Abstract
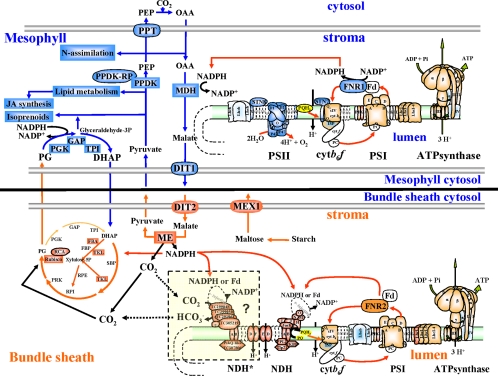
Chloroplasts of maize leaves differentiate into specific bundle sheath (BS) and mesophyll (M) types to accommodate C4 photosynthesis. Chloroplasts contain thylakoid and envelope membranes that contain the photosynthetic machineries and transporters but also proteins involved in e.g. protein homeostasis. These chloroplast membranes must be specialized within each cell type to accommodate C4 photosynthesis and regulate metabolic fluxes and activities. This quantitative study determined the differentiated state of BS and M chloroplast thylakoid and envelope membrane proteomes and their oligomeric states using innovative gel-based and mass spectrometry-based protein quantifications. This included native gels, iTRAQ, and label-free quantification using an LTQ-Orbitrap. Subunits of Photosystems I and II, the cytochrome b6f, and ATP synthase complexes showed average BS/M accumulation ratios of 1.6, 0.45, 1.0, and 1.33, respectively, whereas ratios for the light-harvesting complex I and II families were 1.72 and 0.68, respectively. A 1000-kDa BS-specific NAD(P)H dehydrogenase complex with associated proteins of unknown function containing more than 15 proteins was observed; we speculate that this novel complex possibly functions in inorganic carbon concentration when carboxylation rates by ribulose-bisphosphate carboxylase/oxygenase are lower than decarboxylation rates by malic enzyme. Differential accumulation of thylakoid proteases (Egy and DegP), state transition kinases (STN7,8), and Photosystem I and II assembly factors was observed, suggesting that cell-specific photosynthetic electron transport depends on post-translational regulatory mechanisms. BS/M ratios for inner envelope transporters phosphoenolpyruvate/Pi translocator, Dit1, Dit2, and Mex1 were determined and reflect metabolic fluxes in carbon metabolism. A wide variety of hundreds of other proteins showed differential BS/M accumulation. Mass spectral information and functional annotations are available through the Plant Proteome Database. These data are integrated with previous data, resulting in a model for C4 photosynthesis, thereby providing new rationales for metabolic engineering of C4 pathways and targeted analysis of genetic networks that coordinate C4 differentiation.
In leaves of C4 grasses such as maize (Zea mays), photosynthetic activities are partitioned between two morphologically and biochemically distinct bundle sheath (BS)1 and mesophyll (M) cells. A single ring of BS cells surrounds the vascular bundle followed by a concentric ring of specialized M cells, creating the classical Kranz anatomy. C4 differentiation occurs along a developmental gradient with proplastids at the leaf base and fully differentiated C4 M and BS chloroplasts at the leaf tip. Genetic screens for mutants affected in BS differentiation identified various mutants (1–4). However, the molecular basis for C4 differentiation is still poorly understood but includes transcriptional regulation through DNA regulatory elements, transcription factors, and likely also metabolic signals (5, 6).
Differential protein accumulation and activities between M and the BS chloroplasts of maize and sorghum (both NADP-malic enzyme (ME) type C4) have been studied using various low throughput techniques. This has shown that BS and M cells each accumulate a distinct set of C4 photosynthetic enzymes and has established the general pathway for C4 photosynthesis (5–7). A quantitative analysis of purified M and BS chloroplast stromal proteomes determined BS/M accumulation ratios for 125 proteins, covering a wide range of plastid functions and allowing integration of information from past studies (8).
M thylakoids have a complete linear electron transport chain, containing the Photosystem II (PSII), cytochrome b6f (cytb6f), and PSI complexes, similar to C3 plants. In contrast, fully differentiated BS thylakoids have little functional PSII and normal or increased PSI levels, whereas cytb6f and ATP synthase complexes are quite evenly distributed between BS and M thylakoids. As a consequence, BS thylakoids mostly carry out cyclic electron flow and have low rates of linear electron flow (9–11). After several conflicting reports on protein levels of PSII and light-harvesting complex (LHC) II (11–13), a subsequent study partially clarified both activity and protein accumulation levels of PSI and PSII complexes in BS and M thylakoids (14). However, a more comprehensive overview of accumulation levels and assembly state of the thylakoid-bound photosynthetic apparatus remains to be determined. Thylakoids also contain NAD(P)H dehydrogenase (NDH) complexes of which several chloroplast-encoded and nucleus-encoded subunits were identified in maize (15) with a clear enrichment of NDH complexes in the BS membranes (16–18). However, a better understanding of the composition and assembly state of the NDH complex, as well as interactions with other electron transport components, will be important to understand its contribution to cyclic electron flow.
Proteome analysis of thylakoid membranes (19–22) and associated lipoprotein particles (plastoglobules) (23, 24) as well as the plastid chromosome (the nucleoid) (25) from Arabidopsis chloroplasts also identified many proteins with non-photosynthetic functions (26). In addition forward and reverse genetics studies in Arabidopsis (27, 28) as well as maize (29) identified several nucleus-encoded thylakoid proteins affecting biogenesis and chloroplast functions (30). Finally a number of stromal metabolic enzymes may functionally interact with the thylakoid, e.g. to transfer reducing equivalents to metabolic pathways, such as GcpE/IspG in the methylerythritol phosphate pathways (31). Thus, it is highly likely that numerous thylakoid (-associated) proteins with non-photosynthetic functions accumulate to different levels in the two cell types and may contribute to BS/M chloroplast differentiation; their quantification will help to better understand cell specific differentiation.
Chloroplasts are surrounded by a double envelope membrane containing the protein import apparatus (32, 33) and transporters for exchange of ions and metabolites and also participate in metabolic pathways and plastid-nucleus signaling (34–37). Hundreds of proteins have been experimentally identified in the inner and/or outer envelope proteome of Arabidopsis (38–40). However, no systematic comparison of envelope proteomes of differentiated BS and M maize chloroplasts has been carried out, but major differences can be expected in e.g. translocators of carbohydrates (34, 41).
Gel-based and, in particular, MS-based techniques to determine quantitative differences between proteomes have greatly improved in recent years (42, 43). Some of these techniques (two-dimensional (2D) IEF gels, ICAT, and label-free MS-based quantification using MS ion chromatograms) were applied in our previous comparative analysis of differentiated BS and M stromal proteomes (8). However, quantitative comparison of M and BS chloroplast membrane proteomes represents a significant challenge because of their hydrophobicity (19, 21, 44, 45). Moreover the presence of about 100 abundant photosynthetic proteins in thylakoids representing some 98% of the protein mass provides an additional challenge to quantify the low abundance proteins. In addition, although most maize genes are probably represented in the collection of maize EST and unigenes, no complete and assembled maize genome is yet available, thus making protein identification and quantification more difficult.
The prime objectives of this study were to determine maize BS and M cell-specific differences in (i) assembly state and composition of the four photosynthetic complexes and the NDH complex, (ii) auxiliary functions of the thylakoid proteome, and (iii) metabolic and transport functions of M and BS thylakoids and envelopes. Therefore, we carried out a quantitative proteome analysis of differentiated BS and M membranes using techniques compatible with membrane proteomes and also taking advantage of a new, fast, and highly accurate mass spectrometer, the LTQ-Orbitrap (46–48). This clarified the organization of photosynthetic machineries of BS and M thylakoids, revealed large NDH complexes and associated proteins with unknown function, and determined functional differentiation of transporters and biosynthetic pathways. These results complement our recent comparative analysis of soluble stromal BS and M chloroplast proteomes (8) and will provide new entry points for future studies to unravel cell-specific chloroplast differentiation in maize. Proteomics data and functional annotation are available via the Plant Proteome Database.
EXPERIMENTAL PROCEDURES
Maize Genotype, Plant Growth, and Purification of BS and M Chloroplast Membranes—
WT-T43 maize plants were grown for 12–14 days in a growth chamber (16-h light/8-h dark, 400 μmol of photons·m−2·s−1) until the fourth leaf was emerging. M and BS chloroplasts were purified from the top 4-cm section of the third leaf, harvested about 2 h after the onset of the light period, using several hundreds of leaf tips following procedures described previously (8). Purified M and BS chloroplasts were broken with a Dounce homogenizer, and thylakoid and envelope membranes were collected by 20-min centrifugation at 80,000 × g. The cross-contamination of M and BS chloroplast fractions was assessed from the presence of the M and BS markers (pyruvate, phosphate dikinase (PPDK) and Rubisco, respectively) visualized on stained 1-D SDS-PAGE gels as described previously (8). Cross-contamination between BS and M chloroplast membranes was between 5 and 15% (allowing minimal and maximal BS/M ratio from 0.01–0.15 to 8.5–9.5), whereas contamination with non-chloroplast proteins was estimated to be below 0.1% in terms of protein mass. On average, preparations with less than 15% cross-contamination yielded about 700 μg of protein equivalent to about 150 μg of chlorophyll for each M and BS fraction. Protein concentrations were determined with the bicinchoninic acid assay (49).
2D Blue Native Gel Analysis and MS-based Protein Identification—
After purification of BS and M membranes, identical amounts of BS and M proteins were solubilized in β-n-dodecyl maltoside (β-DM) (Sigma) and separated on blue native gels as described previously (50). The gel lanes were then cut out, and proteins were denatured, reduced, and alkylated (8) and separated by Tricine-SDS-PAGE (51) as described previously. The resulting focused 2D gels were stained with Coomassie Brilliant Blue R-250 (USB Corp.) and scanned (Epson Perfection 4490). Seven independent M and BS chloroplast preparations were resolved on seven pairs of 2D BN-PAGE gels. Image analysis was performed using Phoretix software (Nonlinear Dynamics Ltd.). About 180 spots/gel were detected with spot volumes spanning 4 orders of magnitude. Spot matching was manually verified with a large time investment in spot matching and verification. Ambiguous matches were resolved by protein identification from MS. After background removal and normalization of each spot “volume” to the total gel spot volume, virtual average M and BS gels were created where each spot volume represented the average of volumes of matched spots in M or in BS gels. We included on the average gels only those matched spots that were present at least three times on M or BS gels. BS/M accumulation ratios were obtained for 85 spots. Spot quantification data are shown in supplemental Table 1.
Stained gel spots were excised manually or using a ProPic robot (Genomic Solutions, Ann Arbor, MI). The spots were washed, digested with modified trypsin (Promega), and extracted using a ProGest robot (Genomic Solutions). The extracted peptides were dried and resuspended in 15 μl of 5% formic acid (FA). Protein identification was performed by peptide mass fingerprinting using MALDI-TOF MS in reflectron mode (Voyager DE-STR from Perseptive Biosystems) and on-line LC-ESI-MS/MS (Q-TOF, Micromass). All protein identification data are shown in supplemental Table 1.
The mass spectra were obtained automatically by MALDI-TOF MS in reflectron mode followed by automatic internal calibration using tryptic peptides from autodigestion. Peak lists (.mgf files) from the MALDI data were generated using MoverZ software m/z (freeware edition, Proteometrics, Inc.) using a minimum signal to noise ratio of 3.0 and peak resolution of 5000. A large portion of the spectral annotations (in particular assignments of monoisotopic masses) were manually verified. The resulting peptide mass lists were used to search the Maize EST assembly from TIGR (ZmGI v16.0, 56,364 entries) by Mascot (v2.2) in automated mode using the following search parameter criteria: significant protein Mowse score p < 0.05, no missed cleavages allowed, variable methionine oxidation, fixed carbamidomethylation of cysteines, and minimum mass accuracy of 100 ppm. These search result pages were extracted and further filtered to pass the following criteria for positive identification: (i) at least five or more matching peptides with mass error distribution clustered within 25 ppm and (ii) at least 15% sequence coverage. Only peptides without missed cleavages (by trypsin) were considered with methionine oxidation as a variable modification and carbamidomethylation as a fixed modification. In exceptional cases (i.e. proteins less than 20 kDa and matching gel coordinates) four matching peptides were considered as positive identification. The criteria for positive protein identification used here result in less than 1% false positives as determined by searching the data from these gel spots using the target-decoy database consisting of the ZmGI v16.0 database (56,364 entries) concatenated with a decoy where all the sequences were shuffled. FPR was calculated as follows: 2 × decoy_hits/total_hits. PDF files of annotated spectra with assigned peptide masses are provided in the supplemental data. Data for protein identification by MALDI-TOF peptide mass fingerprinting are listed in supplemental Table 1, supplemental Fig. 1, and associated additional files showing spectral annotations.
The Q-TOF instrument was operated in positive ion mode with a sample cone voltage of 35 kV, capillary voltage of 3.3 kV, and source temperature of 90 °C. The samples were run in data-dependent mode where each full MS scan was followed by three consecutive MS/MS scans. The MS survey scans (m/z 350–1550) had a scan time of 1 s and an interscan time of 0.08 s. MS/MS spectra were automatically acquired when the peak intensity rose above a threshold of 10 counts·s−1. Normalized collision energies for peptide fragmentation were set using the charge state recognition files for 1+, 2+, 3+, and 4+ peptide ions provided by MassLynx (Waters). For tandem MS acquisition we used a scan range from m/z 50 to 1550 with a scan time of 1.92 s, an interscan time of 0.08 s, and a dynamic exclusion window of 240 s. Argon was used as the collision gas. Peak lists (.mgf files) from the Q-TOF data were generated using Mascot Distiller (v2.0).
The peak lists from Q-TOF were searched against the Maize EST assembly from TIGR (ZmGI v16.0, 56,364 entries) by Mascot (v2.2) with p < 0.05, with a maximum mass error of 0.8 and 2 Da for product and precursor ions, respectively, and with a minimum ion score of 20. Mascot results were subsequently filtered to increase the stringency for positive identification by MS/MS as follows. If only one or two matching peptides were found, clear partial Y-ion series and partial complementary B-ion series needed to be present (as determined by manual inspection) with a minimum peptide ion score of 31 for one-peptide identifications and minimum peptide ion scores of 22 for two-peptide identifications. If three or more sequence tags were found, manual inspection was not a requirement, but the protein Mowse score must be over 70 with only peptides with ion scores of 20 or more contributing. These criteria resulted in a protein false positive rate below 1% as determined from target-decoy database searches using the ZmGI v16.0 database (56,364 entries) concatenated with a decoy where all the sequences were shuffled. Protein FPR was calculated as follows: 2 × decoy_hits/total_hits. Protein identification data are displayed in supplemental Table 1, and additional Mascot protein report pages for single peptide identifications are provided as supplemental files.
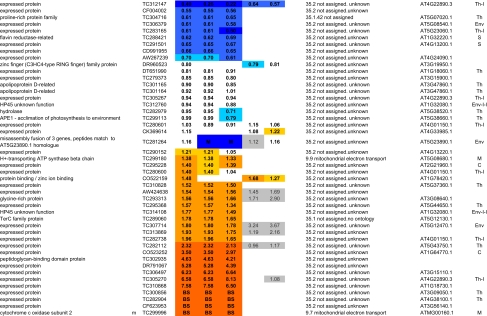
Seven independent biological replicates (pairs of BS and M membranes) were used for this 2D BN analysis. The minimum requirement for spot quantification was spot presence in at least three biological replicates for either M or BS membranes. The average BS/M ratio across the replicates and their coefficients of variation (cvs) for each quantified accession, normalized average spot volume with their cvs for each BS and M sample, and the histograms with spot volume for each of the seven replicates are listed in supplemental Table 1. We report only one representative accession for each group in Table I; redundant accessions are reported in supplemental Table 1.
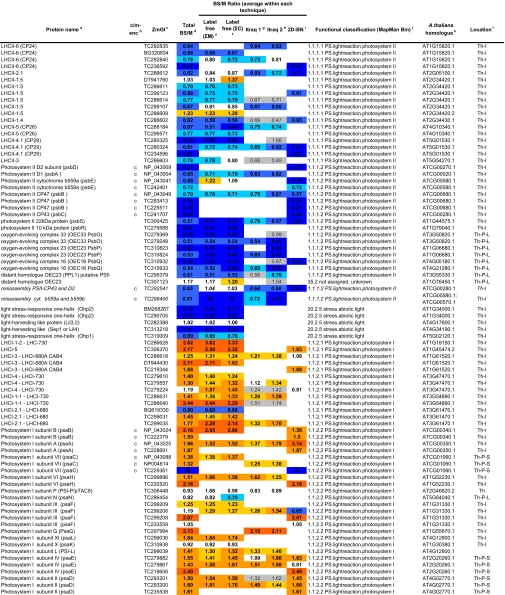
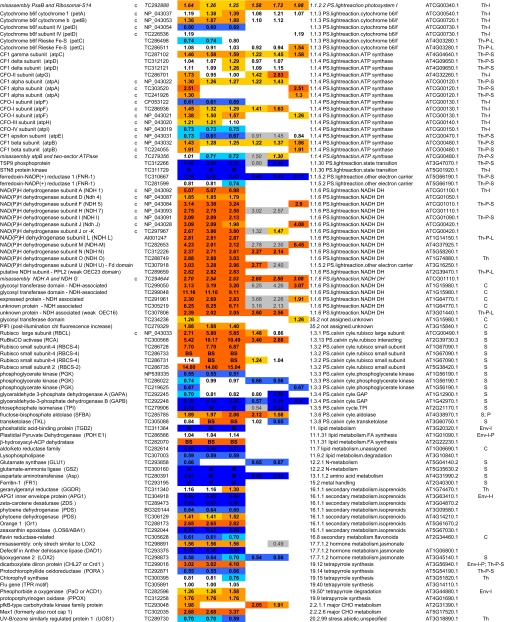
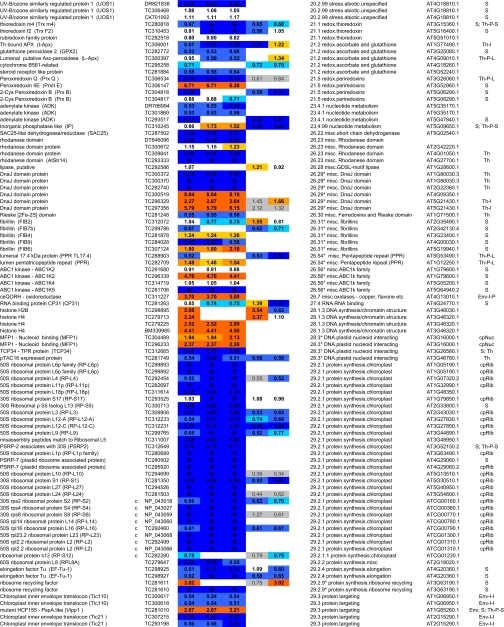
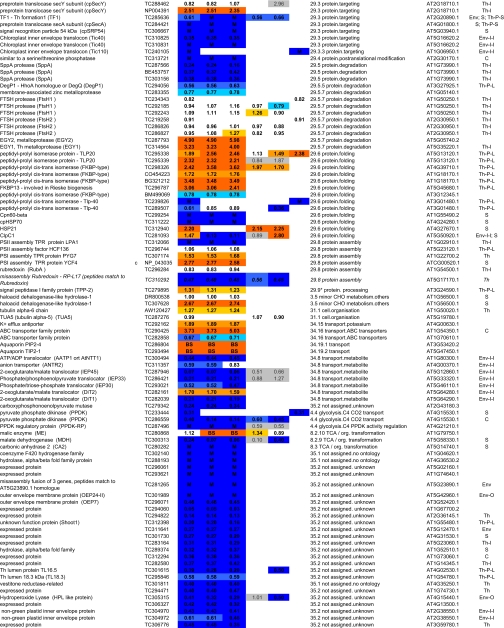
Table I.
