Abstract
A virulent (P18) strain of the Pichinde arenavirus produces a disease in guinea pigs that somewhat mimics human Lassa fever, whereas an avirulent (P2) strain of this virus is attenuated in infected animals. It has been speculated that the composition of viral genomes may confer the degree of virulence in an infected host; the complete sequence of the viral genomes, however, is not known. Here, we provide for the first time genomic sequences of both S and L segments for both the P2 and P18 strains. Sequence comparisons identify three mutations in the GP1 subunit of the viral glycoprotein, one in the nucleoprotein NP, and five in the viral RNA polymerase L protein. These mutations, alone or in combination, may contribute to the acquired virulence of Pichinde infection in animals. The 3 amino acid changes in the variable region of the GP1 glycoprotein subunit may affect viral entry by altering its receptor-binding activity. While NP has previously been shown to modulate the host immune responses to viral infection, we found that the R374K change in this protein does not affect the NP function in suppressing interferon-β expression. Four out of the five amino acid changes in the L protein occur in a small region of the protein that may contribute to viral virulence by enhancing its function on viral genomic RNA synthesis.
Introduction
Arenaviruses are bi-segmented ambisense single-stranded RNA viruses [7]. Five of the viruses (i.e., Lassa, Junin, Machupo, Guanarito, and Sabia) can cause severe viral hemorrhagic fevers (VHFs) in humans [20]. Lassa fever is endemic in certain regions of West Africa [38]. Currently, there are limited effective vaccines and treatment options for VHFs. As a result, highly pathogenic arenaviruses are included in the Category A Pathogen List of the Center for Disease Control and Prevention (CDC). Efforts to understand the basic biology and pathogenesis of these pathogenic arenaviruses in order to develop effective antiviral strategies are hampered by the hazardous nature of these infectious agents and the necessary requirement to work with them in a BSL-4 laboratory.
Pichinde virus (PICV) is a nonpathogenic arenavirus that acquires virulence in guinea pigs upon long-term passaging in the animals in a conventional BSL-2 laboratory [25]. The high-passage, virulent (P18) strain of PICV causes a disease in guinea pigs that mimics human Lassa fever in some aspects, including the correlation between viremia and outcome [1, 26], terminal vascular leakage syndrome [27], and identical distribution of viral antigens within infected hosts [8].
PICV shares a similar genomic organization with all other arenaviruses [7]. Its genome consists of two RNA segments, the large segment (L) of approximately 7.2 kb and the small (S) fragment of ~ 3.4 kb. The L RNA segment encodes the viral polymerase L protein and a small multi-functional Z protein. The S RNA segment encodes the nucleoprotein (N or NP) and the envelope glycoprotein precursor GPC that is post-translationally processed into GP1 and GP2 subunits. The genes are encoded in an ambisense orientation [7], such that the NP and L genes are encoded in the conventional negative sense, whereas the GPC and Z genes are encoded in the positive sense. At both ends of the RNA segments, the terminal 19 nucleotides, conserved among all arenaviruses, are complementary to each other and are predicted to form panhandle structures that are the cis-acting signals required for viral RNA transcription and replication [21, 40]. A unique feature among the arenavirus genomic RNAs is the noncoding intergenic regions (IGRs) located between the two ORFs on each of the segments [7]. The IGRs, ranging from 59 to 217 nts in length, are predicted to form one to three energetically stable stem-loop structures in both the genomic and antigenomic RNAs [50] and are proposed to contribute to transcriptional termination [15, 23, 32, 42]. Both L and NP proteins are essential for viral transcription and replication [41]. The viral glycoprotein GPC is expressed as a single polypeptide with an extended signal peptide and is posttranslational processed by the cellular SKI-1/S1P subtilase into GP1 and GP2 [4, 30]. GP1 is a peripheral membrane protein and GP2 a transmembrane protein. Z protein carries short proline-rich sequences called the L domains, which are known to promote virus particle budding by interacting with cellular proteins, such as Tsg101 and Nedd4 [16]. It has been shown that Lassa fever Z protein alone is sufficient for the formation of virus-like particles [48].
