Abstract
Gene expression studies in humans and animals have shown that elevated stearoyl-CoA desaturase (SCD1) activity is associated with increased fat accumulation and monounsaturation of saturated fatty acids in skeletal muscle. However, results of the two reported association studies in humans are inconsistent. In the present study, we annotated the bovine SCD1 gene and identified 3 single nucleotide polymorphisms (SNPs) in its 3'untranslated region (UTR). Genotyping these SNPs on a Wagyu x Limousin reference population revealed that the SCD1 gene was significantly associated with six fat deposition and fatty acid composition traits in skeletal muscle, but not with subcutaneous fat depth and percent kidney-pelvic-heart fat. In particular, we confirmed that the high stearoyl-CoA desaturase activities/alleles were positively correlated with beef marbling score, amount of monounsaturated fatty acids and conjugated linoleic acid content, but negatively with amount of saturated fatty acids. The inconsistent associations between human studies might be caused by using different sets of markers because we observed that most associated markers are located near the end of 3'UTR. We found that the proximity of the polyadenylation signal site is highly conserved among human, cattle and pig, indicating that the region might contain functional elements involved in posttranscriptional control of SCD1 activity. In conclusion, our cross species study provided solid evidence to support SCD1 gene as a critical player in skeletal muscle fat metabolism.
Keywords: SCD1, SNPs, association, fat deposition, fatty acid composition.
1. Introduction
Stearoyl-CoA desaturase (SCD1) is a rate-limiting enzyme that catalyzes the synthesis of monounsaturated fatty acids, which are components of triglycerides, wax esters, cholesteryl esters and membrane phospholipids 1. The gene is highly expressed in white adipose tissue, brown adipose tissue, meibomian gland, Harderian and preputial gland under normal dietary conditions 2. However, this gene is also highly expressed in skeletal muscle from extremely obese humans 3 and from obese insulin-resistant Zucker diabetic fatty rats 4. In the human study, the authors observed that elevated SCD1 expression is associated with decreased fatty acid oxidation, increased triacylglycerol synthesis and increased monounsaturation of muscle saturated fatty acids. In the rat study, the authors revealed impairments of glucose uptake and of different steps of the insulin signaling cascade in a muscle cell line that overexpresses SCD1 and accumulates lipids. In addition, SCD1-/- mice with a targeted disruption of the SCD1 gene have reduced body adiposity, increased insulin sensitivity, and are resistant to diet-induced weight gain 5. These data suggest that elevated expression of SCD1 in skeletal muscle contributes to abnormal lipid metabolism and progression of obesity and type 2 diabetes.
However, associations of the gene with obesity and type 2 diabetes-related phenotypes in the human subjects have been reported in only two studies; therefore results are inconsistent. In 1143 elderly Swedish men involved in the Uppsala Longitudinal Study of Adult Men, Warensjö and colleagues 6 reported that subjects homozygous for the rare alleles of rs10883463, rs7849, rs2167444, and rs508384 had decreased body mass index (BMI) and waist circumference and improved insulin sensitivity. In particular, the rare allele of rs7849 demonstrated the strongest effect on both insulin sensitivity (regression coefficient (beta)=1.19, p=0.007) and waist circumference (beta=-4.4, p=0.028), corresponding to 23% higher insulin sensitivity and 4 cm less waist circumference. In contrast, Liew and colleagues 7 found that there was no association detected between SCD1 gene (at the allele, genotype or haplotype level) and diabetes-related traits including BMI and waist-to-hip ratio in a primary set of 608 diabetic subjects and 600 control subjects as well as in a replicate set of 350 young-onset diabetic patients and 747 controls. Therefore, we decided to pursue a cross-species study using cattle as a model organism and determine if the SCD1 gene is involved in muscle fat metabolism.
