Abstract
Statistical mining and integration of complex molecular data including metabolites, proteins, and transcripts is one of the critical goals of systems biology (Ideker, T., Galitski, T., and Hood, L. (2001) A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2, 343–372). A number of studies have demonstrated the parallel analysis of metabolites and large scale transcript expression. Protein analysis has been ignored in these studies, although a clear correlation between transcript and protein levels is shown only in rare cases, necessitating that actual protein levels have to be determined for protein function analysis. Here, we present an approach to investigate the combined covariance structure of metabolite and protein dynamics in a systemic response to abiotic temperature stress in Arabidopsis thaliana wild-type and a corresponding starch-deficient mutant (phosphoglucomutase-deficient). Independent component analysis revealed phenotype classification resolving genotype-dependent response effects to temperature treatment and genotype-independent general temperature compensation mechanisms. An observation is the stress-induced increase of raffinose-family-oligosaccharide levels in the absence of transitory starch storage/mobilization in temperature-treated phosphoglucomutase plants indicating that sucrose synthesis and storage in these mutant plants is sufficient to bypass the typical starch storage/mobilization pathways under abiotic stress. Eventually, sample pattern recognition and correlation network topology analysis allowed for the detection of specific metabolite-protein co-regulation and assignment of a circadian output regulated RNA-binding protein to these processes. The whole concept of high-dimensional profiling data integration from many replicates, subsequent multivariate statistics for dimensionality reduction, and covariance structure analysis is proposed to be a major strategy for revealing central responses of the biological system under study.
Metabolomic technologies enable the very rapid non-targeted analysis of metabolites and provide a diagnostic tool for pattern recognition of biological samples (2–5). Typical pattern recognition methods are variance discrimination algorithms such as principal components analysis (PCA)1 or independent component analysis (ICA) (2, 6–9). Independent component analysis is an extension of covariance analysis by looking for kurtosis thresholds or high entropy (8, 10) and thus adds a further value for biological interpretation. Variance discrimination of samples relies strongly on a high biological variability of independent biological replicate analysis (4, 11, 12). Recently, we demonstrated that these covariance matrixes of experimentally determined metabolite levels are connected with the elasticities of pathway reaction networks (13). Consequently, changes in the structure of these covariance networks reveal biochemical regulations (4). This was confirmed by using topology studies of differential metabolite correlation/covariance networks to investigate a silent phenotype sucrose synthase antisense plant and alterations in a starch-deficient Arabidopsis thaliana mutant (9, 14). Further we used a computational kinetic model of the Calvin cycle coupled to sucrose biosynthesis in plant leaf metabolism to demonstrate changes in metabolite correlation/covariance networks as a response to protein phosphorylation and enzymatic regulation (15, 16). The statistical model implies that variance discrimination analysis such as PCA will optimize sample grouping according to differences in biochemical regulation, thus providing for the first time a fundamental relationship between large scale profiling methods such as metabolomics combined with multivariate data analyses, biochemical regulation, and pattern recognition (4, 12) (see Fig. 1). However, although regulatory hubs can be identified in differential metabolite correlation networks, causal relationships in experimental systems are not derivable without integration of additional parameters such as external environmental perturbation and further molecular levels like protein concentrations or RNA expression data (1, 4). Computer simulation of enzymatic activities of a biochemical network enables calculation of corresponding metabolite correlation networks (4, 13, 15–17). This idea has been further substantiated by recent calculations of metabolic networks (18, 19). In these studies the authors identified high variances in gene expression and protein activity as causes for metabolite correlations. Obviously, the model for metabolite correlations can be extended to systemic fluctuations in complex biochemical networks (20). Consequently, the integration of rapid sample classification and metabolic network analysis using metabolomic techniques with quantitative non-targeted protein profiling will add a further dimension for protein function analysis and systems biology. Furthermore, integrated metabolite and protein measurements offer an improved method for distinguishing among phenotypes (i.e. causes for phenotypes) (4, 9, 12, 21). The systematic comparison of mRNA expression levels, enzymatic activities, and protein levels revealed a low correlation in most studies so far indicating that high throughput microarrays are not sufficient to understand genome-wide protein dynamics or biochemical regulation (22, 23). The systematic integration of transcript and metabolite profiling, thus, necessitates time course resolution. A more direct interaction can be expected for proteins and metabolites. However, only a few examples are existing, consequently investigating metabolomics and proteomics data integration. Recent examples demonstrate such an approach (9, 11, 24–26). These studies clearly demonstrate the need for data integration, however, show that several further obstacles have to be addressed: (i) data quality and comprehensiveness; (ii) sample throughput; and (iii) algorithms and statistics to extract significant information and to cope with the high dimensionality structure of the data. All these issues are directly related and dominate the outcome of an integrative study. In the present study a strategy for metabolomic and proteomic phenotype integration is shown coping with these problems. The overall strategy is based on recent work by us for the systematic analysis of the combined covariance structure of metabolites and proteins in a complex systemic response (see Fig. 1) (9, 11). Recent approaches were restricted to only low numbers of individual proteins. In the present work we improved protein identification and quantification rates strongly without limiting the sample throughput, which is a requirement to exploit biological variability for sample classification and biological interpretation as described above (4, 9, 11).
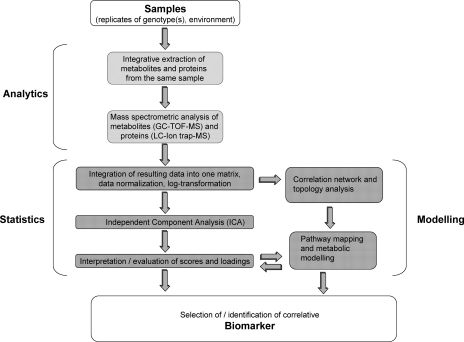
Fig. 1.
Strategy to analyze the combined covariance/correlation matrix of metabolites and proteins using integrative extraction from a multitude of biological replicates versus different experiments. By data integration it is possible to enhance the interpretation of the extracted independent components and assign specific biomarkers. In parallel pathway mapping, correlation network analysis and stochastic metabolic modeling can be linked to the whole process in an iterative manner to improve metabolic models and their predictive power (15, 42).
Molecular responses of temperature acclimation at 4 and 32 °C after 3 days were investigated in a sugar accumulating starch-deficient A. thaliana plant mutant phosphoglucomutase (PGM) and its corresponding wild-type (WT) ancestor. Metabolites and proteins were identified and quantified from the same tissue samples according to Weckwerth et al. (11). Typical metabolite stress markers and novel members of the RNA-binding protein family indicating involvement of post-transcriptional mechanisms were identified with a significant impact on genotype discrimination, temperature treatment, and cold acclimation, respectively. We propose the applicability of the whole process to all kinds of biological systems revealing systemic responses to environmental conditions and correlative sets of biomarkers.
EXPERIMENTAL PROCEDURES
Reagents—
Chemicals were purchased from Sigma (Taufkirchen, Germany), except d-sorbitol-P13PCB6B, dl-leucine-2,3,3-dB3B, and l-aspartic acid-2,3,3-dB3B, which were obtained from Isotech (Miamisburg, OH). Acetonitrile was from J. T. Baker (Deventer, Netherlands), endoproteinase Lys-C from Roche Applied Science, and PoroszymeP®P immobilized trypsin from Applied Biosystems (Foster City, CA).
