Abstract
Recent studies using stable isotope labeling with amino acids in culture (SILAC) in quantitative proteomics have made mention of the problematic conversion of isotope-coded arginine to proline in cells. The resulting converted proline peptide divides the heavy peptide ion signal causing inaccuracy when compared with the light peptide ion signal. This is of particular concern as it can effect up to half of all peptides in a proteomic experiment. Strategies to both compensate for and limit the inadvertent conversion have been demonstrated, but none have been shown to prevent it. Additionally, these methods combined with SILAC labeling in general have proven problematic in their large scale application to sensitive cell types including embryonic stem cells (ESCs) from the mouse and human. Here, we show that by providing as little as 200 mg/liter l-proline in SILAC media, the conversion of arginine to proline can be rendered completely undetectable. At the same time, there was no compromise in labeling with isotope-coded arginine, indicating there is no observable back conversion from the proline supplement. As a result, when supplemented with proline, correct interpretation of “light” and “heavy” peptide ratios could be achieved even in the worst cases of conversion. By extending these principles to ESC culture protocols and reagents we were able to routinely SILAC label both mouse and human ESCs in the absence of feeder cells and without compromising the pluripotent phenotype. This study provides the simplest protocol to prevent proline artifacts in SILAC labeling experiments with arginine. Moreover, it presents a robust, feeder cell-free, protocol for performing SILAC experiments on ESCs from both the mouse and the human.
Large scale identification of proteins using mass spectrometry (MS)-based1 proteomics has become common place in recent years (1). However, the generation of increasingly long lists of proteins now confronts researchers with the more challenging problem of determining the biological significance of particular proteins. For this reason, researchers are exploiting quantitative proteomic strategies to extract more meaningful information from their data sets. By comparing multiple samples and different conditions versus controls, it is possible to discern proteins that are altered in a given set of experimental or biological conditions from the thousands of proteins that are identified (2).
In MS-based proteomics, quantitative strategies typically involve the use of stable isotopic reagents to generate “heavy” and “light” samples, which retain their chemical identity and can then be differentiated and directly compared by MS analysis. Although chemical labeling methods such as ICAT (isotope-coded affinity tags) have demonstrated widespread applicability (1), metabolic incorporation strategies such as stable isotope labeling with amino acids in cell culture (SILAC) are becoming more common for cell types that can be grown for extensive periods of time in vitro. Though there are numerous advantages for using SILAC-based methods, compared with chemical labeling, a major drawback is the unintended metabolic inter-conversion of isotopic amino acids in the labeling process, generating artifacts affecting the quantification. This is a particular problem with arginine, which is a metabolic precursor for proline biosynthesis (Fig. 1). In general, large scale SILAC experiments use both isotope-coded arginine and lysine to obtain labeling of all possible tryptic peptides thereby maximizing quantitative coverage of all potential peptides in a given experiment.
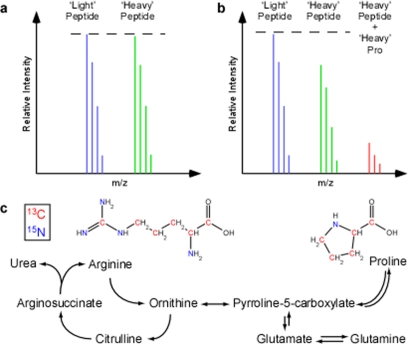
Fig. 1.
Metabolic conversion of isotope-coded arginine to proline in SILAC experiments. The unpredictable conversion of isotopic arginine to proline creates inaccuracy in SILAC-based quantitative proteomic experiments. a, a conceptual mass spectra of a non-proline containing peptide ion from a 1:1 mixture of light and heavy labeled samples. Here the expected Light and Heavy peptide ions have an equivalent signal. b, a spectra from a proline containing peptide where arginine to proline conversion has occurred in the same 1:1 mixture. The resulting heavy proline peptide signal (red) has been subtracted from the expected ‘heavy’ peptide ion signal. c, metabolic pathway outlining the inter-conversion of arginine and proline. Isotope-coded arginine with carbon 13 (red) and nitrogen 15 (green) when used as a synthetic precursor increases the expected mass of proline.
Several studies using SILAC have described the problematic conversion of isotopically coded arginine to labeled proline in cells (3–7) (Fig. 1), which results in an underestimation of the abundance of “heavy” tryptic peptides containing proline in a relative quantification experiment. This is of particular concern because ∼50% of all tryptic peptides between 700 and 6000 Da in the international protein index human data base contain at least one proline. Moreover, in a recent study it was reported that ∼30–40% of all observable proline containing peptides exhibited some level of conversion from heavy arginine (7). Besides acting to skew the relative abundance observed, conversion of “heavy” arginine to “heavy” proline also further complicates the mass spectrometry data by increasing the number of peptide ion peaks. One strategy to minimize this problem is the reduction of the arginine concentration in the SILAC media rendering it metabolically unfavorable as a precursor for proline synthesis (8). Although this does reduce the conversion, it does not prevent it completely (3). The other is to avoid the use of arginine altogether (3). Finally, alternative corrections can be done mathematically, post-analysis, but these are inaccurate and do not solve the problem of increased peptide ion complexity (9).
