Abstract
Influenza A viruses cause significant morbidity and mortality annually, and the threat of a pandemic underscores the need for new therapeutic strategies. Here we briefly discuss novel antiviral agents under investigation, the limitations of current antiviral therapy and stress the importance of secondary bacterial infections in seasonal and pandemic influenza. Additionally, the lack of new antibiotics available to treat increasingly drug resistant organisms such as methicillin-resistant Staphylococcus aureus, pneumococci, Acinetobacter, extended spectrum beta-lactamase producing gram negative bacteria and Clostridium difficile is highlighted as an important component of influenza treatment and pandemic preparedness. Addressing these problems will require a multidisciplinary approach, which includes the development of novel antivirals and new antibiotics, as well as a better understanding of the role secondary infections play on the morbidity and mortality due to influenza infection.
Introduction
Influenza viruses are among the most common causes of respiratory infections in humans [1] and are also associated with high morbidity and mortality, especially in the elderly, in infants and in people with chronic diseases. In the USA alone, influenza results in approximately 200,000 hospitalizations and 36,000 deaths in a typical endemic season [2]. In addition to annual winter outbreaks, antigenically novel strains of influenza virus occasionally emerge causing pandemics, on average three times per century [3,4]. Although the impact of past pandemics has been highly variable, up to 50% or more of a population can be infected in a single pandemic year and [S2] the number of deaths caused by influenza can dramatically exceed what is normally expected in an endemic season [5,6]. For example, in the past 120 years there were pandemics in 1889, 1918, 1957 and 1968 [7]. The 1957 pandemic caused 66,000 excess deaths in the USA [6]. In 1918, the worst pandemic in recorded history caused approximately 675,000 deaths in the USA [4] and killed up to 50 million people worldwide [8].
It is highly likely that influenza will return in pandemic form [4,9]. Concern about a future influenza pandemic caused by human infection with a highly pathogenic avian influenza (HPAI) virus of H5N1 subtype [9–12] has prompted renewed interest in influenza pandemic preparedness planning and in basic and applied influenza virus research.
After its reemergence in 2003, the ongoing H5N1 HPAI epizootic continues to produce human spillover infections [13]. As of December 2007, 336 confirmed cases of human H5N1 infection had been documented, of which 207 were fatal [14], yielding a case fatality rate of 62%. Concerns about the emergence of an H5N1 pandemic virus hinges not only upon sporadic transmission events between infected poultry and exposed humans but also crucially upon its potential for sustained person-to-person transmission. Several small case clusters of H5N1 infections have been reported [15,16]. Although epidemiologic information has been limited, person-to-person transmission of H5N1 has been suggested in a few instances, usually involving family members [17]. It is unknown whether this represents infection associated with particularly intimate or prolonged contact, or shared but unidentified host factors affecting either infection risk or virus transmissibility [18,19].
Challenges of Therapy
Effective pharmacological treatment of influenza virus infection must take into account the rapid time-course of acute viral infection. Influenza A viral replication peaks approximately 48 hours after inoculation into the nasopharynx and declines slowly, with little virus shed after about six days. The virus can replicate in the upper and lower respiratory tract. Even after infectious virus can no longer be recovered, viral antigen can be detected in cells and secretions of infected individuals for several additional days [1]. Human influenza infection is an acute respiratory disease characterized in its full form by the sudden onset of fever, coryza, cough, headache, myalgia, prostration, malaise and inflammation of the upper respiratory tree and trachea. Acute symptoms and fever often persist for 7 to 10 days and weakness and fatigue can linger for weeks. People with chronic pulmonary or cardiac disease, diabetes mellitus or other chronic illnesses have a higher risk of developing severe complications from influenza, which may include hemorrhagic bronchitis, pneumonia (both primary viral and secondary bacterial) and death. Hemorrhagic [S3] bronchitis and pneumonia can develop within hours and fulminant, fatal influenza viral pneumonia, may present with rapid onset of dyspnea, cyanosis, hemoptysis and pulmonary edema. Death can follow in as little as 48 hours after onset of symptoms, but most influenza fatalities occur later, after the development of secondary bacterial pneumonias and other complications [20].
Effective measures currently available against influenza infection include prevention by either vaccination with inactivated or live attenuated vaccines, or administration of antiviral drugs prophylactically or therapeutically [21]. The role of novel vaccines which target highly pathogenic influenza viruses in preparedness for future pandemics has been extensively reviewed elsewhere [22–26]. Given concerns about a deadly new pandemic and the inevitable delays in producing and administering large quantitities of vaccines, pandemic planning initiatives have also focused on the possible use of antiviral drugs as a component of pandemic mitigation and response [27]. To prevent disease, antiviral drugs must be administered promptly and continuously at times of influenza exposure. However, the use of available antiviral drugs in an influenza pandemic can have limitations [28], including development of drug resistance with retention of virulence and transmissibility properties [29].