2. Materials and Methods
Animals
A Wagyu x Limousin reference population was used in the present study, including 6 F1 bulls, 113 F1 dams and ~250 F2 progeny. The breeding program, calf management and feeding regime were described previously by Jiang et al. 8, Kunej et al. 9 and Alexander et al. 10. Evidence has shown that the Japanese Wagyu (Black) breed of cattle has greater muscle fat deposition and softer fat than the Limousin breed (4.5% vs. 3.1% intramuscular fat content measured for the former trait and 40.2°C vs. 37.8°C melting point measured for the latter trait) 11. In particular, Wagyu fat has considerably less saturated and more unsaturated fatty acids resulting in much higher unsaturated/saturated ratios (1.9) compared with the other breeds (1.0) 12. Certainly, such a Wagyu x Limousin reference population should be a unique resource to examine association between SCD1 gene and fat metabolism phenotypes that might provide new insights into understanding genetic aspects of obesity and type 2 diabetes. As indicated above, fat accumulation and monounsaturation of saturated fatty acids in skeletal muscle are pathobiological contributors to insulin resistance in patients with these conditions.
Fat deposition measurements
Beef marbling score (BMS), subcutaneous fat depth (SFD) and percent kidney-pelvic-heart fat (KPH) were assessed by a trained evaluator. BMS was determined at the interface of the 12th and 13th ribs and was evaluated by subjective comparison of the amount of fat within the longissimus muscle with photographic standards (National Livestock and Meat Board 1981). SFD was recorded at the 12th rib at a point three-fourths the width of the longissimus muscle from its chine bone end. The amount of kidney, pelvic, and heart (KPH) fat was estimated and recorded as a percentage of carcass weight.
Fatty acid analysis
The fatty acid composition was determined using a method described previously by Rule et al. 13. Briefly, the entire core of the longissimus dorsi was sampled (i.e., devoid of trim fat and extraneous muscles) by dicing the muscle into 1.0-cm cubes, freeze-dried (Genesis 25 freeze dryer; The VirTis Co.) and then ground and homogenized using a home-style electric coffee grinder. Approximately 150 mg of dried muscle was weighed, in duplicate, into 16 × 125-mm screw-capped tubes that contained 1.0 mg of tridecanoic acid as an internal standard, and then subjected to direct saponification 13. Samples were reacted with 4.0 ml of 1.18 M KOH in ethanol at 90 °C along with frequent vortex-mixing until the sample was completely dissolved. After about 45 min, tubes were cooled, 2.0 ml of water was added and cholesterol (CHOL) was extracted with 2.0 ml of hexane that contained 0.1 mg/ml of stigmasterol as an internal standard for the cholesterol assay; the hexane phase was transferred to gas liquid chromatography (GLC) vials and sealed. One millilitre of concentrated HCl was added to the original tubes and fatty acids were extracted in 2.0 ml of hexane for fatty acid methyl ester (FAME) preparation using methanolic HCl as a catalyst. The amount of conjugated linoleic acid (CLA) was measured using acid catalysts.
Based on these measurements, we calculated: saturated fatty acids (SFA) = myristic + pentadecanoic + palmitic + heptadecanoic + stearic, monounsaturated fatty acids (MUFA) = myristoleic + pentadecenoic + palmitoleic + heptadecenoic + oleic + vaccenic and polyunsaturated fatty acids (PUFA) = linoleic + linolenic. The relative amount of SFA, MUFA and PUFA was defined as rSFA = (SFA/total fat in 100 g dry meat) x 100%, rMUFA = (MUFA/total fat in 100 g dry meat) x 100% and rPUFA = (PUFA/total fat in 100 g dry meat) x 100%, respectively. Three stearoyl-CoA desaturase activities were estimated as R1 = (14:1/14:0) x 100%, R2 = (16:1/16:0) x 100% and R3 = (18:1/18:0) x 100%.