Plant Material and Harvest—
A. thaliana plants Col-0 (wild-type) and a plastidic PGM mutant (27) were cultivated simultaneously under identical phytotron conditions set as follows: The light conditions were 160 μE for 8 h followed by 16 h at 0 μE (darkness). Relative humidity and temperature conditions were set to 70% and 20 °C during the light and dark period, respectively.
Plants were harvested at the developmental stage 5.10 (28) after 3 days at 4, 20 (control), and 32 °C, with 12 different plants per treatment and genetic background, respectively. Enzymatic activity was quenched by immediately freezing the plants in liquid nitrogen. The material of two plants per experiment was pooled to give six samples per treatment and genetic background. Tissues were stored at −80 °C until further analysis.
Extraction Procedure and Sample Preparation for Metabolite and Protein Analysis from One Sample—
Frozen leaf tissue was individually homogenized under liquid nitrogen using a pre-chilled mortar and pestle. Approximately 50 mg of powdered material was used for analysis. Simultaneous extraction of metabolites and proteins from individual plants was performed as described in Weckwerth et al. (11, 29) with modifications. For metabolite extraction, 1 ml of the extraction mixture containing methanol:chloroform:water (2.5:1:0.5 v:v:v) and 10 μl of an internal standard solution containing 2 mg/ml each d-sorbitol-P13PCB6B, dl-leucine-2,3,3-dB3B, and l-aspartic acid-2,3,3-dB3B was added. Soluble metabolites were extracted by mixing the solution at 4 °C for 10 min. After centrifugation for 6 min at 20,000 rpm, the supernatant was separated into chloroform and water/methanol phases. The aqueous phase was used for metabolite analysis.
Samples were derivatized by dissolving the dried metabolite pellet in 20 μl of methoxyamine hydrochloride (40 mg/ml pyridine) and shaking the mixture for 90 min at 30 °C. After the addition of 180 μl of N-methyl-N-trimethylsilyltrifluoroacetamid, the mixture was incubated at 37 °C for 30 min with vigorous shaking. A solution of even numbered fatty acid methylesters, methylcaprylate (C8-ME), methylcaprate (C10-ME), methyllaurate (C12-ME), methylmyristate (C14-ME), methylpalmitate (C16-ME), methylstearate (C18-ME), methyleicosanoate (C20-ME), methyldocosanoate (C22-ME), lignoceric acid methylester (C24-ME), methylhexacosanoate (C26-ME), methyloctacosanoate (C28-ME), and triacontanoic acid methylester (C30-ME) (each 0.8 mg/ml CHClB3B) was spiked into the derivatized sample prior to injection into the gas chromatography (GC). The remaining proteins pellets were dissolved in 200 μl of protein extraction buffer (50 mm HEPES-KOH, 40% sucrose (w/v), 1% β-mercaptoethanol, pH 7.5) per 50 mg of fresh weight. 600 μl of (3 volumes) TE-buffer (10 mm Tris, 1 mm EDTA-Na2)-equilibrated phenol were added and shaken for 30 min at 4 °C. After centrifugation at 4,000 × g and 4 °C for 8 min, the soluble proteins were dissolved in the upper phenolic phase (the high sucrose concentration causes a phase inversion). The phenolic phase was separated and the proteins precipitated out of the phenolic phase overnight in 5 volumes of ice-cold acetone. After centrifugation at 4,000 × g and 4 °C for 8 min the pellets were washed 3 times with ice-cold methanol and stored at −80 °C until further use. The dried protein pellets were then digested in two steps using endoproteinase Lys-C (1:100) first and then PoroszymeP®P immobilized trypsin according to the manufacturer's instructions (buffer 1: Lys-C digestion buffer (50 mm Tris, 8 m urea, 100 mm methylamine, pH 7.5); buffer 2: trypsin digestion buffer (50 mm Tris, 10% acetonitrile, 10 mm CaCl2, pH 7.5), after Lys-C digestion the sample is 1:4 diluted to have an end concentration of 2 m urea). Protein content was determined using the Bradford assay employing ovalbumin as the standard protein. The protein digest was desalted with SPECP®P C18 columns. After lyophilization the pellet was stored at −20 °C until use.
GC-TOF-MS Analysis—
The GC-TOF-MS analysis was performed on an HP 5890 gas chromatograph with deactivated standard spit/splitless liners containing glasswool (Agilent, Böblingen, Germany). One-μl sample was injected in the splitless mode at 230 °C injector temperature. GC was operated on an MDN-35 capillary, 30 m × 0.32 mm inner diameter, 25-μm film (SUPELCO, Bellefonte, PA), at constant flow of 2-ml/min helium. The temperature program started with 2 min isocratic at 85 °C, followed by temperature ramping at 15 °C/min to a final temperature of 360 °C, which was held for 8 min. Data acquisition was performed on a Pegasus II TOF mass spectrometer (LECO, St. Joseph, MI) with an acquisition rate of 20 scans sP−1P in the mass range of m/z = 85–600.
The obtained data were analyzed at first by defining a reference chromatogram with the maximum number of detected peaks over a signal/noise threshold of 50. Afterward all chromatograms were matched against the reference with a minimum match factor of 800. Compounds were annotated by retention index and mass spectra comparison to a user defined spectra library. Selected unique fragment ions specific for each individual metabolite were used for quantification.
LC-MS Shotgun Protein Analysis—
Prior to MS analysis, pellets of protein digests were dissolved in 5% formic acid. 10 μg per sample were concentrated on a pre-column and subsequently loaded onto a 50 cm silica-based C18 RP monolithic column (50 μm inner diameter) (30). Elution of the peptides was performed using a 4 h gradient from 100% solvent A (5% acetonitrile, 0.1% formic acid in water) to 100% solvent B (90% acetonitrile, 0.1% formic acid in water) using the Agilent nano high pressure liquid chromatography (HPLC) system (Agilent, Böblingen, Germany) with a flow rate of 400 nL per min. Eluting peptides were analyzed with a linear ion trap mass spectrometer (Thermo Electron, San Jose, CA) operated in a data-dependent mode. Each full MS scan was followed by three MS/MS scans in which the three most abundant peptide molecular ions were dynamically selected for collision-induced dissociation using a normalized collision energy of 35%. The temperature of the heated capillary and electrospray voltage was 150 °C and 1.9 kV, respectively. After MS analysis, DTA files were created from raw files and searched against a database using Bioworks 3.1. With DTASelect, a list of identified proteins was obtained using the following criteria: Xcorr: −1 2.0, −2 2.0, −3 3.3 (31) for hits with at least 2 different peptides. For quantitative analysis, Contrast was used to compare and align identified proteins and peptides from different runs and to determine the ion count per protein (32). Only proteins were included in the list, which at least appeared in five of the six independent replicates of one experimental treatment thereby ensuring reproducibility of the analysis. According to Liu et al. (2004) (33) the cumulative sum of recorded peptides per protein called spectral count was applied as a quantitative measure (33). All the identified peptide product ion spectra can be downloaded from ProMEX to reveal identification criteria and to judge the quality of the spectra. ProMEX is a mass spectral reference library for plant proteomics and can be searched also with unknown samples (34).
Statistical Data Analysis—
All data were normalized to mg fresh weight and stable isotope-labeled standard compounds. Statistical tests were performed in Matlab® 7.0 (Mathworks, Natick, MA) on the basis of log-transformed data.