Summary of all quantifications of maize chloroplast BS and M membrane proteomes
ZmGI accessions were quantified by one or more of the following techniques: 2D BN gels, iTRAQ, and 1D BN gels followed by LC-MS-based quantificatins of unlabeled peptides (label-free method). BS/M ratios for the label-free methods are calculated based on normalized protein Mowse score and on normalized spectral count; only proteins identified in three replicate runs with a minimum ΣCount of 10 were included. BS/M ratios determined by iTRAQ are based on pairwise reporter ion quantifications (Only quantifications based on at least two peptides are included; however, when the protein was quantified by one or both other methods, then iTRAQ quantifications based on one peptide were included but are marked in gray). The BS/M ratios for the 2D BN analysis are based on normalized spot volumes. Spots were only included in the analysis if they were present in at least three biological replicates for either M or BS membranes. The “heat map” scale of BS/M ratios is as follows: M to 0.5, dark blue; 0.5–0.7, medium blue; 0.7–0.8. light blue; 0.8–1.2, white; 1.2–1.5, yellow; 1.5–2, light orange; 2 to BS, dark orange; grey, only one peptide.
aAssigned protein name based on information from BLAST alignments.
bOrganelle-encoded genes are indicated: c, chloroplast-encoded, m, mitochondria-encoded.
cZmGI accession number or maize chloroplast genome accession number (obtained from NCBI) identified by MS.
dAverage BS/M protein accumulation ratio across all experiments.
eAverage BS/M protein accumulation ratio based on sum of Mowse scores obtained in the label-free quantitication experiment.
fAverage BS/M protein accumulation ratio based on sum spectral counts obtained in the label-free quantification experiment.
gQuantification from iTRAQ labeling experiment 1.
hQuantification from iTRAQ labeling experiment 2.
iAverage BS/M ratio based on spots quantification on 2D-BN-PAGE.
jFunctional assignment based on the MapManBin system.
kBest A. thaliana homologues as judged by BLAST E-value.
lCurated chloroplast sub-proteome localizations: plastid (C), stroma (S), thylakoid integral (Th-I), thylakoid peripheral stromal side (Th-P-S), thylakoid lumenal side (Th-P-L), thylakoid integral (Th-I), thylakoid (Th), Envelope (Env), Envelope integral (Env-I), Envelope inner integral (Env-I-I), Envelope inner peripheral (Env-I-P), Envelope outer (Env-O), Mitochondria (M), plastoglobule (cpP), plastid nucleoids (cpNuc), plastid ribosome (cpRib).
1D Blue Native Gel Analysis and Quantification by Nano-LC-LTQ-Orbitrap MS—
700 μg of protein from BS and M chloroplast membranes were solubilized as described above. Each gel lane was cut into 27 bands followed by reduction, alkylation, and in-gel digestion with trypsin (52). The peptides extracted from these bands were analyzed in triplicate by on-line LC-LTQ-Orbitrap MS. Peptides extracts were loaded on a guard column (LC Packings MGU-30-C18PM) followed by separation on a PepMap C18 reverse-phase nanocolumn (LC Packings nan75-15-03-C18PM) using 90-min gradients with 95% water, 5% ACN, 0.1% FA (solvent A) and 95% ACN, 5% water, 0.1% FA (solvent B) at a flow rate of 200 nl/min. Each sample injection and analysis were followed by two blank injections to prevent carryover. The acquisition cycle consisted of a survey FT MS scan at the highest resolving power (100,000) followed by five data-dependent MS/MS scans acquired in the LTQ. Dynamic exclusion was used with the following parameters: exclusion size, 500; repeat count, 2; repeat duration, 30 s; exclusion time, 180 s; and exclusion window, ±6 ppm. Target values were set at 5 × 105 and 104 for the survey and tandem MS scans, respectively, and the maximum ion accumulation times were set at 200 ms in both cases. Peak lists (.mgf format) were generated using DTA supercharge software and searched with Mascot v2.2 (Matrix Science). For off-line calibration, first a preliminary search was conducted with a broad precursor tolerance window set at ±30 ppm. Peptides with ion scores above 45 were chosen as benchmarks to determine the offset for each LC-MS/MS run. This offset was then applied to adjust precursor masses in the peak lists of the respective .mgf file for recalibration using a Perl script.2
The recalibrated peak lists were searched against the ZmGI v16.0 database (56,364 entries) concatenated with a decoy where all the sequences were shuffled. Each of the peak lists was searched using Mascot v2.2 (maximum p value of 0.01) for full tryptic peptides using a precursor ion tolerance window set at ±6 ppm, variable methionine oxidation, fixed cysteine carbamidomethylation, and a minimal ion score threshold of 42 that yielded a peptide false discovery rate below 1%. Peptide FPR was calculated as follows: 2 × decoy_hits/total_hits. The false protein identification rate of protein identified with two or more peptides was 0. To reduce the false protein identification rate of proteins identified by one peptide, the Mascot search results were further filtered as follows. The ion score threshold was increased to 45, and mass accuracy on the precursor ion was required to be within ±3 ppm. All filtered results were uploaded into the PPDB. Protein identification data are also provided in supplemental Table 2.
To assess the reproducibility between technical replicates, we used the G-test of independence (53). Recently this test has been used to assess significance of expression changes in label-free comparative proteomics experiments for eukaryotes and prokaryotes (54, 55). We applied the G-test to three technical replicates of each of the MS and BS preparations. In both of the cases, about 2% of all proteins were determined to be significantly changed between the three replicates, showing that the technical replicates were highly reproducible.
To determine protein accumulation ratios between BS and M, we calculated two parameters for quantification: (i) normalized number of matched spectra for each protein across all 27 gel slices per replicate (ΣCounts) and (ii) normalized total Mowse protein score for each protein across all 27 gel slices per replicate (ΣMowse). Only those proteins for which the sum of ΣCounts across the three replicates was higher than 10 in either cell type were used for quantification. G-tests were used to help determine this cutoff as explained above. The BS/M protein ratios were then calculated based on averaged (across the three replicates) ΣMowse score or ΣCounts for each protein per cell type (Table I and supplemental Table 7). We report only one representative accession for each group in Table I; redundant accessions and all peptide sequences are reported in supplemental Table 2.
iTRAQ Analysis—
Two independent biological pairwise replicates of BS and M cells were isolated, and 30 μg protein from each of the four samples were precipitated and digested as described previously (23). The peptides were dissolved with 30 μl of 0.5 m triethylammonium bicarbonate and mixed with the four iTRAQ reagents (Applied Biosystems). The samples BS1, M1, BS2, and M2 were labeled for 90 min with the four iTRAQ reagents 114, 115, 116, and 117 and then pooled. The sample was then diluted 10-fold to a final concentration of 25% ACN and 1% FA, and half of the final volume was separated using strong cation exchange chromatography on a PolySULFOETHYL A™ column (200 × 2.1 mm, 5 μm, 300 Å) from PolyLC Inc. connected to an Agilent 1100 HPLC system. Solution A was 25% ACN, and solution B was 25% ACN, 0.5 m ammonium formate (pH 3.0, set with FA). 500 μl of the sample were injected and eluted at a flow rate of 200 μl/min as follows: 3% B, 0–10 min; 3–10% B, 10–15 min; 10–60% B, 15–25 min; 60–100% B, 25–35 min; 100% B, 35–45 min; 100–3% B, 45–47 min; and 3% B, 47–55 min. The fractions were collected in microtiter plates, lyophilized, resuspended with 15 μl of 5% FA, and analyzed by MALDI-TOF MS. The 10 fractions containing the majority of the labeled peptides were selected for analysis by Q-TOF MS. The data were processed with Mascot Distiller v2.1 (Matrix Science) resulting in .mgf files, and the areas of the reporter ions were extracted from the .mgf files and searched against ZmGI v16.0 (56,364 entries) using Mascot (v2.2) with p < 0.05 using the following criteria: one missed cleavage allowed, variable methionine oxidation, cysteine carbamidomethylation and iTRAQ label as fixed modifications, and a minimum peptide ion score threshold of 20.
Search results were extracted from the XLM pages and further filtered for overlap with other accessions using an in-house program written by Dr. Qi Sun. All peptides where the difference in score between the highest and the second highest interpretations of the same query was less than 10 points were manually checked, and if it was not possible to distinguish which was the best interpretation, both were removed. Areas from spectra derived from the same sequence (e.g. repetitive fragmentation of the same precursor, different charge states, or variable modifications) were pooled. The areas were normalized by adding the areas from all peptides with all four reporter ions. Spectra lacking one or two reporter ions were kept if both ions from the same biological replicate (e.g. BS1 and M1 or BS2 and M2) were present and used to calculate the protein average. Peptides were grouped by accession. In the case of seven proteins quantified with six to 20 peptides, one or two peptides showing a ratio clearly different from the average of the majority of the peptides were removed from the calculation of the average protein accumulation.
In total, 183 accessions were identified of which 116 were assigned an average BS/M accumulation ratio based on quantification of at least two peptides, and the corresponding S.D. and cv are reported (Table I and supplemental Table 7). Proteins exclusively identified in M or BS proteomes were removed from the quantification analysis. The average cv across the quantified peptides per protein for all protein ratios was 0.26. In a number of cases, the MS data matched two or more closely related proteins, particular in the LHC family and ribosomal proteins. We report only one representative accession for each group in Table I; redundant accessions and all peptide sequences used for iTRAQ quantification are reported in supplemental Table 5. All sequences used for iTRAQ quantification are listed in supplemental Table 6.
The Plant Proteome Database and Functional Assignment of Identified Proteins—
The Mascot output files were automatically processed by in-house software, 3 and a number of output parameters were uploaded into the PPDB. Because many of the maize ZmGI accessions lack functional annotation, we functionally annotated all identified ZmGI accessions using a combination of best BLAST hits in the predicted rice proteome (Oryza sativa Gene Index) and the predicted Arabidopsis thaliana proteome, ATHv6 (from The Arabidopsis Information Resource). Pairwise BLAST search results between ATHv6, O. sativa Gene Index v4, and ZmGI v16.0 are available via PPDB. Each identified protein was assigned to a molecular function using the hierarchical, non-redundant classification system developed for MapMan (56), adjusted after manual verification and information from the literature, and incorporated into the PPDB. In some cases ZmGI accessions numbers were identified as a set of accessions sharing entirely the set of identified peptides; this was essentially due to a lack of good assembly of ZmGI accessions within small gene families (e.g. LHCs). In these situations we cite only one accession number in quantification tables; the subset accessions are listed in tables containing the corresponding identification information (supplemental Tables 1, 2, and 5).
RESULTS
Purification of M and BS Chloroplast Membranes from Maize Leaf Tips—
The aim of this study was to identify and compare differentiated M and BS chloroplast membrane proteomes with associated proteins within the final stages of C4 maize leaf differentiation. Therefore, chloroplasts were isolated from the tip of the third leaf of 12–14-day-old seedlings. After purification, M and BS chloroplasts were each lysed, and chloroplast envelope and thylakoid membranes were collected together by ultracentrifugation. These BS- and M-specific membrane fractions were deliberately not further “stripped” of associated, non-integral proteins as they are of biological interest and relevance to BS/M differentiation. Furthermore although envelope and thylakoid membranes can be separately purified (see the Introduction), we decided to collect envelopes and thylakoid together because this reduced the number of samples and also avoided variability resulting from additional fractionation. The purity of each preparation was systematically “prescreened” for quality based on the presence of M and BS cytosolic and chloroplast markers using 1D SDS-PAGE analysis (see “Experimental Procedures”). The purity of M/BS preparations was further assessed based on the quantification of 23 marker proteins chosen for their established BS/M relative accumulation and chloroplast localization (see “The Identified and Quantified Proteomes and Consistency”). For this study, we used 10 prescreened and independent preparations of BS/M pairs.
Overview of the Comparative Proteome Analysis—
To overcome the challenges of comparative membrane proteomics in maize, we used a combination of three comparative proteomics methods that are compatible with membrane proteins and that are based on different principles for quantification as follows: (i) quantification based on spot image analysis of 2D gels using blue native gels (50) as the first dimension and SDS-PAGE as the second dimension (further referred to as “2D BN”), (ii) MS-based quantification using label-free analysis by nano-reverse phase LC-LTQ-Orbitrap MS (further referred to as the “label-free” method), and (iii) MS-based quantification using differential peptide labeling with an amine-reactive isobaric tagging reagent (iTRAQ).
The mass spectral data were searched against the assembled transcript sequences of maize TIGR ZmGI v16.0 supplemented with complete chloroplast and mitochondria genome sequences obtained from the National Center for Biotechnology Information (NCBI). The collective data set was functionally annotated based on manual annotation for about 400 accessions and BLAST alignments to rice and Arabidopsis and the MapMan functional classification system (56). Mass spectral information extracted from the Mascot search pages, functional annotation, and the highest scoring BLAST results against Arabidopsis and rice are available through the PPDB. Matched peptides projected on translated reading frames can also be viewed via PPDB. A number of ZmGI unigenes were identified that appeared to be assembled incorrectly; this is indicated in the annotations.
2D BN Analysis—
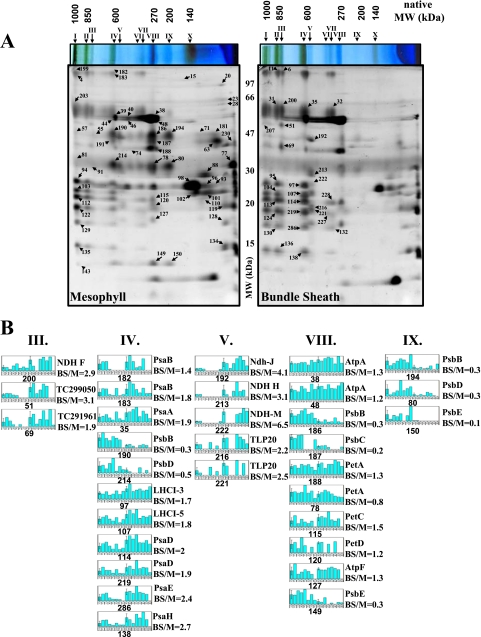
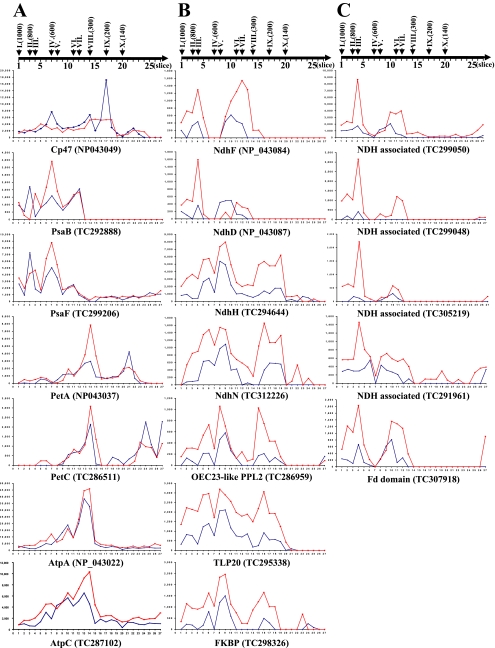
2D BN has been used to analyze protein composition and oligomeric states of chloroplast envelope and thylakoid membranes (57, 58). Following similar procedures, the purified BS and M membranes were solubilized in β-DM and run on BN gels followed by separation of each gel lane by SDS-PAGE. Seven independent biological replicates (pairs of BS and M membranes) were used. The minimum requirement for spot quantification was spot presence in at least three biological replicates for either M or BS membranes. Representative 2D BN gels of M and BS chloroplast membranes with annotation of the major complexes and proteins are shown in Fig. 1A. The differential BS/M accumulation ratio was determined for 85 spots (supplemental Table 1), and examples of quantifications are shown as bar diagrams (Fig. 1B).
Fig. 1.