Mutation detection
Two pairs of primers (forward, 5'-AAC TCC TAG AGA TTC CCC CAC AGG-3'/reverse: 5'-ACC TCC ATT CCA GTC CTG ACT CAC-3' and forward, 5'-GCA GAT CTA CAC ATC AGC GCA TCT-3'/reverse, 5'-TTC TCC TCG GCT TCT CTT ACA TCG-3') were designed to target the promoter region and two pairs of primers (forward, 5'- GAG CAG GGC AGT CCT AAA ACT CAA/reverse, 5'-GGC AAC CTG GCT AAT TCT TCC TCT-3' and forward, 5'- CTA CCA CTG TGC CAC TGA CTT GCT-3'/reverse, 5'-AAA AAG CTC AGA CAC AGG GCA ATC-3') to target the 3'UTR of the bovine SCD1 gene. Approximately 50 ng of genomic DNA each from six Wagyu x Limousin F1 bulls was amplified in a final volume of 10 μl that contained 12.5 ng of each primer, 150 μM dNTPs, 1.5 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl and 0.25 U of Platinum Taq polymerase (Invitrogen, Carlsbad, CA). The PCR conditions were carried out as follows: 94oC for 2 min, 35 cycles of 94oC for 30 sec, 60oC for 30 sec and 72oC for 30 sec, followed by a further 5 min extension at 72oC. PCR products were then sequenced on an ABI 3730 sequencer in the Laboratory for Biotechnology and Bioanalysis (Washington State University) using a standard protocol and polymorphisms were identified.
Genotyping and Data analysis
Three SNPs in the 3'UTR of the bovine SCD1 gene were genotyped on a Sequenom iPLEX assay using services provided by the Children's Hospital Oakland Research Institute, Oakland, California. The HAPLOVIEW program was used to determine the degrees of Hardy-Weinberg equilibrium within each marker and linkage disequilibrium among different markers 14. The phenotypic data for all measurements described above were analyzed in a fixed effects model that included the effects of year, gender, age at harvest (linear) and the genotype for each marker using the GLM (general linear model) procedure of SAS v9.1 (SAS Institute Inc., Gary, NC). Pair-wise comparisons of least squares means were performed using a protected t-test. Additionally, quantitative transmission disequilibrium test (QTDT) 15 was performed to further examine the association between markers and adjusted obesity-related phenotype data. P value <0.05 was considered statistically significant.
3. Results and Discussion
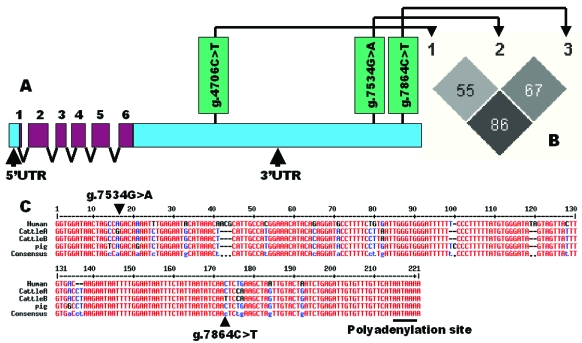
A full-length cDNA sequence of the bovine SCD1gene was retrieved from the GenBank database with a total of 5,108 bp including a 5'UTR of 144 bp, a reading frame of 1,080 bp and a long 3'UTR of 3,884 bp (NM_173959.4), respectively. However, an EST sequence (DY209631) further extends the 5'UTR to 323bp, thus producing a full-length cDNA sequence of 5,287 bp for the bovine gene. A BLAST search using this full-length cDNA sequence as a query obtained three genomic DNA sequences (AAFC02116637.1, AAFC02151622.1 and AAFC02181333.1) from the GenBank database. Alignment between cDNA and these genomic DNA sequences indicated that, like human, the bovine gene has six exons (Figure 1A). Four pairs of primers were designed to target both the promoter and 3' UTR for mutation detection, but direct sequencing of PCR products on 6 F1 bulls revealed three SNPs only in the 3'UTR (AAFC02181333.1:g.4706C>T, g.7534G>A and g.7864C>T). Interestingly, Keating and colleagues 16 also failed to identify any sequence polymorphisms in the SCD1 promoter region of Holstein Friesian, Montbeliarde, Normande, Norwegian Red, Charlois, Limousin and Kerry breeds of cattle. HAPLOVIEW analysis revealed that these three SNPs in the 3'UTR of the bovine SCD1 gene are still segregating in the population with a r2 value of 55% - 86% (Figure 1B).
Figure 1.