For ICA an in-house Matlab script was used (10). The covariance of the data was first analyzed by PCA giving a restricted set of principal components covering 95% of variance. ICA was then applied to these new components, and new independent components were ranked by the kurtosis measure. The contributions of each metabolite/protein to an independent component can be obtained by combining the transformation matrix W of PCA with the transformation matrix V of ICA to a direct transformation U = W*V. The elements of the i-th vector in U represent the individual contributions; the loading (see Fig. 3A) to the i-th independent component. For more details see Scholz and Selbig (10). The ICA algorithm used in this study is CuBICA4 (35).
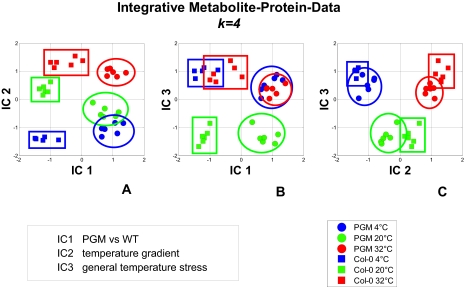
Fig. 3.
Sample pattern recognition in a combined metabolite-protein covariance matrix. For data dimensionality, reduction and extraction of the most significant biological information in the data, independent component analysis was performed (9, 10). In the experimental setup two different genotypes, a starch-deficient (PGM) mutant plant and the corresponding WT plant, were adapted to temperature at 4, 20, and 32 °C for 3 days. For each experiment six independent biological replicates (two pooled plants per sample) were measured (see under “Experimental Procedures” and supplemental material). The 20 °C treatments serve as controls to distinguish between cold and high temperature adaptation. The first 4 principal components (k = 4) used for the subsequent ICA explain 95% of the total variance. However, for simplicity only the 3 most important independent components are used for pattern recognition. A, visualization of independent components 1 and 2 (IC1 and IC2); B, visualization of independent components 1 and 3 (IC1 and IC3); C, visualization of independent components 2 and 3 (IC2 and IC3). The following biological information was extracted from these plots: (i) IC1 separates WT and PGM (see A); (ii) IC2 separates the temperature gradient shared by both genotypes WT and PGM; thus distinguishes 4, 20, and 32 °C (see A and B); (iii) IC3 separates temperature effects similar in 4C and 32 °C, shared by both genotypes WT and PGM (see B and C). By plotting IC2 and IC3 (see C) only temperature effects are observed, completely resolved from the genotype influence.
To test for differences in the median concentrations of metabolites and proteins between stressed and unstressed plants we used Kruskal-Wallis one-way analysis (ANOVA) by ranks implemented in Matlab® 7.0. Differences were considered statistically significant at p < 0.05.
RESULTS AND DISCUSSION
Parallel Metabolite and Protein Analysis by Combining an Integrative Extraction Protocol with GC-TOF-MS and LC-Ion Trap-MS Analysis—
Plant material was extracted using an extraction protocol for sequential isolation of metabolites and proteins from one sample to minimize technical standard deviation, increase sample throughput, and exploit metabolite-protein covariance for sample classification (9, 11, 29) (see Fig. 1). The data matrix consists of 36 independent biological replicates of different experiments with 332 variables for each experiment in the form of relative levels of proteins (160) and metabolites (172). In contrast to the former extraction protocol protein recovery was improved 2-fold by adding 40% sucrose into the protein extraction buffer and reducing degradation because of high temperature and long extraction times (29). For metabolite analysis a standard operation protocol consisting of chemical derivatization and subsequent analysis with gas chromatography coupled to time-of-flight mass spectrometry (GC-TOF-MS) was performed (9, 11, 14, 36). GC-TOF-MS metabolite profiling resulted in a list of 172 reproducible identifiable mass spectra including known carbohydrates, amino acids, and organic acids (for a complete list of identified metabolites, see supplemental Table S1). For protein analysis we used a label-free non-gel-based approach analyzing tryptic peptides of the complex protein sample on reversed phase liquid chromatography coupled to ion trap mass spectrometry (LC/MS) (9, 11, 30, 37, 38). Recently, it was demonstrated that the cumulative sum of recorded peptides called spectral count in such data-dependent LC/MS analyses correlates with protein abundance in the samples (33, 39, 40). This was also proven in a combination with stable isotope labeling (39). In a recent study we compared spectral count and peak integration from complex total ion chromatograms of a complex protein sample in combination with multivariate data mining (41). Dynamic range and limits of detection were studied by spiking a protein with known concentrations into the complex protein matrix. Both methods gave similar sample pattern recognition; however, spectral count was advantageous with respect to the total number of identified and quantified proteins. A typical protein analysis using peak integration showed high abundance proteins such as Calvin cycle enzymes and protein of the photosystem I and II (9, 11, 30). Using spectral counts the list of identified proteins exceeded significantly these functional protein classes (for a complete list see supplemental Table S2). Low abundance proteins as well as proteins known to be involved in temperature adaptation (At5g15970 and At5g52310) were observed.
All the identified proteins and their corresponding peptide product ion spectra can be downloaded from ProMEX site, a mass spectral reference library for plant proteomics (34). This library can also be used to search with unknown samples for protein identification. All the entries in the data base indicate the experimental conditions under which the protein was detected (34).
Correlation Network Topology Analysis and Sample Pattern Recognition Reveal the Structure of the Metabolite-Protein Covariance Matrix—
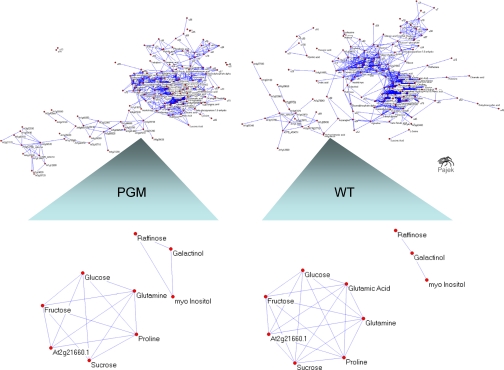
In recent studies we have proposed that the differential correlation between two components of a data matrix, say a specific metabolite and a protein, reflects the underlying biochemical regulation (4, 9, 14, 15). Following this line we analyzed the correlation network topology of the starch-deficient mutant PGM versus the corresponding wild-type under different temperature regimes 4, 20, and 32 °C (see Fig. 2). Correlation networks and their visualization using multidimensional scaling was generated with Pajek. PGM showed within the complete data set of all temperature treatment, a total of 1990 correlations, and after Bonferroni correction 1020 significant correlations. The wild-type plants exhibited 1535 correlations and 1101 significant correlations after Bonferroni correction. As expected both genotypes exhibited different network topologies (see Fig. 2). A refined analysis revealed that a component of the circadian regulation, named AtGRP7 (At2g21660), was strongly up-regulated under cold and had a strong correlation with proline and glutamine but not with raffinose and galactinol. To further reveal the covariance/correlation structure of the variables, the metabolites and proteins, and to rank the major information content of the data we applied ICA according to Morgenthal et al. and Scholz and Selbig (9, 10) (see Fig. 1). The chemometric analysis of the covariance/correlation structure of a complex data matrix is typically performed with PCA. PCA is applied to the high-dimensional data set first to extract the variables with the highest variance and to reduce data dimensionality. Subsequently, the principal components are used for independent component analysis (for detailed explanation, see Ref. 10). ICA is optimized to detect the inherent differences and ignore differences introduced globally by placing more emphasis on the independence of variables than their variances. The primary feature of the independent components is their kurtosis measure, a negative value indicating flatter or more uniform distribution throughout the data set and so a strong factor for data separation. Furthermore, ICA is an unsupervised statistical method, thereby guaranteeing that no bias for sample pattern recognition is introduced. Based on sample discrimination it is possible to assign the covariance/correlation structure of the metabolites and proteins to specific biological processes and to identify a ranked list of corresponding correlative biomarkers.