Although reducing the concentration of available arginine in SILAC labeling media seems to be the method of choice for preventing its conversion to proline, this approach could be problematic for particular cells types like those with rapid cellular metabolism (10). Moreover, certain sensitive cell types, such as human embryonic stem cells (hESCs) die or differentiate under restrictive metabolic conditions (7, 11). To this end, Van hoof et al. (7) sought to compensate for arginine to proline conversion in SILAC experiments by advocating the use of isotopically labeled arginine (15N4-Arg) in light SILAC media as well as (15N4-13C6-Arg) in the heavy media. With the assumption that arginine to proline conversion would be the same under both conditions, the monoisotopic peak of light and heavy proline containing peptide ions could be compared with correctly determined relative abundance, as both would be reduced proportionately. This method does provide a solution but it further complicates the procedure by generating additional converted proline peptide ions in the MS spectra and by adding the cost of expensive isotopic reagents. Also, there is no guarantee that the rate of arginine to proline conversion will be the same when comparing different cell types or treatments. Adding to these difficulties is the fact that SILAC labeling of both hESCs (7) and ESCs from the mouse (mESC) (12) requires the use of mouse embryonic fibroblast (MEF) feeder cells, which precludes the attainment of high cell numbers necessary for large scale, SILAC-based proteomic applications (8, 13–16). Faced with these challenges, we sought a simple solution, for arginine to proline conversion in SILAC experiments that would be compatible with a robust hESC culture protocol free of feeder cells.
In this study, we demonstrate that by supplementing SILAC media with standard l-proline, the conversion of isotope-coded arginine to labeled proline can be prevented. In addition, when providing sufficient amounts of arginine, no back conversion from the increased proline supplement was observed. When proline supplementation of SILAC media was adapted to both mouse and human ESC culture reagents complete labeling with isotope-coded lysine and arginine can be routinely achieved using standard procedures in the absence of feeder cells without compromising the ESC phenotype.
EXPERIMENTAL PROCEDURES
SILAC Media Formulations—
All SILAC media formulations and labeling procedures were modified from previously described protocols (5, 17). For all SILAC labeling experiments embryonic stem cell qualified Dulbecco's Modified Eagle's Medium (DMEM; 4500 mg/liter glucose, with sodium pyruvate, and no l-glutamine) was custom ordered without lysine or arginine (Specialty Media, Millipore). Before use, media was re-supplemented with 2.5 mm l-glutamine (Invitrogen), 0.798 mm isotope-coded l-lysine (13C6, 15N2), and 0.398 mm isotope-coded l-arginine (either 13C6 or 13C6, 15N4). In the case of light media, standard l-lysine and l-arginine were used. For HeLa cell culture the DMEM was supplemented with 10% (v/v) dialyzed fetal bovine serum (FBS), 1% (v/v) penicillin-streptomycin (both Invitrogen), and 0–800 mg/liter l-proline (Sigma). The DMEM for ESC cell culture was supplemented with 0.1 mm 2-mercaptoethanol, 1% non-essential amino acids (Invitrogen), and 20% or 15% Knockout Serum Replacement (KOSR; Invitrogen) reagent for hESC or mESC culture, respectively. Note that ESC media with 1% non-essential amino acids will have 11.5 mg/liter proline whereas KOSR at 20 and 15% (v/v) will contribute an additional 800 and 600 mg/liter l-proline, respectively (18). All media were filter-sterilized at 0.2 μm following formulation.
Preparation and Digestion of Cell Samples—
To monitor isotopic amino acid incorporation and conversion, trypsin digests were prepared from whole cell lysates for LC-MS analysis. Cell culture media was removed, and cells were washed with phosphate-buffered saline. Cells were then mechanically removed from the plate, lysed by dissociation in 8 m urea, and 50 mm ammonium bicarbonate and frozen at −80 °C. The protein content was determined by Bradford assay, and where indicated, light and heavy isotopic samples were mixed (1:1) based on total protein concentration prior to digestion. For trypsin digestion, samples were reduced with 10 mm dithiothreitol, alkylated with 30 mm iodoacetamide, diluted 1:4 with 50 mm NH4CO3, and treated with modified trypsin (1:25 enzyme/substrate ratio; Promega) at 37 °C overnight. The resulting tryptic peptides were extracted using a 1-ml C18 solid phase extraction cartridge (Waters), eluted with 50% (v/v) acetonitrile, 0.1% (v/v) formic acid to remove urea and salts and were re-concentrated in a vacuum centrifuge. Dried fractions were reconstituted in 10% formic acid for LC-MS/MS analysis.
Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Analysis—
Between 1.0 and 0.5 μg of each original sample was injected on a NanoAcquity UPLC (ultra performance liquid chromatography) (Waters) equipped with a 15 cm × 75 μm C18 reverse phase column employing a 90-min LC gradient (5–40% ACN, 0.1% formic acid) and detected in a data-dependent acquisition mode by tandem MS (Q-ToF Ultima; Waters). The MS was directed to use the following data-dependent acquisition parameters: survey scans range 400–1800 m/z, 1 s scans, 1–4 precursors selected based on charge state (+2, +3 and, +4 ions). MS/MS fragmentation was then performed on the ions using the charge-state collision energy profile function.
Analysis of Isotopic Amino Acid Labeling and Conversion—
Raw data files were processed using Protein Lynx Global Server 2.2.5 and summarized to peak list files (pkl). The processed data was searched against the human or mouse international protein index data base version 3.40 (appended with common protein contaminants including human keratin, bovine albumin, and trypsin) using the built in Protein Lynx Global Server search engine. The search parameters were: 100 ppm for MS and 0.1 Da for MS/MS, 2 missed cleavages on tryptic peptides, carbamidomethyl Cys as a fixed modification, and oxidized Met as a variable modification. For isotopic labeling, variable modifications of 5 or 6 Da for proline, 6 or 10 Da for arginine, and 8 Da for lysine were included in the search where appropriate. Quantification of peak areas was performed manually using MassLynx software version 4.1 on peptides indicated. MS spectra containing peptide ions of interest were summed, and background was subtracted using a polynomial order of 5 at 10% below the curve. The peak area was determined by integration using the built-in algorithm in MassLynx 4.1 to output peak area values. Peak area of the monoisotopic ions was used for comparison and determination of intensity ratios of peptide candidates.