The molecular determinants of Pichinde virulence in guinea pigs have not been characterized due in part to the lack of sequence information of both of the viral genomic segments and a molecular clone for the virus. Published sequences of the S segment between the P2 and P18 strains have identified several sequence variations within the GPC and NP genes, suggesting that viral genomic sequence variations may dictate the differential behaviors of different strains of the virus in an infected host [51]. Reassortment experiments further suggest that genes encoded on both the S and L segments may contribute to the acquired virulence [52]. To our knowledge, the full-length sequences of the L segment for the P2 and P18 viruses are not yet available. Here, we report for the first time the complete sequences of both L and S segments of P2 and P18 viruses. Our data have confirmed the previously identified sequence variations in coding sequences of the S segments and revealed novel sequence changes in the L segments that may help define the molecular determinants of PICV virulence in an infected animal.
Materials and Methods
Viruses and cells
Both P2 and P18 viruses are derived from the Pichinde Munchique strain CoAn4763 (the 2 and 18 indicate passage numbers). The Pichinde CoAn4763 virus was passaged once in guinea pigs in Dr. Peter Jahrling’s laboratory (USAMRID). This strain was subsequently passaged once more in a guinea pig in order to produce the P2 strain. Virulent, high guinea pig-passage Pichinde virus, derived from Jahrling’s original passage-adapted CoAn4763 adPIC virus, was obtained from Dr. Dorian Coppenhaver (UTMB) as spleen stock of the 15th guinea pig passage, and was successively passaged further in inbred guinea pigs in order to obtain the P18 strain [25]. Both BHK-21 and Vero cells were maintained in modified Eagle’s medium (MEM) supplemented with 10% fetal bovine serum and 50 μg per ml of penicillin and streptomycin.
Viral RNA isolation
The P2 and P18 viruses were amplified once in BHK-21 cells at low multiplicity of infection (MOI=0.1). At 4 days post infection (dpi), viral RNA was extracted from viral particles using the Qiagen viral RNA extraction kit (Qiagen).
cDNA synthesis and PCR amplification of viral sequences
Viral RNA samples extracted from virus supernatants were cleared of possible DNA contamination by DNase I treatment for 30 min at 37°C and then at 85 °C for 15 min. Reverse transcription was conducted using specific primers for PICV S and L segments (5′ CGCACCGGGGATCCTAGG 3′, 5′ CGCACCGAGGATCCTAGGC 3′, and 5′ CGCACAGTGGATCCTAGGC 3′) according to the protocol outlined in the SuperScript III first-strand synthesis kit (Invitrogen).
PCR amplification of L or S segment was conducted using the Phusion Hot Start High-Fidelity DNA Polymerase in GC-rich buffer (New England BioLabs) with paired primers, 5′ CGCACCGGGGATCCTAGG 3′ and 5′ CGCACCGAGGATCCTAGGC 3′ for the L segment, 5′ CGCACCGGGGATCCTAGG 3′ and 5′ CGCACAGTGGATCCTAGGC 3′ for the S segment. The PCR reaction conditions were 30 cycles of 98 °C for 30 sec, 59 °C for 20 sec, and 72 °C for 3.5 min (L segment) or 1.5 min (S segment). PCR reaction was performed with the Pfu Turbo Taq polymerase (Stratagene) with paired primers targeting the flanking sequences of the IGR to amplify either a 360-bp IGR of the S segment or a 455-bp IGR of the L segment. The PCR reaction conditions were 30 cycles of 98 °C for 30 sec, 56 °C for 30 sec, and 72 °C for 30 sec.
Cloning and Sequencing
A minimum of five plasmids for each of the PCR products was sequenced in order to obtain a consensus sequence. All sequence analyses were conducted using either the Vector NTI Advance 10.0 software from InforMax (Invitrogen) or the MacVector 7.2.3 software (MacVector Inc.). GenBank accession numbers for the reported sequences are: P2 S segment, EF529744; P2 L segment, EF529745; P18 S segment, EF529746; P18 L segment, EF529747.