Characterization of single nucleotide polymorphisms in the bovine SCD1 gene. A: Genomic organization and three SNPs detected in the 3'UTR of the bovine SCD1 gene; B: Pairwise linkage disequilibrium relationship for these three SNPs mutations illustrated based on r2 measurements; and C: The proximity region of polyadenylation site is highly conserved among human, cattle and pig, where two SNPs were located in the bovine gene.
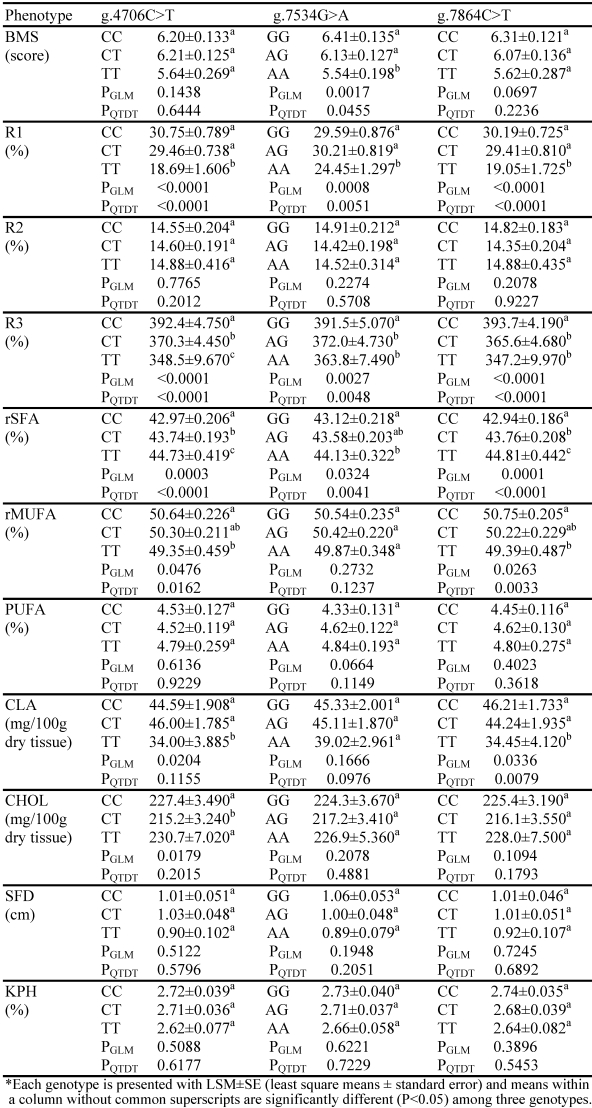
As shown in Table 1, SCD1 genotypes had no significant effects on both SFD and KPH. Rather, the gene was significantly associated with skeletal muscle fat deposition and fatty acid composition measurements. Overall, both GLM analysis and QTDT test revealed that AAFC02181333.1:g.4706C>T was significantly associated with R1, R3, rSFA and rMUFA; g.7534G>A with BMS, R1, R3 and rSFA; and g.7864C>T with R1, R3, rSFA, rMUFA and CLA (P<0.05) (Table 1), respectively. Doran and colleagues 17 investigated the effect of a reduced protein diet on the activity and expression of SCD1 as well as on the level of total lipids and the fatty acid composition of muscle and subcutaneous adipose tissue in pigs. The authors observed that a significant effect of the diet on SCD1 activity and SCD1 protein expression in muscle, but not in subcutaneous adipose tissue. These data indicate that the expression of SCD1 might be tissue-specific under a different regulatory mechanism between skeletal muscle and subcutaneous fat tissues.
Table 1.