New therapeutic strategies are needed to lessen the impact of seasonal influenza as well as to prepare for another influenza pandemic. Particularly in developed nations, the population is aging and persons are living longer and with higher rates of chronic diseases that put them into a high-risk category for influenza complications and fatality [2,30]. Immunosuppressive therapies and immunomodulating drugs are being used to treat many such patients. Neither the population impact of a pandemic nor the effectiveness of current therapies can be predicted. The relative paucity of intensive care unit beds, invasive positive pressure ventilation systems, emergency room facilities and staff are all problems that could have a major impact during a pandemic. The emergence of multi-drug resistant bacteria, both nosocomial and community acquired, including methicillin-resistant Staphylococcus aureus (MRSA), resistant pneumococci, Acinetobacter, extended spectrum beta-lactamase (ESBL)-producing gram negative organisms and Clostridium difficile, could all be major factors in the morbidity and mortality of a future pandemic. It is clear that a multidisciplinary approach is required in preparing for an influenza pandemic, and that prevention and mitigation strategies must address many issues.
Current Pharmacotherapy and Drug Development
Most influenza pandemic plans consider drug therapy as a key part of the initial response. In the event of a pandemic, not only could primary viral pneumonia, respiratory distress and other syndromes secondary to the influenza virus itself play a major part in morbidity and mortality but it is also highly likely that secondary infections such as pneumonias and other community acquired and nosocomial infections will contribute as well. Anti-influenza drug availability is limited[S5], and there is obviously a need for development of new classes of antivirals. In addition, drug therapies targeting the wide array of complications associated with influenza infection and hospitalization, such as secondary pneumonias, healthcare associated infections, ventilator associated lung injury and others, are also important.
Currently there are two major classes of drugs that target the influenza virus itself: matrix 2 ion channel inhibitors; and neuraminidase (NA) inhibitors. These drugs have been extensively evaluated [28] but their efficacy in a pandemic or highly pathogenic influenza situation is still unknown. Matrix 2 ion channel blockers (amantadine and rimantadine) are only effective against influenza A viruses. Resistant viral strains develop rapidly and have been recognized in 15–92% of patients in a given year [31–33]. Interestingly, a dramatic increase in the frequency of resistance to adamantanes by human influenza A (H3N2) viruses has occurred in recent years, now up to 90%, associated with a single S31N amino acid replacement in the viral matrix M2 protein [34]. The more recently developed NA inhibitors, zanamivir and oseltamivir, are effective against influenza A and B viruses[S6]. Both classes of drugs are effective in preventing influenza when administered prophylactically [28,29]. To be clinically effective, early administration within the first 12 hours of disease onset of each of these drugs is important [28,35]. Drug resistance to NA inhibitors has also been reported [36]. Prevalence of drug resistant strains of seasonal influenza viruses could be increasing as suggested by European reports that 14% of H1N1 influenza A isolates were resistant to oseltamivir during the 2007–2008 season [37]. Recent reports have also documented the development of resistance mutations in H5N1 strains following treatment with NA inhibitors [39].
Unfortunately, neither class of drug has been proven to be of value in pandemic mitigation nor has been shown to be efficacious for highly pathogenic influenza viruses, such as H5N1 [38,39]. A third class of drug being assessed is the inosine monophosphate (IMP) dehydrogenase inhibitors, such as ribavirin. Small controlled studies have shown some efficacy in decreasing fever and influenza A virus shedding when IMP inhibitors are given by aerosol, but no efficacy when given orally [40]. Further study needs to be undertaken to examine the possibility that combinations of the available drugs would increase efficacy.
Research efforts are also focusing on development of new drug classes that target unique aspects of influenza virus replication and infection. Antiviral therapies currently under investigation include viral receptor blockers, viral release inhibitors, viral polymerase inhibitors, RNA interference, among others [41]. Some examples are highlighted below.