2D BN gels from BS and M chloroplast membranes and examples of quantifications. A, BS and M membranes were solubilized with β-DM and first separated based on native mass by BN-PAGE. The focused gel lanes were then denatured, reduced, and alkylated, and proteins were separated based on denatured mass by SDS-PAGE. The resulting 2D-BN gels were stained with Coomassie Blue, and spots were detected, matched, quantified, and normalized against the total spot volume. The analysis was carried out with seven (independent) biological replicates. Proteins in the spots were identified by peptide mass fingerprinting using MALDI-TOF MS and/or by on-line nano-LC-ESI-MS/MS. The mass spectral data were searched against the maize EST assembly from TIGR (ZmGI v16.0). BS/M ratios are listed in Table I. Details of spot quantifications, their estimated 1D and 2D molecular weights, and protein content are shown in supplemental Table 1. Major complexes are indicated by Roman numerals with their BS/M ratio and S.D. as follows: I (mixture of PSI and PSII “supercomplexes”), II (PSI dimer, BS/M = 1.9 ± 0.5), III (complex of unknown function, BS/M = 3.1 ± 0.57), IV (PSI, BS/M = 2 ± 0.38; and PSII dimer, BS/M = 0.4 ± 0.14), V (NDH and Tlp20, BS/M = 3.7 ± 1.7), VI (partially assembled PSI), VII (partially assembled PSI, BS/M = 1.6 ± 0.37), VIII (ATP synthase, BS/M = 1.3 ± 0.05; cytb6f, BS/M = 1.2 ± 0.3; and PSII monomer, BS/M = 0.3 ± 0.05), IX (partially assembled PSII, BS/M = 0.2 ± 0.11), and X (LHCII trimer, BS/M = 0.6). B, examples of quantifications of the major thylakoid complexes: PSI, PSII, cytb6f, ATP synthase, the NDH complex, and the high molecular weight complexes of unknown function (complexes III, IV, V, VIII, and IX). Spot numbers, protein identities, and BS/M spot ratios are indicated. Bar diagrams indicate the following. Bar 1 shows the M average spot volume and S.D. calculated from normalized spots volumes in seven M gels shown in bars 2–8, and bar 9 shows BS average spot volume and S.D. calculated from normalized spots volumes in seven BS gels shown in bars 10–16.
The 2D BN analysis provides a BS/M comparison of the oligomeric state and relative abundance of the most abundant proteins. As expected, PSII content was reduced in the BS membranes. In M thylakoids, three major PSII complexes (dimer at 600 kDa, monomer at ∼270 kDa, and a partial core at ∼200 kDa) were observed. Accumulation of the PSII monomer and a partially assembled PSII core was respectively 3- and 5-fold reduced in BS membranes, whereas the high molecular weight PSII complexes observed in M thylakoid were below detection in BS membranes. Interestingly the accumulation of LHCII trimers (∼140-kDa complex X) was only 1.6-fold reduced in the BS membranes suggesting that a subset of LHCII proteins do function in the BS consistent with previous findings (13) (Fig. 1B). Furthermore BS thylakoids were characterized by a 40% higher accumulation of PSI and its antennae LHCI subunits in particular within the high molecular mass forms of PSI-LHCI oligomeric assemblies (>700 kDa). A 30% increased BS accumulation of the ATP synthase complex (300 kDa) was also observed. NDH complex (complex V) was detected at ∼550–600 kDa and was 400% enriched in the BS membranes (Fig. 1B). Interestingly thylakoid lumenal protein isomerase TLP20 was identified with complex V, suggesting a possible association with the NDH complex. Finally an abundant and novel complex of ∼800 kDa (complex III) was found in BS membranes; it includes two homologous proteins (TC299048 and TC299050) of unknown function that co-migrate with NdhF on 2D BN (spot 200; Fig. 1B) and show a strong co-migration pattern with the NdhD and -F subunits (see below for 1D BN profile analysis). FNR1 showed a strong M accumulation (BS/M = 0.2), the FtsH thylakoid protease accumulated with a BS/M ratio of 0.9, and a strong M expression was determined for the Tic110 subunit of the inner envelope translocon.
1D BN-PAGE and Label-free Quantitative Analysis by LTQ-Orbitrap MS—
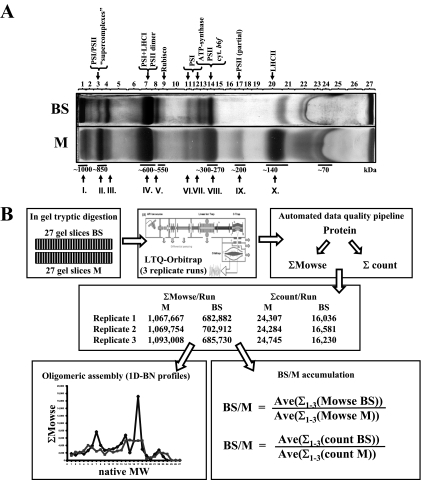
The 2D BN shows distinct differences between BS and M membranes, but proteins of lower abundance are not visible on the gels and thus cannot be quantified. Moreover, spots containing more than one protein do not provide reliable quantification. Therefore, we separated the BS and M proteomes by BN-PAGE only and then directly analyzed and quantified the proteins by MS using the fast and accurate LTQ-Orbitrap. Each BN-PAGE lane was cut in 27 bands, proteins were cleaved by in-gel digestion with trypsin, and extracted peptides were analyzed by MS/MS (Fig. 2, A and B). The on-line chromatography and MS acquisition were optimized for label-free quantification (59). MS data were analyzed against ZmGI v16.0 followed by additional filtering (see “Experimental Procedures”). Each sample was analyzed in triplicate to reduce variation due to chromatography and MS acquisition. The chromatography was very reproducible (not shown), which was also reflected by similar total numbers of matched spectra (counts) and total Mowse scores between the replicate runs (Fig. 2B).
Fig. 2.
Comparative analysis of BS and M chloroplast membranes by 1D BN-PAGE followed by LC-MS-based quantification of unlabeled peptides. A, BS and M membranes were solubilized with β-DM and separated based on native mass by 1D BN-PAGE. Approximate native molecular mass (below the gel) and the identities of the most abundant complexes (above the gel) are indicated. Roman numerals indicate the most abundant complexes observed in the first dimension of the 2D BN in Fig. 1. Each BN-PAGE lane was cut into 27 bands (indicated above the gel). B, schematic overview of the identification and quantification process. Proteins were denatured, reduced, alkylated, and cleaved by in-gel digestion with trypsin. Extracted peptides were analyzed by data-dependent acquisition nano-LC-ESI-MS/MS. The high accuracy precursor ion masses (in MS) were determined in the Orbitrap (maximum mass error in database search <6 ppm) followed by data-dependent MS/MS in the LTQ part of the instrument. Each sample was analyzed in three replicates. Spectral data were searched against ZmGI v16.0 using Mascot followed by additional filtering and extraction of relevant information. The sum of the number of matched MS/MS spectra (ΣCount) or Mowse scores (ΣMowse) obtained per protein was calculated for each replicate. The BS/M protein ratios were then calculated based on averaged (Ave; across the three replicates) ΣMowse or ΣCounts for each protein per cell type.
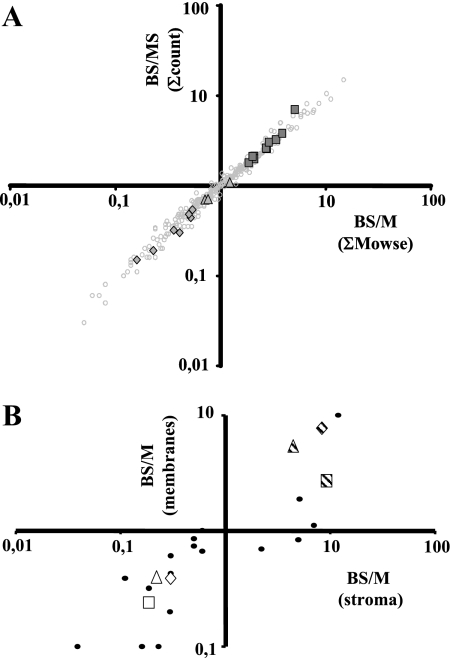
We identified 543 proteins (supplemental Table 2) of which 333 were identified in both M and BS; 60 and 150 proteins were only identified in BS and M membranes, respectively (supplemental Table 7). To determine protein accumulation ratios between BS and M, we calculated two possible parameters for quantification: (i) normalized number of matched spectra for each protein across all 27 gel slices per replicate (ΣCounts) and (ii) normalized total Mowse protein score for each protein across all 27 gel slices per replicate (ΣMowse). For information on the average cv, see “Experimental Procedures.” To increase the “robustness” of the data set for quantification, we discarded those proteins for which the sum of ΣCounts across the three replicates was lower than 10 in either cell type (corresponding to a minimum ΣMowse of ∼420). G-tests were used to help determine this cutoff (see “Experimental Procedures”). This reduced the number of proteins only found in BS or M cells to 11 and 51, respectively; and 289 proteins were found in both BS and M. The BS/M protein ratios were then calculated based on averaged (across the three replicates) ΣMowse score or ΣCounts for each protein per cell type (Table I and supplemental Table 7). Cross-correlation of the BS/M protein ratios calculated by the two parameters showed a strong linear correlation (r2 = 0.97), indicating that the two parameters yielded comparable results (Fig. 3A).
Fig. 3.
Cross-correlation of quantifications of BS/M ratios. A, cross-correlation of the BS/M protein ratios for 290 proteins calculated based on averaged (across the three replicates) ΣMowse score or ΣCounts for each protein per cell type. The BS/MS ratios for the subunits of the NDH complex (squares) and the PSII core complex of chloroplast-encoded (triangles) and nucleus-encoded (diamonds) subunits are highlighted in the overall population of quantified accessions (circles). B, cross-correlation of the BS/M protein ratios for proteins characterized in the comparative analysis of BS and M chloroplast stromal proteomes (8) and our current thylakoid data set. A direct comparison was obtained for 23 proteins (circles). Average BS/M ratios across the methods were used. Examples of markers of the C4 carbon fixation pathway are highlighted for the following: PPDK, BS/M = 0.4 (open triangle); MDH, BS/M = 0.24 (open square); GAPB, BS/M = 0.39 (open diamond); RBCL, BS/M = 2.71 (striped square); RBCS, BS/M = 7.69 (striped diamond); and RCA, BS/M = 5.42 (striped triangle). Proteins that were only identified in M or BS were assigned a ratio of 0.1 and 10, respectively.
To further evaluate the quality of the quantifications, we took advantage of established, biological internal controls: many protein complexes are known to accumulate with strict stoichiometry between the subunits due to transcriptional and/or post-transcriptional control mechanisms. The BS/M ratios of the subunits within PSI (including LHCI), the cytb6f complex, and ATP synthase complex were compared and showed good consistency (Table I). As an example, the BS/MS ratios for the subunits of the NDH complex and the PSII core complex are highlighted in the overall population of quantified accessions (Fig. 3A).
The use of the 1D BN as a protein separation method prior to the MS analysis also provided information about oligomeric state and was complementary to the 2D BN analysis. We calculated the normalized ΣMowse score for each protein per gel slice (supplemental Table 3) and created profiles of their oligomeric assembly state in BS and M membranes. Examples of profiles for subunits of photosynthetic complexes are shown in Fig. 4; profiles corresponded well to these observed from gel image analysis on 2D BN. These native mass profiles will be used further below to characterize BS/M differentiation.
Fig. 4.
Profiles of oligomeric state of selected proteins determined by 1D BN-PAGE and MS analysis. For each protein the ΣMowse per gel slice for BS (in red) and M (in blue) are represented. Slices were numbered (as indicated by the scale on top of each panel) following a decreasing native molecular weight. Roman numerals indicate the major thylakoid complexes as described in Fig. 1 with approximate native molecular weight indicated in parentheses. A, examples of 1D BN profiles for subunits of PSII (CP47), PSI (PsaB,F), cytb6f (PetA,C), and ATP synthase (AtpA,C). B, 1D BN profiles of two membrane-embedded (NdhD and -F), two connecting complex subunits (NdhH and -N) of the NDH complex, and potential interacting partners co-migrating with the NDH-connecting complex subunits (PPL2, TLP20, and FKBP). C, 1D BN profiles for four unknown proteins co-migrating in a BS-localized 800-kDa high molecular mass complex and potentially interacting with Fd-like or NdhU and NDH membrane-embedded subunits.
iTRAQ—
To further quantify relative protein accumulation between BS and M membranes, we used the differential stable isotope labeling technique named iTRAQ and analysis by a nano-LC-Q-TOF MS instrument. The structure of this isobaric tagging reagent, the labeling principle, and quantification have been explained previously (60, 61). Two independent biological replicates of pairs of BS and M membrane proteomes were in-solution digested using a procedure that we established earlier (23). The peptides were labeled with four iTRAQ tags (BS1 (114), M1 (115), BS2 (116), and M2 (117)) and pooled. The protein mixture was separated using strong cation exchange chromatography followed by LC-ESI-MS/MS of selected fractions. Examples of MS/MS spectra from our Q-TOF instrument show the well resolved reporter ions (61).
In total, 183 accessions were identified of which 116 were assigned an average BS/M accumulation ratio based on quantification of at least two peptides, and the corresponding S.D. and cv are reported (Table I and supplemental Table 7). Proteins exclusively identified in M or BS proteomes were removed from the quantification analysis. In a number of cases, the MS data matched two or more closely related proteins, particularly in the LHC family and ribosomal proteins. We report only one representative accession for each group of homologues in Table I; redundant accessions and all peptide sequences used for iTRAQ quantification are reported in supplemental Table 6.
The Identified and Quantified Proteomes and Consistency—
In total 610 proteins were identified with the highest number (543 proteins) obtained by the 1D BN analysis. Proteins were functionally classified using the MapMan functional classification system developed for Arabidopsis (56) (supplemental Table 7). 398 proteins passed the minimal criteria for quantification (described above) by at least one of the three quantification methods and were assigned a BS/M ratio (summarized in Table I). An additional 192 proteins that did not pass the quantification quality filters are only listed in supplemental Table 7. Generally the three methods gave similar BS/M ratios with an average cv across the three methods of 0.28, but we did note that the BS/M ratios determined by iTRAQ appeared to be generally closer to unity than the ratios determined by the other two methods (Table I). This suggests that the label-free quantification (also named spectral counting) has a better dynamic resolution than iTRAQ. Furthermore the quantification from the 2D BN gel image analysis of the more abundant proteins showed similar or slightly more extreme BS/M ratios than the label-free quantification.
A direct comparison of the BS/M ratios of 23 proteins with quantifications from our previous stromal analysis (8) showed good consistency and also confirmed that the BS/M cross-contamination was well below 15% (Fig. 3B and supplemental Table 8). Examples of abundant markers of the C4 carbon fixation pathway (average ratios across three methods) were: PPDK, BS/M = 0.4; malate dehydrogenase (MDH), BS/M = 0.24; glyceraldehyde dehydrogenase subunit B (GAPB), BS/M = 0.39; Rubisco large subunit (RBCL), BS/M = 2.71; Rubisco small subunit (RBCS), BS/M = 7.69; and Rubisco activase (RCA), BS/M = 5.42. Furthermore only a handful of non-chloroplast proteins were identified, and none passed the criteria for quantification (see supplemental Table 7), indicating that the chloroplasts were of high purity.
Differential Accumulation of Photosystem II Subunits and Subcomplexes—
PSII can be viewed as the assembly of the PSII reaction center surrounded by the core subunits, the oxygen-evolving complex (OEC16,23,33), and the major and minor LHCII antennae proteins. Fifteen proteins of the reaction center and core are encoded by the plastid genome, whereas the other subunits are encoded by the nuclear genome; in the context of BS/M differentiation this might be important because transcriptional and translational controls of these two groups are different.
The PSII Core and OEC—
The chloroplast-encoded PSII core subunits D1 (NP_043004), CP47 (NP_043049; TC283413; TC225511), CP43 (TC241707), and cytb559α (NP_043041; TC242401) showed average BS/M ratios between 0.25 and 0.72 (Table I). The nucleus-encoded core subunit PsbR (TC279588) showed a BS/M ratio of 0.41 (Table I), whereas nucleus-encoded subunits PsbTn-1 (TC305359), PsbW (TC282454), and PsbH phosphoprotein (NP_043052) were only identified in M thylakoids (supplemental Table 7). OEC16, -23, and -33 proteins represented by six ZmGI accessions showed a BS/M accumulation ratio between 0.22 and 0.54, which was on average lower than the rest of the PSII core (Table I). In contrast, two distantly related OEC16-like proteins with unknown function (AI001247 and TC307806) each showed much higher BS/M accumulation ratios of 2.39 and 2.81, respectively, strongly suggesting that they do not belong to PSII. In fact AI001247 showed a weak similarity to the NdhL subunit of Synechocystis elongatus PCC6803, and both proteins TC307806 and AI001247 co-migrated with the NDH complexes (see below). We also identified three distant homologues of OEC23, namely TC307123, TC295379, and TC289659, with BS/M ratios of 1.17, 0.61, and 2.82. Interestingly Arabidopsis homologues named PPL1 (AT3G553330) and PPL2 (AT2G39470) for TC295379 and TC289659, respectively, were recently characterized in Arabidopsis (62). PPL1 was suggested to be required for PSII stability and repair, whereas PPL2 was suggested to be required for accumulation of the NDH complex. This does fit very well with the BS/M ratios for the two maize proteins. Moreover bona fide maize OEC subunits were identified essentially in the low molecular weight fraction in 1D BN analysis probably because they were released from the membrane upon detergent treatment. In contrast, AI001247 (distant OEC16 and putative NdhL), TC289659 (PPL2 homologue and putative NDH subunit), and TC307806 (distant OEC23) were all found in high molecular mass complexes between 250 and 750 kDa (supplemental Table 3). The PPL2 and “distant OEC16” homologues showed a clear co-migration pattern and similar BS/M expression ratios with different oligomeric assemblies of the NDH complex, suggesting that they might be functionally related with the NDH complex.