Associations of SNPs in the bovine SCD1gene with fat deposition and fatty acid composition*
All three SNPs in the SCD1 gene were significantly associated with the estimated stearoyl-CoA desaturase activities for R1 and R3 (Table 1). In particular, the g.4706C>T polymorphism showed the largest effect on R1, being 64.53% higher in CC animals than in TT homozygotes (P<0.05); while the g.7864C>T polymorphism led to the largest difference by 13.39% in R3 between the CC and TT homozygotes (P<0.05). For g.7534G>A, the differences between GG and AA animals were 21.02% in R1 and 7.61% in R3, respectively (Table 1). Interestingly, alleles g.4706C, g.7534G and g.7864C had dominant effects on the estimated SCD1 activity R1 in comparsion to g.4706T, g.7534A and g.7864T, because the heterozygotes were almost equal to the homozygotes with the former alleles (P=0.9836). However, the inheritance mode between two alleles for each of these three polymorphic sites turned to the additive fashion for R3, because the performance of heterozygotes was close to the average between two homozygotes (P=0.9287). Unfortunately, none of these SNPs were associated with R2 (PGLM=0.2078–0.7765 and PQTDT=0.2012–0.9227) (Table 1).
Furthermore, the alleles associated with high stearoyl-CoA desaturase activities were also associated with increased beef marbling scores and increased amounts of monounsaturated fatty acids regardless of their statistical significance levels (Table 1). Among these three polymorphic sites, g.7534G>A showed the largest effect on the former trait, being 0.87 scores higher in GG animals than in AA (P<0.05); while g.7864C>T had the biggest impact on the latter phenotype, being 1.36 units higher in CC animals than in TT animals (P<0.05). However, all these alleles associated with high SCD1 activities were negatively associated with amount of saturated fatty acid in skeletal muscle (Table 1). The differences in rSFA between homozygotes with the high SCD1 active allele and those with the low SCD1 active allele ranged from –1.87 units at g.7864C>T to –1.01 units at g.7534G>A (P<0.05).
Two polymorphic sites, g.4706C>T and g.7864C>T were significantly associated with conjugated linoleic acid in the skeletal muscle based on either GLM analysis or QTDT test. The difference in the trait between CC and TT was 10.59 mg/100 g dry muscle sample for the former SNP (P<0.05) and 11.76 mg/100 g dry muscle sample for the latter SNP (P<0.05) (Table 1). Two additional features were also observed: first, alleles associated with high SCD1 activities had a high amount of CLA in muscle and second, the highly active alleles showed almost completely dominant effects on the phenotypes, because both CC and CT animals had very similar performances (P=0.8018) (Table 1). Although GLM analysis indicated significant association between g.4706C>T and CHOL, QTDT test failed to confirm the strong association (Table 1).
Our cross species study using cattle as a model organism confirmed significant associations of SCD1 gene with fat metabolism in skeletal muscle, thus supporting the finding reported by Warensjö and colleagues 6 that the SCD1 gene is related to obesity in humans. The reason for failed associations between the gene and diabetes-related traits including BMI and waist-to-hip ratio reported by Liew and colleagues 7 could be simple: the selection of SNPs for the association study. In the former study 6, markers rs10883463, rs7849, rs2167444, and rs508384 with the significant associations are 11692bp, 15341 bp, 17482 bp and 17499 bp apart from ATG (+1) site, while SNPs used in the latter study 7 are located in a range of –1057 bp to 14555 bp in relation to the ATG site of the human SCD1 gene. Certainly, none of the markers used in both studies overlapped or were in common. Interestingly, Liew and colleagues 7 reported that genotype frequencies at the +14301 A>C SNP in the 3' UTR showed borderline association (p~0.06) when evidence for linkage was taken into account. Therefore, SNPs near the 3'UTR had a stronger likelihood to show significant associations.
In our present study, these three SNPs are located in the 3'UTR of the bovine gene and in particular, two are in the proximity of polyadenylation signal site (AATAAA) (NM_173959.4). In human (NM_005063.4), cattle (NM_173959.4), pig (NM_213781.1), mouse (NM_009127.3) and rat (NM_139192.2), the SCD1 gene has a long 3'UTR of 3350 bp – 3903 bp in comparison with a coding sequence of 1068 bp to 1080 bp. Generally speaking, untranslated regions are usually less conserved than coding sequences across species. However, highly conserved untranslated regions might contribute significantly to gene function in posttranscriptional control. For example, the 3' UTRs may contain sequence elements that play crucial roles in transcript cleavage, polyadenylation and nuclear export, and in regulating the level of transcript and the stability of transcripts 18. Indeed, the 3' UTR that contains g.7534G>A and g.7864C>T in the bovine gene is also highly conserved in human and pig (Figure 1C).