Titration of Proline—
HeLa cells were passaged twice at a ratio of 1:8 in SILAC media containing the indicated amount of l-proline to ensure maximum incorporation of the isotopic arginine label (either 13C6 or 13C6, 15N4-Arg). SILAC media was changed every 3 days or as needed based on phenol red indicator. Whole cell lysates were digested and subject to LC-MS/MS in the initial sample analysis. Both potential and confirmed (by MS/MS) proline containing peptides resulting from the conversion of isotopic arginine were noted. All samples were then re-run in triplicate using the “include only” function to target previously detected/suspected proline containing peptides across all the samples. The candidate list of proline containing peptides was then used to follow the conversion of arginine to proline over varying proline supplement concentrations. Peptides containing labeled proline derived from isotope-coded arginine were confirmed by MS/MS in the 0 and 50 mg/liter proline samples. 13 proline-converted peptides observed in the replicate analysis and confirmed by MS/MS were used to monitor isotopic arginine to proline conversion in these experiments. These were: VAPEEHPVLLTEAPINPK, LILPGELAK, PMFIVNTNVPR, NTGIICTIGPASR, MSVQPTVSLGGFEITPPVVLR, PPYTVVYFPVRGR, YSSLVPIEK, AVFPSLVGRPR, VAPEEHPVLLTEAPINPK, SSGPYGGGGQYFAKPR, QYPKVPR, LAVNMVPFPR, and SSRAGLQFPVGR.
To investigate the possible back conversion of proline to arginine, a similar process was repeated except the arginine containing peptide candidates did not have a proline in their sequence, so that proline conversion could not interfere with arginine incorporation calculations. Based on replicate analysis, those peptides used for arginine incorporation that were consistently observed and confirmed MS/MS were: STELLIR, AGTGVDNVDLEAATR, VIGSGCNLDSAR, TIAQDYGVLKADEGISFR, GKVKVGVDGFGR, and GITINAAHVEYSTAAR.
SILAC Labeling of Human and Mouse ESCs—
For SILAC labeling of both hESC and mESC cultures measures were taken to remove all non-isotopic Arg and Lys from the system. All cells were washed with either phosphate-buffered saline or DMEM lacking Arg and Lys prior to exposure to SILAC media. Any reagents requiring dilution with DMEM were prepared in DMEM lacking Arg and Lys as well. hESC lines H1 and H9 (19) were maintained in feeder cell-free culture on 1:15 Matrigel-coated plates (BD Biosciences). hESC SILAC media was supplemented with 4 ng/ml basic fibroblast growth factor (bFGF; Invitrogen) and pre-conditioned on irradiated mouse embryonic fibroblasts (MEFs) according to previously established protocols (20). The MEF conditioned hESC SILAC media was further supplemented with 8 ng/ml bFGF prior to addition to hESC cultures and changed daily. Cells were passaged every 5–7 days through dissociation with 200 units/ml collagenase IV (Invitrogen). hESCs were cultured for two passages (>12 days) in SILAC media to achieve maximum isotopic amino acid incorporation. Analysis of hESC culture integrity and expression of pluripotent cell surface markers was performed as described previously (21). E14K mESC cultures from 129/Ola mice were also maintained in the absence of feeder cells on 0.1% (v/v) gelatin-coated plates (Sigma) as described previously (22). mESC SILAC media was supplemented with 1000 units/ml leukemia inhibitory factor (ESGROW, Millipore) prior to addition to cultures. Cells were passaged 1:3 every 2 days through dissociation in 0.25% trypsin, 2.21 mm EDTA (Wisent). mESCs were cultured for 4 passages (>8 days) in SILAC media to achieve maximum isotopic amino acid incorporation.
To monitor isotope-coded amino acid incorporation and determine the optimum times for SILAC adaptation a list of both Lys and Arg containing candidate peptides for both human and mouse ESCs was generated. Those candidate peptides that were consistently identified by MS/MS across all samples during the labeling time course were: NLPQYVSNELLEEAFSVFGQVER, IHFPLATYAPVISAEK, SLQDIIAILGMDELSEEDKLTVSR, LISWYDNEFGYSNR, GVDEVTIVNILTNR, SSGPTSLFAVTVAPPGAR, ALMLQGVDLLADAVAVTMGPK, EVAAFAQFGSDLDAATQQLLSR, LLEDGEDFNLGDALDSSNSMQTIQK, EKPYFPIPEEYTFIQNVPLEDR, VGKDELFALEQSCAQVVLQAANER, GFGFVTYATVEEVDAAMNARPHK, VVLAYEPVWAIGTGK, and YPIEHGIITNWDDMEK for hESC; and KATGPPVSELITK, ALLFVPR, GLFIIDDK, YQAVTATLEEK, ALMLQGVDLLADAVAVTMGPK, SGETEDTFIADLVVGLCTGQIK, MVNHFIAEFK, IGYPAPNFK, IAIYELLFK, ALYETELADARR, SLMDEVVKATSR, ERPPNPIEFLASYLLK, TKVHAELADVLTEAVVDSILAIR for mESCs.
RESULTS
In mammalian cells, the metabolism of arginine and proline are related, and each can serve as precursors for one another through slightly different but related pathways (Fig. 1c; reviewed in (23)). In a related pathway, both glutamate and glutamine can be precursors for arginine biosynthesis and products of arginine metabolism (Fig. 1c). However, only artifacts of proline synthesis from isotopic arginine have been reported in SILAC experiments. This observation raised the question of availability of amino acids in vitro cell culture media, particularly in SILAC formulations because previous in vivo mammalian studies have demonstrated that the inter-conversion of amino acids in this pathway (Fig. 1c) is primarily dependent on their bioavailability (24). Because l-proline is a non-essential amino acid, its concentration in the two main basal media used in standard SILAC experiments, RPMI and DMEM, is only 20 and 0 mg/L, respectively. At the same time, the typical concentration of arginine, a metabolic precursor of proline (Fig. 1b), is 200 and 84 mg/liter, respectively. Based on this we surmised that the arginine to proline conversion artifacts in SILAC experiments were a direct result of the low or absent concentration of proline in standard SILAC labeling formulations.