Sialylmimetics are compounds that mimic the molecular structure of cell receptor sialic acid residues involved in binding influenza viruses. Extensive study of these compounds led to the development of NA inhibitors, which are monomeric sialylmimetics [42]. Further study of a number of new sialyloligosaccharides and sialylmimetics that inhibit virus attachment by the hemagglutinin (HA), or inhibit viral detachment through the activity of the NA, has led to the development of two new drugs. One, multimeric zanamavir, is in the preclinical stages and the other, DAS181(Fludase), is in Phase I clinical trials [41,42]. Novel entry blocker proteins, such as the 20 amino-acid peptide derived from fibroblast growth factor 4 that blocks viral entry by binding to the HA and exhibits antiviral activity against influenza in tissue culture and in mice are also under investigation[43].
Novel amino acid entry blockers, sialylmimetics, and related compounds that target entry of the virus into the cell might eventually expand the armamentarium against influenza, but these types of agents can be subject to some of the same limitations as current drugs in their requirement for early or prophylactic administration for maximum benefit. Further study is also needed to determine their efficacy and safety in humans.
A number of groups have been exploring compounds that inhibit the influenza virus RNA-dependant RNA polymerase [41]. Important and unique influenza polymerase properties, such as activation induced by binding of viral genomic RNA to specific amino acids, and the catalytic activity of the cap-dependant endonuclease, have been suggested as drug targets. A number of compounds that target polymerase function, particularly its catalytic activity, have already been identified [44,45]. Li et al. have suggested that this approach is too nonspecific and could adversely effect host cell enzymes [45]. Further testing of these compounds continues, and a non-peptide small molecule that targets the viral polymerase function [46,47] is currently in Phase I clinical trials [41].
RNA interference as a mechanism for viral inhibition has been reported for a number of viruses including influenza A in vitro [41]. Double stranded RNA molecules can cause sequence-specific translational inhibition, while synthetic small interfering RNAs (siRNAs) have inhibited influenza virus replication when transfected into cells before or after viral challenge [41,48]. In a recent publication, Zhou et al. developed M2- and NA-specific siRNAs that not only inhibited influenza A in transfected cells but also produced a 16–50 fold reduction in mouse lung titers following intravenous injection [48]. This strategy has been tested with promising results against pathogenic avian influenza A viruses of the H5 and H7 subtypes in mice using an intranasal delivery system with a cationic transfection reagent [49]. Questions of unwanted induction of interferon responses and toxic side effects have been raised, and further animal studies using intranasal and intravenous routes of delivery are needed [41].
Therapeutics for Secondary and Nosocomial Infections
The cause of fatal influenza has been studied scientifically in each of the four most recent pandemics, as well as in seasonal epidemics spanning the past 175 years. The accumulated data, which represent autopsies with gross pathological description since 1837, autopsies with histologic and bacteriologic examination since 1889 and correlational studies of series of fatal cases autopsied with standardized protocols since 1918, demonstrate that most pandemic influenza deaths are associated with severe secondary bacterial pneumonia. Even after the 1933 isolation of the first human influenza virus, most bacteriologists and pathologists were of the opinion that primary influenza virus pneumonia was rarely fatal. That opinion changed somewhat during the next pandemic (1957), when occasional cases of alleged primary viral pneumonia were reported [50], although as the first pandemic to occur during the antibiotic era it was difficult to rule out antibiotic suppressive effects conclusively. Moreover, whereas pneumococci and streptococci had predominated in 1918 and probably also in 1889 [51–53] (Figure 1), Staphylococcus aureus, a bacterium more often resistant to then-common antibiotics such as penicillin, stremptomycin and chloramphenicol, was the principal infecting organism in 1957 pneumonia cases [50]. Whatever the role of primary viral pneumonia in fatal influenza, however, pathogenic “models” for fatal influenza put forth in 1918 remain relevant today. In the 10 early 20th Century, views of leading researchers like William MacCallum and Eugene Opie [53–55] were that the influenza virus initiates a process of lung injury by inducing necrosis of the respiratory epithelium (necrosis and sloughing of the columnar epithelium down to the basal layer). If this injury proceeds downward to the bronchi, and especially in the presence in the upper respiratory tract of Haemophilus influenzae, which seems to play a facilitative role, pathogenic organisms like pneumococci and streptococci can gain direct access to the lungs via the bronchial tree, thereby causing bronchopneumonia. This hypothesis is still reasonable today. During the 1918 pandemic severe bronchopneumonia had a very high fatality rate, often approaching 40% [56]. Even during the most recent pandemic (1968), patients with secondary staphylococcal pneumonia had a 33% case fatality rate [57], reflecting the inherent difficulty in treating severe secondary bronchopneumonia caused by known pathogens even in the antibiotic era.
Figure 1. Hematoxylin and Eosin-stained section of lung from a 1918 ‘Spanish’ influenza victim.