A missense mutation that causes an amino acid replacement from valine (type V) to alanine (type A) was also identified in the fifth exon of bovine SCD1 gene 19. The authors genotyped this marker on 1003 Japanese Black steers and found that the SCD1 type A gene contributes to higher MUFA percentage and lower melting point in intramuscular fat. Mele and colleagues 20 also tested the same marker on dairy cows and found that milk of AA cows had a greater content of cis-9 C18:1 and total monounsaturated fatty acids and a higher C14:1/C14 ratio than did milk of VV cows. The relative contribution of SCD1 genotype to variation of monounsaturated fatty acids, cis-9 C18:1, and cis-9 C14:1 was 5, 4, and 7.7%, respectively. These animal data further confirmed that the SCD1 gene is a critical player in fatty acid metabolism.
Beef is the number one protein source in American diets and the demand for beef continues to grow. Beef is a highly nutritious and valued food, but it also contains high amounts of saturated fat as a result of biohydrogenation in the rumen 21. For many years, fats in meat have received considerable interest because in many people's minds they not only make you fat, but they also increase the risk of a number of health problems such as heart disease, stroke, diabetes, obesity, and some cancers 22. In fact, a diet high in saturated fats tends to increase blood cholesterol levels while diets high in unsaturated fats tend to lower blood cholesterol levels. Cholesterol is carried in the bloodstream as lipoproteins. Low-density lipoprotein (LDL) cholesterol is the "bad" cholesterol because elevated LDL levels are associated with an increased risk of coronary artery (heart) disease. In contrast, high-density lipoprotein (HDL) cholesterol is the "good" cholesterol since high HDL levels are associated with less coronary disease. Therefore, heart diseases are favorably affected by the consumption of certain unsaturated fatty acids because they lower plasma total chlolesterol and “bad” cholesterol levels 23. No doubt, our present study provides evidence that it is possible to produce beef with high marbling, high amount of monounsaturated fatty acids and conjugated linoleic acid content, but with low amount of saturated fatty acids. Therefore, our findings can help beef breeders and producers produce nutritionally healthy food, thus reducing the burden of health care in the nation.
Acknowledgments
This work was supported by Merial Ltd. Animal Genomics Research Fund to Z.J. This activity was also funded, in part, with an Emerging Research Issues Internal Competitive Grant from the Washington State University, College of Agricultural, Human, and Natural Resource Sciences, Agricultural Research Center to Z.J.
References
- 1.Miyazaki M, Ntambi JM. Role of stearoyl-coenzyme A desaturase in lipid metabolism. Prostaglandins Leukot Essent Fatty Acids. 2003;68:113–121. doi: 10.1016/s0952-3278(02)00261-2. [DOI] [PubMed] [Google Scholar]
- 2.Dobrzyń A, Dobrzyń P. Stearoyl-CoA desaturase--a new player in skeletal muscle metabolism regulation. J Physiol Pharmacol. 2006;57(Suppl 10):31–42. [PubMed] [Google Scholar]
- 3.Hulver MW, Berggren JR, Carper MJ et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2:251–261. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voss MD, Beha A, Tennagels N et al. Gene expression profiling in skeletal muscle of Zucker diabetic fatty rats: implications for a role of stearoyl-CoA desaturase 1 in insulin resistance. Diabetologia. 2005;48:2622–2630. doi: 10.1007/s00125-005-0025-2. [DOI] [PubMed] [Google Scholar]
- 5.Ntambi JM, Miyazaki M, Stoehr JP et al. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci U S A. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warensjö E, Ingelsson E, Lundmark P et al. Polymorphisms in the SCD1 gene: associations with body fat distribution and insulin sensitivity. Obesity (Silver Spring) 2007;15:1732–1740. doi: 10.1038/oby.2007.206. [DOI] [PubMed] [Google Scholar]