Proline Availability Effects on Isotopic Arginine Conversion to Proline—
In standard DMEM-based SILAC media, in the absence of proline supplement, cells readily convert isotopic arginine to proline, which is then incorporated into newly synthesized proteins. This conversion divides the MS signal of the heavy proline containing peptide in a complex fashion, resulting in false relative abundance values when compared with an equal amount of light sample (Fig. 1, a and b). To eliminate this artifact in arginine-based SILAC procedures we investigated whether increased l-proline supplementation could prevent the conversion in a concentration-dependent manner.
Equal amounts of cell lysates extracted from HeLa cells cultured in SILAC media (13C6-Arg) containing 0–800 mg/liter proline were digested with trypsin and subjected to LC-MS/MS analysis. Detected proline conversion peptides were used to build a list of peptide candidates for quantification. By using +2 or +3 charge states, the selection of the peptide candidates was based on whether they were observed in all samples, the number of proline residues, the extent of conversion observed, and whether the sequence contained arginine. Based on the triplicate analysis of all lysates, a list of thirteen peptide candidates was chosen and quantified across the 0–800 mg/liter proline concentrations.
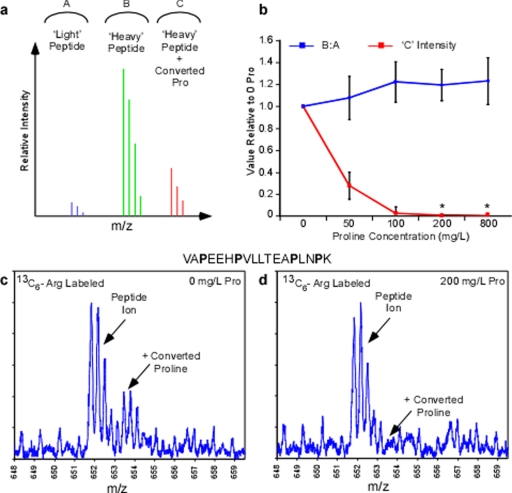
Throughout the titration of l-proline in SILAC media both signals of the converted proline containing peptides was monitored (Fig. 2, a and b). In addition, the ratio of heavy labeled Arg peptide to light peptide was measured to detect whether a decrease in the converted proline peptide signal resulted in a subsequent increase in the expected heavy peptide (Fig. 2, a and b). Within the set of peptide candidates proline conversion consumed an average of 28% of the monoisotopic signal of the heavy arginine peptide in the 0 mg/liter proline sample (standard SILAC media) (5). As the proline concentration was increased to 50, 100, and 200 mg/liter, the monoisotopic heavy proline peak occupied 9, 3, and 2%, respectively. However, within this list of proline-containing peptides no peaks corresponding to a converted proline could be detected in the MS spectrum beyond 100 mg/liter proline. As such, 2% of the total signal seen at 200 mg/liter proline was a result of unsubtracted background in the MS spectrum where there was low signal-to-noise. Moreover, maximum relative signal of heavy arginine-labeled peptide (∼20% increase versus 0 mg/liter Pro) was seen at 200 mg/liter and beyond (Fig. 2b). Consequently, even in some of the worst cases of proline conversion (Fig. 2c) no proline conversion could be observed in cells supplemented with 200 mg/liter proline or greater (d). Taken together, these data not only demonstrate that supplementation of standard SILAC media can eliminate the proline conversion, but also return the “lost signal” to the expected peak within the heavy SILAC sample. Most surprisingly, this critical improvement in arginine-based SILAC methods can be achieved by supplementing cell culture medium with as little as 200 mg/liter l-proline.
Fig. 2.
Titration of proline during SILAC labeling with isotope-coded arginine. Candidate proline containing tryptic peptides were monitored in whole cell extracts in 13C6-arginine samples with 0–800 mg/liter l-proline. a and b, conceptual mass spectra indicating the possible peptide ion peaks observed in the arginine labeling experiment. Both the ratio of labeled to unlabeled peptide as well as relative intensity of converted proline peptide ion was monitored in the experiment across 0–800 mg/liter proline (b) where * indicates no converted proline peptide ions were detectable from background. c, a mass spectrum from 0 mg/liter proline sample representing the peptide VAPEEHPVLLTEAPINPK at m/z 652 demonstrating a prevalent converted proline peak at +1.7 m/z from the expected peptide ion. d, mass spectrum from the 200 mg/liter proline sample depicting the same peptide ion as shown in c and the converted proline ion is notably absent.
Proline Availability Effects on Isotopic Arginine Labeling—
One concern with supplementing an arginine-based SILAC experiment with proline is that, by a similar biosynthetic mechanism (Fig. 1c), the excess proline might be a precursor for arginine synthesis thereby decreasing the efficiency of labeling by the isotope-coded arginine provided. Although proline is known to be one of the main substrates in the formation of citrulline, a direct precursor for arginine biosynthesis, it has been demonstrated that this process is only favorable when there is a lack of available arginine (24, 25). Moreover, to date, the presence of other citrulline precursors, such as glutamine in standard SILAC media, has had no reported detrimental effect on SILAC procedures.
To prevent back conversion of proline into arginine and to increase the compatibility of the medium with nutritionally sensitive cell types like ESCs (7), we re-supplemented the DMEM-based SILAC media with its standard complement of isotope-coded arginine (0.398 mm). This was in direct contrast to the reported reduction of the concentration of isotope-coded arginine to prevent its availability for conversion to proline (5, 8). At the end point in the same proline titration experiments we monitored the heavy arginine labeling efficiency by tracking the ratio of heavy to light peptides, which contained arginine (Fig. 3a). To minimize proline conversion artifacts that would interfere with these calculations, we avoided peptides with proline in their sequences.