Fig. 2.
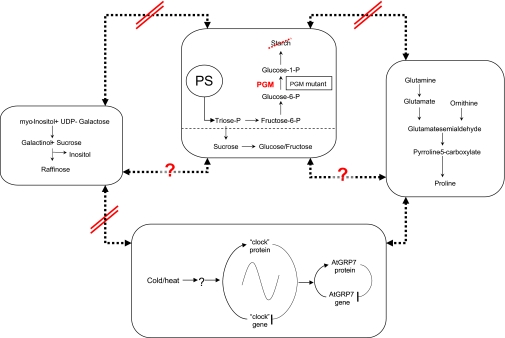
Correlation network analysis of an integrated metabolite-protein data set. Responses to 4, 20, and 32 °C treatment are combined in a data matrix for a genotype-dependent correlation analysis. Differential correlation networks for topology analysis were generated according to recent studies (4, 9, 14). The correlation network of the starch-deficient mutant PGM and the corresponding wild-type show different network topologies according to our stochastic metabolic modeling approach based on different data covariance structures and thus different biochemical regulation (4, 13, 15, 18). In the figure enlargement below, a sub-fraction of the correlations is shown between the RNA-binding protein ATGRP7 and temperature marker proline, glutamine, raffinose, and galactinol. A positive correlation is observed with proline and glutamine but not with galactinol and raffinose suggesting that galactinol and raffinose respond differently to temperature stress. For further discussion, see the text and Fig. 4.
ICA of the metabolites alone gave almost complete separation of sample groups (data not shown). The extracted transformation vectors IC1–IC3 indicated the occurrence of specific metabolites giving similar relative metabolite level responses for different processes because of a time-lag effect or analogous biochemical regulation. This was indeed recently demonstrated in an analysis of temperature-treated plants (43) and the diurnal rhythm of a plant (9). In contrast, the sample pattern of the proteins in ICA revealed a sample pattern according to the performed experiment, showing the genotype separation on the one hand and the temperature gradient on the other. No further biological characteristics were observed using the protein data alone. In Fig. 1 the strategy for the combined analysis of the metabolite-protein covariance matrix is shown. Following this strategy and decomposing the combined metabolite and protein data matrix into independent components revealed additional information, especially for protein marker identification. First, the mutation (PGM) and the WT plants (WT) were separated on IC1 (see Fig. 3A). The second component (IC2) depicts the temperature gradient response in both the mutant and the wild-type plants (see Fig. 3A) and the third component (IC3) shows temperature stress responses similar in 4 °C and 32 °C treatments, respectively (see Fig. 3B). Eventually, by plotting IC2 and IC3 only differences of temperature treatment were resolved independent from the genotype (see Fig. 3C). Thus, response effects of different genotypes to temperature treatment and general temperature acclimation mechanisms were completely resolved by combining the distinct features of metabolite and protein data.
Biomarker Identification Based on Combined Metabolite-Protein Covariance Analysis—
Because of a clear sample discrimination by ICA in Fig. 3 it is possible to assign roles to the detected proteins, which are correlated with a network of significant metabolite marker. This process is based on the biological interpretation of the transformation vectors IC1-IC3, the new independent components IC1 to IC3, allowing for a ranked assignment of metabolite and protein sets involved in the observed phenomena.
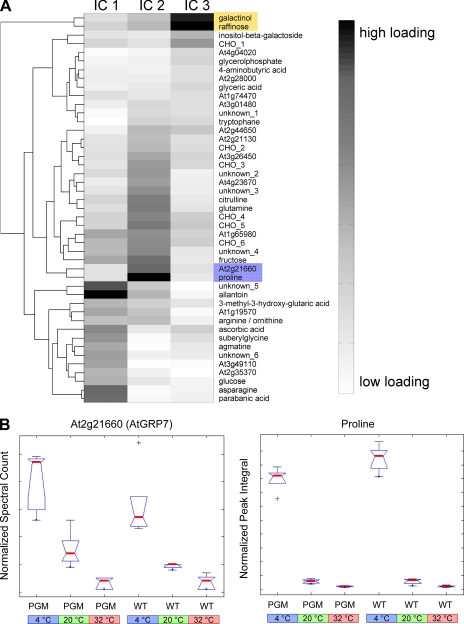
In Fig. 4A the loadings of different independent components IC1, IC2, and IC3 are visualized in a biclustering diagram. The loadings of ICA are proportional to the influence of a corresponding metabolite or protein on the observed sample discrimination along the transformation vectors (IC). If the transformation vectors (or independent components) can reasonably be interpreted, the loadings of the different metabolites and proteins are directly related to their importance on this biological phenomenon. It becomes clear that specific metabolites and proteins have different response effects to IC1, IC2, and IC3. Low temperature (IC2) strongly triggers osmolyte biosynthesis including proline, glutamine, and raffinose family oligosaccharides (RFO) like raffinose and galacinol. These compounds are accumulated during cold acclimation (43–45). However, from studies with galactinol synthase mutants, it has been shown that raffinose is not essential for basic freezing tolerance or for cold acclimation of A. thaliana (46). Furthermore, a comparison of heat- and cold-shock response patterns revealed that the majority of heat-shock responses on the metabolite level were shared with cold-shock responses (43, 47). These observations coincide with our study visible by IC3 in Fig. 3, which extracts effects similar in cold and heat adaptation. Here, raffinose and galactinol have the highest weights for 4 °C and 32 °C samples in the PGM and the wild-type and have similar mutant/WT ratios in 4 °C and 32 °C samples (see Fig. 4A and supplemental Table S1). Thus, there is evidence that these intermediates were involved in general temperature stress in contrast to proline and glutamine, which showed different loadings on IC2 for 4, 20, and 32 °C (see Fig. 4A; IC2) whereas having conserved levels in PGM and WT (see boxplot for proline in Fig. 4B). Unexpectedly, the primary effect of starch deficiency and channeling of triosephosphate into sucrose, glucose, and fructose in the PGM mutant has no strong impact on typical metabolite marker for cold acclimation processes. The observation is a stress-induced increase of RFO levels in temperature-treated PGM plants. Thus, starch storage and mobilization is not essential for the typical accumulation of these stress marker metabolites. This indicates that sucrose accumulation and storage in the starch-deficient mutant plants are sufficient to bypass the typical starch storage/mobilization pathways under abiotic stress (48). Furthermore the increase of RFO is not directly correlated with ATGRP7, proline, glutamine, and sucrose (see Fig. 2) for 20, 4, and 32 °C conditions in both the mutant and the wild-type plants indicating that those processes might work independently. This suggests a very complex up/down-regulation of several pathways and metabolic storage pools simultaneously and a high potential of metabolic flexibility (14, 49). However, these processes remain elusive and whether the increased RFO levels origin from storage pools or from de novo synthesis will be tested in future with stable isotope labeling techniques and metabolic flux measurements.
Fig. 4.
Clustering of the ICA loadings. A, in the graph the weight distribution on IC1–IC3 for the metabolites and proteins based on the combined covariance data matrix are shown. The weights (also called “loadings”) are proportional to the importance or significance of a metabolite/protein for a corresponding independent component, in other words, the observed biological phenomenon. The interpretations of IC1, IC2, and IC3 correspond to Fig. 3 (see also text for more details). For this diagram the weights are normalized and visualized in a biclustering diagram. Only the first 50 ranked loadings for the corresponding IC1, IC2, and IC3 were selected for clarity of visual inspection. B, boxplots of identified cold stress biomarkers for IC2. Based on the inspection of the loadings for IC2, ATGRP7, and proline show a similar pattern dependent on the temperature treatment and independent on the genotype.