LHCII Antennae—
The major and minor LHCII antennae were represented by 18 different accessions with an average BS/M ratio of 0.68 (cv = 0.33) (Table I). Six ZmGI accessions matched best to LHCII1.5; within this cluster two accessions appeared to have higher BS/M ratios than the other LHCII proteins, which may indicate that these proteins function in both compartments. Indeed although Schuster et al. (11) observed the absence of LHCII polypeptides in BS chloroplasts in 2–3-week-old maize seedlings, others determined that LHCII gene family members showed different cell-specific expression patterns (measured at the mRNA level) in 14-day-old seedlings (63). These results were supported by the observation of different ultrastructures of the LHCII in freeze fracture experiments between maize M and BS thylakoids (13), suggesting that complexes of LHCII proteins in the M and BS are of different composition.
Light Stress Proteins with Chlorophyll Binding Domains—
PsbS (TC300425) was identified with a very high ΣMowse score (20,612) in the label-free experiment and a BS/M ratio of 0.5, similar to the PSII core, underscoring the involvement of PsbS in dissipation of the excess energy from PSII (64). PsbS 1D BN profiles show large peaks (at ∼650, ∼350, and ∼200 kDa) encompassing PSII assemblies (supplemental Table 3). However, these interactions are relatively weak because most PsbS was found in low molecular weight fraction 25 consistent with observations that PsbS links LHCII antennae complexes with the PSII core rather than being a stable core subunit (65). We also identified and quantified maize homologues of small light stress proteins with chlorophyll binding domains, Ohp1 (TC310009), Ohp2 (BM268267 and TC290705), Lil3.2 (TC282388), and Lil4/Sep1 (TC313218) that was only found in M thylakoids. The two Ohp2 accessions were strongly enriched in M thylakoids (BS/M ratio, 0.18 and 0.35), whereas the Ohp1 BS/M ratio was 0.69. 1D BN protein profiles for Ohp2 (BM268267) showed a co-migration with the PSI and PSII dimers suggesting possible interaction and its contribution in PSII and/or PSI stress response (supplemental Table 3). In contrast, Lil3.2 was equally distributed over the two cell types (BS/M = 1), suggesting a role in PSI stress response.
PSI, LHCI, and FNR—
The PSI and LHCI proteins, represented by 25 and 13 quantified accessions, respectively, showed preferential BS expression with average BS/M ratios of 1.57 (cv = 0.28) and 1.72 (cv = 0.43), respectively (Table I and supplemental Table 7). These observations are consistent with previously suggested higher accumulation of PSI in BS chloroplasts of C4 plants based on an immunoblot against the PsaA protein (12). The exception was LHCI-680B (BQ619330), which showed higher M accumulation. PSI-associated PsaP (TC306448) protein was expressed equally in both compartments. Strong differential BS/M expression was observed by all thee methods for FNR1 in agreement with our previous observation for the soluble stromal proteomes (8). FNR1, represented by two accessions, was either strongly expressed in the M (TC310667) or equally distributed over both compartments (TC281599).
Cytb6f and ATP Synthase Complexes—
The three quantified subunits of cytb6f (cytf, Rieske Fe-S, and subunit IV), represented by six accessions, showed very little BS/M differentiation with an average BS/M ratio of 1.03 (cv = 0.26). This is logical because cytb6f is needed for both linear and cyclic electron flow. Some variability was observed within two ZmGI accessions corresponding to the Rieske subunit (TC286498 and TC286511), suggesting that two different isoforms are differentially expressed between M and BS (Table I and supplemental Table 7). All nine subunits of the ATP synthase complex (represented by 15 ZmGI accessions) were quantified with an average BS/M ratio of 1.33 (cv = 0.37), indicating a fairly equal distribution of the ATP synthase over BS and M thylakoids. The somewhat higher cv is most likely due to redundant and incorrect ZmGI unigene assemblies (Table I).
The NDH Complex, New Interaction Partners, and IMMUTANS—
Thylakoid NDH complex is involved in one of the two pathways for cyclic electron flow around PSI and also in chlororespiration. In C3 plants, the NDH complex is of much lower abundance than the four major photosynthetic complexes (estimated at 0.2% of total thylakoid protein (66)). The plastid genome encodes for 11 putative NDH subunits (A–K), and several have been identified in both C3 (67) and C4 (15, 18, 68) plants. Recently five nucleus-encoded Ndh subunits (M, N, and O and CRR3 and -7) were identified in Arabidopsis (69–71).
Here we quantified seven chloroplast-encoded (A, D, F, H, J, I, and K) and three nucleus-encoded maize NDH subunits (M, N, and O), all showing strong BS expression with an average BS/M ratio of 3.0 (cv = 0.32) (Table I). In addition we identified NdhB (CF039487) with low scores in complex V (spot 213; data not shown) in 2D BN and quantified the maize homologue of PPL2 (BS/M = 2.8) as mentioned above. A misassembled accession (TC294644) containing fragments of NdhA and -G and an accession (TC297967) containing fragment sequences similar to NDH-J and -K annotated here as NdhJorK were both quantified with similar BS/M ratios (2.67 and 2.70). Preferential accumulation of the NDH complex was described in BS chloroplasts of sorghum (68) and maize with BS/M ratios between 2.5 and 3 (16, 17), which is consistent with our observations. We did not detect any of the CRR proteins, involved in NDH biogenesis or NDH subunits (67, 70, 71), but we note that the maize homologue of soluble CRR1 (TC242930) was found in our previous stromal analysis (8).
Analysis of the 1D BN label-free profiles showed that all identified NDH subunits assemble in high molecular weight complexes with the exception of NdhA, which was only found as a monomer (band 24). Detachment of NdhA was also observed previously using either sucrose gradient fractionation (15) or affinity purification (69), suggesting that it is located at the periphery of the complex. Based on peaks detected for each of the NDH subunits in the 1D BN profiles (and by 2D BN analysis for the ∼800- and ∼550-kDa complexes), we conclude that the membrane-embedded and -connecting NDH subunits co-migrate in two assemblies at ∼1000 and ∼550 kDa (Figs. 4B and 5 and supplemental Table 4). Although we did not identify all membrane-embedded NDH subunits (because of their small size and hydrophobicity), it is likely that these ∼550- and ∼1000-kDa complexes represent a monomer and a dimer, respectively, of a fully assembled NDH complex (Fig. 5, A and C). The 1D BN profiles showed additional NDH assemblies with a highly abundant and well defined peak at ∼800 kDa (containing NdhD and -F) and at ∼650 kDa (containing NdhJorK, -M, -N, and -O) and a less abundant complex at ∼320 kDa (NdhD and -F), at ∼220 kDa (NdhH, -J, -JorK, -M, and -N), and at ∼170 kDa (NdhH, -I, -J, -JorK, -M, -N, and -O). These are likely the result of destabilization due to the detergent, and they do suggest a modular organization of the NDH complex. Consistently the NDH complex in tobacco and maize was purified as an assembly of 550 kDa (15, 72). Using 2D BN-PAGE and four NDH-directed antibodies, the Ndh complex in maize was found at 550 and ∼1000 kDa, and subcomplexes were found at 300 and 250 kDa (18).
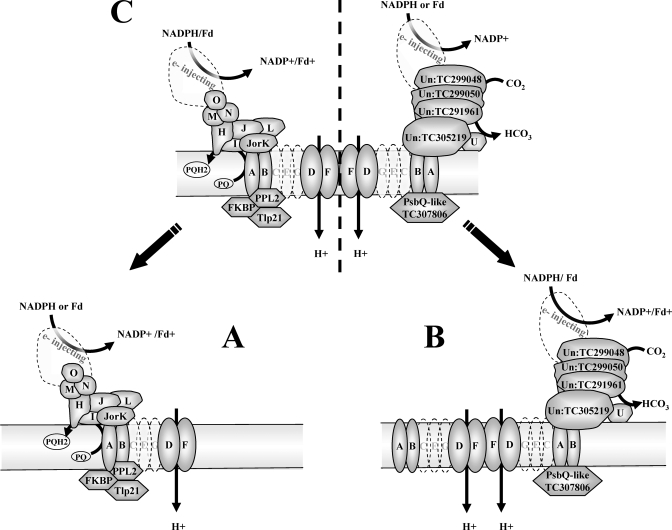
Fig. 5.
Schematic representation of the suggested oligomeric organization of the NDH complex and potential interacting proteins. Co-migration of proteins was deduced from peaks observed on 1D BN profiles (see also supplemental Tables 3 and 4 and examples of profiles in Fig. 4C). A, the ∼550-kDa complex represents the NDH monomer with the connecting complex (the classical L-shaped complex observed for mitochondrial complex I) and three associated lumenal proteins (PPL2 (TC289659), Tlp20 (TC295339), and FKBP (TC298326)). B, the ∼800-kDa complex represents a dimeric membrane-embedded complex with the associated new complex postulated to be involved in CCM. C, the ∼1000-kDa complex represents the U-shaped NDH complex with both the connecting complex and the newly associated complex. Possible electron donors, CO2 hydration, and proton translocation are indicated. Un, unknown
We observed that a number of additional BS enriched proteins co-migrated with NDH oligomeric assemblies (in both 1D and 2D BN). In particular, putative NdhL protein (AI0012470), Tlp20 (TC295339), and FKBP (TC298326) co-migrated with the NDH complexes at 1000, 650, and 550 kDa. The major accumulation peak for these three proteins was found at ∼550 kDa (Figs. 3 and 4 and supplemental Table 4). The 2D BN gel analysis showed TLP20 co-migrating with the ∼550-kDa NDH complex, whereas FKBP was found in the same spot (spot 192) as NdhJ and PPL2.
Interestingly a second set of four unknown proteins (TC291961, TC299050, TC299048, and TC305219) and a ferredoxin (Fd) domain protein (TC307918) showed 1D BN profiles very similar to those of NdhD and -F with peaks at ∼1000, ∼800, and ∼320 kDa with a predominant and well defined peak at ∼800 kDa (band 4) (Fig. 4, B and C, and supplemental Tables 3,4). Surprisingly abundant, this ∼800-kDa complex was also observed on 2D BN gels (Fig. 1, complex III). An additional ZmGI accession (TC234236) similar to TC299050 was identified on 2D BN at ∼1000 kDa (Fig. 1A, spot 207) also preferentially expressed in the BS (Table I). TC299050 and TC299048 are similar to Arabidopsis proteins AT1G15980 and AT1G64770. These Arabidopsis proteins were identified in the chloroplast (73) and thylakoids (59) but have no known function and no clear predicted functional domains. The strong clustering of BS/M ratios between these proteins and the NDH subunits and their co-migration in three different native assemblies suggest that they constitute interacting partners of the NDH complex (Fig. 5, B and C).
A recent study in Arabidopsis proposed that the PIFI protein (AT3G15840) (postillumination fluorescence increase protein) was a new and essential component of Ndh (74). We identified a maize homologue of PIFI (TC279329) with a BS/M ratio of 1.88; however, it was not associated with NDH complex assemblies. A maize homologue (TC307307) of the Arabidopsis IMMUTANS (IM) protein was identified only once in this study and only in BS membranes. IM is a chloroplast alternative oxidase and is likely to be the elusive terminal oxidase in chlororespiration. IM appears to be a versatile electron sink especially early in chloroplast development, and its inactivation in Arabidopsis leads to leaf variegation (75). Our data suggest surprisingly low accumulation levels as compared with NDH.
Regulation of Light Harvesting Capacity—
Reversible phosphorylation of light-harvesting antennae LHCII and subunits of PSII regulate the state transitions (76, 77) and the PSII repair cycle (57), respectively. State transitions involve reversible interactions of LHCII with PSII and PSI, thus changing the balance of the light harvesting capacity of the two photosystems, and involve lateral movement of LHCII antennae proteins. Two Arabidopsis thylakoid serine-threonine kinases, STN8 and STN7 (78–80), have been identified. Whereas STT7 and STN7 phosphorylate LHCII and are required for state transitions, STN8 specifically phosphorylates the N-terminal threonine residues of PSII core subunits D1 and D2 and CP43 proteins and Thr-4 in the PsbH protein (80).
We identified maize homologues of both STN7 (TC305194) and STN8 (TC311729) kinases. Both proteins were only identified in the M proteome suggesting enrichment in the M thylakoid (we note that STN8 was identified in all three replicates with good scores; STN7 was only identified in one replicate; see supplemental Table 7). In addition, 1D BN profiles indicated that STN7 migrates as a dimer (∼110 kDa) or associated as a monomer with its target, the LHCII trimer (supplemental Table 3). STN8 migrated in two complexes (∼180–200 and ∼100 kDa).
A soluble thylakoid phosphoprotein TSP9 was shown to interact with PSII antennae as well as the PSI and PSII cores, and it was suggested to play a role in regulation of light harvesting capacity (81). We identified a maize homologue of TSP9 (TC312266) that was more highly expressed in M thylakoids (BS/M = 0.45), and its 1D BN profile showed peaks in bands 10 (∼500 kDa) and 13 (∼300 kDa) with the majority found as monomers (in band 24). We also identified homologues of Arabidopsis PGRL1 protein (82) involved in NDH-independent cyclic electron flow, one accumulating with a very pronounced ratio in the M thylakoid (TC312147; BS/M = 0.26) and the other in the BS membranes (TC305270; BS/M = 6.58).
The Chloroplast Expression Apparatus—
The plastid chromosome binds to the inner envelope and thylakoid membranes (83). We quantified four maize homologues of Arabidopsis proteins found to interact with the chloroplast chromosome. The Arabidopsis matrix attachment filament protein 1 (MFP1; AT3G16000) is a coiled coil protein that is believed to anchor the plastid chromosome to thylakoids (84). We quantified MFP1-like proteins (TC304489 and TC296233) with BS/M ratios of 1.9 and 2.4, respectively. These proteins are present in high molecular mass complexes of ∼700, ∼500, and ∼250 kDa. We also quantified maize homologues of TCP34 (TC312665) and pTAC16 (TC281749) with BS/M ratios of 0.43 and 0.54, respectively. TCP34 is a tetratricopeptide repeat (TPR) protein found in association with a transcriptionally active protein-DNA complex (TAC) from chloroplasts; a regulatory function of TCP34 in plastid gene expression was proposed (85). pTAC16 was identified in TACs from A. thaliana and mustard chloroplasts and was shown to be involved in plastid gene expression (25).
We identified 27 chloroplast ribosomal subunits (mostly 50 S subunits) either only identified in M membranes or identified with a low BS/M accumulation ratio (average 0.5). Also plastid ribosome-associated proteins (PSRPs) showed M preferential accumulation, including PSRP-7 (TC295920 and TC280502), PSRP-2 (TC312649), and elongation factor Tu (TC298927 and TC298925). Two accessions (TC281611 and TC281610) corresponding to the ribosome recycling factor yielded opposite expression ratios (3.0 and M only, respectively), suggesting specialization according to cell type. Chloroplast-encoded membrane proteins are synthesized on ribosomes docked onto the thylakoid membrane (86). It is the 50 S rather than the 30 S ribosomal particle that makes the interaction to the thylakoid membrane (via the Sec translocon); this is consistent with the higher number of identified 50 S subunits.
Chloroplast Protein Import, Membrane Biogenesis, and Protein Targeting—
Nuclear-encoded chloroplast proteins are imported via the Toc-Tic complex, consisting of several outer and inner membrane proteins (33). Components of the Tic complex consistently showed preferential M accumulation (average BS/M of 0.46 or only in M), namely Tic110 (TC300617 and TC300618), Tic21 (TC307215 and TC293198), and Tic40 (TC310831 and TC310825) (Table I). Tic110 was identified in a 170-kDa complex with its Tic40 and Tic21 partners in both 2D BN and label-free native profiles. An additional complex of lower intensity also was observed for these three Tic subunits around 500 kDa. The major outer envelope preprotein receptor Toc159 (TC300201) was also identified, but there was not enough information for quantification (supplemental Table 7). Assuming a similar envelope/thylakoid ratio, the enrichment for Tic components in the M chloroplast suggests more active protein import as compared with BS chloroplasts. It is important to note that other inner envelope proteins, not involved in protein import, are clearly enriched in the BS samples, e.g. chloroplast envelope quinone oxidoreductase (TC311227) (87) with a BS/M ratio of 3.7, HP45 unknown function (TC314108), and the 2-oxoglutarate/malate translocator (DIT2; TC282161), indicating that the low BS/M ratio for Tic components cannot be explained by the amount of envelopes in the purified membranes.
We identified two maize proteins that are homologues to Arabidopsis proteins TF1 (thylakoid formation 1) (88) and VIPP1 (vesicle-inducing protein in plastids) (89), both involved in thylakoid membrane formation. Loss of VIPP1 expression is deleterious to thylakoid membrane formation (89), and VIPP1 appears to be needed for formation of the thylakoid bilayer rather than the functional assembly of thylakoid protein complexes (90). Maize TF1 (TC285636) was only identified with a low BS/M ratio (0.61), whereas maize VIPP1 (TC281010) protein was identified with a high BS/M ratio (2.67) in very high molecular mass complexes (band 1) and as monomers.
Several pathways for insertion and translocation of thylakoid proteins have been discovered in C3 and C4 plants (91, 92). The Sec pathway involving cpSecA,Y,E is used for insertion of both chloroplast- and nucleus-encoded proteins (92, 93). Two accessions for cpSecY (TC288462 and NP004391) were quantified with quite different BS/M ratios of 0.82 and 2.51, respectively (Table I). Other intrachloroplast targeting components such as TatC (TC300579) and two accessions for cpSecA were identified but could not be quantified (supplemental Table 7), whereas cpSRP54 (TC306667) was identified only in the M. The expression of the thylakoid signal peptidase TPP2 (TC279895) involved in cleavage of lumenal transit peptides showed a BS/M of 1.3.
DnaJ Domain Proteins and Immunophilins—
DnaJ proteins are best known as co-chaperones and interacting partners of Hsp70 (DnaK) and GrpE chaperones. The Arabidopsis genome encodes about 90 DnaJ domain proteins (94), and several have been identified in Arabidopsis thylakoid membranes (19). The function of some DnaJ domain proteins in chloroplasts have been determined and include ARC6 involved in plastid division (95), maize BSD2 involved in biogenesis of the Rubisco complex (2), and the Orange 1 protein (Or1) in Brassica oleracea required for carotenoid accumulation in chromoplasts (96).