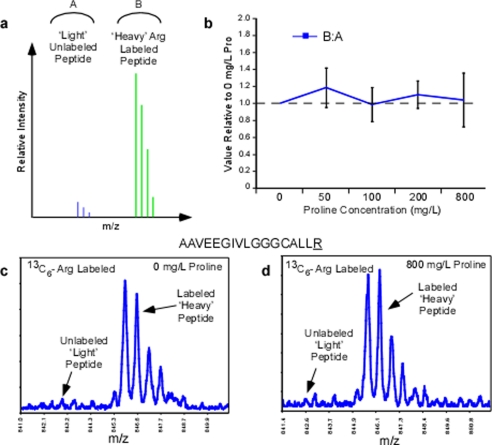
Fig. 3.
Isotope-coded arginine labeling efficiency across the titration of proline. Candidate arginine containing tryptic peptides lacking proline was monitored in whole cell extracts in 13C6-arginine samples with 0–800 mg/liter l-proline. a, conceptual mass spectra indicating the possible peptide ion peaks observed in the arginine labeling experiment. The peak area ratio between the heavy and light arginine monoisotopic peaks was monitored across the 0–800 mg/liter proline samples and plotted relative to 0 mg/liter proline (b). c, a mass spectrum representing the peptide AAVEEGIVLGGGCALLR at m/z 845 from 0 mg/liter proline sample. d, mass spectra of the same peptide ion in the 800 mg/liter proline sample demonstrating no compromise in the isotope-coded arginine labeling efficiency or proline to arginine back conversion. Isotopic amino acids are underlined.
In comparison to 0 mg/liter proline (standard SILAC media), the average relative peak area for the 50–800 mg/liter proline samples exhibited only slight deviations but overall remained unchanged (Fig. 3b). Larger deviations were only observed for peptides with lower signal-to-noise ratios and did not have a noticeable trend as a function of proline concentration. None of the arginine peptides examined, in the 0 mg/liter proline sample (Fig. 3c) exhibited an increase in unlabeled light peptide signal, even in samples that were supplemented with 800 mg/liter of proline (Fig. 3d). These data provide evidence that our SILAC media formulation (standard arginine in combination with proline supplementation) is effective in minimizing arginine to proline conversion artifacts without compromising the efficiency of isotope-coded arginine labeling. Thus, in the cell types examined here, an arginine concentration of 0.398 mm, even in the presence of high exogenous proline (up to 800 mg/liter), is sufficient to prevent observable proline to arginine back conversion that would interfere with SILAC labeling efficiency.
Adaptation of SILAC Labeling to Feeder Cell-free hESC Culture—
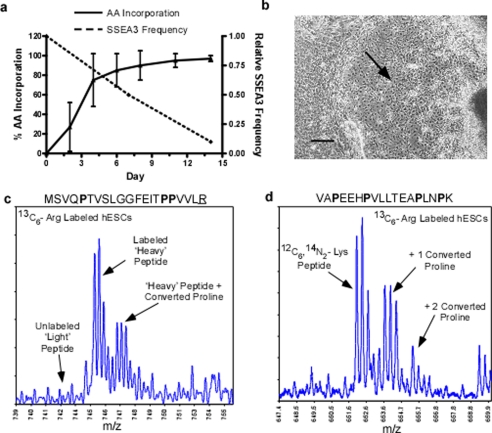
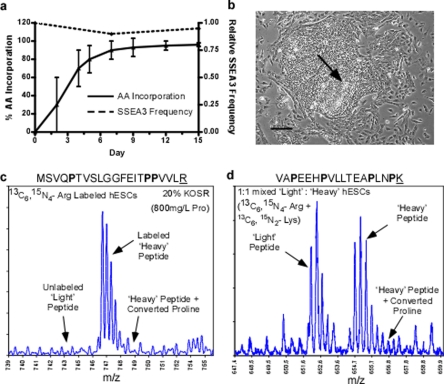
To perform a wide variety of large scale SILAC-based proteomic experiments with hESCs the ability to generate large numbers of labeled cells has to be achieved. It became clear that SILAC methods compatible with feeder cell-free culture systems would be necessary to attain these high cell numbers (reviewed in (11)). In an attempt to extend standard SILAC labeling procedures to feeder cell-free hESC culture we confirmed incompatibilities previously highlighted using a feeder-cell-based culture system (7). Using standard SILAC media with 13C6-arginine and supplemented with 36 ng/ml bFGF (26), hESC cultures achieved maximum levels of isotope-coded arginine incorporation after only 2 passages or after more than 12 days under SILAC culture conditions (Fig. 4a). By day 15 in SILAC medium, hESCs had reached 96.5% average efficiency in isotope-coded amino acid labeling. However, in the same period of time the hESCs rapidly differentiated as demonstrated by the loss of SSEA3 expression (Fig. 4a), a marker of pluripotent stem cells. We also observed the loss of defined hESC undifferentiated colony structure and tight cell morphology indicating the presence of differentiated cell types (Fig. 4b). Moreover, manual inspection of the MS data showed that the conversion of arginine to proline was exacerbated in these cells. Although it was clear that arginine incorporation was complete in day 14 samples the proline-converted peptide ions were prevalent (Fig. 4c). In some extreme cases, converted proline peptide ions consumed greater than 50% of the expected peptide ion signal (Fig. 4d). The instability of the hESC phenotype combined with the pronounced arginine to proline conversion artifacts indicated that the standard SILAC media formulation was not a suitable protocol to adapt to feeder cell-free hESC culture.
Fig. 4.
Labeling feeder cell-free hESCs in standard SILAC medium. hESCs culture on Matrigel was grown in standard SILAC medium (13C6-Arg, dialyzed FBS, 0 mg/liter proline) for 2 weeks. Arginine incorporation, conversion to proline, and hESC phenotype were monitored. a, the relative SSEA3 frequency and percent heavy arginine incorporation over the SILAC labeling time course. b, differentiated phenotype of hESC after 14 days in standard SILAC medium as observed by light microscopy. Scale bars equal 250 μm. c, a mass spectrum of heavy arginine peptide ion MSVQPTVSLGGFEITPPVLR at 745 m/z. Indicated are the prominent converted proline peptide ion shifted +1.7 m/z and the absence of the unlabeled light arginine peptide ion corresponding to the high level of isotopic amino acid incorporation. d, a mass spectrum of the peptide ion VAPEEHPVLLTEAPLNPK at 652 m/z. Indicated are the peptide ions shifted +1.7 m/z and +3.4 m/z corresponding to the incorporation of 1 and 2 converted heavy proline and consuming more than half of the expected peptide ion intensity. Isotopic amino acids are underlined.