Fructose is highly accumulated in PGM (see supplemental Table S1), thus PGM and WT separate based on the loadings for fructose (see Fig. 4A). However, allantoin, asparagine, and an oxidative derivative of urea parabanic acid, all involved in the urea cycle metabolism, are more discriminatory for PGM and WT based on the loadings for IC1 (see Fig. 4A). The reason is that fructose has also strong influence in temperature adaptation in the plants, thus has high loadings also on IC2. Urea and asparagine metabolism, in contrast, are strongly impaired in the PGM mutant plant compared with the WT plants according to studies analyzing global transcriptional activity in the PGM mutant plant (50). Consequently, intermediates of urea metabolism are unique biomarkers for the PGM mutation probably because of severely retarded growth (see also our recent study (9)). This interaction of metabolic mutation and temperature response is nicely visible in the presented work here, thereby indicating that the proposed integration of different genotypes, experimental conditions, and molecular levels reveal novel insights on the systemic behavior, eventually leading to a new route of functional studies (see Fig. 1).
Because of an improved recognition of sample pattern, which is demonstrated with “proof of concept-metabolite markers” like proline, raffinose, and galactinol (see above), it is possible to assign specific proteins to these processes. In a recent study we investigated the proteins separating PGM and WT (9). The loadings of proteins with respect to their differentiating capability can be seen in the biclustering diagram in Fig. 4A. The ranking is found to be the same for chloroplastidic GAPDH (At3g26650) and cytosolic GAPDH (At1g13440) indicating that the chloroplastidic isoform has a stronger impact on PGM-WT discrimination (see also (9). However, in this study, we considerably increased the number of quantified proteins. Consequently, new protein markers were identified with higher rankings than GAPDH. Interestingly, proteins involved in redox-stress At3g49110 and At1g19570 have high loadings for IC1 in separating the PGM mutant and the WT.
High protein loadings for IC3, general temperature response similar for 4 and 32 °C, is observed for At2g44650 (see Fig. 4A and supplemental Table S1), a novel chloroplast chaperonin (CPn10) with unknown function (51–53). Recently, a homolog of CPn10 was implicated as having a very specific role in temperature stress adaptation in other species (54, 55). CPn10 shows a mixed-type behavior and is consequently found with high loadings for components, IC2 (temperature gradient marker) and IC3 (general temperature stress marker). Other proteins with high loadings for IC3 are typical temperature stress markers, like cyclophilins or proteins involved in oxidative stress (At3g01480 and At1g65980, respectively).
IC2 encodes differences between 4 and 32 °C temperature acclimation (see Fig. 3A). Accordingly, proteins identified as having high loadings for this component belong to the cold responsive proteins such as At5g52310 low temperature-induced protein 78 (sp Q06738) and At5g15970 cold-regulated protein COR6.6 (KIN2) (see Fig. 3A). At2g37220 and At3g53460 are nuclear encoded, targeted to the chloroplast, and have a consensus sequence-type RNA-binding domain originally isolated in tobacco (56). In our study, these proteins have slightly higher loadings for component IC2 than the cold-regulated proteins. In supplemental Table S2 the PGM mutant and WT ratios to the 20 °C control samples are shown. The ratios of these two RNA-binding proteins decrease strongly under higher temperature. In contrast, the ratios of the cold-regulated proteins increase under 4 °C treatment. Thus, it is very important that biomarkers identified by covariance analysis (multivariate statistics) have to be further compared on their median levels (univariate statistics, multiple means testing, analysis of variance (ANOVA)) (see supplemental Tables S1 and S2).
Other protein markers with very high loadings on IC2 were also RNA-binding proteins, which is also in agreement with recent studies (53, 57–70). Kim et al. (71) demonstrated that over-expression of a glycine-rich RNA-binding protein resulted in enhanced cold-shock resistance in Escherichia coli. A novel candidate At2g21660 (ATGRP7) is a homologue of this protein family. It was identified in our study as the strongest cold treatment marker increasing under cold and decreasing under heat (highest loadings on IC2; see Fig. 4, A and boxplot in B). At2g21660 (ATGRP7) was subjected to genetic analysis and suggested to be stress-related (72, 73) and a circadian output gene (74). In a recent whole-genome analysis of transcript levels under cold stress this gene showed increasing expression after 2 days, which correlates with our experimental conditions and quantitative protein data. However, the direct correlation of RNA expression and protein levels has to be further studied. In a recent investigation of circadian clock-regulated gene expression a feedback loop between ATGRP7 gene and protein abundance was proposed (75). In a very recent study ATGRP7 was shown to complement cold-sensitive E. coli mutants lacking cold-shock proteins (76). ATGRP7 is also a homologue of so-called cold-inducible RNA-binding proteins identified in human, mouse, and rat (77) mediating cold-induced growth suppression. Last, a strong covariance of ATGRP7 is found with the cold-acclimation marker proline (see Figs. 2 and 4A and boxplots in Fig. 4B). The relations of the different biomarkers are summarized in Fig. 5. Biochemical building blocks represent the different metabolic compartments in which the individual marker is involved. Based on the parallel analysis of the starch-deficient mutant versus wild type and the analysis of the metabolite-protein covariance structure a decoupling of starch synthesis and mobilization from temperature compensation mechanisms driven by circadian genes/proteins are observed. These observations together support a functional role of ATGRP7, other RNA-binding proteins, and post-transcriptional control in plant cold adaptation indicating that these processes in plants are closely related and share mechanisms with the mammalian system (77–79).
Fig. 5.
Proposed interaction of biochemical modules as a response to cold/heat. A cold-induced increase in ATGRP7 and a down-regulation at moderate heat correlates with the increase/decrease of the metabolite markers proline and glutamine (see also supplemental Tables S1 and S2), whereas the raffinose and galactinol of RFO show increase in both temperature treatments, 4 °C and 32 °C. This effect is also observed in the starch-deficient mutant thereby indicating that starch deficiency and resulting changes in starch mobilization and raffinose/galactinol synthesis are not necessarily linked in response to temperature stress and ATGPP7 as long as the sucrose synthesis pathway is functioning. However, these observations have to be investigated in further detail, also according to a recent discussion on plant temperature stress and metabolic effects by Guy et al. (47). A clear correlation of sucrose with RFO is not observed. Here, other methods like conditional correlation analysis are planned in future. In summary, the parallel investigation of the starch-deficient mutant and the corresponding WT and the identification of correlative metabolite-protein biomarkers uncover the separation of starch storage/mobilization and other metabolic processes such as the raffinose/galactinol pathway, proline/glutamine pathways, and cold temperature compensation mechanisms driven by circadian output genes like the RNA-binding protein At2g21660 (ATGRP7).