Interferon-β reporter assay
293T cells were cotransfected by calcium phosphate with 0.5 μg of a vector that expresses the firefly luciferase (Fluc) reporter gene from a known functional promoter of the IFN-β gene (pIFNβ-LUC), and 2 μg of the individual protein expression plasmid pCAGGS (‘empty’ control vector), pCAGGS-P2NP (expressing the P2 NP protein), pCAGGS-P18NP (expressing the P18 NP), or pcDNA3.1-NS1 (expressing the influenza A virus NS1 protein that is known to inhibit the IFN expression [17]). A β-gal expressing plasmid was included in each of the transfection reactions for normalization purpose. Six hours post transfection, cells were washed with phosphate-buffered saline and supplemented with fresh DMEM medium. At 24 h post transfection, cells were infected with Sendai virus (at moi =1) in order to induce the IFN-β expression. At 24 hpi, cell lysates were prepared for luciferase and β-gal assays using the appropriate kits that were purchased from Promega. Fluc activities obtained from the transfection reactions were normalized to the β-gal values and compared.
Results & Discussion
Cloning and sequence analysis of the S segments of the P2 and P18 viruses
PICV P2 and P18 viruses were amplified once in BHK-21 cells in order to extract enough viral RNAs for further manipulations. A previous study [52] has shown that limited passages of viruses on cultured cells required for plaque purification did not change viral pathogenicity in vivo. We, therefore, believe that a single round of virus amplification should not greatly alter the native genomic sequences of the P2 and P18 viruses. Viral RNAs were reverse transcribed using primers specific to the PICV viral terminal sequences that were deposited in GenBank. The S segment was amplified using a high-fidelity enzyme and cloned into the pCR-XL-TOPO vectors using the TA cloning technique. Consensus sequences of most of the S segments, except for the IGRs, were assembled from multiple sequences of the respective P2 and P18 strains. We chose to subclone the PCR products prior to sequencing because sequence data obtained by direct sequencing of the PCR products were usually suboptimal in quality and hard to interpret, especially around the GC-rich IGR region. In order to obtain accurate IGR consensus sequences for P2 and P18, we amplified short sequences spanning only the IGRs of the respective strains and cloned into the pGEM-T-easy vector. Multiple independent clones reveal identical sequences of the IGR for P2 and P18 as have previously been reported [51]. All in all, we believe that we have successfully assembled the consensus sequence for the full-length S segments of P2 and P18 strains that only show minor sequence variations from those that have been published previously [51]. The P2 S segments differ at 17 positions (81, 135, 187, 405, 471, 567, 699, 1014, 1158, 1206, 1242, 1365, 2466, 2712, 3222, 3374, 3398), whereas the P18 S segments differ at 5 positions (1336, 1713, 1860, 3236, 3387) from the published sequences [51]. The differences in sequence variations of the S segments obtained from the current and the previous study might result from the different sequencing methods employed. Regardless, both studies revealed the same set of amino acid changes in the S segment, except for one in the NP protein.
A published report compared sequences of the S segments between the different guinea-pig passaged PICVs and found a total of 43 nt changes [51]. However, only 19 nt changes between the avirulent strains P1 and P2 and the virulent strains P17 and P18 appeared to be associated with the degree of viral virulence. Interestingly, these nucleotide substitutions are located within the GPC and NP protein coding regions and most of which are silent mutations. Five substitution mutations (G407C, A469G, A541G, C2218T, and C3236T) result in three nonsynonymous amino acid changes (S119T, K140E, and I164V) in the GP1 subunit of the viral glycoprotein and two residue changes (R374K and A35T) in the NP protein. In the current study, a total of 33 nt differences, all of which are located within protein coding regions were found between the P2 and P18 viruses (Table 1). Consistent with the published data, most of these changes produced silent mutations. We have not formally ruled out the possibility that any of these nucleotide substitutions can alter the synthesis and/or stability of the viral S RNA segment. Four of the nt changes in the S segment (G407C, A469G, A541G, and C2218T) result in 3 amino acid substitutions in the GP1 subunit of the viral glycoprotein (S119T, K140E, and I164V) and a residue change in the NP protein (R374K). Unlike the published report, however, we did not observe the A35T change at position 3236 of the NP gene [51]. Taken together, our study has mostly confirmed data reported previously and suggests that sequences of the avirulent PICVs differ from the virulent ones at positions 119, 140, and 164 of the GPC gene and at position 374 of the NP gene (Table 1).
Table 1.