We identified and quantified the BS/M accumulation patterns for several maize DnaJ domain proteins. Accessions TC292740, TC300370, and TC300372 were enriched in M membranes, whereas TC296329 and TC297356 were strongly enriched in BS membranes. 1D BN profiles indicated that all of these DnaJ proteins are present in high molecular mass complexes at ∼580 kDa (band 8) and ∼650 kDa (band 6) situated just above and below the abundant ∼600-kDa (band 7) band containing PSI-LHCI and PSII dimers. A maize homologue of Or1 (TC288173) was found preferentially expressed in the BS (BS/M = 2.65) and was only found as a monomer (or small complex).
We identified nine thylakoid-localized peptidyl-prolyl isomerases, corresponding to five known Arabidopsis thylakoid lumen proteins. Two of these Arabidopsis proteins, TLP20 and TLP40, are believed to hold most of the isomerase activity (97). In particular the TLP20 homologues appear to be relatively abundant in the maize thylakoid proteome based on their high cumulative Mowse scores (up to 14,000) and spectral counts (up to 600). Two maize homologues (TC295339 and TC295338) of the isomerase protein TLP20 in Arabidopsis had clear preferential BS accumulation (BS/M = 2.3 and 1.9), whereas the maize homologues of TLP40 (TC298326 and TC289507) showed preferential accumulation in the M thylakoid. This M enrichment for TLP40 is in line with its proposed role in light-mediated protein phosphorylation of PSII subunits (98). Also FKBP-type protein (BM499069) showed preferential accumulation in M (BS/M = 0.78), but its role is unknown. Peptidyl-prolyl cis-trans isomerases CO454223, BG321212, and TC296787 homologous to FKBP13 involved in the biogenesis of the Rieske cytb6f protein (99) all showed high BS/M ratios (1.7, 3.5, and 3.1, respectively).
PSII and PSI Assembly Factors—
We identified a maize homologue of the Arabidopsis “low PSII accumulation 1 protein” (LPA1) (TC312066) with good scores in all three replicates of M thylakoids but not in BS thylakoids. LPA1 has two TPR repeats and is implicated in the synthesis of the chloroplast-encoded D1 protein and assembly of PSII in A. thaliana (100). The observation of LPA1 in M thylakoids and not in BS thylakoids is consistent with its proposed role as a specific factor in PSII biogenesis. We also identified the maize homologue of Arabidopsis protein HCF136, shown to be involved in assembly of the PSII reaction center assembly factor (101) and also in maize (4). Maize HCF136 (TC296744) distributed equally between M and BS. The equal distribution of HCF136 suggests that HCF136 is not specific for PSII or that it is a remnant of the PSII synthesis and assembly machinery that was not (yet) lost during BS differentiation and PSII down-regulation. It is interesting to note that HCF136 in M membranes showed a predominant accumulation in a complex of ∼180 kDa (band 18), whereas HCF136 predominantly accumulated at ∼100 kDa (band 24) in BS membranes. This difference in oligomeric organization might indicate a partial loss of function in the absence of high levels of active PSII in the BS.
We identified two PSI assembly factors, YCF3 (TC293155) and YCF4 (NP_043035). YCF3 was identified in all three replicates in M (albeit with low scores and thus did not pass the filter for Table I) but not in BS, whereas YCF4 accumulated preferentially in the BS (BS/M = 2.7) and was identified with good scores. Both proteins are essential for the stability and accumulation, but not synthesis, of PSI in tobacco and Chlamydomonas reinhardtii (Refs. 102–104; for a review, see Ref. 30). Sucrose gradient fractionation and immunoblots indicate that YCF4 is part of a high molecular weight complex in C. reinhardtii that does not co-sediment with PSI subunits, whereas YCF3 is monomeric (103). Consistently we found that YCF4 accumulated in a ∼500 kDa complex (band 10, above PSI monomer) as well as at ∼220 kDa (band 16) and in low molecular weight fractions (regions below LHCIIs, bands 21–25). In contrast, maize YCF3 was found only in low molecular weight fractions, similar to that observed in Chlamydomonas (103). The contrasting BS/M accumulation patterns and oligomeric state of YCF3 and YCF4 suggest that they have different roles in PSI assembly.
We also quantified the expression of a maize homologue of Arabidopsis pale yellow green7 protein (PYG7; TC307174) with BS/M ratio of 1.53, which is similar to ratios observed for PSI subunits. PYG7 in A. thaliana is required for a specific accumulation of PSI subunits without affecting the accumulation of other photosynthetic complexes and was qualified as a PSI assembly factor (105). The protein contains a TPR motif, and it was only identified in low molecular weight complexes on 1D BN. Thylakoid-bound Rubredoxin A (RubA) was proposed to be specifically required for the assembly of the F(X) iron-sulfur cluster in PSI but was not needed for iron-sulfur clusters in other thylakoid proteins (e.g. Rieske protein). Homologues of RubA were detected in thylakoids of spinach and C. reinhardtii (106, 107). We identified a maize homologue of RubA (TC296284) expressed in both compartments (BS/M = 0.8).
A maize homologue of the Arabidopsis protein APE1 (acclimation of photosynthesis to environment 1) (TC299113) showed an equal distribution between BS and M membranes. A mutant in APE1 was isolated in a chlorophyll fluorescence screen for light acclimation mutants (108), and the protein was identified by proteomics in the thylakoid membrane of Arabidopsis (19).
Protein Degradation—
Protein degradation has a fundamental role in chloroplast development and maintenance as evidenced by strong phenotypes of chloroplast protease mutants in Arabidopsis (109, 110), and it is thus likely that proteases also play a role in BS/M differentiation. Chloroplast proteases in Arabidopsis include soluble ATP-dependent ClpP protease, thylakoid ATP-dependent FtsH metalloproteases, thylakoid SppA and EGY1, and members of the DegP family (109, 110).
Three maize accessions (TC287566, BE453757, and TC303156) with homology to thylakoid-localized SppA protease (111) accumulated preferentially in the M membranes (average BS/M ratio = 0.33). It has been suggested that this high light-inducible protease might target PSII and LHCII proteins in A. thaliana (111). We identified maize SppA mainly in a ∼500-kDa high molecular mass complex that could correspond to a dimeric form of a previously observed Arabidopsis SppA complex of 270 kDa (111). EGY1 was identified as a metalloprotease required in early chloroplast development (112), and we observed two EGY accessions (matching to EGY1 and -2) with high BS/M ratios (TC314564 and TC287793) of 3.23 and 4.9, respectively, suggesting substrates specifically enriched in BS thylakoids.
Various thylakoid-associated DegP protease are likely involved in degradation of the D1 protein (113–115). We identified lumenal DegP1 (TC294056) in low molecular weight fractions with a BS/M ratio of 0.56. Six accessions for FtsH proteases quantified by label-free analysis, iTRAQ, and 2D BN showed that FtsH protease is present in both BS and M chloroplasts with equal distribution (TC292185, TC292243, TC234343, TC286826, TC286827, and TC219258). FtsH is mainly present in a high molecular mass complex of ∼750 kDa and in ∼650- and ∼500-kDa complexes.
We did not identify any components of the ClpP protease complex, such as ClpP,R or ClpS (110). However, we did observe ClpC (TC281093), one of three closely related Clp chaperones, in a distinct major peak at ∼650 kDa (band 5) that could correspond to the predicted hexameric assembly. A putative oligopeptidase (TC294060) was identified with a high Mowse score (2200 in M and 103 in BS) and a BS/M ratio of 0.05. This is one of the most extreme ratios observed in our data set, suggesting a very specific function in M chloroplasts.
Redox and ROS Response Proteins—
Chloroplasts have an elaborate enzymatic system to detoxify ROS (116–119). M chloroplasts have high rates of linear photosynthetic electron transport, whereas BS chloroplasts mainly carry out cyclic electron flow. It is thus quite likely that there are substantial differences in the quantity and “quality” of ROS produced in these two chloroplast types. Indeed the majority of ROS defense proteins quantified in our study showed higher accumulation in M membranes than in BS membranes, similar to those observed for stromal M and BS maize proteins (8). For example, the thylakoid-bound APX (TC306001), the lumenal Peroxiredoxin Q (Prx-Q; TC306534), and two accessions for peripheral thylakoid protein 2-Cys Prx-B (TC304817 and TC304818) have BS/M ratios of 0.67, 0.14, 0.8, and 0.45, respectively. The stromal glutathione peroxidase 2 (GPX2; TC282772) had a similar BS/M ratio of 0.53. Interestingly PrxII-E (TC306147) seems to have a specialized BS function as indicated by its very high BS/M of 6.7. Neither the precise function nor the electron donor of PrxII-E is known in Arabidopsis (119).
The expression of ferritins in maize is regulated by abscisic acid in the case of Ferritin 2 (ZmFer2) and by oxidative stress in the case of ZmFer1 (for review, see Ref. 120). The preferential accumulation of Ferritin 1 (TC293195) in the M membrane is consistent with these observations and supports a role in iron chelation to reduce formation of reactive hydroxyl radicals by interaction of O2 with ferrous ions as suggested previously (120).
Calvin Cycle and C4 Markers—
Because the BS and M membrane fractions analyzed here were deliberately not washed with salts, abundant stromal enzymes involved in carbon fixation and C4-specific pathways could be identified and quantified in this study. As we mentioned earlier (Fig. 3B), these proteins provided an excellent test set to determine the consistency between the current study and our previous study regarding the BS and M maize stromal proteomes (8).
The specific Calvin cycle enzymes RBCL (NP_043033), RBCS (TC286735, TC286728, and TC286733), RCA (TC300568), and fructose-bisphosphate aldolase-2 (SFBA; TC285785) all showed strong BS accumulation. In agreement with our previous BS/M stromal analysis, we observed that the Calvin cycle enzymes involved in triose phosphate reduction, (phosphoglycerate kinase, NP539335, TC286022, and TC219625; glyceraldehyde-3-phosphate dehydrogenase A,B, TC292245 and TC292248; and triose-phosphate isomerase, TC279906) preferentially localized to M or were equally distributed over BS and M chloroplasts (Table I). Within the malate-pyruvate C4 shuttle, we observed strong M expression for PPDK (TC233444 and TC286559) and MDH (TC300313). Carbonic anhydrase (TC280282) was only identified in M membranes; this is consistent with activity assays (121). As expected the NADP-malic enzyme (TC280868) was only identified in the BS.
The activity of PPDK is regulated by a bifunctional kinase/phosphatase PPDK-regulatory protein (PPDK-RP) (122). This unusual enzyme uses ADP as source of Pi (123). Consistently we observed that adenylate kinase (ADK; DR786994, TC301869 and TC293517), which phosphorylates the AMP produced in this PPDK phosphorylation reaction, preferentially accumulated in the M. In agreement with our stromal analysis, we identified PPDK-RP (TC287496 in ZmGI v16.0) in M chloroplasts and not in BS chloroplasts consistent with the localization of its substrate PPDK. 1D BN profiles showed a co-migration of PPDK and PPDK-RP in high molecular mass complexes at >1000 kDa (band 1), ∼650 kDa (band 5), and ∼500 kDa (band 10) (not shown). The relative stoichiometry of PPDK and PPDK-RP appears to be conserved, strongly suggesting stable interaction. An additional ∼180-kDa (band 18) complex was observed for PPDK only, possibly representing homodimeric PPDK.
Translocation of Triose Phosphates and Starch—
M chloroplasts have to maintain high rates of export of phosphoenolpyruvate (PEP) (generated by PPDK) into the cytosol for oxaloacetate production by PEP carboxylase. Consistently we observed that the PEP/Pi translocator (PPT), located in the inner chloroplast envelope membrane, accumulated with a low BS/M ratio of 0.21 (TC286421). PPT homologues in Arabidopsis were shown to be expressed specifically in the mesophyll cells (PPT1) or veins (PPT2) and are required for formation of mesophyll tissue (124).
Interestingly we also identified two close maize homologues of inner envelope 2-oxoglutarate/malate translocator (Dit). TC282039 was closer to A. thaliana Dit1 (AT5G64290), TC282161 was closer to Dit2 (AT5G64280), and they preferentially accumulated in the M envelopes (BS/M = 0.2) and BS envelopes (BS/M = 1.7), respectively. This observation is consistent with previous observations of cell-specific transcript accumulation of Dit1,2 in sorghum (125) and maize (41). Consequently a two-translocator model for Dit1,2 was proposed (126) in which M-enriched Dit1 imports 2-oxoglutarate into chloroplast in exchange for malate, whereas BS enriched Dit2 imports malate and exports glutamate (for a review, see Ref. 127). In C3 plants, Dit1,2 serve to facilitate the photorespiratory pathway, and indeed the Dit2-null mutant is a classic photorespiratory mutant in Arabidopsis (125). Because photorespiration is strongly suppressed in maize (and other C4 species), the Dit translocators in maize essentially serve to drive malate from the M cell to the BS chloroplast.
The maize inner envelope maltose transporter Mex1 (TC302035) accumulated strongly in the BS membranes (BS/M ratio of 2.7). Arabidopsis Mex1 was shown to function as the main exporter of maltose to the cytosol (128) and is essential for carbohydrate export in Arabidopsis leaves at night. Its BS localization is consistent with previous localization of starch synthesis and degradation mechanisms in maize BS chloroplasts (see Ref. 8 for a discussion and references). Clearly, differentiation of envelope proteomes of BS and M chloroplasts participates in the adaptation to C4 photosynthesis.
Isoprenoid and Tetrapyrrole Pathways (Carotenoid, Plastoquinone, and Chlorophyll Biosynthesis)—
Within the initial steps of the carotenoid biosynthetic pathway, we found two isoforms of phytoene desaturase (TC306129 (BS/M = 1.41) and BG320144 (BS/M = 0.64)), a ζ-carotene desaturase (TC289473; BS/M = 0.42), and, further downstream, zeaxanthin epoxidase (TC292044; BS/M = 0.23). A β-hydroxylase (DT944642), a homologue of A. thaliana LUT5 (129), was identified in the M thylakoid. A maize homologue of Arabidopsis inner envelope membrane protein APG1 (TC304918) accumulated at higher levels in the M membranes. APG1 likely functions as an S-methionine methyltransferase in the methylation step of plastoquinone (PQ) biosynthesis (130).
We identified seven enzymes participating in the chlorophyl biosynthetic pathway. The initial steps in the pathway are carried out by soluble stromal enzymes and were not identified in the membrane samples in our current study. The most upstream enzyme in our study was envelope-associated Mg2+-protoporphyrin IX chelatase subunit H (CF244872 and TC287014) (CHLH, also named Gun5 in Arabidopsis), only identified in the M fraction with low scores. Mg-protoporphyrin IX monomethyl ester cyclase (TC299016), two steps further downstream, showed preferential BS accumulation (BS/M = 3.0). Homologues of this enzyme in Arabidopsis and C. reinhardtii are thylakoid- and envelope-associated CHL27 (131) and CRD1 (132), respectively. Two more steps further down in the pathway, we observed NADPH-protochlorophyllide oxidoreductase A (PORA; TC292871) with a BS/M ratio of 0.55. Geranylgeranyl reductase (TC311340) and chlorophyll synthase (TC300395) showed a BS/M ratio of 1.2 and 0.8, respectively. TC305891, a close homologue of the Arabidopsis (AT3G14110) FLU protein and responsible for negative feedback regulation within the tetrapyrrole pathway (133), accumulated with a BS/M ratio of 1. Finally maize lethal leaf spot protein (134), a pheophorbide a oxygenase (PaO; TC282596) involved in chlorophyll degradation, had a BS/M ratio of 1.26. Thus with exception of the CRD1/CHL27 homologue (see “Discussion”), enzymes in the chlorophyll metabolism were quite evenly distributed.
Jasmonic Acid Synthesis—
Jasmonic acid (JA) is an important hormone in plant development and defense, and we quantified the first two committed steps of JA synthesis. Phospholipase (Defective in Anther Dehiscence 1 protein or DAD1; TC293375), which performs the initial step in JA synthesis, showed a BS/M ratio of 0.38, whereas Lipoxygenase 2 (LOX2; TC298873), which carries out the second step in the JA biosynthetic pathway, also preferentially accumulated in M thylakoids (BS/M = 0.58).
The Plastoglobular Proteome—
Chloroplast plastoglobules (PGs) are thylakoid-associated lipoprotein particles; their size and number vary greatly during chloroplast differentiation, senescence, and abiotic stress (for a review, see Ref. 135). Analyses of PGs isolated from Arabidopsis thylakoids show that they have a unique proteome and are a transient storage site for e.g. prenyl quinones and tocopherol (vitamin E) (135). We quantified 11 maize accessions with homology to these Arabidopsis PG proteins. Two structural lipid coat proteins of the plastoglobulin family were expressed in the M (TC299786 and TC284028), whereas two others were preferentially expressed in the BS (TC281878 and TC307124). This family of proteins is believed to control PG size and stability, and in Arabidopsis the family members respond differentially to various environmental conditions and developmental stage (136). Additionally we identified four ABC1 kinases with ABC1K5 (TC281706) only found in M samples and ABC1K2 (TC296339) strongly expressed in the BS (BS/M = 4.8). The function of the kinases is unclear, but homologues in yeast mitochondria and in E. coli are believed to regulate quinone synthesis (for a brief discussion, see Ref. 23). The BS/M ratio of some other plastoglobuli components could be quantified and included a flavin reductase-like protein (TC305628) and an aldo/keto reductase-like protein (TC282614), both with preferential expression in M membranes (Table I).
Additional Quantified Proteins with Unknown Roles—
Numerous additional proteins without a known function or predicted functional domain were quantified, showing a wide expression between BS and M (Table I). Arabidopsis homologues for many of these proteins have been localized previously in the thylakoid or chloroplast envelope by proteomics (see the PPDB). The proteins with strong BS/M cell-specific accumulation patterns likely play a role in functional BS/M differentiation and represent new entry points to study C4 differentiation.