There are two main factors responsible for the incompatibility standard SILAC medium with hESC culture, the first being the dialyzed FBS and the second, the limited growth factor microenvironment (problems reviewed in (11)). To address these issues we revisited the original development of feeder cell-free hESC culture protocols (20). Xu and co-workers formulated a DMEM-based hESC medium using a serum replacement reagent (18) that is now commercially available as knockout serum replacement (KOSR, Invitrogen). The hESC medium was then pre-conditioned on MEF cell feeder layers to supply the appropriate complement of signaling molecules and growth factors to maintain hESC homeostasis. This method has since become the most widely adopted form of feeder cell-free hESC culture. Coincidentally, the KOSR formulation contains no free lysine or arginine, making it compatible with most popular SILAC applications. Moreover, as we demonstrate to be important here, KOSR contributes 800 mg/liter proline when used at 20% (v/v) in hESC medium (18).
An hESC SILAC medium with 20% KOSR and 13C6, 15N4-Arg and 13C6, 15N2-Lys were formulated taking into account the SILAC compatibility of the media (see under “Experimental Procedures”). The hESC SILAC medium was then pre-conditioned on irradiated MEFs according to previously established protocols (20). The resulting medium, when supplemented with 8 ng/ml bFGF, allowed hESCs to incorporate the isotope-coded amino acids at the same rate as observed with standard SILAC medium where after 2 passages and more than 12 days in SILAC culture hESC had reached their maximum levels of incorporation (Fig. 5a). Consistent with the preliminary proline titration data, there was no difficulty encountered in isotopic amino acid labeling because the cells were on average 96.1% labeled by day 14. Moreover, in the new medium formulation the cells were now able to maintain SSEA3 expression (Fig. 5a) and continue to form tight, undifferentiated colony structures (B). Inspection of the MS spectra (Fig. 5c) of the hESC lysates labeled using the new formulation demonstrated the same level of isotopic amino acid incorporation as standard SILAC medium when examining the same peptides (Fig. 4c). Additionally, for the same peptide there was no longer any arginine to proline conversion artifacts observed (Fig. 5c), even when examining peptides with the highest proportion of converted proline peptide ions (Fig. 5d versus Fig. 4d). Consequently, when a 14-day SILAC-labeled (13C6, 15N4-Arg and 13C6, 15N2-Lys) hESC extract was mixed (1:1) with a light hESC lysate, equivalent levels of the expected light and heavy peptides were observed even in sequences most susceptible to proline conversion artifacts (Fig. 5d). These observations clearly demonstrate that combining KOSR as an alternative serum supplement with both a SILAC compatible hESC media formulation and pre-conditioning on feeder cells offers a reliable solution for large scale hESC applications in quantitative proteomics without compromising data quality.
Fig. 5.
Labeling feeder cell-free hESCs in conditioned SILAC medium with KOSR. hESCs cultured on Matrigel were grown in MEF-conditioned ESC optimized SILAC media (13C6 15N4-Arg and 13C6, 15N2-Lys, 20% KOSR providing 800 mg/liter l-proline) for 2 weeks. Amino acid incorporation, conversion of arginine to proline, and hESC phenotype were monitored. a, the relative SSEA3 frequency and percent heavy amino acid incorporation over the SILAC labeling time course. b, undifferentiated phenotype maintained by hESCs after more than 15 days in optimized SILAC media as observed by light microscopy. Scale bars equal 250 μm. c, a mass spectrum of heavy arginine peptide ion MSVQPTVSLGGFEITPPVLR at ∼747 m/z. Indicated are the absence of both a converted proline peptide ion and the unlabeled light arginine peptide ion. The absent light peptide ion at 743 m/z also indicates no back conversion of proline to unlabeled arginine. d, a mass spectrum of the 1:1 mixed labeled and unlabeled hESC lysates showing light and heavy peptide ion VAPEEHPVLLTEAPLNPK at ∼652 m/z and 655 m/z. The prominent converted proline peptide ions are now almost undetectable and do not effect the observed 1:1 ratio. Isotopic amino acids are underlined.
SILAC Procedures with Feeder Cell-free mESC Cultures—
Recently, a landmark analysis of mESCs using a SILAC approach was reported by Graumann et al. (12) in which a total of 5111 proteins were identified. However, this study was also routed in a feeder-cell-based culture system, which by the authors’ own admission contributed to contamination in the analysis. To circumvent this issue these authors performed a cumbersome transition from feeder cell to feeder cell-free culture during the labeling process. As these authors noted, the use of dialyzed FBS, prescribed in standard SILAC medium (17) made feeder cell independent culture of mESCs difficult. Moreover, having to culture mESCs on feeder layers and/or transition those to feeder-cell-independent conditions could negatively impact both the scalability of this procedure and the integrity of the mESC phenotype. Because KOSR was originally designed and used as an alternative to FBS for the culture of mESCs (18) we hypothesize that the formulation that we successfully applied to hESCs could also be used in creating a protocol to perform SILAC on completely feeder-cell-independent mESC cultures.
