Spectrom. 20, 933–936 [DOI] [PubMed] [Google Scholar]
- 28.Al Younes, H. M., Rudel, T., Brinkmann, V., Szczepek, A. J., and Meyer, T. F. ( 2001) Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell. Microbiol. 3, 427–437 [DOI] [PubMed] [Google Scholar]
- 29.van Helden, J., Andre, B., and Collado-Vides, J. ( 2000) A web site for the computational analysis of yeast regulatory sequences. Yeast 16, 177–187 [DOI] [PubMed] [Google Scholar]
- 30.van Helden, J., del Olmo, M., and Perez-Ortin, J. E. ( 2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 28, 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakanari, J. A., Nadler, S. A., Chan, V. J., Engel, J. C., Leptak, C., and Bouvier, J. ( 1997) Leishmania major: comparison of the cathepsin L- and B-like cysteine protease genes with those of other trypanosomatids. Exp. Parasitol. 85, 63–76 [DOI] [PubMed] [Google Scholar]
- 32.Schneider, P., Rosat, J. P., Bouvier, J., Louis, J., and Bordier, C. ( 1992) Leishmania major: differential regulation of the surface metalloprotease in amastigote and promastigote stages. Exp. Parasitol. 75, 196–206 [DOI] [PubMed] [Google Scholar]
- 33.Benz, C., Nilsson, D., Andersson, B., Clayton, C., and Guilbride, D. L. ( 2005) Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143, 125–134 [DOI] [PubMed] [Google Scholar]
- 34.Gopal, S., Awadalla, S., Gaasterland, T., and Cross, G. A. ( 2005) A computational investigation of kinetoplastid trans-splicing. Genome Biol. 6, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp, P. M., and Li, W. H. ( 1987) The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol. Biol. Evol. 4, 222–230 [DOI] [PubMed] [Google Scholar]
- 36.Carbone, A., Zinovyev, A., and Kepes, F. ( 2003) Codon adaptation index as a measure of dominating codon bias. Bioinformatics 19, 2005–2015 [DOI] [PubMed] [Google Scholar]
- 37.Szafron, D., Lu, P., Greiner, R., Wishart, D. S., Poulin, B., Eisner, R., Lu, Z., Anvik, J., Macdonell, C., Fyshe, A., and Meeuwis, D. ( 2004) Proteome Analyst: custom predictions with explanations in a web-based tool for high-throughput proteome annotations. Nucleic Acids Res. 32, W365–W371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiller, K., Schobert, M., Hundertmark, C., Jahn, D., and Munch, R. ( 2003) JVirGel: calculation of virtual two-dimensional protein gels. Nucleic Acids Res. 31, 3862–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medina-Acosta, E., Karess, R. E., Schwartz, H., and Russell, D. G. ( 1989) The promastigote surface protease (gp63) of Leishmania is expressed but differentially processed and localized in the amastigote stage. Mol. Biochem. Parasitol. 37, 263–273 [DOI] [PubMed] [Google Scholar]
- 40.Souza, A. E., Waugh, S., Coombs, G. H., and Mottram, J. C. ( 1992) Characterization of a multi-copy gene for a major stage-specific cysteine proteinase of Leishmania mexicana. FEBS Lett. 311, 124–127 [DOI] [PubMed] [Google Scholar]
- 41.Mottram, J. C., Frame, M. J., Brooks, D. R., Tetley, L., Hutchison, J. E., Souza, A. E., and Coombs, G. H. ( 1997) The multiple cpb cysteine proteinase genes of Leishmania mexicana encode isoenzymes that differ in their stage regulation and substrate preferences. J. Biol. Chem. 272, 14285–14293 [DOI] [PubMed] [Google Scholar]
- 42.Clayton, C. E. ( 2002) Life without transcriptional control? From fly to man and back again. EMBO J. 21, 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayho, M., Fenn, K., Craddy, P., Crosthwaite, S., and Matthews, K. ( 2006) Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 34, 5312–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNicoll, F., Muller, M., Cloutier, S., Boilard, N., Rochette, A., Dube, M., and Papadopoulou, B. ( 2005) Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 280, 35238–35246 [DOI] [PubMed] [Google Scholar]
- 45.Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., and Bumann, D. ( 2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307 [DOI] [PubMed] [Google Scholar]
- 46.Wolff, S., Antelmann, H., Albrecht, D., Becher, D., Bernhardt, J., Bron, S., Buttner, K., van Dijl, J. M., Eymann, C., Otto, A., Tam, L. T., and Hecker, M. ( 2007) Towards the entire proteome of the model bacterium Bacillus subtilis by gel-based and gel-free approaches. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 849, 129–140 [DOI] [PubMed] [Google Scholar]
- 47.Foucher, A. L., Papadopoulou, B., and Ouellette, M. ( 2006) Prefractionation by digitonin extraction increases representation of the cytosolic and intracellular proteome of Leishmania infantum. J. Proteome Res. 5, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 48.Domon, B., and Aebersold, R. ( 2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 49.Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H., and Aebersold, R. ( 1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 50.Mann, M. ( 2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 51.Opperdoes, F. R., and Coombs, G. H. ( 2007) Metabolism of Leishmania: proven and predicted. Trends Parasitol. 23, 149–158 [DOI] [PubMed] [Google Scholar]
- 52.Cutter, A. D., Wasmuth, J. D., and Blaxter, M. L. ( 2006) The evolution of biased codon and amino acid usage in nematode genomes. Mol. Biol. Evol. 23, 2303–2315 [DOI] [PubMed] [Google Scholar]