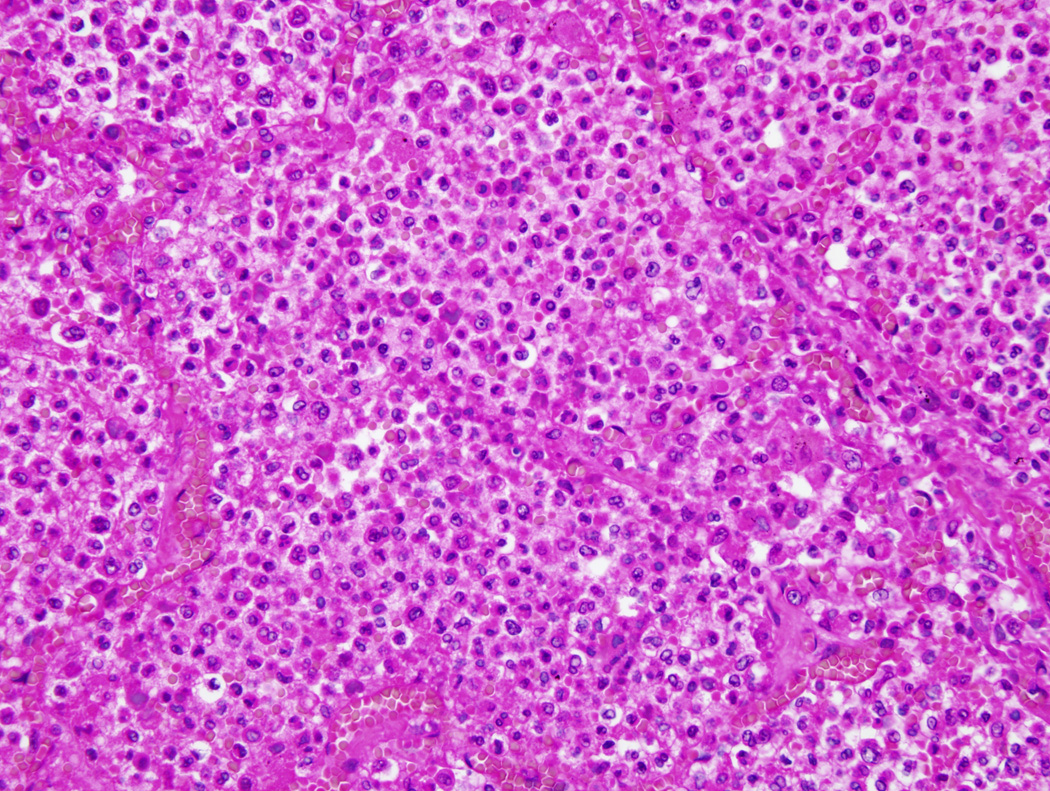
Slide shows massive infiltrate of neutrophils that fills the alveolar air spaces in early bacterial bronchopneumonia. Alveolar capillary congestion is also prominent (original magnification 200×).
Given the impact of secondary pneumonias observed during previous influenza pandemics, and associated with seasonal influenza each year [6], it seems likely that during a new pandemic secondary bacterial infections will again be a major cause of morbidity and mortality. In addition to secondary pneumonias, nosocomial infections associated with hospitalization, the use of positive pressure ventilation and increased host susceptibility to infection would also likely play a significant part in morbidity and mortality. These secondary and nosocomial infections would probably include strains of antibiotic resistant bacteria that have recently become more prevalent including MRSA, vancomycin resistant Enterococcus (VRE), C. difficile, Pseudomonas, Acinetobacter and drug resistant pneumococci.