DISCUSSION
C4 photosynthesis has evolved multiple times among the angiosperms and involves partitioning of photosynthetic activities between morphologically and biochemically distinct BS and mesophyll M cells (5, 137). Although the basic principles and central players of C4 photosynthesis are established, a full characterization of BS- and M-specific chloroplast membrane proteomes is lacking but is needed to test hypotheses regarding the cell-specific BS and M chloroplast membrane functions and metabolic exchange. This study fills this void, determining the BS and M cell-specific differences in thylakoid and envelope proteome composition and assembly state and identifying many new maize membrane proteins. This shows that the evolution of C4 photosynthesis strongly affects the differentiation of M and BS chloroplast proteomes, adapting each proteome to their specialized roles.
To integrate the quantitative and qualitative data obtained in this study, we summarized several of our findings in Fig. 6. This provides a schematic representation of the differential expression of photosynthetic and auxiliary thylakoid functions between the M and the BS as well as an integrated organization of the C4 carbon fixation and main chloroplast metabolic pathways. In the following section we discuss and conceptualize our findings particularly in the context of follow-up experiments to address mechanisms of BS/M differentiation and with relevance to molecular engineering of C4 photosynthesis (138, 139).
Fig. 6.
Simplified overview of the differential expression of major thylakoid and envelope functions between the M and BS chloroplasts. Photosynthetic complexes as well as number of auxiliary photosynthetic and metabolic functions associated with the thylakoids and envelopes are listed. Protein BS/M accumulation ratios are represented as color-coding for M (BS/M < 0.5, dark blue; 0.6 < BS/M < 0.8, light blue) and for BS preferential accumulation (1.2 < BS/M < 2, light orange; BS/M > 2, purple). Proteins that did not show differential accumulation are represented in white (0.8 < BS/M < 1.2), and those that were not quantified are shown as transparent (dashed lines). Abbreviations are listed in Table I except for sedoheptulose-1,7-bisphosphatase (SBP), ribulose-5-phosphate isomerase (RPI), fructose bisphosphatase (FBP), dihydroxyacetone (DHAP), phosphoglycerate (PG), and oxaloacetate (OAA). The solid line indicates the cells walls of adjacent BS and M cells. The reduced amounts of PSII and NDH complexes in BS and M thylakoids, respectively, are not shown. NDH* (yellow-shaded area) represents a suggested organization of the U-shaped NDH complex containing a symmetric novel arm subcomplex (NDH*) that we speculate is involved in hydration of CO2 into HCO3−.
Differential Accumulation of PSI, II, cytb6f, and ATP Synthase and Regulation of PSII Levels—
When compared with M thylakoids, the BS thylakoids have a 55% reduced PSII content, unchanged ATP synthase content, and a 65% increased PSI and 33% increased cytb6f contents. The BS/M ratios for levels of PSII and PSI correspond well to BS/M ratios observed with Western blots against D1 (0.2–0.4), CP47 (0.35–0.8), and PsaA/B (1.1–1.4) (14). In terms of complex assembly, we did not observe any obvious differences for PSI, ATP synthase, and the cytb6f complex, but large PSII complexes were strongly underrepresented in the BS thylakoids and were below detection in the 2D BN gels. These large PSII complexes likely represent the active PSII complexes. We also note that accumulation of oxygen-evolving complex proteins was more strongly reduced to about 15% (12, 14). Based on some of the older literature (see the Introduction), one could have expected that the observed PSII protein levels in BS thylakoids would be even more reduced than observed here. Our data suggest that accumulation of PSII core proteins in BS thylakoids did not result in a proportional amount of active PSII complexes. This explains the strongly reduced linear electron transport rates in BS thylakoids as has been consistently shown in many previous studies (9–11). Thus the PSII activity is down-regulated in the BS thylakoids through reduced expression of the nucleus-encoded OEC proteins, whereas accumulation of the chloroplast-encoded proteins is less tightly controlled.
Many proteins are members of (small) gene families, and similar to our previous analysis of the BS- and M-specific stromal proteomes (8), we observed that different paralogues (e.g. for phosphoglycerate kinase) often show cell-specific accumulation. For instance, two LHCII members accumulated with higher BS/M ratios than the other 16 LHCII accessions. These BS enriched LHCII proteins could represent LHCII proteins preferentially interacting with PSI. In addition, several subunits of PSI (e.g. PsaE and PsaH) were represented by more than one accession, some of which showed differences in BS/M ratios. Based on observations of neofunctionalization between paralogues of PsaE and PsaH in Arabidopsis (140), this may suggest subtle differences in PSI composition between BS and M thylakoids, for instance to adapt to higher rates of cyclic electron flow in BS membranes. A high quality annotated maize genome sequence and additional experiments will be required to sort out these subtle but interesting differences.
Characterization of NDH Complexes and Associated Proteins: a Role for NDH in CO2 Concentration in Addition to Cyclic Electron Flow?—
Thylakoid-localized Ndh complex is proposed to play a role in chlororespiration and in cyclic electron flow around PSI in both C3 and C4 plants (Refs. 72, 141, and 142; for discussions, see Refs. 16 and 143). During chlororespiration in the dark (or light), the NDH complex oxidizes stromal metabolites and transfers these electrons to PSI (or IMMUTANS) in a process coupled to proton translocation to help generate ATP. The role of NDH in the light-driven cyclic electron flow is to accept electrons (likely via Fd rather than FNR (see Ref. 69)) from PSI and then cycle the electrons back to PSI via intersystem components in a process coupled to proton translocation to generate ATP (144). An alternative cyclic electron flow route does not involve NDH but requires PGR5, PGRL1, Fd, and PQ as well as the cytb6f complex (16, 82, 145, 146); all components were detected in maize thylakoids.
Our analysis identified eight of 11 chloroplast-encoded NDH subunits as well as homologues of four (putative) nucleus-encoded NDH subunits (NdhM, -N, and -O and PPL2) recently discovered in Arabidopsis (62, 69) and two putative maize homologues of Thermosynechococcus elongatus NdhL and NdhU. These 14 subunits accumulated with an average BS/M ratio of 3.0, which is in good agreement with Western blot data for NDH-H (16) and BS/M ratios observed for several other subunits (17, 18). These subunits likely form the L-shaped complex observed in cyanobacteria and complex I in mitochondria (Ref. 147 and references therein). In addition, we consistently found associated lumen-localized isomerases TLP20 and FKPB; together they likely function in protein folding, and the linkage to NDH possibly facilitates oxidation or reduction of lumenal proteins (148). The observed native masses of the NDH complex (550 kDa for the monomer and 1000 kDa for the dimer) and the various subcomplexes are consistent with published analyses (15, 17, 18). Three additional Arabidopsis proteins, CRR1, -4, and -6 (67, 149, 150), were shown to be involved in NDH biogenesis but were not considered bona fide NDH subunits, and we did not identity their homologues in the maize samples.
In cyanobacteria, specialized low CO2-induced thylakoid-localized NDH complexes (D3/F3 type) participate in carbon-concentrating mechanisms (CCMs) (151, 152). Recent analysis of their structural organization by single particle electron microscopy in T. elongatus (147) showed (i) an “L”-shaped complex composed of the connecting hydrophilic arm and the membrane-embedded subunits, ii) a “U”-shaped configuration with an additional hydrophilic arm, and (iii) a smaller complex on the lumenal side. The presence of the U-shaped NDH form is unlikely to be an artifact because milder purification conditions dramatically increased the population of U-shaped particles compared with L-shaped particles (147). In fact, the authors suggested that the U-shaped particle is the functional form of the NDH-1 in T. elongatus. The composition of the extra arm in the U-shaped NDH-1 complex is unknown, but genetics showed that ChpY (also named CupA) is an essential component in thylakoid-associated CCM (152). Surprisingly we identified an additional subcomplex in BS thylakoids with four identified proteins that was consistently found in higher molecular mass assemblies containing NDH subunits. Based on the presence of the U-shaped complex discussed above, we speculate that the 1000-kDa thylakoid NDH complex represents a functional homologue of the U-shaped NDH-1 complex in T. elongatus. Thus the additional abundant (peripheral) complex in the BS that we clearly observed on the 2D BN-PAGE gels as well as in the 1D BN analysis may be the functional equivalent of the extra “arm” involved in CO2 hydration in cyanobacteria. We speculate that this novel complex may be involved in hydration of CO2 into HCO3−, thus strongly reducing diffusion rates of inorganic carbon through chloroplast and cellular membranes and providing a reserve/buffer of inorganic carbon within the BS chloroplast (Fig. 6). This would prevent wasteful leakage of CO2 from the BS chloroplast, in particular when ME decarboxylation rates are higher than Rubisco carboxylation rates. A likely source of NADPH to perform this function would be the decarboxylation of malate by ME furnishing CO2 and NADPH in a 1:1 ratio. The benefits for C4 photosynthesis are clear, and this hypothesis is consistent with the low BS/M ratio for carbonic anhydrase (TC280282; only identified in the M with good scores). The preferential location of carbonic anhydrase in M chloroplasts may further be enhanced by the preferential location of lipid biosynthesis in M chloroplasts with a much lower internal CO2 concentration because HCO3− is needed as a co-substrate for acetyl-CoA carboxylase (153). Thus we suggest that maize may have retained a thylakoid-associated CMM that is best understood in cyanobacteria (152, 154), the progenitor of plant plastids.
Control and Regulation of Excitation Energy and ROS Defense—
The redox state, partial pressures of molecular oxygen, and the quality and quantity of the light absorption cross-sections are different between M and BS chloroplasts. Hence there must be BS- and M-specific differentiation to avoid light stress and optimize light harvesting capacity. This includes balancing excitation of PSI and PSII reaction centers through (de)phosphorylation-driven state transitions, non-photochemical quenching of excess excitation energy in the antennae, detoxification of triplet chlorophyll, and detoxification of the various ROS using enzymatic and non-enzymatic systems (116–119). We were able to quantify several membrane (-associated) proteins involved in these processes, and these data were complementary to findings of the BS and M stromal proteomes (8). In particular, the very low BS/M ratio for lumenal Prx-Q suggests a specific role in ROS defense in the M thylakoid lumen, whereas the very high BS/M ratio for PrxII-E on the stromal side of the thylakoid membrane suggests involvement with metabolic redox regulation in BS chloroplasts. The strong M enrichment of maize homologues of light stress proteins Ohp1,2, Lil4/Sep1, and PsbS suggests that their functions are related to PSII and/or that they are generally up-regulated under conditions of high linear electron transport. The preferential M accumulation of thylakoid kinases STN7 and STN8 and phosphoprotein TSP9 as well as lumenal isomerase TLP40 strongly suggests that M thylakoids retained phosphorylation-driven regulation of LHCII state transitions and PSII repair, whereas BS thylakoids have down-regulated these functions in the absence of significant levels of functional PSII.
Control of Protein Homeostasis in BS and M Chloroplast Differentiation—
To accommodate protein homeostasis in the two cell types, it is expected that the BS and M chloroplasts also display differences in the protein import and sorting, folding, assembly, and degradation machineries. Inspection of BS/M accumulation ratios for proteins involved in these different processes suggested indeed a number of divergences. High levels of import machinery in the inner envelope (Tic110,21,40) of M chloroplasts suggested an increased protein flux. Consistently, cpSRP45 involved in targeting of LCHC and chloroplast-encoded proteins (92) was enriched in M thylakoids. The low BS/M ratios for ribosomal and ribosome-associated proteins strongly suggest higher rates of translation (of thylakoid proteins) in M chloroplasts; this would be consistent with higher accumulation levels of PSII and high rates of damage and resynthesis of the D1 protein. This hypothesis is supported by protein 35S pulse labeling in the C4 plant Digitaria sanguinalis showing high rates of synthesis of D1 and D2 PSII subunits in the M, whereas only labeled RBCL is visible in BS chloroplasts (155).
Finally several thylakoid proteins were quantified that are believed to be involved in maturation or assembly of thylakoid complexes, such as lumenal isomerases. Of particular interest were maize homologues for Arabidopsis LPA1 and HCF136. Although we observed enrichment in the M membrane of LPA1 consistent with its role in synthesis of the D1 protein (100), HCF136 clearly involved in assembly of PSII reaction centers in Arabidopsis (101) and maize (4) showed a BS/M ratio close to 1, suggesting additional roles. Also interesting to note is that although PSII assembly factors Pyg7 and RubA have BS/M ratios similar to that of PSI itself, YCF3 and YCF4, have very different ratios. Thus the comparative analysis of BS and M thylakoids provides a good system to study the functions and specificities of various assembly factors.
Numerous studies in Arabidopsis indicated that thylakoid FtsH and Deg proteases are involved in the degradation of the D1 protein (113, 156, 157). However, we did not observe that the FtsH family was particularly enriched in either cell type, whereas lumenal DegP1 was nearly 2-fold higher in M thylakoids. Two maize homologues of Arabidopsis thylakoid protease EGY1 (112) and uncharacterized EGY2 showed more than 3–5-fold higher accumulation in BS membranes, whereas a putative oligopeptidase (TC294060) showed one of the lowest BS/M ratios observed in our data set, suggesting very specific functions in BS and M chloroplasts, respectively. Studies that follow cell type-specific accumulation of proteases along the developmental gradient could further explore the role of proteolysis in BS/M differentiation.
Cell-specific Differentiation of Chlorophyll, Carotenoid, and Quinone Biosynthetic Pathways—
The thylakoid membrane and its proteome contain a high number of various carotenoids and quinones (plastoquinone and phylloquinone) as well as tocopherol. The final steps in their biosynthesis occur at the thylakoids and/or PGs (135), and therefore one would expect that the abundance and/or activity of the enzymes in the BS or M membrane is proportional to demand. Demand is determined by the composition of the electron transport chain which suggests higher amounts of plastoquinone, carotenoids, and tocopherol in the M thylakoid. Indeed relevant biosynthetic enzymes were either only found in M membranes or found with low BS/M ratios. Although we quantified a number of PG proteins, it was particularly interesting to observe very divergent BS/M ratios in the family of ABC1 kinases. In particular ABC1K2 showed a very high BS/M ratio, whereas ABC1K5 was only found in M membranes with good scores. The function of the ABC1 kinase family is unknown, but homologues in E. coli and yeast are involved in regulation of (ubi)quinone synthesis (see Ref. 23). The observed BS/M differentiation provides an excellent opportunity to further analyze ABC1K and PG function.
In contrast, enzymes in chlorophyll synthesis and degradation were equally distributed over the two cell type with the exception of BS enriched CHL27, which requires molecular oxygen and NADPH in the cyclase reaction. Because the O2 concentration is lower in BS than in M, this higher BS accumulation may serve to compensate for slower reaction kinetics and is consistent with enhanced expression of homologue CRD1 in C. reinhardtii when grown under low O2 (158).
Regulation of BS/M Differentiation—
The molecular basis for C4 differentiation is still poorly understood, but it has been established that it includes nuclear transcriptional regulation through DNA regulatory elements, transcription factors, histone modifications, and likely also metabolic signals (5, 159). Nuclear gene expression is also influenced by signals from the plastid, but the actual signals have not been identified conclusively (160, 161). However, it is clear that multiple components within the plastid contribute to plastid-nucleus signaling (in both C3 and C4 plants), possibly integrated in a single pathway (via pentatricopeptide repeat protein GUN1) when the signal is “leaving” the plastid (162). These plastid components/contributions have been suggested to include (i) the redox state of the PQ pool and possible other redox-active components in the thylakoid and stroma, (ii) translational activity within the plastid, and (iii) intermediates of the tetrapyrrole pathway. It is conceivable that intraplastid signaling also involves phosphorylation cascades, various ROS species, and other small molecules.
We explored our data for possible evidence of differences in BS and M plastid-nucleus signaling, and indeed we did observe several components and processes that have been implicated in signaling. Within the tetrapyrrole/chlorophyll synthesis in particular CHL27 stands out for its high BS/M ratio. Clearly the BS and M differences in linear electron flow rates, tightly intertwined with differences in the ROS defense system, possibly provide a good starting point for differential plastid-nucleus signaling. Also redox equivalents imported via the malate shuttle and possibly redistributed via chlororespiration (involving the NDH complex) are likely to contribute to plastid-nucleus signaling. Although these components may help to complete the BS/M differentiation process and to enforce the differentiated state (the status quo), they are not the primary event in the differentiation process. The search for components involved in the primary steps in BS/M differentiation should involve leaf zones closer to the base of the leaves. It is interesting to note that when the plastid PsbA gene encoding for the D1 protein was deleted in tobacco the NDH complex was up-regulated 4-fold (163). This suggests that parts of regulatory networks or signal transduction chains involved in C4 chloroplast differentiation maybe be similar to those in C3 plants, also underlining the notion that genes necessary for C4 photosynthesis are present in C3 species (137). Finally two isoforms of haloacid dehalogenase were identified, one with no differential expression and the other with strong expression in the BS. In recent years, metabolism of the disaccharide trehalose in plants has received a lot of attention in particular because trehalose 6-phosphate is involved in the regulation of developmental and metabolic processes (164–167). A possible function of the trehalose pathway in BS and M differentiation has yet to be explored.
Conclusions and Challenges—
The current study complements our previous stromal analysis, completes a first quantitative overview of the differentiated state of M and BS chloroplasts, and shows a fascinating number of differences to meet the demand for carbon and energy as well as (re)distribution of metabolites between C4 photosynthetic compartments. The identification and quantification of hundreds of chloroplast proteins integrates observations from many studies over the last few decades. Moreover our study provides many new entry points to study the mechanisms and consequences of C4 differentiation in maize and proposes a new function for the NDH complex in C4 plants. A logical next step will be to study the kinetics of BS/M differentiation along the leaf gradient from base to tip; this is needed to establish the sequence of events. Our study also demonstrated the strength of label-free quantification using a highly sensitive, fast MS instrument with high mass accuracy, the LTQ-Orbitrap. Finally a completely sequenced maize genome, high quality assembly, and gene discovery will be needed to refine this analysis. Our MS/MS data set can help to annotate gene models as we demonstrated earlier on a small scale for Arabidopsis (20) and as demonstrated in large scale studies in other organisms such as Drosophila melanogaster (168).