Sequence comparison of the S segment of different Pichinde virus strains (sequences are reported by Zhang et al. and in this paper)
| avirulent | avirulent | virulent | virulent | avirulent | virulent | ||
|---|---|---|---|---|---|---|---|
| nt # | P1 (Zhang et al.) | P2 (Zhang et al.) | P17 (Zhang et al.) | P18 (Zhang et al.) | P2 (this paper) | P18 (this paper) | codon |
| 81 | C (Ser) | C (Ser) | T (Ser) | T (Ser) | T (Ser) | T (Ser) | |
| 99 | A (Gln) | A (Gln) | G (Gln) | G (Gln) | A (Gln) | G (Gln) | |
| 135 | C (Thr) | C (Thr) | C (Thr) | C (Thr) | T (Thr) | C (Thr) | |
| 136 | C (Leu) | T (Leu) | C (Leu) | C (Leu) | T (Leu) | C (Leu) | |
| 187 | C (Leu) | T (Phe) | C (Leu) | C (Leu) | C (Leu) | C (Leu) | |
| 225 | C (Asp) | T (Asp) | T (Asp) | T (Asp) | T (Asp) | T (Asp) | |
| 226 | A (Ser) | G (Gly) | G (Gly) | G (Gly) | G (Gly) | G (Gly) | |
| 243 | A (Arg) | A (Arg) | G (Arg) | G (Arg) | A (Arg) | G (Arg) | |
| 405 | T (Thr) | T (Thr) | T (Thr) | T (Thr) | C (Thr) | T (Thr) | |
| 407 | G (Ser) | G (Ser) | C (Thr) | C (Thr) | G (Ser) | C (Thr) | GPC (S119T) |
| 469 | A (Lys) | A (Lys) | G (Glu) | G (Glu) | A (Lys) | G (Glu) | GPC (K140E) |
| 471 | A (Lys) | G (Lys) | A (Lys) | A (Lys) | A (Lys) | A (Lys) | |
| 541 | A (Ile) | A (Ile) | G (Val) | G (Val) | A (Ile) | G (Val) | GPC (I164V) |
| 567 | A (Glu) | A (Glu) | G (Glu) | G (Glu) | G (Glu) | G (Glu) | |
| 699 | C (Asn) | T (Asn) | T (Asn) | T (Asn) | C (Asn) | T (Asn) | |
| 897 | C (Asp) | C (Asp) | T (Asp) | C (Asp) | C (Asp) | C (Asp) | |
| 1014 | G (Glu) | A (Glu) | A (Glu) | A (Glu) | G (Glu) | A (Glu) | |
| 1053 | A (Gln) | G (Gln) | G (Gln) | G (Gln) | G (Gln) | G (Gln) | |
| 1158 | C (Leu) | C (Leu) | C (Leu) | C (Leu) | T (Leu) | C (Leu) | |
| 1206 | C (Tyr) | C (Tyr) | C (Tyr) | C (Tyr) | T (Tyr) | C (Tyr) | |
| 1242 | A (Pro) | G (Pro) | G (Pro) | G (Pro) | A (Pro) | G (Pro) | |
| 1336 | C (Leu) | C (Leu) | C (Leu) | G (Val) | C (Leu) | C (Leu) | |
| 1365 | C (Gly) | T (Gly) | T (Gly) | T (Gly) | C (Gly) | T (Gly) | |
|
| |||||||
| 1625 | C | T | T | T | T | T | IGR |
|
| |||||||
| 1698 | G (Pro) | G (Pro) | A (Pro) | A (Pro) | G (Pro) | A (Pro) | |
| 1703 | G (Leu) | G (Leu) | A (Leu) | A (Leu) | G (Leu) | A (Leu) | |
| 1713 | T (Pro) | T (Pro) | G (Pro) | T (Pro) | T (Pro) | C (Pro) | |
| 1716 | C (Lys) | C (Lys) | T (Lys) | C (Lys) | C (Lys) | C (Lys) | |
| 1728 | G (Val) | G (Val) | A (Val) | A (Val) | G (Val) | A (Val) | |
| 1860 | A (Tyr) | A (Tyr) | A (Tyr) | A (Tyr) | A (Tyr) | G (Tyr) | |
| 2067 | G (Leu) | G (Leu) | A (Leu) | A (Leu) | G (Leu) | A (Leu) | |
| 2218 | C (Arg) | C (Arg) | T (Lys) | T (Lys) | C (Arg) | T (Lys) | NP (R374K) |
| 2300 | C (Asp) | T (Asn) | T (Asn) | T (Asn) | T (Asn) | T (Asn) | |
| 2328 | C (Lys) | T (Lys) | C (Lys) | C (Lys) | T (Lys) | C (Lys) | |