CONCLUSION
A method is presented combining high throughput metabolite and protein profiling for the investigation of systemic responses of A. thaliana to abiotic stress. The integration clearly benefits from the heterogeneity of the data, thus, improves sample pattern recognition and therefore biological interpretation and identification of potential correlative metabolite-protein biomarker. However, a principle drawback of the presented profiling methods is its unbiased nature. For instance, the coverage of metabolic enzymes is comparatively low, as is the overlap between metabolites and their corresponding enzymes. This agrees with the observation that sample pattern recognition is indeed complementary for both of the molecular fractions, metabolites, and proteins (9, 80). Consequently, the integration of metabolite and protein data adds a further level of complementary information resulting in a better sample pattern recognition. However, for a detailed analysis of the interaction between metabolic enzymes and their corresponding metabolites, targeted approaches are much more feasible (81–84). Also, the quantitative pathway activity information captured in the metabolic network can be compared at the system level with metabolic fluxes estimated by metabolic flux analysis that uses only the metabolic data set (85). In future work system responses to abiotic temperature stress can be compared based on such modeling approaches and by the integration of metabolic and proteomic data sets.
In summary, metabolite profiling using GC-TOF-MS provides a very rapid and comprehensive technique for characterizing biological samples based on identification and quantification of hundreds of compounds. However, sample classification generally relies on covariance between metabolites. Integration of proteomics data from the same sample introduces a further level of causality and reveals an increased information extraction based on complementary sample patterns. Consequently, correlated metabolites and proteins can be assigned to distinct biological processes, thereby generating new hypothesis about the interaction of different biochemical building blocks. Besides transcript profiling the integration of enzyme activities represents an important complement to the described mass spectrometry-based protein profiling method. Especially, high throughput platforms for measuring many different enzymatic activities at the same time are very useful (86). Another very important aspect is flux measurement. Especially in the case of abiotic stress it will be interesting to reveal metabolic fluxes between central sugar metabolism and the RFO because these RFO were identified in our study as rather independent general stress markers. The whole concept of high-dimensional data integration from many replicates and multivariate statistics for covariance structure analysis is proposed to be a unique way to reveal systemic responses of the biological system under study, which is a prerequisite for gene/protein function discovery in the genome/systems biology era.
Supplementary Material
Acknowledgments
We thank Megan McKenzie for revising the manuscript.
Footnotes
Published, MCP Papers in Press, April 28, 2008, DOI 10.1074/mcp.M700273-MCP200
Author's Choice—Final version full access.
The abbreviations used are: PCA, principal components analysis; ICA, independent component analysis; WT, wild-type; GC, gas chromatography; TOF-MS, time-of-flight mass spectrometry; RFO, raffinose family oligosaccharides; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PGM, phosphoglucomutase.
This work was supported by the Max Planck Society. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Tables S1 and S2.
REFERENCES
- 1.Ideker, T., Galitski, T., and Hood, L. ( 2001) A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2, 343–372 [DOI] [PubMed] [Google Scholar]
- 2.Fiehn, O., Kopka, J., Dormann, P., Altmann, T., Trethewey, R. N., and Willmitzer, L. ( 2000) Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 3.Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G., and Kell, D. B. ( 2004) Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252 [DOI] [PubMed] [Google Scholar]
- 4.Weckwerth, W. ( 2003) Metabolomics in systems biology. Annu. Rev. Plant Biol. 54, 669–689 [DOI] [PubMed] [Google Scholar]
- 5.Fernie, A. R., Trethewey, R. N., Krotzky, A. J., and Willmitzer, L. ( 2004) Innovation: Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5, 763–769 [DOI] [PubMed] [Google Scholar]
- 6.Liebermeister, W. ( 2002) Linear modes of gene expression determined by independent component analysis. Bioinformatics 18, 51–60 [DOI] [PubMed] [Google Scholar]
- 7.Zamboni, N., and Sauer, U. ( 2004) Model-independent fluxome profiling from H-2 and C-13 experiments for metabolic variant discrimination. Genome Biol. 5, [DOI] [PMC free article] [PubMed]
- 8.Scholz, M., Gatzek, S., Sterling, A., Fiehn, O., and Selbig, J. ( 2004) Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20, 2447–2454 [DOI] [PubMed] [Google Scholar]
- 9.Morgenthal, K., Wienkoop, S., Scholz, M., Selbig, J., and Weckwerth, W. ( 2005) Correlative GC-TOF-MS-based metabolite profiling and LC-MS-based protein profiling reveal time-related systemic regulation of metabolite-protein networks and improve pattern recognition for multiple biomarker selection. Metabolomics 1, 109–121 [Google Scholar]
- 10.Scholz, M., and Selbig, J. ( 2007) Visualization and analysis of molecular data. Methods Mol. Biol. 358, 87–104 [DOI] [PubMed] [Google Scholar]
- 11.Weckwerth, W., Wenzel, K., and Fiehn, O. ( 2004) Process for the integrated extraction identification, and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4, 78–83 [DOI] [PubMed] [Google Scholar]
- 12.Weckwerth, W., and Morgenthal, K. ( 2005) Metabolomics: from pattern recognition to biological interpretation. Drug Discov. Today 10, 1551–1558 [DOI] [PubMed] [Google Scholar]
- 13.Steuer, R., Kurths, J., Fiehn, O., and Weckwerth, W. ( 2003) Observing and interpreting correlations in metabolomic networks. Bioinformatics 19, 1019–1026 [DOI] [PubMed] [Google Scholar]
- 14.Weckwerth, W., Loureiro, M. E., Wenzel, K., and Fiehn, O. ( 2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc. Natl. Acad. Sci. U. S. A. 101, 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgenthal, K., Weckwerth, W., and Steuer, R. ( 2006) Metabolomic networks in plants: Transitions from pattern recognition to biological interpretation. Biosystems 83, 108–117 [DOI] [PubMed] [Google Scholar]
- 16.Weckwerth, W., and Steuer, R. ( 2005) Metabolic networks from a systems perspective: from experiment to biological interpretation. (Vaidyanathan, S., Harrigan, G. G., and Goodacre, R., eds) pp. 265–289, Springer, NY
- 17.Steuer, R., Kurths, J., Fiehn, O., and Weckwerth, W. ( 2003) Interpreting correlations in metabolomic networks. Biochem. Soc. Trans. 31, 1476–1478 [DOI] [PubMed] [Google Scholar]
- 18.Camacho, D., Fuente, A., and Mendes, P. ( 2005) The origin of correlations in metabolomics data. Metabolomics 1, 53–63 [Google Scholar]
- 19.Mendes, P., Camacho, D., and de la Fuente, A. ( 2005) Modelling and simulation for metabolomics data analysis. Biochem. Soc. Trans. 33, 1427–1429 [DOI] [PubMed] [Google Scholar]