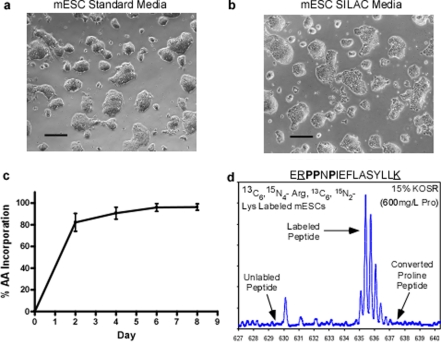
Therefore a mESC SILAC medium containing 15% KOSR was formulated (18) and the media supplemented with 1000 units/ml of leukemia inhibitory factor prior to addition to feeder cell-free mESC cultures. After 4 passages in our mESC SILAC medium the pluripotent culture morphology was indiscernible from those mESCs cultured under standard conditions (Fig. 6, a and b). After the same period of time mESCs also achieved maximum incorporation of isotopic amino acids (13C6, 15N4-Arg and 13C6, 15N2-Lys) (Fig. 6c) with an average labeling of 96.3%. Moreover, visual inspection of SILAC-labeled mESC MS spectra demonstrates similar results as observed with hESCs samples where there is no compromise with levels of isotopic arginine incorporation and no detectable incidence of proline conversion artifacts even in peptide ions with multiple proline residues in the sequence (Fig. 6d). In accordance with the observations in the hESC system, the application of a KOSR-based SILAC medium to a feeder cell-free mouse ESC culture, now provides a robust protocol for routine, large scale quantitative proteomic experiments using SILAC in mESCs as well.
Fig. 6.
Labeling feeder cell-free mESCs in conditioned SILAC media with KOSR. mESCs cultured on gelatin were grown in ESC-optimized SILAC media (13C6, 15N4-Arg and 13C6, 15N2-Lys, 15% KOSR providing 600 mg/liter l-proline) for 4 passages (8 days). Amino acid incorporation, conversion of arginine to proline, and mESC phenotype were monitored. The undifferentiated phenotype of mESCs grown under standard feeder cell-free conditions (a) compared with those grown in optimized SILAC medium for 8 days (b) as observed by light microscopy. Scale bars equal 250 μm. c, the average heavy Lys and Arg incorporation over the SILAC labeling time course. d, a mass spectrum of peptide ERPPNPIEFLASYLLK at 635.6 m/z from mESCs in ESC-optimized SILAC media. The heavy Lys and Arg peptide ions are prominent whereas the unlabeled light peptide or converted heavy proline peptide ions are absent. The absent light peptide ion at m/z 633 also indicates no back conversion of proline to unlabeled arginine. Isotopic amino acids are underlined.
DISCUSSION
General SILAC procedures take a reductionist approach in controlling amino acid content in cell culture to achieve the desired labeling outcome. These include the use of dialyzed FBS, reduced concentration or total removal of many amino acids (i.e. Arg and Pro), and overall culture conditions optimized using immortalized tissue culture. We believed these methodological aspects precluded the extension of SILAC procedures to ESCs and were at the heart of amino acid conversion artifacts seen using the general labeling procedure. In this study, we demonstrate that through supplementation of isotope-coded arginine-based SILAC experiments with as little as 200 mg/liter l-proline, isotopic arginine to proline conversion artifacts can be virtually eliminated in a variety of cell types. In addition, using KOSR, which provides 800 mg/liter l-proline at 20% as a serum supplement in lysine and arginine-based SILAC experiments instead of dialyzed FBS creates a robust media formulation for completely feeder cell-free SILAC experiments using ESCs from both the mouse and the human. We also tested the compatibility of KOSR-based SILAC media on a number of traditional tissue culture cell types including HeLa and MCF7 cells with no observable differences compared with standard SILAC media, except the absence of arginine to proline conversion (data not shown). As such, because of its consistent, reagent-like nature KOSR might act as a more nutrient rich replacement for dialyzed FBS in SILAC experiments in general, especially in cell types, which are sensitive to dialyzed FBS.
Based on the cellular pathways of proline and arginine metabolism (Fig. 1c), the addition of proline is an obvious solution to the conversion issue; however the concern has always been the potential back conversion to arginine, which would interfere with isotope-coded arginine labeling (8). Some studies, such as the recent one using SILAC in mESCs stress that all reagents are prepared without proline for this very reason (12). To our knowledge, no study to date has demonstrated that the presence of proline actually induces significant back conversion to arginine in a concentration-dependent manner during a SILAC procedure. In fact, we demonstrate here that between 0 and 800 mg/liter proline there is no significant change detectable in the labeling of cells with isotope-coded arginine (Fig. 3).
Proline is a preferred precursor for arginine biosynthesis in neonates in vivo just as arginine is for proline. However it has been clearly demonstrated in a number of studies that the de novo biosynthesis of either arginine or proline using the other as a precursor is most strongly influenced by their bioavailability (24, 25, 27). In other words, if there is sufficient free proline present to maintain cellular homeostasis the endogenous production of proline will not be favored, regardless of the concentration of its available precursors. The same statement can be made in the opposite direction for arginine. With respect to energy and metabolism this observation makes sense as it would be the most efficient. Even though a diminished arginine concentration may be sufficient in certain cell types, it has been reported that this is not completely efficient at preventing proline conversion artifacts (3, 8). Our study suggests that this was due to the absence of proline in the standard SILAC media formulation. Although the inter-conversion of proline and arginine may still exist in cells labeled with isotope-coded arginine using our media formulation, we were unable to detect it with any level of significance. More importantly, our strategy of supplying additional or high concentrations of amino acids appears to be more effective at eliminating the problem and makes our strategy the most applicable to biologically important in vitro cell types like embryonic stem cells.
Acknowledgments
We thank James Duncan and David Litchfield for facilitating HeLa cell labeling, Monica Graham for MEF cell culture and conditioning, and Zheng “John” Ye for calculations of proteomic distribution of proline.
Footnotes
Published, MCP Papers in Press, DOI 10.1074/mcp.M800113-MCP200
The abbreviations used are: MS, mass spectrometry; bFGF, basic fibroblast growth factor; DMEM, Dulbecco's Modified Eagle's Medium; hESC, human embryonic stem cell; FBS, fetal bovine serum; hESC, human embryonic stem cell; KOSR, knockout serum replacement; LC, liquid chromatography; MEF, mouse embryonic fibroblast; mESC, mouse embryonic stem cell; SILAC, stable isotope labeling with amino acids in culture; LC-MS, liquid chromatography mass spectrometry; LC-MS/MS, liquid chromatography tandem mass spectrometry.