| 2343 | C (Val) | T (Val) | C (Val) | T (Val) | T (Val) | T (Val) | |
| 2355 | G (Asp) | A (Asp) | G (Asp) | A (Asp) | A (Asp) | A (Asp) | |
| 2435 | G (Leu) | G (Leu) | A (Leu) | A (Leu) | G (Leu) | A (Leu) | |
| 2466 | A (Thr) | A (Thr) | A (Thr) | A (Thr) | G (Thr) | A (Thr) | |
| 2712 | G (Phe) | G (Phe) | G (Phe) | G (Phe) | A (Phe) | G (Phe) | |
| 2757 | G (Val) | G (Val) | A (Val) | A (Val) | G (Val) | A (Val) | |
| 2805 | C (Leu) | C (Leu) | T (Leu) | T (Leu) | C (Leu) | T (Leu) | |
| 2844 | G (Asn) | G (Asn) | A (Asn) | A (Asn) | G (Asn) | A (Asn) | |
| 2945 | G (Leu) | G (Leu) | A (Leu) | A (Leu) | G (Leu) | A (Leu) | |
| 2946 | T (Gln) | T (Gln) | C (Gln) | C (Gln) | T (Gln) | C (Gln) | |
| 3078 | A (Leu) | G (Leu) | A (Leu) | A (Leu) | G (Leu) | A (Leu) | |
| 3213 | G (Phe) | G (Phe) | A (Phe) | A (Phe) | G (Phe) | A (Phe) | |
| 3222 | A (Ala) | A (Ala) | A (Ala) | A (Ala) | G (Ala) | A (Ala) | |
| 3236 | C (Ala) | C (Ala) | T (Thr) | T (Thr) | C (Ala) | C (Ala) | |
|
| |||||||
| 3374 | G | A | G | G | G | G | 3′UTR |
| 3387 | A | A | A | G | A | A | |
| 3398 | G | A | G | G | G | G | |
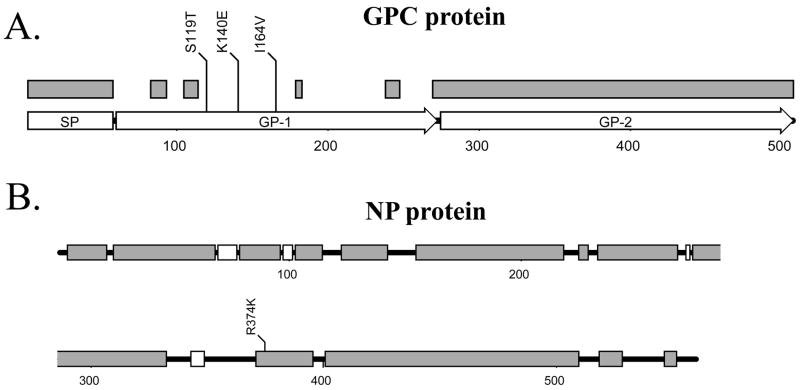
Arenaviral GPC precursor protein contains a long conserved 58-aa signal peptide (SP) and is cleaved into GP-1 and GP-2 by the host SKI-1/S1P subtilase [4, 30]. Sequence alignment of all known arenaviral GPC proteins suggests that GP-1 is highly variable among the arenaviruses with only a few short stretches of conserved sequences (Fig. 1A, gray boxes), whereas sequences of GP-2 and the signal peptide (SP) are highly conserved. Sequence variations in GP1 may correlate with its functions: GP-1 forms the receptor-binding site and is recognized by host neutralizing antibodies, which may drive the evolution of this viral glycoprotein subunit. GP-2, on the other hand, is a transmembrane anchored subunit of the glycoprotein, which is usually not exposed to neutralizing antibodies and, therefore, can maintain its conservative sequence and structure. Accordingly, the 3 mutations (S119T, K140E, and I164V) identified in our study are located within the highly variable regions of the GP-1 subunit (Fig. 1A). These residue changes may affect virus-receptor binding specificity and may contribute to pathogenesis in an infected host. Many viral surface glycoproteins play a major role in determining viral tropism and pathogenic processes. For example, influenza virus hemagglutinin (HA) is one of the virulence determinants of the H5N1 viruses in mammals. Sequence changes in the viral HA glycoprotein have been shown to alter its receptor binding specificity that allows the virus to “jump” host species [19, 45, 47]. The Ebola GP glycoprotein is another example of a major determinant of viral tropism and virulence [12, 13, 49].