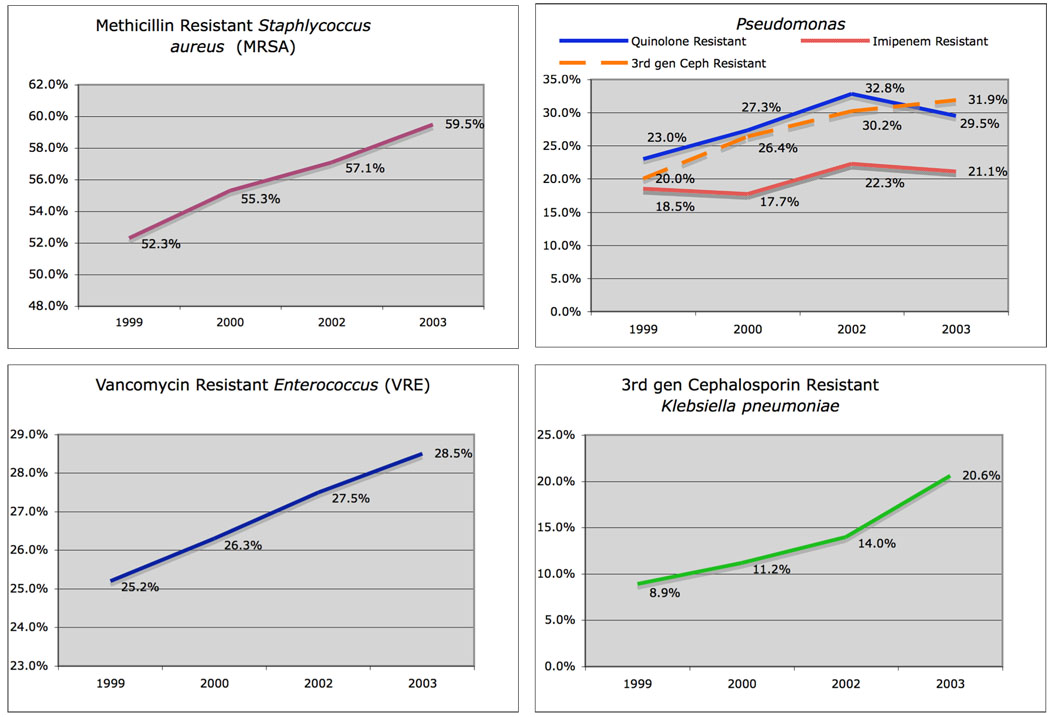
As a corollary to the challenges that could be faced in a future influenza pandemic, it is worth noting that currently over 2 million people acquire infections in US hospitals each year, resulting in 90,000 deaths and a cost of at least US$5 billion annually in the USA alone [58]. About 70% of these infections are caused by organisms resistant to at least one antibiotic drug [58]. Patients at highest risk for resistant infections are those in intensive care units, those who are immunocompromised and those who reside in chronic care facilities. The rates of antibiotic resistant organisms found in intensive care unit and nursing home patients have been increasing (Figure 2). The 2004 CDC National Nosocomial Infections Surveillance report found increases in antibiotic resistance rates of up to 47% against common organisms found in nosocomial infections in intensive care unit patients [59]. As of 2005, data collected by the CDC showed that, out of 250,000 reported infections in nursing home patients, 27,000 infections were caused by antibiotic resistant bacteria [60].
Figure 2. Antimicrobial resistance rates of select pathogens associated with nosocomial infections in intensive care unit patients in the United States 1999–2003.
Data collected from hospitals around the United States by the CDC National Nosocomial Infections Surveillance System (NNIS) [46] over a four year period showed increases in antimicrobial resistance amongst common nosocomial pathogens: A. rates of methicillin resistance of Staphylococcus aureus isolates, B. resistance rates of pseudomonas species to multiple drugs commonly used to treat pulmonary and nosocomial infections, C. vancomycin resistance rates in enterococcal isolates, D. rates of resistance to 3rd generation cephlosporins amongst Klebsiella pneumoniae isolates.
Antibiotic resistant infections are not only increasing in hospitals and chronic care facilities but also in the community and in younger, previously healthy people [58]. The problem of community acquired MRSA demonstrates this clearly.
MRSA has been directly associated with severe community acquired necrotizing pneumonia [61,62] and, during the 2003–2004 influenza season, was found to be a cause of severe secondary bacterial pneumonia in patients with influenza infection. Seventeen cases were reported from nine states in the USA. The median age of these patients was only 21 years old, and only five patients had a previous underlying illness. Of these 17 patients, 81% required intensive care, 62% required positive pressure ventilation and 29% died [63].
The emergence of MRSA as a secondary pathogen in seasonal influenza was not an isolated phenomenon during 2003–2004. The CDC reports that in the only two states evaluated from January 2006 to December 2007 (Louisiana and Georgia), 15 cases of severe MRSA community acquired pneumonia associated with influenza infection were identified. The patients were young (median age 17.5 years) and all but one had been previously healthy, the exception being a patient with chronic hepatitis C infection and hypertension. Death occurred in 60% of these patients, with a median time to death of 3.5 days from illness onset [64].
Currently, no formal surveillance has determined the full impact of MRSA pneumonia secondary to seasonal influenza infections. The lessons of the past, and the current CDC reports, underscore the importance of learning more about how MRSA and other resistant bacterial secondary infections affect the morbidity and mortality of influenza, and what this might mean in the event of a new pandemic.