- 53.Carbone, A., Kepes, F., and Zinovyev, A. ( 2005) Codon bias signatures, organization of microorganisms in codon space, and lifestyle. Mol. Biol. Evol. 22, 547–561 [DOI] [PubMed] [Google Scholar]
- 54.Breitling, R., Klingner, S., Callewaert, N., Pietrucha, R., Geyer, A., Ehrlich, G., Hartung, R., Muller, A., Contreras, R., Beverley, S. M., and Alexandrov, K. ( 2002) Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 25, 209–218 [DOI] [PubMed] [Google Scholar]
- 55.Glaser, T. A., Baatz, J. E., Kreishman, G. P., and Mukkada, A. J. ( 1988) pH homeostasis in Leishmania donovani amastigotes and promastigotes. Proc. Natl. Acad. Sci. U. S. A. 85, 7602–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pimenta, P. F., and de Souza, W. ( 1983) Leishmania mexicana amazonensis: surface charge of amastigote and promastigote forms. Exp. Parasitol. 56, 194–206 [DOI] [PubMed] [Google Scholar]
- 57.Jungblut, P. R., Bumann, D., Haas, G., Zimny-Arndt, U., Holland, P., Lamer, S., Siejak, F., Aebischer, A., and Meyer, T. F. ( 2000) Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol. 36, 710–725 [DOI] [PubMed] [Google Scholar]
- 58.Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., Ward, N. L., Tettelin, H., Davidsen, T. M., Beanan, M. J., Deboy, R. T., Daugherty, S. C., Brinkac, L. M., Madupu, R., Dodson, R. J., Khouri, H. M., Lee, K. H., Carty, H. A., Scanlan, D., Heinzen, R. A., Thompson, H. A., Samuel, J. E., Fraser, C. M., and Heidelberg, J. F. ( 2003) Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 100, 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nandan, D., and Reiner, N. E. ( 2005) Leishmania donovani engages in regulatory interference by targeting macrophage protein tyrosine phosphatase SHP-1. Clin. Immunol. 114, 266–277 [DOI] [PubMed] [Google Scholar]
- 60.Burchmore, R. J., Rodriguez-Contreras, D., McBride, K., Merkel, P., Barrett, M. P., Modi, G., Sacks, D., and Landfear, S. M. ( 2003) Genetic characterization of glucose transporter function in Leishmania mexicana. Proc. Natl. Acad. Sci. U. S. A. 100, 3901–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naderer, T., Ellis, M. A., Sernee, M. F., De Souza, D. P., Curtis, J., Handman, E., and McConville, M. J. ( 2006) Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc. Natl. Acad. Sci. U. S. A. 103, 5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hart, D. T., and Coombs, G. H. ( 1982) Leishmania mexicana: energy metabolism of amastigotes and promastigotes. Exp. Parasitol. 54, 397–409 [DOI] [PubMed] [Google Scholar]
- 63.Schwartz, R., Ting, C. S., and King, J. ( 2001) Whole proteome pI values correlate with subcellular localizations of proteins for organisms within the three domains of life. Genome Res. 11, 703–709 [DOI] [PubMed] [Google Scholar]
- 64.Ueda-Nakamura, T., Attias, M., and de Souza, W. ( 2001) Megasome biogenesis in Leishmania amazonensis: a morphometric and cytochemical study. Parasitol. Res. 87, 89–97 [DOI] [PubMed] [Google Scholar]
- 65.Alberio, S. O., Dias, S. S., Faria, F. P., Mortara, R. A., Barbieri, C. L., and Freymuller, H. E. ( 2004) Ultrastructural and cytochemical identification of megasome in Leishmania chagasi. Parasitol. Res. 92, 246–254 [DOI] [PubMed] [Google Scholar]
- 66.De Gaudenzi, J., Frasch, A. C., and Clayton, C. ( 2005) RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryot. Cell 4, 2106–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Sayed, N. M., Myler, P. J., Blandin, G., Berriman, M., Crabtree, J., Aggarwal, G., Caler, E., Renauld, H., Worthey, E. A., Hertz-Fowler, C., Ghedin, E., Peacock, C., Bartholomeu, D. C., Haas, B. J., Tran, A. N., Wortman, J. R., Alsmark, U. C., Angiuoli, S., Anupama, A., Badger, J., Bringaud, F., Cadag, E., Carlton, J. M., Cerqueira, G. C., Creasy, T., Delcher, A. L., Djikeng, A., Embley, T. M., Hauser, C., Ivens, A. C., Kummerfeld, S. K., Pereira-Leal, J. B., Nilsson, D., Peterson, J., Salzberg, S. L., Shallom, J., Silva, J. C., Sundaram, J., Westenberger, S., White, O., Melville, S. E., Donelson, J. E., Andersson, B., Stuart, K. D., and Hall, N. ( 2005) Comparative genomics of trypanosomatid parasitic protozoa. Science 309, 404–409 [DOI] [PubMed] [Google Scholar]
- 68.Bringaud, F., Muller, M., Cerqueira, G. C., Smith, M., Rochette, A., El Sayed, N. M., Papadopoulou, B., and Ghedin, E. ( 2007) Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 3, 1291–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aebischer, T., Wolfram, M., Patzer, S. I., Ilg, T., Wiese, M., and Overath, P. ( 2000) Subunit vaccination of mice against new world cutaneous leishmaniasis: comparison of three proteins expressed in amastigotes and six adjuvants. Infect. Immun. 68, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sabarth, N., Hurwitz, R., Meyer, T. F., and Bumann, D. ( 2002) Multiparameter selection of Helicobacter pylori antigens identifies two novel antigens with high protective efficacy. Infect. Immun. 70, 6499–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freiwald, A. ( 2006) Establishing of ICPL labeling for proteome studies of the micro-organism Leishmania mexicana. M.Sc. thesis, Technische Fachhochschule, Berlin
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.