Fig. 1.
Location of various natural mutations in viral GPC and NP proteins. (A) A schematic representation of GPC protein, which consists of a long signal peptide (SP), GP-1, and GP-2. The conserved regions among GPC proteins of arenaviruses are shown as gray boxes. The three natural mutations identified between the P2 and P18 viral variants are also shown. (B) A schematic representation of the NP protein. The conserved domains among all arenaviruses are shown as gray bars, while the conserved regions only among the New World arenaviruses are shown as white bars. The single R374K mutation identified between the P2 and P18 viral variants is also shown.
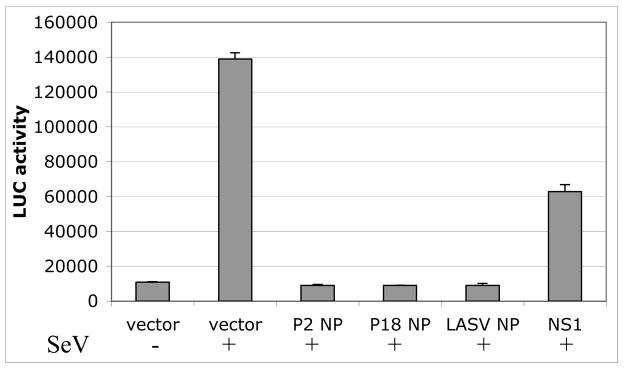
The NP protein is indispensable for viral RNA replication and transcription as it intimately interacts with viral genomic RNA. Its primary sequence is, therefore, generally highly conserved among the arenaviruses (Fig. 1B). A single residue change identified between P2 and P18 viruses turns out to be a conserved amino acid substitution (i.e., R374K). Although this amino acid residue is located within one of the conserved domains of the protein (Fig. 1B), the residue itself is not conserved among the arenaviruses. Recently, a novel function for the NP protein of the LCMV virus in down-regulating interferon expression of the infected cells has been revealed [35]. This implicates NP as a potential virulence factor. We, therefore, have undertaken a study to determine whether the R374K substitution can inhibit the IFN-β production. To do this, 293T cells were cotransfected by calcium phosphate with 0.5 μg of a vector that expresses the firefly luciferase (Fluc) reporter gene from a known functional promoter of the IFN-β gene, and 2 μg of the individual protein-expression plasmid pCAGGS (‘empty’ control vector), pCAGGS-P2NP (expressing the P2 NP protein), pCAGGS-P18NP (expressing the P18 NP), or pcDNA3.1-NS1 (expressing the flu-A NS1 protein, which has been shown to strongly inhibit the IFN-β gene). A β-gal expressing plasmid (at 0.1 μg) was included in each of the transfection reactions for normalization purpose. Six hours post transfection, cells were washed with phosphate-buffered saline and supplemented with fresh DMEM medium. At 24 h post transfection, cells were infected with Sendai virus (at moi =1) in order to induce the IFN-β expression. At 24 hpi, cell lysates were prepared for luciferase and β-gal assays using the appropriate kits and protocols supplied by Promega. Fluc activities obtained from the transfection reactions were normalized by the β-gal values and compared. Our data showed that both the P2 NP and P18 NP inhibited IFN-β expressions, as judged by the decreased levels of the Fluc reporter expression, to a similar extent (Fig. 2), suggesting that a single conservative amino acid change in the NP gene between the P2 and P18 virus does not impact its role in IFN-β inhibition. This is consistent with recent findings that NP proteins from Pichinde virus and a variety of other arenaviruses, except for the Tacaribe virus, inhibit activation of the promoters of IFN-β and interferon regulatory factor 3 (IRF3), as well as the nuclear translocation of the IRF3 factor [36].
Fig. 2.
Both P2 and P18 NP proteins inhibit IFN-β expression. 293T cells were transfected with an IFNβ-LUC reporter plasmid, together with either empty vector or vector expressing P2 NP, P18 NP, Lassa virus NP, or influenza A virus NS1. A β-gal expression plasmid was included in each reaction for normalization purpose. Cells were infected with or without Sendai virus to induce IFN-β expression. Luciferase activity was measured at 24 hpi and normalized against β-gal activity.