This work was supported by grants from the Ontario Research and Development Challenge Fund (ORDCF) (to G. L.), CIHR and Stem Cell Network (to M. B.), and by Canada Research Chair positions (to B. D. and M. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2.Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 3.Hwang, S. I., Lundgren, D. H., Mayya, V., Rezaul, K., Cowan, A. E., Eng, J. K., and Han, D. K. ( 2006) Systematic characterization of nuclear proteome during apoptosis: a quantitative proteomic study by differential extraction and stable isotope labeling. Mol. Cell Proteomics 5, 1131–1145 [DOI] [PubMed] [Google Scholar]
- 4.Ong, S. E., Kratchmarova, I., and Mann, M. ( 2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 5.Ong, S. E., and Mann, M. ( 2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 6.Schmidt, F., Strozynski, M., Salus, S. S., Nilsen, H., and Thiede, B. ( 2007) Rapid determination of amino acid incorporation by stable isotope labeling with amino acids in cell culture (SILAC). Rapid Commun. Mass Spectrom. 21, 3919–3926 [DOI] [PubMed] [Google Scholar]
- 7.Van Hoof, D., Pinkse, M. W., Oostwaard, D. W., Mummery, C. L., Heck, A. J., and Krijgsveld, J. ( 2007) An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat. Methods 4, 677–678 [DOI] [PubMed] [Google Scholar]
- 8.Blagoev, B., and Mann, M. ( 2006) Quantitative proteomics to study mitogen-activated protein kinases. Methods 40, 243–250 [DOI] [PubMed] [Google Scholar]
- 9.Gruhler, A., Olsen, J. V., Mohammed, S., Mortensen, P., Faergeman, N. J., Mann, M., and Jensen, O. N. ( 2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 10.Scott, L., Lamb, J., Smith, S., and Wheatley, D. N. ( 2000) Single amino acid (arginine) deprivation: rapid and selective death of cultured transformed and malignant cells. Br. J. Cancer 83, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman, L. M., and Carpenter, M. K. ( 2005) Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 23, 699–708 [DOI] [PubMed] [Google Scholar]
- 12.Graumann, J., Hubner, N. C., Kim, J. B., Ko, K., Moser, M., Kumar, C., Cox, J., Schoeler, H., and Mann, M. ( 2007) SILAC-labeling and proteome quantitation of mouse embryonic stem cells to a depth of 5111 proteins. Mol. Cell Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 13.Andersen, J. S., Lam, Y. W., Leung, A. K., Ong, S. E., Lyon, C. E., Lamond, A. I., and Mann, M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed]
- 14.Ong, S. E., Mittler, G., and Mann, M. ( 2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 [DOI] [PubMed] [Google Scholar]
- 15.Selbach, M., and Mann, M. ( 2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 16.Yocum, A. K., Gratsch, T. E., Leff, N., Strahler, J. R., Hunter, C. L., Walker, A. K., Michailidis, G., Omenn, G. S., O'Shea, K. S., and Andrews, P. C. ( 2008) Coupled global and targeted proteomics of human embryonic stem cells during induced differentiation. Mol. Cell Proteomics 7, 750–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 18.Price, P. J., Goldsborough, M. D., and Tilkin, M. L. ( 1998) Embryonic stem cell serum replacement. International patent Application WO98/30679 p. 28
- 19.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M. ( 1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 20.Xu, C., Inokuma, M. S., Denham, J., Golds, K., Kundu, P., Gold, J. D., and Carpenter, M. K. ( 2001) Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19, 971–974 [DOI] [PubMed] [Google Scholar]
- 21.Bendall, S. C., Stewart, M. H., Menendez, P., George, D., Vijayaragavan, K., Werbowetski-Ogilvie, T., Ramos-Mejia, V., Rouleau, A., Yang, J., Bosse, M., Lajoie, G., and Bhatia, M. ( 2007) IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature 448, 1015–1021 [DOI] [PubMed] [Google Scholar]
- 22.Doble, B. W., Patel, S., Wood, G. A., Kockeritz, L. K., and Woodgett, J. R. ( 2007) Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 12, 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris, S. M., Jr. ( 2004) Enzymes of arginine metabolism. J. Nutr. 134, 2743S–2747S; discussion 2765S–2767S [DOI] [PubMed] [Google Scholar]
- 24.Urschel, K. L., Rafii, M., Pencharz, P. B., and Ball, R. O. ( 2007) A multitracer stable isotope quantification of the effects of arginine intake on whole body arginine metabolism in neonatal piglets. Am. J. Physiol. Endocrinol. Metab. 293, E811–E818 [DOI] [PubMed] [Google Scholar]
- 25.Urschel, K. L., Evans, A. R., Wilkinson, C. W., Pencharz, P. B., and Ball, R. O. ( 2007) Parenterally fed neonatal piglets have a low rate of endogenous arginine synthesis from circulating proline. J. Nutr. 137, 601–606 [DOI] [PubMed] [Google Scholar]
- 26.Wang, L., Li, L., Menendez, P., Cerdan, C., and Bhatia, M. ( 2005) Human embryonic stem cells maintained in the absence of mouse embryonic fibroblasts or conditioned media are capable of hematopoietic development. Blood 105, 4598–4603 [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson, D. L., Bertolo, R. F., Brunton, J. A., Shoveller, A. K., Pencharz, P. B., and Ball, R. O. ( 2004) Arginine synthesis is regulated by dietary arginine intake in the enterally fed neonatal piglet. Am. J. Physiol. Endocrinol. Metab. 287, E454–E462 [DOI] [PubMed] [Google Scholar]