The challenge of drug resistant bacterial infections was extensively reviewed by the Infectious Disease Society of America in 2004 in a report entitled “Bad Bugs, No Drugs”[58]. Although drug development continues at a rapid pace, antibiotic research and development has slowed dramatically in the past 20 years due to the risk and high cost of research and development, combined with lower profits than generated by drugs to treat chronic conditions. Between 1983 and 1987 16 new antibacterials were introduced; and between 2003 and 2007 only 4 were introduced [58]. As reported by the CDC, this decline has been ongoing since 1980 despite the steady increase in infections with antibiotic resistant organisms such as MRSA, VRE and Pseudomonas [59]. Of the 10 new antibacterial agents introduced between 1998 and 2004, only 2, daptomycin and linezolid, were of novel classes [65].
The global community has also recognized this problem and in 2002 increasing antibiotic resistance prompted the World Health Organization to develop the “WHO Global Strategy for Containment of Antimicrobial Resistance” [66,67]. This includes improving worldwide access to antibiotics, improving the appropriate use of antibiotics to limit resistance, strengthening healthcare systems and surveillance capabilities, and encouraging development of new drugs [66].
The problem of antimicrobial resistance is directly related to influenza pandemic preparedness. In the event of an influenza pandemic, the number of patients at high risk for acquiring secondary pneumonias and nosocomial infections could greatly increase because of widespread influenza infection as well as increased need for hospitalization, ICU care, positive pressure ventilation and exposure to medical care facilities. These effects will not only be seen in the USA but also around the world where access to antimicrobials could be even further limited. Efforts to increase antimicrobial development, improve access to drugs and to judiciously use antimicrobials to limit resistance are extremely important in preparing for a pandemic. We must consider the development of new antibacterials for resistant organisms imperative for influenza pandemic preparedness as well as a means of reducing the morbidity and mortality of seasonal influenza. Careful evaluation and management of antibiotic stockpiles worldwide may also be necessary. Through legislation, education and cooperation, further progress must be made in these areas as part of the global strategy in pandemic influenza preparedness.
Summary and Conclusions
The challenges we face in developing better therapeutic strategies for seasonal and pandemic influenza are great and include the paucity of effective antiviral therapies, the lack of new antibiotic development in the face of increasing antibacterial resistance, limitations of influenza drug therapy secondary to the rapid infection and progression of influenza infection, the time lag in vaccine production and the lack of availability of these therapies worldwide. Fortunately, many avenues of research are now being explored in addition to those discussed here. Currently, widely available drugs such as statins, which have been hypothesized to reduce influenza mortality, have been studied with controversial results [68–70]. Other inexpensive preventive strategies such as intranasal gels are under development [71]. Further research in the areas of sepsis and systemic inflammatory response syndrome (SIRS) might also be of benefit.
The intimately related problems of seasonal influenza and of pandemic influenza preparedness will require multidisciplinary and coordinated efforts by healthcare workers and scientists around the world. Infection control, access to healthcare and therapies, drug development and vaccination programs all need to play major parts in pandemic planning strategies. Broad efforts to understand the basic virology underlying seasonal as well as pandemic and highly pathogenic avian influenza viruses must continue, but we must not forget the many other secondary bacterial complications that an influenza pandemic will cause.