Cloning and sequence analysis of the L segments of the P2 and P18 viruses
To clone the L segment, we first attempted to amplify the entire segment in a single PCR reaction using primers that annealed to the conserved ends. We successfully amplified a 7-kb PCR product, which is the approximate size of the full-length L segment, but failed to clone it into vector. Since no published sequences of the L segment for P2 or P18 viruses were available, we could not design PCR primers to amplify shorter fragments for subcloning purpose. Instead, we decided to digest the 7-kb PCR product with BamH I and Bgl II restriction enzymes and subcloned a 1.4-kb DNA fragment representing the middle region of the L segments into the pBluescript vector. Using the consensus sequences of this fragment, we designed primers to amplify the rest of the viral L segments from viral genomic RNA. At least 5 independent clones for each of the fragments, representing the 5′, middle, and 3′ regions of the L segments were sequenced in order to obtain the consensus sequences. The IGR sequence of the L segment was obtained similarly as described above for that of the S segment.
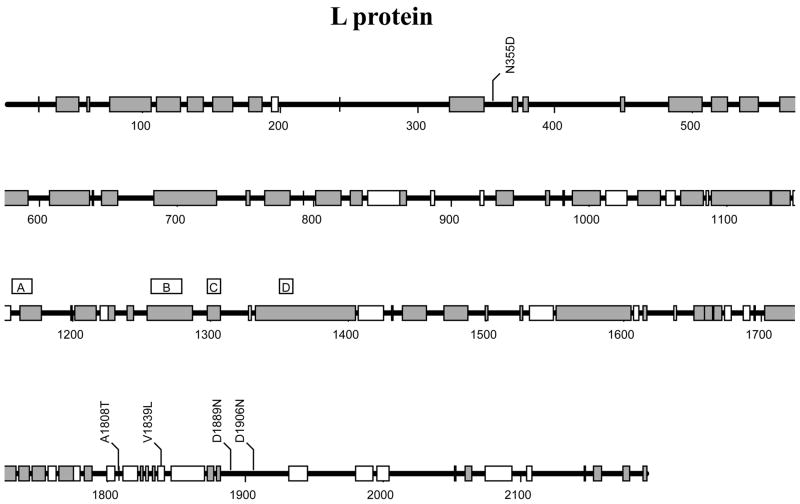
The full-length sequences of the L segment of the P2 and P18 viruses have a length of 7057 nts and carry 15 nucleotide differences between these two virus strains. The sequences of the Z protein between P2 and P18 viruses are absolutely conserved, suggesting that no differences in its known multitudes of activity, such as virus assembly and budding [11, 29, 39, 48] and regulation of viral RNA replication [9, 10, 18, 24, 31], are expected between the viruses. All changes on the L segments are localized to the L polymerase protein-coding region, five of which cause amino acid changes (N355D, A1808T, V1839L, D1889N, and D1906N) (Table 2). These changes are predicted to affect the function of the L polymerase protein and thus to potentially alter the viral replicative potential in vivo. The rest of the sequence changes (at nts #473, 860, 1178, 1529, 2972,3173, 3485, 3611, 6149, and 6812) produce silent mutations (Table 2). Again, we have not determined whether these silent mutations can alter the synthesis and/or stability of the L RNA segment.
Table 2.
Sequence comparison of the L segment from avirulent P2 and virulent P18 Pichinde viruses.