Supplementary Material
Footnotes
Published, MCP Papers in Press, May 2, 2008, DOI 10.1074/mcp.M800016-MCP200
The abbreviations used are: BS, bundle sheath; M, mesophyll; ME, NADP-malic enzyme; PS, Photosystem; cyt, cytochrome; LHC, light-harvesting complex; NDH, NAD(P)H dehydrogenase; FNR, ferredoxin-NAD(P)+ reductase; Fd, ferredoxin; ROS, reactive oxygen species; LTQ, linear ion trap triple quadrupole; PPDB, Plant Proteomics Database; BN, blue native; β-DM, β-n-dodecyl maltoside; FA, formic acid; cv, coefficient of variation; PPDK, pyruvate, phosphate dikinase; MDH, malate dehydrogenase; GAPB, glyceraldehyde dehydrogenase subunit B; RBCL, Rubisco large subunit; RBCS, Rubisco small subunit; RCA, Rubisco activase; PG, plastoglobule; PEP, phosphoenolpyruvate; PQ, plastoquinone; TPR, tetratricopeptide repeat; TAC, transcriptionally active chromosome; PSRP, plastid ribosome-associated protein; Rubisco, ribulose-bisphosphate carboxylase/oxygenase; iTRAQ, isobaric tags for relative and absolute quantification; 2D, two-dimensional; 1D, one-dimensional; EST, expressed sequence tag; TIGR, The Institute for Genomic Research; Mowse, molecular weight search; ZmGI, Z. mays Gene Index; FPR, false positive rate; BLAST, Basic Local Alignment Search Tool; OEC, oxygen-evolving complex; FKBP, FK506-binding protein; CRR, chlororespiratory reduction; PIFI, postillumination fluorescence increase; IM, IMMUTANS; MFP1, matrix attachment filament protein 1; LPA1, low PSII accumulation 1 protein; PYG7, pale yellow green7 protein; RubA, Rubredoxin A; APX, ascorbate peroxidase; Prx, Peroxiredoxin; PPDK-RP, PPDK-regulatory protein; ADK, adenylate kinase; PPT, PEP/Pi translocator; JA, jasmonic acid; CCM, carbon-concentrating mechanism.
B. Zybailov, unpublished data.
Q. Sun, B. Zybailov, and K. J. van Wijk, unpublished data.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Hall, L. N., Rossini, L., Cribb, L., and Langdale, J. A. ( 1998) GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10, 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roth, R., Hall, L. N., Brutnell, T. P., and Langdale, J. A. ( 1996) bundle sheath defective2, a mutation that disrupts the coordinated development of bundle sheath and mesophyll cells in the maize leaf. Plant Cell 8, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brutnell, T. P., Sawers, R. J., Mant, A., and Langdale, J. A. ( 1999) BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11, 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covshoff, S., Majeran, W., Liu, P., Kolkman, J. M., van Wijk, K. J., and Brutnell, T. P. ( 2008) Deregulation of maize C4 photosynthetic development in a mesophyll cell-defective mutant. Plant Physiol. 146, 1469–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheen, J. ( 1999) C-4 gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 187–217 [DOI] [PubMed] [Google Scholar]
- 6.Edwards, G. E., Franceschi, V. R., Ku, M. S., Voznesenskaya, E. V., Pyankov, V. I., and Andreo, C. S. ( 2001) Compartmentation of photosynthesis in cells and tissues of C4 plants. J. Exp. Bot. 52, 577–590 [PubMed] [Google Scholar]
- 7.Langdale, J. A. ( 1998) Cellular differentiation in the leaf. Curr. Opin. Cell Biol. 10, 734–738 [DOI] [PubMed] [Google Scholar]
- 8.Majeran, W., Cai, Y., Sun, Q., and van Wijk, K. J. ( 2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17, 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo, K. C., Anderson, J. M., Boardman, N. K., Downton, W. J., Osmond, C. B., and Thorne, S. W. ( 1970) Deficient photosystem II in agranal bundle sheath chloroplasts of C4 plants. Proc. Natl. Acad. Sci. U. S. A. 67, 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardt, H., and Kok, B. ( 1978) Comparison of photosynthetic activities of spinach chloroplasts with those of corn mesophyll and corn bundle sheath tissue. Plant Physiol. 62, 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuster, G., Ohad, I., Martineau, B., and Taylor, W. C. ( 1985) Differentiation and development of bundle sheath and mesophyll thylakoids in maize. Thylakoid polypeptide composition, phosphorylation, and organization of photosystem II. J. Biol. Chem. 260, 11866–11873 [PubMed] [Google Scholar]
- 12.Oswald, A., Streubel, M., Ljungberg, U., Hermans, J., Eskins, K., and Westhoff, P. ( 1990) Differential biogenesis of photosystem-II in mesophyll and bundle-sheath cells of ‘malic’ enzyme NADP+-type C4 plants. A comparative protein and RNA analysis. Eur. J. Biochem. 190, 185–194 [DOI] [PubMed] [Google Scholar]
- 13.Bassi, R., and Simpson, D. J. ( 1986) Differential expression of LHCII genes in mesophyll and bundle sheath cells of maize. Carlsberg Res. Commun. 51, 363–370 [Google Scholar]
- 14.Meierhoff, K., and Westhoff, P. ( 1993) Differential biogenesis of photosystem II in mesophyll and bundle sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: the non-stoichiometric abundance of the subunits of photosystem II in the bundle sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 191, 23–33 [Google Scholar]
- 15.Funk, E., Schäfer, E., Steinmüller, K. ( 1999) Characterization of the complex I-homologous NAD(P)H-plastoquinone-oxidoreductase (NDH-complex) of maize chloroplasts. J. Plant Physiol. 154, 16–23 [Google Scholar]
- 16.Takabayashi, A., Kishine, M., Asada, K., Endo, T., and Sato, F. ( 2005) Differential use of two cyclic electron flows around photosystem I for driving CO2-concentration mechanism in C4 photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 102, 16898–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darie, C. C., De Pascalis, L., Mutschler, B., and Haehnel, W. ( 2006) Studies of the Ndh complex and photosystem II from mesophyll and bundle sheath chloroplasts of the C4-type plant Zea mays. J. Plant Physiol. 163, 800–808 [DOI] [PubMed] [Google Scholar]
- 18.Darie, C. C., Biniossek, M. L., Winter, V., Mutschler, B., and Haehnel, W. ( 2005) Isolation and structural characterization of the Ndh complex from mesophyll and bundle sheath chloroplasts of Zea mays. FEBS J. 272, 2705–2716 [DOI] [PubMed] [Google Scholar]
- 19.Peltier, J. B., Ytterberg, A. J., Sun, Q., and van Wijk, K. J. ( 2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279, 49367–49383 [DOI] [PubMed] [Google Scholar]
- 20.Peltier, J. B., Emanuelsson, O., Kalume, D. E., Ytterberg, J., Friso, G., Rudella, A., Liberles, D. A., Soderberg, L., Roepstorff, P., von Heijne, G., and van Wijk, K. J. ( 2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., and Wijk, K. J. ( 2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert, M., Petersson, U. A., Haas, B. J., Funk, C., Schröder, W. P., and Kieselbach, T. ( 2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277, 8354–8365 [DOI] [PubMed] [Google Scholar]
- 23.Ytterberg, A. J., Peltier, J. B., and van Wijk, K. J. ( 2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts; a surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140, 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidi, P. A., Kanwischer, M., Baginsky, S., Austin, J. R., Csucs, G., Dormann, P., Kessler, F., and Brehelin, C. ( 2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281, 11225–11234 [DOI] [PubMed] [Google Scholar]
- 25.Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K. J., and Oelmuller, R. ( 2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Wijk, K. J. ( 2004) Plastid proteomics. Plant Physiol. Biochem. 42, 963–977 [DOI] [PubMed] [Google Scholar]
- 27.Meurer, J., Meierhoff, K., and Westhoff, P. ( 1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198, 385–396 [DOI] [PubMed] [Google Scholar]
- 28.Leister, D. ( 2003) Chloroplast research in the genomic age. Trends Genet. 19, 47–56 [DOI] [PubMed] [Google Scholar]
- 29.Stern, D. B., Hanson, M. R., and Barkan, A. ( 2004) Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends Plant Sci. 9, 293–301 [DOI] [PubMed] [Google Scholar]
- 30.Rochaix, J. D. ( 2004) Genetics of the biogenesis and dynamics of the photosynthetic machinery in eukaryotes. Plant Cell 16, 1650–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann, M., Tse Sum Bui, B., Wolff, M., Miginiac-Maslow, M., and Rohmer, M. ( 2006) Isoprenoid biosynthesis in plant chloroplasts via the MEP pathway: direct thylakoid/ferredoxin-dependent photoreduction of GcpE/IspG. FEBS Lett. 580, 1547–1552 [DOI] [PubMed] [Google Scholar]
- 32.Soll, J., and Schleiff, E. ( 2004) Protein import into chloroplasts. Nat. Rev. Mol. Cell. Biol. 5, 198–208 [DOI] [PubMed] [Google Scholar]
- 33.Kessler, F., and Schnell, D. J. ( 2006) The function and diversity of plastid protein import pathways: a multilane GTPase highway into plastids. Traffic 7, 248–257 [DOI] [PubMed] [Google Scholar]
- 34.Weber, A. P., Schwacke, R., and Flugge, U. I. ( 2005) Solute transporters of the plastid envelope membrane. Annu. Rev. Plant Biol. 56, 133–164 [DOI] [PubMed] [Google Scholar]
- 35.Joyard, J., Teyssier, E., Miege, C., Berny-Seigneurin, D., Marechal, E., Block, M. A., Dorne, A. J., Rolland, N., Ajlani, G., and Douce, R. ( 1998) The biochemical machinery of plastid envelope membranes. Plant Physiol. 118, 715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarvis, P. ( 2003) Intracellular signalling: the language of the chloroplast. Curr. Biol. 13, R314–R316 [DOI] [PubMed] [Google Scholar]
- 37.Block, M. A., Douce, R., Joyard, J., and Rolland, N. ( 2007) Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth. Res. 92, 225–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferro, M., Salvi, D., Riviere-Rolland, H., Vermat, T., Seigneurin-Berny, D., Grunwald, D., Garin, J., Joyard, J., and Rolland, N. ( 2002) Integral membrane proteins of the chloroplast envelope: Identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. U. S. A. 99, 11487–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferro, M., Salvi, D., Brugiere, S., Miras, S., Kowalski, S., Louwagie, M., Garin, J., Joyard, J., and Rolland, N. ( 2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2, 325–345 [DOI] [PubMed] [Google Scholar]
- 40.Froehlich, J. E., Wilkerson, C. G., Ray, W. K., McAndrew, R. S., Osteryoung, K. W., Gage, D. A., and Phinney, B. S. ( 2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Proteome Res. 2, 413–425 [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi, Y., Nagasaki, J., Kawasaki, M., Miyake, H., Sugiyama, T., and Taniguchi, M. ( 2004) Differentiation of dicarboxylate transporters in mesophyll and bundle sheath chloroplasts of maize. Plant Cell Physiol. 45, 187–200 [DOI] [PubMed] [Google Scholar]
- 42.Goshe, M. B., and Smith, R. D. ( 2003) Stable isotope-coded proteomic mass spectrometry. Curr. Opin. Biotechnol. 14, 101–109 [DOI] [PubMed] [Google Scholar]
- 43.Domon, B., and Aebersold, R. ( 2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 44.Rolland, N., Ferro, M., Ephritikhine, G., Marmagne, A., Ramus, C., Brugiere, S., Salvi, D., Seigneurin-Berny, D., Bourguignon, J., Barbier-Brygoo, H., Joyard, J., and Garin, J. ( 2006) A versatile method for deciphering plant membrane proteomes. J. Exp. Bot. 57, 1579–1589 [DOI] [PubMed] [Google Scholar]
- 45.Komatsu, S., Konishi, H., and Hashimoto, M. ( 2007) The proteomics of plant cell membranes. J. Exp. Bot. 58, 103–112 [DOI] [PubMed] [Google Scholar]
- 46.Olsen, J. V., de Godoy, L. M., Li, G., Macek, B., Mortensen, P., Pesch, R., Makarov, A., Lange, O., Horning, S., and Mann, M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 47.Makarov, A., Denisov, E., Kholomeev, A., Balschun, W., Lange, O., Strupat, K., and Horning, S. ( 2006) Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 [DOI] [PubMed] [Google Scholar]
- 48.Scigelova, M., and Makarov, A. ( 2006) Orbitrap mass analyzer—overview and applications in proteomics. Proteomics 6, Suppl. 2, 16–21 [DOI] [PubMed] [Google Scholar]
- 49.Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J., and Klenk, D. C. ( 1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 50.Schagger, H., Cramer, W. A., and von Jagow, G. ( 1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 51.Schagger, H., and von Jagow, G. ( 1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 52.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 53.Sokal, R., and Rohlf, F. J. ( 1995) Biometry, pp. 685–793, W. H. Freeman and Co., New York
- 54.Zhang, B., VerBerkmoes, N. C., Langston, M. A., Uberbacher, E., Hettich, R. L., and Samatova, N. F. ( 2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 5, 2909–2918 [DOI] [PubMed] [Google Scholar]
- 55.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A., and Ahn, N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 56.Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L. A., Rhee, S. Y., and Stitt, M. ( 2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 57.Aro, E. M., Suorsa, M., Rokka, A., Allahverdiyeva, Y., Paakkarinen, V., Saleem, A., Battchikova, N., and Rintamaki, E. ( 2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56, 347–356 [DOI] [PubMed] [Google Scholar]
- 58.Heinemeyer, J., Eubel, H., Wehmhoner, D., Jansch, L., and Braun, H. P. ( 2004) Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry 65, 1683–1692 [DOI] [PubMed] [Google Scholar]
- 59.Zybailov, B., Rutschow, H., Friso, G., Rudella, A., Emanuelsson, O., Sun, Q., and van Wijk, K. J. ( 2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zieske, L. R. ( 2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J. Exp. Bot. 57, 1501–1508 [DOI] [PubMed] [Google Scholar]
- 61.Rudella, A., Friso, G., Alonso, J. M., Ecker, J. R., and van Wijk, K. J. ( 2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18, 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishihara, S., Takabayashi, A., Ido, K., Endo, T., Ifuku, K., and Sato, F. ( 2007) Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145, 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sheen, J. Y., and Bogorad, L. ( 1986) Differential expression of six light-harvesting chlorophyll a/b binding-protein genes in maize leaf cell-types. Proc. Natl. Acad. Sci. U. S. A. 83, 7811–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niyogi, K. K., Li, X. P., Rosenberg, V., and Jung, H. S. ( 2005) Is PsbS the site of non-photochemical quenching in photosynthesis? J. Exp. Bot. 56, 375–382 [DOI] [PubMed] [Google Scholar]
- 65.Holt, N. E., Fleming, G. R., and Niyogi, K. K. ( 2004) Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43, 8281–8289 [DOI] [PubMed] [Google Scholar]
- 66.Sazanov, L. A., Burrows, P., and Nixon, P. J. ( 1996) Detection and characterization of a complex I-like NADH-specific dehydrogenase from pea thylakoids. Biochem. Soc. Trans. 24, 739–743 [DOI] [PubMed] [Google Scholar]
- 67.Shimizu, H., and Shikanai, T. ( 2007) Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52, 539–547 [DOI] [PubMed] [Google Scholar]
- 68.Kubicki, A., Funk, E., Westhoff, P., and Steinmuller, K. ( 1996) Differential expression of plastome-encoded ndh genes in mesophyll and bundle-sheath chloroplasts of the C4 plant Sorghum bicolor indicates that the complex I-homologous NAD(P)H-plastoquinone oxidoreductase is involved in cyclic electron transport. Planta 199, 276–281 [Google Scholar]
- 69.Rumeau, D., Becuwe-Linka, N., Beyly, A., Louwagie, M., Garin, J., and Peltier, G. ( 2005) New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 17, 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muraoka, R., Okuda, K., Kobayashi, Y., and Shikanai, T. ( 2006) A eukaryotic factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Physiol. 142, 1683–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamruzzaman Munshi, M., Kobayashi, Y., and Shikanai, T. ( 2005) Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J. 44, 1036–1044 [DOI] [PubMed] [Google Scholar]
- 72.Burrows, P. A., Sazanov, L. A., Svab, Z., Maliga, P., and Nixon, P. J. ( 1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kleffmann, T., Russenberger, D., Von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., and Baginsky, S. ( 2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362 [DOI] [PubMed] [Google Scholar]
- 74.Wang, D., and Portis, A. R., Jr. ( 2007) A novel nucleus-encoded chloroplast protein, PIFI, is involved in NAD(P)H dehydrogenase complex-mediated chlororespiratory electron transport in Arabidopsis. Plant Physiol. 144, 1742–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aluru, M. R., and Rodermel, S. R. ( 2004) Control of chloroplast redox by the IMMUTANS terminal oxidase. Physiol Plant. 120, 4–11 [DOI] [PubMed] [Google Scholar]
- 76.Tikkanen, M., Piippo, M., Suorsa, M., Sirpio, S., Mulo, P., Vainonen, J., Vener, A. V., Allahverdiyeva, Y., and Aro, E. M. ( 2006) State transitions revisited—a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 62, 779–793 [DOI] [PubMed] [Google Scholar]
- 77.Rochaix, J. D. ( 2007) Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett. 581, 2768–2775 [DOI] [PubMed] [Google Scholar]
- 78.Bellafiore, S., Barneche, F., Peltier, G., and Rochaix, J. D. ( 2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433, 892–895 [DOI] [PubMed] [Google Scholar]
- 79.Bonardi, V., Pesaresi, P., Becker, T., Schleiff, E., Wagner, R., Pfannschmidt, T., Jahns, P., and Leister, D. ( 2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437, 1179–1182 [DOI] [PubMed] [Google Scholar]
- 80.Vainonen, J. P., Hansson, M., and Vener, A. V. ( 2005) STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J. Biol. Chem. 280, 33679–33686 [DOI] [PubMed] [Google Scholar]
- 81.Hansson, M., Dupuis, T., Stromquist, R., Andersson, B., Vener, A. V., and Carlberg, I. ( 2007) The mobile thylakoid phosphoprotein TSP9 interacts with the light-harvesting complex II and the peripheries of both photosystems. J. Biol. Chem. 282, 16214–16222 [DOI] [PubMed] [Google Scholar]