Acknowledgements
This work was supported by the Intramural Research Program of the NIH and the NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wright PF, et al. Orthomyxoviruses. In: Knipe DM, Howley PM, et al., editors. Fields Virology. Lippincott: Williams & Wilkins; 2005. pp. 1691–1740. [Google Scholar]
- 2.Thompson WW, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 3.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12(1):15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beveridge W. Influenza: The Last Great Plague, an Unfinished Story. Prodist; 1977. [Google Scholar]
- 6.Simonsen L. The global impact of influenza on morbidity and mortality. Vaccine. 1999;17 Suppl 1:S3–S10. doi: 10.1016/s0264-410x(99)00099-7. [DOI] [PubMed] [Google Scholar]
- 7.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195(7):1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 8.Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 "Spanish" influenza pandemic. Bull Hist Med. 2002;76(1):105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 9.Taubenberger JK, et al. The next influenza pandemic: can it be predicted? JAMA. 2007;297:2025–2027. doi: 10.1001/jama.297.18.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris JS, et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363(9409):617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302(5650):1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 12.Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355(21):2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 13.Webster RG, et al. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 2006;12(1):3–8. doi: 10.3201/eid1201.051024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO. World Health Organization; Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. 2007;Vol. 2007
- 15.Ungchusak K, et al. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005;352(4):333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, et al. Human influenza A (H5N1) cases, urban areas of People's Republic of China, 2005–2006. Emerg Infect Dis. 2007;13(7):1061–1064. doi: 10.3201/eid1307.061557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, et al. Detecting human-to-human transmission of avian influenza A (H5N1) Emerg Infect Dis. 2007;13(9):1348–1353. doi: 10.3201/eid1309.07-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdel-Ghafar AN, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358(3):261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 19.Maines TR, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103(32):12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taubenberger JK, Morens DM. The Pathology of Influenza Virus Infections. Annu Rev Pathol. 2007 doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couch RB. Prevention and treatment of influenza. N Engl J Med. 2000;343(24):1778–1787. doi: 10.1056/NEJM200012143432407. [DOI] [PubMed] [Google Scholar]
- 22.Subbarao K, Luke C. H5N1 viruses and vaccines. PLoS Pathog. 2007;3(3):e40. doi: 10.1371/journal.ppat.0030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Sahly HM, Keitel WA. Pandemic H5N1 influenza vaccine development: an update. Expert Rev Vaccines. 2008;7(2):241–247. doi: 10.1586/14760584.7.2.241. [DOI] [PubMed] [Google Scholar]
- 24.Haque A, et al. Confronting Potential Influenza A (H5N1) Pandemic with Better Vaccines. Emerg Infect Dis. 2007;13(10):1512–1518. doi: 10.3201/eid1310.061262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mossad SB. Influenza update 2007–2008: vaccine advances, pandemic preparation. Cleve Clin J Med. 2007;74(12):889–894. doi: 10.3949/ccjm.74.12.889. [DOI] [PubMed] [Google Scholar]
- 26.Tosh PK, Poland GA. Emerging vaccines for influenza. Expert Opin Emerg Drugs. 2008;13(1):21–40. doi: 10.1517/14728214.13.1.21. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson NM, et al. Strategies for mitigating an influenza pandemic. Nature. 2006;442(7101):448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden FG, Pavia AT. Antiviral management of seasonal and pandemic influenza. J Infect Dis. 2006;194 Suppl 2:S119–S126. doi: 10.1086/507552. [DOI] [PubMed] [Google Scholar]
- 29.Hayden FG. Antiviral resistance in influenza viruses--implications for management and pandemic response. N Engl J Med. 2006;354(8):785–788. doi: 10.1056/NEJMp068030. [DOI] [PubMed] [Google Scholar]
- 30.Neuzil KM, et al. Influenza-associated morbidity and mortality in young and middle-aged women. JAMA. 1999;281(10):901–907. doi: 10.1001/jama.281.10.901. [DOI] [PubMed] [Google Scholar]
- 31.Bright RA, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366(9492):1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 32.Deyde VM, et al. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis. 2007;196(2):249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 33.Belshe RB, et al. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J Virol. 1988;62(5):1508–1512. doi: 10.1128/jvi.62.5.1508-1512.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonsen L, et al. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol. 2007;24(8):1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 35.Treanor JJ, et al. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. US Oral Neuraminidase Study Group. JAMA. 2000;283(8):1016–1024. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 36.Kiso M, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364(9436):759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 37.Dyer O. European agencies find 14% of flu isolates are resistant to oseltamivir. BMJ. 2008;336(7639):298. [Google Scholar]