- 20.Rao, C. V., Wolf, D. M., and Arkin, A. P. ( 2002) Control, exploitation and tolerance of intracellular noise. Nature 420, 231–237 [DOI] [PubMed] [Google Scholar]
- 21.Patterson, S. D., and Aebersold, R. H. ( 2003) Proteomics: the first decade and beyond. Nat. Genet. 33, 311–323 [DOI] [PubMed] [Google Scholar]
- 22.Gibon, Y., Usadel, B., Blaesing, O. E., Kamlage, B., Hoehne, M., Trethewey, R., and Stitt, M. ( 2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 7, R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. ( 1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey, I. M., Rubio-Aliaga, I., Siewert, A., Sailer, D., Drobyshev, A., Beckers, J., de Angelis, M. H., Aubert, J., Hen, A. B., Fiehn, O., Eichinger, H. M., and Daniel, H. ( 2007) Profiling at mRNA, protein, and metabolite levels reveals alterations in renal amino acid handling and glutathione metabolism in kidney tissue of Pept2−/− mice. Physiol. Genomics 28, 301–310 [DOI] [PubMed] [Google Scholar]
- 25.Clish, C. B., Davidov, E., Oresic, M., Plasterer, T. N., Lavine, G., Londo, T., Meys, M., Snell, P., Stochaj, W., Adourian, A., Zhang, X., Morel, N., Neumann, E., Verheij, E., Vogels, J., Havekes, L. M., Afeyan, N., Regnier, F., Van Der Greef, J., and Naylor, S. ( 2004) Integrative biological analysis of the APOE*3-Leiden transgenic mouse. OMICS 8, 3–13 [DOI] [PubMed] [Google Scholar]
- 26.Perroud, B., Lee, J., Valkova, N., Dhirapong, A., Lin, P. Y., Fiehn, O., Kultz, D., and Weiss, R. H. ( 2006) Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol. Cancer 5, [DOI] [PMC free article] [PubMed]
- 27.Caspar, T., Huber, S. C., and Somerville, C. ( 1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 79, 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyes, D. C., Zayed, A. M., Ascenzi, R., McCaskill, A. J., Hoffman, N. E., Davis, K. R., and Gorlach, J. ( 2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgenthal, K., Wienkoop, S., Wolschin, F., and Weckwerth, W. ( 2006) Integrative profiling of metabolites and proteins: improving pattern recognition and biomarker selection for systems level approaches. Methods Mol. Biol. 358, 57–76 [DOI] [PubMed] [Google Scholar]
- 30.Wienkoop, S., Glinski, M., Tanaka, N., Tolstikov, V., Fiehn, O., and Weckwerth, W. ( 2004) Linking protein fractionation with multidimensional monolithic RP peptide chromatography/mass spectrometry enhances protein identification from complex mixtures even in the presence of abundant proteins. Rapid Commun. Mass Spectrom. 18, 643–650 [DOI] [PubMed] [Google Scholar]
- 31.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J., and Gygi, S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 32.Tabb, D. L., McDonald, W. H., and Yates, J. R. ( 2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, H., Sadygov, R. G., and Yates, J. R., III ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 34.Hummel, J., Niemann, M., Wienkoop, S., Schulze, W., Steinhauser, D., Selbig, J., Walther, D., and Weckwerth, W. ( 2007) ProMEX: a mass spectral reference database for proteins and protein phosphorylation sites. BMC Bioinformatics 8, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaschke, T., and Wiskott, L. ( 2004) CuBICA: independent component analysis by simultaneous third- and fourth-order cumulant diagonalization. Ieee Trans. Signal Process. 52, 1250–1256 [Google Scholar]
- 36.Weckwerth, W., Tolstikov, V., and Fiehn, O. ( 2001) Metabolomic characterization of transgenic potato plants using GC/TOF and LC/MS analysis reveals silent metabolic phenotypes, in Proceedings of the 49th ASMS Conference on Mass spectrometry and Allied Topics, American Society of Mass Spectrometry, Chicago, USA
- 37.Wienkoop, S., Zoeller, D., Ebert, B., Simon-Rosin, U., Fisahn, J., Glinski, M., and Weckwerth, W. ( 2004) Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry 65, 1641–1649 [DOI] [PubMed] [Google Scholar]
- 38.Wienkoop, S., and Weckwerth, W. ( 2006) Relative and absolute quantitative shotgun proteomics: targeting low-abundance proteins in Arabidobsis thaliana. J. Exp. Bot. 57, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 39.Zybailov, B., Coleman, M. K., Florens, L., and Washburn, M. P. ( 2005) Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 77, 6218–6224 [DOI] [PubMed] [Google Scholar]
- 40.Cox, B., Kislinger, T., and Emili, A. ( 2005) Integrating gene and protein expression data: pattern analysis and profile mining. Methods 35, 303–314 [DOI] [PubMed] [Google Scholar]
- 41.Wienkoop, S., Larrainzar, E., Niemann, M., Gonzalez, E. M., Lehmann, U., and Weckwerth, W. ( 2006) Stable isotope-free quantitative shotgun proteomics combined with sample pattern recognition for rapid diagnostics. J. Sep. Sci. 29, 2793–2801 [DOI] [PubMed] [Google Scholar]
- 42.Weckwerth, W. ( 2008) Integration of metabolomics and proteomics in molecular plant physiology - coping with the complexity by data-dimensionality reduction. Physiol. Plant. 132, 176–189 [DOI] [PubMed] [Google Scholar]
- 43.Kaplan, F., Kopka, J., Haskell, D. W., Zhao, W., Schiller, K. C., Gatzke, N., Sung, D. Y., and Guy, C. L. ( 2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomashow, M. F. ( 1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Phys. 50, 571–599 [DOI] [PubMed] [Google Scholar]
- 45.Cook, D., Fowler, S., Fiehn, O., and Thomashow, M. F. ( 2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 101, 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuther, E., Buchel, K., Hundertmark, M., Stitt, M., Hincha, D. K., and Heyer, A. G. ( 2004) The role of raffinose in the cold acclimation response of Arabidopsis thaliana. Febs Lett. 576, 169–173 [DOI] [PubMed] [Google Scholar]
- 47.Guy, C., Kaplan, F., Kopka, J., Selbig, J., and Hincha, D. K. ( 2008) Metabolomics of temperature stress. Physiol. Plant. 132, 220–235 [DOI] [PubMed] [Google Scholar]
- 48.Kaplan, F., and Guy, C. L. ( 2005) RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 44, 730–743 [DOI] [PubMed] [Google Scholar]
- 49.Ishii, N., Nakahigashi, K., Baba, T., Robert, M., Soga, T., Kanai, A., Hirasawa, T., Naba, M., Hirai, K., Hoque, A., Ho, P. Y., Kakazu, Y., Sugawara, K., Igarashi, S., Harada, S., Masuda, T., Sugiyama, N., Togashi, T., Hasegawa, M., Takai, Y., Yugi, K., Arakawa, K., Iwata, N., Toya, Y., Nakayama, Y., Nishioka, T., Shimizu, K., Mori, H., and Tomita, M. ( 2007) Multiple high throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 [DOI] [PubMed] [Google Scholar]
- 50.Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L. A., Rhee, S. Y., and Stitt, M. ( 2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 51.Hill, J. E., and Hemmingsen, S. M. ( 2001) Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 6, 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koumoto, Y., Shimada, T., Kondo, M., Hara-Nishimura, I., and Nishimura, M. ( 2001) Chloroplasts have a novel Cpn10 in addition to Cpn20 as co-chaperonins in Arabidopsis thaliana. J. Biol. Chem. 276, 29688–29694 [DOI] [PubMed] [Google Scholar]
- 53.Levy-Rimler, G., Viitanen, P., Weiss, C., Sharkia, R., Greenberg, A., Niv, A., Lustig, A., Delarea, Y., and Azem, A. ( 2001) The effect of nucleotides and mitochondrial chaperonin 10 on the structure and chaperone activity of mitochondrial chaperonin 60. Eur. J. Biochem. 268, 3465–3472 [DOI] [PubMed] [Google Scholar]