- 7.Liew CF, Groves CJ, Wiltshire S et al. Analysis of the contribution to type 2 diabetes susceptibility of sequence variation in the gene encoding stearoyl-CoA desaturase, a key regulator of lipid and carbohydrate metabolism. Diabetologia. 2004;47:2168–2175. doi: 10.1007/s00125-004-1575-4. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Z, Kunej T, Michal JJ et al. Significant associations of the mitochondrial transcription factor A promoter polymorphisms with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Biochem Biophys Res Commun. 2005;334:516–523. doi: 10.1016/j.bbrc.2005.06.120. [DOI] [PubMed] [Google Scholar]
- 9.Kunej T, Wang Z, Michal JJ, Daniels TF, Magnuson NS, Jiang Z. Functional UQCRC1 polymorphisms affect promoter activity and body lipid accumulation. Obesity (Silver Spring) 2007;15:2896–2901. doi: 10.1038/oby.2007.344. [DOI] [PubMed] [Google Scholar]
- 10.Alexander LJ, Macneil MD, Geary TW, Snelling WM, Rule DC, Scanga JA. Quantitative trait loci with additive effects on palatability and fatty acid composition of meat in a Wagyu-Limousin F2 population. Anim Genet. 2007;38:506–513. doi: 10.1111/j.1365-2052.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 11.Pitchford WS, Deland MP, Siebert BD, Malau-Aduli AE, Bottema CD. Genetic variation in fatness and fatty acid composition of crossbred cattle. J Anim Sci. 2002;80:2825–2832. doi: 10.2527/2002.80112825x. [DOI] [PubMed] [Google Scholar]
- 12.Yang A, Larsen TW, Powell VH, Tume RK. A comparison of fat composition of Japanese and long-term grain-fed Australian steers. Meat Science. 1999;51:1–9. doi: 10.1016/s0309-1740(98)00065-5. [DOI] [PubMed] [Google Scholar]
- 13.Rule DC, Broughton KS, Shellito SM, Maiorano G. Comparison of muscle fatty acid profiles and cholesterol concentrations of bison, beef cattle, elk, and chicken. J Anim Sci. 2002;80:1202–1211. doi: 10.2527/2002.8051202x. [DOI] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keating AF, Stanton C, Murphy JJ, Smith TJ, Ross RP, Cairns MT. Isolation and characterization of the bovine Stearoyl-CoA desaturase promoter and analysis of polymorphisms in the promoter region in dairy cows. Mamm Genome. 2005;16:184–193. doi: 10.1007/s00335-004-2325-0. [DOI] [PubMed] [Google Scholar]
- 17.Doran O, Moule SK, Teye GA et al. A reduced protein diet induces stearoyl-CoA desaturase protein expression in pig muscle but not in subcutaneous adipose tissue: relationship with intramuscular lipid formation. Br J Nutr. 2006;95:609–617. doi: 10.1079/bjn20051526. [DOI] [PubMed] [Google Scholar]
- 18.Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5:105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi M, Utsugi T, Oyama K et al. Genotype of stearoyl-CoA desaturase is associated with fatty acid composition in Japanese Black cattle. Mamm Genome. 2004;15:142–148. doi: 10.1007/s00335-003-2286-8. [DOI] [PubMed] [Google Scholar]
- 20.Mele M, Conte G, Castiglioni B et al. Stearoyl-coenzyme A desaturase gene polymorphism and milk fatty acid composition in Italian Holsteins. J Dairy Sci. 2007;90:4458–4465. doi: 10.3168/jds.2006-617. [DOI] [PubMed] [Google Scholar]
- 21.De Smet S, Raes K, Demeyer D. Meat fatty acid composition as affected by fatness and genetic factors: a review. Anim Res. 2004;53:81–98. [Google Scholar]
- 22.Wood JD, Enser M, Fisher AV et al. Manipulating meat quality and composition. Proc Nutr Soc. 1999;58:363–370. doi: 10.1017/s0029665199000488. [DOI] [PubMed] [Google Scholar]
- 23.Human Health Effects of Fatty Acids in Beef. Whetsell MS, Rayburn EB, Lozier JD; http://www.wvu.edu/~agexten/forglvst/humanhealth.pdf. [Google Scholar]