- 82.Dalcorso, G., Pesaresi, P., Masiero, S., Aseeva, E., Schunemann, D., Finazzi, G., Joliot, P., Barbato, R., and Leister, D. ( 2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132, 273–285 [DOI] [PubMed] [Google Scholar]
- 83.Sato, N., Terasawa, K., Miyajima, K., and Kabeya, Y. ( 2003) Organization, developmental dynamics, and evolution of plastid nucleoids. Int. Rev. Cytol. 232, 217–262 [DOI] [PubMed] [Google Scholar]
- 84.Jeong, S. Y., Rose, A., and Meier, I. ( 2003) MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31, 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weber, P., Fulgosi, H., Piven, I., Muller, L., Krupinska, K., Duong, V. H., Herrmann, R. G., and Sokolenko, A. ( 2006) TCP34, a nuclear-encoded response regulator-like TPR protein of higher plant chloroplasts. J. Mol. Biol. 357, 535–549 [DOI] [PubMed] [Google Scholar]
- 86.van Wijk, K. J., Bingsmark, S., Aro, E. M., and Andersson, B. ( 1995) In vitro synthesis and assembly of photosystem II core proteins. The D1 protein can be incorporated into photosystem II in isolated chloroplasts and thylakoids. J. Biol. Chem. 270, 25685–25695 [DOI] [PubMed] [Google Scholar]
- 87.Miras, S., Salvi, D., Piette, L., Seigneurin-Berny, D., Grunwald, D., Reinbothe, C., Joyard, J., Reinbothe, S., and Rolland, N. ( 2007) Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J. Biol. Chem. 282, 29482–29492 [DOI] [PubMed] [Google Scholar]
- 88.Wang, Q., Sullivan, R. W., Kight, A., Henry, R. L., Huang, J., Jones, A. M., and Korth, K. L. ( 2004) Deletion of the chloroplast-localized thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kroll, D., Meierhoff, K., Bechtold, N., Kinoshita, M., Westphal, S., Vothknecht, U. C., Soll, J., and Westhoff, P. ( 2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. U. S. A. 98, 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aseeva, E., Ossenbuhl, F., Sippel, C., Cho, W. K., Stein, B., Eichacker, L. A., Meurer, J., Wanner, G., Westhoff, P., Soll, J., and Vothknecht, U. C. ( 2007) Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 45, 119–128 [DOI] [PubMed] [Google Scholar]
- 91.Jarvis, P., and Robinson, C. ( 2004) Mechanisms of protein import and routing in chloroplasts. Curr. Biol. 14, R1064–R1077 [DOI] [PubMed] [Google Scholar]
- 92.Schunemann, D. ( 2004) Structure and function of the chloroplast signal recognition particle. Curr. Genet. 44, 295–304 [DOI] [PubMed] [Google Scholar]
- 93.Mori, H., and Cline, K. ( 2001) Post-translational protein translocation into thylakoids by the Sec and ΔpH-dependent pathways. Biochim. Biophys. Acta 1541, 80–90 [DOI] [PubMed] [Google Scholar]
- 94.Miernyk, J. A. ( 2001) The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vitha, S., Froehlich, J. E., Koksharova, O., Pyke, K. A., van Erp, H., and Osteryoung, K. W. ( 2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15, 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu, S., Van Eck, J., Zhou, X., Lopez, A. B., O'Halloran, D. M., Cosman, K. M., Conlin, B. J., Paolillo, D. J., Garvin, D. F., Vrebalov, J., Kochian, L. V., Kupper, H., Earle, E. D., Cao, J., and Li, L. ( 2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18, 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Edvardsson, A., Shapiguzov, A., Petersson, U. A., Schroder, W. P., and Vener, A. V. ( 2007) Immunophilin AtFKBP13 sustains all peptidyl-prolyl isomerase activity in the thylakoid lumen from Arabidopsis thaliana deficient in AtCYP20–2. Biochemistry 46, 9432–9442 [DOI] [PubMed] [Google Scholar]
- 98.Fulgosi, H., Vener, A. V., Altschmied, L., Herrmann, R. G., and Andersson, B. ( 1998) A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17, 1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gupta, R., Mould, R. M., He, Z., and Luan, S. ( 2002) A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc. Natl. Acad. Sci. U. S. A. 99, 15806–15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peng, L., Ma, J., Chi, W., Guo, J., Zhu, S., Lu, Q., Lu, C., and Zhang, L. ( 2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18, 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meurer, J., Plucken, H., Kowallik, K. V., and Westhoff, P. ( 1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17, 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ruf, S., Kossel, H., and Bock, R. ( 1997) Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Boudreau, E., Takahashi, Y., Lemieux, C., Turmel, M., and Rochaix, J. D. ( 1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16, 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Naver, H., Boudreau, E., and Rochaix, J. D. ( 2001) Functional studies of Ycf3: its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13, 2731–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stockel, J., Bennewitz, S., Hein, P., and Oelmuller, R. ( 2006) The evolutionarily conserved tetratricopeptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen, G., Zhao, J., Reimer, S. K., Antonkine, M. L., Cai, Q., Weiland, S. M., Golbeck, J. H., and Bryant, D. A. ( 2002) Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277, 20343–20354 [DOI] [PubMed] [Google Scholar]
- 107.Shen, G., Antonkine, M. L., van der Est, A., Vassiliev, I. R., Brettel, K., Bittl, R., Zech, S. G., Zhao, J., Stehlik, D., Bryant, D. A., and Golbeck, J. H. ( 2002) Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of F(X) in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277, 20355–20366 [DOI] [PubMed] [Google Scholar]
- 108.Walters, R. G., Shephard, F., Rogers, J. J., Rolfe, S. A., and Horton, P. ( 2003) Identification of mutants of Arabidopsis defective in acclimation of photosynthesis to the light environment. Plant Physiol. 131, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakamoto, W. ( 2006) Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57, 599–621 [DOI] [PubMed] [Google Scholar]
- 110.Adam, Z., Rudella, A., and van Wijk, K. J. ( 2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9, 234–240 [DOI] [PubMed] [Google Scholar]
- 111.Lensch, M., Herrmann, R. G., and Sokolenko, A. ( 2001) Identification and characterization of SppA, a novel light-inducible chloroplast protease complex associated with thylakoid membranes. J. Biol. Chem. 276, 33645–33651 [DOI] [PubMed] [Google Scholar]
- 112.Chen, G., Bi, Y. R., and Li, N. ( 2005) EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 41, 364–375 [DOI] [PubMed] [Google Scholar]
- 113.Sun, X., Peng, L., Guo, J., Chi, W., Ma, J., Lu, C., and Zhang, L. ( 2007) Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 19, 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spetea, C., Hundal, T., Lohmann, F., and Andersson, B. ( 1999) GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proc. Natl. Acad. Sci. U. S. A. 96, 6547–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haussuhl, K., Andersson, B., and Adamska, I. ( 2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 20, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Apel, K., and Hirt, H. ( 2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 [DOI] [PubMed] [Google Scholar]
- 117.Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. ( 2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 [DOI] [PubMed] [Google Scholar]
- 118.Foyer, C. H., and Noctor, G. ( 2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dietz, K. J., Jacob, S., Oelze, M. L., Laxa, M., Tognetti, V., de Miranda, S. M., Baier, M., and Finkemeier, I. ( 2006) The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57, 1697–1709 [DOI] [PubMed] [Google Scholar]
- 120.Briat, J. F., Lobreaux, S., Grignon, N., and Vansuyt, G. ( 1999) Regulation of plant ferritin synthesis: how and why. CMLS Cell. Mol. Life Sci 56, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Poincelot, R. P. ( 1972) The distribution of carbonic anhydrase and ribulose diphosphate carboxylase in maize leaves. Plant Physiol. 50, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burnell, J. N., and Chastain, C. J. ( 2006) Cloning and expression of maize-leaf pyruvate, Pi dikinase regulatory protein gene. Biochem. Biophys. Res. Commun. 345, 675–680 [DOI] [PubMed] [Google Scholar]
- 123.Ashton, A. R., Burnell, J. N., and Hatch, M. D. ( 1984) Regulation of C4 photosynthesis: inactivation of pyruvate, Pi dikinase by ADP-dependent phosphorylation and activation by phosphorolysis. Arch. Biochem. Biophys. 230, 492–503 [DOI] [PubMed] [Google Scholar]
- 124.Knappe, S., Lottgert, T., Schneider, A., Voll, L., Flugge, U. I., and Fischer, K. ( 2003) Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis—AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 36, 411–420 [DOI] [PubMed] [Google Scholar]
- 125.Renne, P., Dressen, U., Hebbeker, U., Hille, D., Flugge, U. I., Westhoff, P., and Weber, A. P. ( 2003) The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J. 35, 316–331 [DOI] [PubMed] [Google Scholar]
- 126.Woo, K. C., Flugge, U. I., and Heldt, H. W. ( 1987) A two-translocator model for the transport of 2-oxoglutarate and glutamate in chloroplasts during ammonia assimilation in the light. Plant Physiol. 84, 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weber, A., and Flugge, U. I. ( 2002) Interaction of cytosolic and plastidic nitrogen metabolism in plants. J. Exp. Bot. 53, 865–874 [DOI] [PubMed] [Google Scholar]
- 128.Niittyla, T., Messerli, G., Trevisan, M., Chen, J., Smith, A. M., and Zeeman, S. C. ( 2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303, 87–89 [DOI] [PubMed] [Google Scholar]
- 129.Kim, J., and DellaPenna, D. ( 2006) Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. U. S. A. 103, 3474–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Motohashi, R., Ito, T., Kobayashi, M., Taji, T., Nagata, N., Asami, T., Yoshida, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. ( 2003) Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34, 719–731 [DOI] [PubMed] [Google Scholar]
- 131.Tottey, S., Block, M. A., Allen, M., Westergren, T., Albrieux, C., Scheller, H. V., Merchant, S., and Jensen, P. E. ( 2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. U. S. A. 100, 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moseley, J. L., Page, M. D., Alder, N. P., Eriksson, M., Quinn, J., Soto, F., Theg, S. M., Hippler, M., and Merchant, S. ( 2002) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., op den Camp, R., and Apel, K. ( 2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 98, 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gray, J., Close, P. S., Briggs, S. P., and Johal, G. S. ( 1997) A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89, 25–31 [DOI] [PubMed] [Google Scholar]
- 135.Brehelin, C., Kessler, F., and van Wijk, K. J. ( 2007) Plastoglobules: versatile lipoprotein particles in plastids. Trends Plant Sci. 12, 260–266 [DOI] [PubMed] [Google Scholar]
- 136.Laizet, Y., Pontier, D., March, R., and Kuntz, M. ( 2004) Subfamily organization and phylogenetic origin of genes encoding plastid-lipid-associated proteins of the fibrillin type. J. Genome Sci. Technol. 3, 19–28 [Google Scholar]
- 137.Brown, N. J., Parsley, K., and Hibberd, J. M. ( 2005) The future of C4 research—maize, Flaveria or Cleome? Trends Plant Sci. 10, 215–221 [DOI] [PubMed] [Google Scholar]
- 138.Matsuoka, M., Furbank, R. T., Fukayama, H., and Miyao, M. ( 2001) Molecular Engineering of C4 Photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 297–314 [DOI] [PubMed] [Google Scholar]
- 139.Mitchell, P. L., and Sheehy, J. E. ( 2006) Supercharging rice photosynthesis to increase yield. New Phytol. 171, 688–693 [DOI] [PubMed] [Google Scholar]
- 140.Jensen, P. E., Bassi, R., Boekema, E. J., Dekker, J. P., Jansson, S., Leister, D., Robinson, C., and Scheller, H. V. ( 2007) Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta 1767, 335–352 [DOI] [PubMed] [Google Scholar]
- 141.Sazanov, L. A., Burrows, P. A., and Nixon, P. J. ( 1998) The plastid ndh genes code for an NADH-specific dehydrogenase: isolation of a complex I analogue from pea thylakoid membranes. Proc. Natl. Acad. Sci. U. S. A. 95, 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kofer, W., Koop, H. U., Wanner, G., and Steinmuller, K. ( 1998) Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Gen. Genet. 258, 166–173 [DOI] [PubMed] [Google Scholar]
- 143.Ivanov, B., Asada, K., and Edwards, G. E. ( 2007) Analysis of donors of electrons to photosystem I and cyclic electron flow by redox kinetics of P700 in chloroplasts of isolated bundle sheath strands of maize. Photosynth. Res. 92, 65–74 [DOI] [PubMed] [Google Scholar]
- 144.Guedeney, G., Corneille, S., Cuine, S., and Peltier, G. ( 1996) Evidence for an association of ndh B, ndh J gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 378, 277–280 [DOI] [PubMed] [Google Scholar]
- 145.Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. ( 2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110, 361. [DOI] [PubMed] [Google Scholar]
- 146.Nandha, B., Finazzi, G., Joliot, P., Hald, S., and Johnson, G. N. ( 2007) The role of PGR5 in the redox poising of photosynthetic electron transport. Biochim. Biophys. Acta 1767, 1252–1259 [DOI] [PubMed] [Google Scholar]
- 147.Arteni, A. A., Zhang, P., Battchikova, N., Ogawa, T., Aro, E. M., and Boekema, E. J. ( 2006) Structural characterization of NDH-1 complexes of Thermosynechococcus elongatus by single particle electron microscopy. Biochim. Biophys. Acta 1757, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 148.Buchanan, B. B., and Luan, S. ( 2005) Redox regulation in the chloroplast thylakoid lumen: a new frontier in photosynthesis research. J. Exp. Bot. 56, 1439–1447 [DOI] [PubMed] [Google Scholar]
- 149.Munshi, M. K., Kobayashi, Y., and Shikanai, T. ( 2006) Chlororespiratory reduction 6 is a novel factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant. Physiol. 141, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Okuda, K., Nakamura, T., Sugita, M., Shimizu, T., and Shikanai, T. ( 2006) A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281, 37661–37667 [DOI] [PubMed] [Google Scholar]
- 151.Battchikova, N., and Aro, E. M. ( 2007) Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition. Physiol. Plant. 131, 22–32 [DOI] [PubMed] [Google Scholar]
- 152.Price, G. D., Badger, M. R., Woodger, F. J., and Long, B. M. ( 2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59, 1441–1461 [DOI] [PubMed] [Google Scholar]
- 153.Hoang, C. V., and Chapman, K. D. ( 2002) Regulation of carbonic anhydrase gene expression in cotyledons of cotton (Gossypium hirsutum L.) seedlings during post-germinative growth. Plant Mol. Biol. 49, 449–458 [DOI] [PubMed] [Google Scholar]
- 154.Badger, M. R., and Price, G. D. ( 2003) CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot 54, 609–622 [DOI] [PubMed] [Google Scholar]
- 155.Potter, J. W., and Black, C. C. ( 1982) Differential protein composition and gene expression in leaf mesophyll cells and bundle sheath cells of the C4 plant Digitaria sanguinalis (L.) Scop. Plant Physiol. 70, 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bailey, S., Thompson, E., Nixon, P. J., Horton, P., Mullineaux, C. W., Robinson, C., and Mann, N. H. ( 2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277, 2006–2011 [DOI] [PubMed] [Google Scholar]
- 157.Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A. B., Adam, Z., and Andersson, B. ( 2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Moseley, J., Quinn, J., Eriksson, M., and Merchant, S. ( 2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Offermann, S., Danker, T., Dreymuller, D., Kalamajka, R., Topsch, S., Weyand, K., and Peterhansel, C. ( 2006) Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phosphoenolpyruvate carboxylase promoter in maize. Plant Physiol. 141, 1078–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nott, A., Jung, H. S., Koussevitzky, S., and Chory, J. ( 2006) Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759 [DOI] [PubMed] [Google Scholar]
- 161.Leister, D. ( 2005) Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene ( Amst.) 354, 110–116 [DOI] [PubMed] [Google Scholar]
- 162.Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., Lim, J., Mittler, R., and Chory, J. ( 2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 [PubMed] [Google Scholar]
- 163.Baena-Gonzalez, E., Allahverdiyeva, Y., Svab, Z., Maliga, P., Josse, E. M., Kuntz, M., Maenpaa, P., and Aro, E. M. ( 2003) Deletion of the tobacco plastid psbA gene triggers an upregulation of the thylakoid-associated NAD(P)H dehydrogenase complex and the plastid terminal oxidase (PTOX). Plant J. 35, 704–716 [DOI] [PubMed] [Google Scholar]
- 164.Paul, M. ( 2007) Trehalose 6-phosphate. Curr. Opin. Plant Biol. 10, 303–309 [DOI] [PubMed] [Google Scholar]
- 165.Rolland, F., Baena-Gonzalez, E., and Sheen, J. ( 2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709 [DOI] [PubMed] [Google Scholar]
- 166.Eastmond, P. J., and Graham, I. A. ( 2003) Trehalose metabolism: a regulatory role for trehalose-6-phosphate? Curr. Opin. Plant Biol. 6, 231–235 [DOI] [PubMed] [Google Scholar]
- 167.Satoh-Nagasawa, N., Nagasawa, N., Malcomber, S., Sakai, H., and Jackson, D. ( 2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441, 227–230 [DOI] [PubMed] [Google Scholar]
- 168.Brunner, E., Ahrens, C. H., Mohanty, S., Baetschmann, H., Loevenich, S., Potthast, F., Deutsch, E. W., Panse, C., de Lichtenberg, U., Rinner, O., Lee, H., Pedrioli, P. G., Malmstrom, J., Koehler, K., Schrimpf, S., Krijgsveld, J., Kregenow, F., Heck, A. J., Hafen, E., Schlapbach, R., and Aebersold, R. ( 2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.