- 54.Strocchi, M., Ferrer, M., Timmis, K. N., and Golyshin, P. N. ( 2006) Low temperature-induced systems failure in Escherichia coli: insights from rescue by cold-adapted chaperones. Proteomics 6, 193–206 [DOI] [PubMed] [Google Scholar]
- 55.Ferrer, M., Lunsdorf, H., Chernikova, T. N., Yakimov, M., Timmis, K. N., and Golyshin, P. N. ( 2004) Functional consequences of single: double ring transitions in chaperonins: life in the cold. Mol. Microbiol. 53, 167–182 [DOI] [PubMed] [Google Scholar]
- 56.Ohta, M., Sugita, M., and Sugiura, M. ( 1995) 3 Types of nuclear genes encoding chloroplast RNA-binding proteins (Cp29, Cp31 and Cp33) are present in Arabidopsis thaliana - presence of Cp31 in chloroplasts and its homolog in nuclei/cytoplasms. Plant Mol. Biol. 27, 529–539 [DOI] [PubMed] [Google Scholar]
- 57.Savitch, L. V., Allard, G., Seki, M., Robert, L. S., Tinker, N. A., Huner, N. P. A., Shinozaki, K., and Singh, J. ( 2005) The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant Cell Physiol. 46, 1525–1539 [DOI] [PubMed] [Google Scholar]
- 58.Mastrangelo, A. M., Belloni, S., Barilli, S., Ruperti, B., Di Fonzo, N., Stanca, A. M., and Cattivelli, L. ( 2005) Low temperature promotes intron retention in two e-cor genes of durum wheat. Planta 221, 705–715 [DOI] [PubMed] [Google Scholar]
- 59.Zhang, X., Fowler, S. G., Cheng, H. M., Lou, Y. G., Rhee, S. Y., Stockinger, E. J., and Thomashow, M. F. ( 2004) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 39, 905–919 [DOI] [PubMed] [Google Scholar]
- 60.Rohde, P., Hincha, D. K., and Heyer, A. G. ( 2004) Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 38, 790–799 [DOI] [PubMed] [Google Scholar]
- 61.Jia, Y., del Rio, H. S., Robbins, A. L., and Louzada, E. S. ( 2004) Cloning and sequence analysis of a low temperature-induced gene from trifoliate orange with unusual pre-mRNA processing. Plant Cell Rep. 23, 159–166 [DOI] [PubMed] [Google Scholar]
- 62.Viachonasios, K. E., Thoirnashow, M. F., and Triezenberg, S. J. ( 2003) Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim, J. C., Lee, S. H., Cheong, Y. H., Yoo, C. M., Lee, S. I., Chun, H. J., Yun, D. J., Hong, J. C., Lee, S. Y., Lim, C. O., and Cho, M. J. ( 2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 25, 247–259 [DOI] [PubMed] [Google Scholar]
- 64.Dickson, R., Weiss, C., Howard, R. J., Alldrick, S. P., Ellis, R. J., Lorimer, G., Azem, A., and Viitanen, P. V. ( 2000) Reconstitution of higher plant chloroplast chaperonin 60 tetradecamers active in protein folding. J. Biol. Chem. 275, 11829–11835 [DOI] [PubMed] [Google Scholar]
- 65.Li, Q. B., Haskell, D. W., and Guy, C. L. ( 1999) Coordinate and non-coordinate expression of the stress 70 family and other molecular chaperones at high and low temperature in spinach and tomato. Plant Mol. Biol. 39, 21–34 [DOI] [PubMed] [Google Scholar]
- 66.Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. ( 1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442 [DOI] [PubMed] [Google Scholar]
- 67.Suehiro, T., Boros, P., Emre, S., Sheiner, P., Guy, S., Schwartz, M. E., and Miller, C. M. ( 1997) Assessment of liver allograft function by hyaluronic acid and endothelin levels. J. Surg. Res. 73, 123–128 [DOI] [PubMed] [Google Scholar]
- 68.Anderson, J. V., Li, Q. B., Haskell, D. W., and Guy, C. L. ( 1994) Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold-acclimation. Plant Physiol. 104, 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neven, L. G., Haskell, D. W., Hofig, A., Li, Q. B., and Guy, C. L. ( 1993) Characterization of a spinach gene responsive to low-temperature and water-stress. Plant Mol. Biol. 21, 291–305 [DOI] [PubMed] [Google Scholar]
- 70.Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. ( 2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781 [DOI] [PubMed] [Google Scholar]
- 71.Kim, Y. O., Kim, J. S., and Kang, H. ( 2005) Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 42, 890–900 [DOI] [PubMed] [Google Scholar]
- 72.Wilkins, M. B., and Holowins, Aw ( 1965) Occurrence of an endogenous circadian rhythm in a plant tissue culture. Plant Physiol. 40, 907– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smallwood, M., and Bowles, D. J. ( 2002) Plants in a cold climate. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 831–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heintzen, C., Nater, M., Apel, K., and Staiger, D. ( 1997) AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 94, 8515–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Staiger, D., Zecca, L., Wieczorek Kirk, D. A., Apel, K., and Eckstein, L. ( 2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 33, 361–371 [DOI] [PubMed] [Google Scholar]
- 76.Kim, J. S., Park, S. J., Kwak, K. J., Kim, Y. O., Kim, J. Y., Song, J., Jang, B., Jung, C. H., and Kang, H. ( 2007) Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 35, 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng, Y., Kok, K. H., Xu, R. H., Kwok, K. H. H., Tay, D., Fung, P. C. W., Kung, H. F., and Lin, M. C. M. ( 2000) Maternal cold inducible RNA binding protein is required for embryonic kidney formation in Xenopus laevis. FEBS Lett. 482, 37–43 [DOI] [PubMed] [Google Scholar]
- 78.Burd, C. G., and Dreyfuss, G. ( 1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 79.Pan, F., Zarate, J., Choudhury, A., Rupprecht, R., and Bradley, T. M. ( 2004) Osmotic stress of salmon stimulates up-regulation of a cold inducible RNA-binding protein (CIRP) similar to that of mammals and amphibians. Biochimie (Paris) 86, 451–461 [DOI] [PubMed] [Google Scholar]
- 80.Hoehenwarter, W., Van Dongen, J. T., Wienkoop, S., Steinfath, M., Hummel, J., Erban, A., Sulpice, R., Regierer, B., Kopka, J., Geigenberger, P., and Weckwerth, W. ( 2008) A rapid approach for phenotype-screening and database independent detection of cSNP/protein polymorphism using mass accuracy precursor alignment. Proteomics, in press [DOI] [PubMed]
- 81.Gerber, S. A., Rush, J., Stemman, O., Kirschner, M. W., and Gygi, S. P. ( 2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U. S. A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wienkoop, S., and Weckwerth, W. ( 2006) Relative and absolute quantitative shotgun proteomics: targeting low-abundance proteins in Arabidopsis thaliana. J. Exp. Bot. 57, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 83.Mallick, P., Schirle, M., Chen, S. S., Flory, M. R., Lee, H., Martin, D., Ranish, J., Raught, B., Schmitt, R., Werner, T., Kuster, B., and Aebersold, R. ( 2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]
- 84.Brunner, E., Ahrens, C. H., Mohanty, S., Baetschmann, H., Loevenich, S., Potthast, F., Deutsch, E. W., Panse, C., de Lichtenberg, U., Rinner, O., Lee, H., Pedrioli, P. G., Malmstrom, J., Koehler, K., Schrimpf, S., Krijgsveld, J., Kregenow, F., Heck, A. J., Hafen, E., Schlapbach, R., and Aebersold, R. ( 2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 85.Stephanopoulos, G. ( 1998) Metabolic Engineering: principles and methodologies. Academic Press, London
- 86.Gibon, Y., Blaesing, O., Hannemann, J., Carillo, P., Hohne, M., Hendriks, J., Palacios, N., Cross, J., Selbig, J., and Stitt, M. ( 2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16, 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.