- 38.de Jong MD, et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med. 2005;353(25):2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- 39.Le QM, et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature. 2005;437(7062):1108. doi: 10.1038/4371108a. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert BE, et al. Ribavirin small-particle aerosol treatment of infections caused by influenza virus strains A/Victoria/7/83 (H1N1) and B/Texas/1/84. Antimicrob Agents Chemother. 1985;27(3):309–313. doi: 10.1128/aac.27.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torrence PF. Combating the Threat of Pandemic Influenza Drug Discovery Approaches. John Wiley & Sons; 2007. [Google Scholar]
- 42.Sun XL. Recent anti-influenza strategies in multivalent sialyloligosaccharides and sialylmimetics approaches. Curr Med Chem. 2007;14(21):2304–2313. doi: 10.2174/092986707781696582. [DOI] [PubMed] [Google Scholar]
- 43.Jones JC, et al. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J Virol. 2006;80(24):11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai CH, et al. Antiviral therapy targeting viral polymerase. Curr Pharm Des. 2006;12(11):1339–1355. doi: 10.2174/138161206776361156. [DOI] [PubMed] [Google Scholar]
- 45.Li ML, et al. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 2001;20(8):2078–2086. doi: 10.1093/emboj/20.8.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuta Y, et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother. 2002;46(4):977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furuta Y, et al. Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother. 2005;49(3):981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, et al. Effective small interfering RNAs targeting matrix and nucleocapsid protein gene inhibit influenza A virus replication in cells and mice. Antiviral Res. 2007;76(2):186–193. doi: 10.1016/j.antiviral.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Tompkins SM, et al. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101(23):8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Louria DB, et al. Studies on influenza in the pandemic of 1957–1958. II. Pulmonary complications of influenza. J Clin Invest. 1959;38(1 Part 2):213–265. doi: 10.1172/JCI103791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodhart J. Influenza. In: Allbutt T, Rolleston H, editors. System of Medicine. Vol. I. MacMillan & Company; 1905. pp. 942–967. [Google Scholar]
- 52.Lucke B. Pathology and bacteriology of pneumonia. Pennsylvania Med J. 1920;23:369–371. [Google Scholar]
- 53.MacCallum W. Pathological Studies in the recent epidemics of pneumonia. Tr South Surg Assoc. 1919;31:180–192. [Google Scholar]
- 54.MacCallum W. Pathological anatomy of pneumonia associated with influenza. Johns Hopkins Hosp Rep. 1920;20:149–249. [Google Scholar]
- 55.Opie E, et al. The pathology and bacteriology of pneumonia following influenza. In: Opie E, et al., editors. Epidemic Respiratory Disease, The Pneumonias and Other Infections of the Respiratory Tract Accompanying Influenza and Measles. C.V. Mosby; 1921. pp. 107–281. [Google Scholar]
- 56.Lamb F, Brannin E. The epidemic respiratory infection at Camp Cody, N.M. J. Am Med Assoc. 1919;72:1056–1062. [Google Scholar]
- 57.Schwarzmann SW, et al. Bacterial pneumonia during the Hong Kong influenza epidemic of 1968–1969. Arch Intern Med. 1971;127(6):1037–1041. [PubMed] [Google Scholar]
- 58.White Paper: Bad bugs, No Drugs. Infectious Disease Society of America; 2004. As Antibiotic Discovery Stagnates …A Public Health Crisis Brews. [Google Scholar]
- 59.National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32(8):470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 60.Campaign to prevent Antimicrobial Resistance in Healthcare Settings. Centers for Disease Control; 2007. Burden of Infections Among U.S. Nursing Home Residents. [Google Scholar]
- 61.Francis JS, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40(1):100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 62.Johnston BL. Methicillin-resistant Staphylococcus aureus as a cause of community-acquired pneumonia--a critical review. Semin Respir Infect. 1994;9(3):199–206. [PubMed] [Google Scholar]
- 63.Hageman JC, et al. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003–04 influenza season. Emerg Infect Dis. 2006;12(6):894–899. doi: 10.3201/eid1206.051141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Severe methicillin-resistant Staphylococcus aureus community-acquired pneumonia associated with influenza--Louisiana and Georgia, December 2006–January 2007. MMWR Morb Mortal Wkly Rep. 2007;56(14):325–329. [PubMed] [Google Scholar]
- 65.Spellberg B, et al. Trends in antimicrobial drug development: implications for the future. Clin Infect Dis. 2004;38(9):1279–1286. doi: 10.1086/420937. [DOI] [PubMed] [Google Scholar]
- 66.Executive Summary. World Healh Organization; WHO Global Strategy for Containment of Antimicrobial Resistance. 2001
- 67.Schunemann HJ, et al. WHO Rapid Advice Guidelines for pharmacological management of sporadic human infection with avian influenza A (H5N1) virus. Lancet Infect Dis. 2007;7(1):21–31. doi: 10.1016/S1473-3099(06)70684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fedson DS. Statin protection against influenza and COPD mortality. Chest. 2007;132(4):1406. doi: 10.1378/chest.07-0992. author reply 1406–1407. [DOI] [PubMed] [Google Scholar]
- 69.Grijalva CG, et al. Statins and influenza/COPD mortality. Chest. 2007;132(4):1407. doi: 10.1378/chest.07-0952. author reply 1407–1408. [DOI] [PubMed] [Google Scholar]
- 70.Vos E, Mascitelli L. Statins have no role in pulmonary disease mortality. Chest. 2007;132(4):1408. doi: 10.1378/chest.07-1157. author reply 1408–1409. [DOI] [PubMed] [Google Scholar]
- 71.Rennie P, et al. Low pH gel intranasal sprays inactivate influenza viruses in vitro and protect ferrets against influenza infection. Respir Res. 2007;8:38. doi: 10.1186/1465-9921-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]