| avirulent | virulent | ||
|---|---|---|---|
| nt # | P2 | P18 | codon |
| 473 | G (Phe) | A (Phe) | |
| 860 | T (Val) | A (Val) | |
| 1178 | C (Lys) | T (Lys) | |
| 1312 | C (Asp) | T (Asn) | L (D1906N) |
| 1363 | C (Asp) | T (Asn) | L (D1889N) |
| 1513 | C (Val) | G (Leu) | L (V1839L) |
| 1529 | G (Tyr) | A (Tyr) | |
| 1606 | C (Ala) | T (Thr) | L (A1808T) |
| 2972 | T (Gly) | C (Gly) | |
| 3173 | G (Asn) | A (Asn) | |
| 3485 | C (Leu) | T (Leu) | |
| 3611 | A (Asn) | G (Thr) | |
| 5965 | T (Asn) | C (Asp) | L (N355D) |
| 6149 | A (Cys) | G (Cys) | |
| 6812 | G (Leu) | A (Leu) | |
The L polymerase is a large protein of more than 2100 residues that functions as an RNA-dependent RNA polymerase. Due to its enormous size, no structural information is currently available for this protein. Sequence alignments of all known arenaviral L proteins have provided some insights into the possible functional domains of this protein, however. L protein contains, in the central region, the four conserved motifs (A, B, C, and D) identified in all RNA-dependent polymerases, which constitute the “polymerase module” implicated in template recognition and polymerizing activity [43] (Figs. 3 and 4A). In particular, the highly conserved SDD sequence in the motif C of the protein (Fig. 4A) is predicted to play a central role in the enzyme’s polymerizing activity. Alignment of all available sequences of arenaviral L proteins also reveals multiple conserved domains across this protein (Fig. 3, gray boxes), suggesting that the mechanism for L protein to recognize, replicate, and transcribe the ambisense segmented RNA genome is conserved among the arenaviruses. Among the 5 identified residue changes in the L protein between the P2 and P18 viruses, four (N355D, A1808T, D1889N, and D1906N) are localized to the variable regions, whereas the V1839L change is located within a region conserved among the new world arenaviruses (Fig. 3). This residue always encodes a valine among the new world arenaviruses (Fig. 4B), but nevertheless is variable among the old world arenaviruses (data not shown). We have yet to determine whether the V1839L substitution has an effect on L polymerase functions.
Fig. 3.
A schematic representation of the viral polymerase L protein. The conserved domains of this protein among arenaviruses are shown as gray bars, whereas the conserved regions only among the New World arenaviruses are shown as white bars. The four conserved domains (A, B, C, D) among all RNA-dependent polymerases are indicated [43]. The six mutations identified between P2 and P18 viral variants are also shown.
Fig. 4.
(A) Sequence alignment of the four conserved domains (A, B, C, and D) of RNA-dependent polymerases [43] in the arenavirus L proteins, with the invariant D, G, DD, and K residues in each domain boxed. (B) Sequence alignment of a conserved region among the New World arenavirus near the V1839L mutation.
Mutagenesis of the LCMV L gene has produced various viral mutants that affect plaque morphology [28], induce lethality in infected guinea pigs [44], or suppress the cytotoxic T-cell response in order to initiate persistent viral infection [46]. It is, therefore, reasonable to suggest that any of the five amino acid substitutions in the L gene identified in our study, either individually or in combination (Fig. 3), may specify determinants of viral virulence of the PICV P18 virus in an infected animal. Although we do not know how these mutations affect the function of L polymerase in viral RNA synthesis, we speculate that some of these changes are significant as they represent amino acid substitutions that can potentially alter the overall folding of the protein. Interestingly, four of the mutations (A1808T, V1839L, D1889N, and D1906N) are localized within a small region of 99 amino acid residues, implying that this domain may affect the efficiency of viral RNA synthesis and/or replication. It is not uncommon that the molecular determinant of viral virulence is localized to the polymerase enzyme. A single amino acid substitution in one subunit of the influenza viral polymerase (e.g., PB2 E627K) has been shown to enhance the degree of virulence of the H5N1 virus in mice [22] that may explain the high lethality observed with the 1918 pandemic flu virus. It has also been documented that a single amino acid change (K1079Q) in the L gene of the LCMV virus increases virus production and infection of macrophages, which may possibly be responsible for viral persistence and immune suppressive effect in an infected animal [37].
In summary, our sequence analyses of both the S and L segments of the P2 and P18 viruses have uncovered 3 amino acid changes in the GPC protein, 1 in the NP protein, and 5 in the L protein that may be responsible for their observed phenotypic differences in infected animals. We hypothesize that these mutations, either acting alone or in combination, are potential determinants of viral virulence in the animals. Previous studies have suggested that aberrant cytokine production and suppressed immune signaling are the underlying mechanisms for the high virulence of P18 virus in guinea pigs [2, 5, 6, 14]. These observations are consistent with those made for other hemorrhagic fever-causing viruses. For example, Lassa fever virus does not activate macrophages following infection [3, 33]. The immuno-regulatory function of dendritic cell are known to be inhibited by Ebola or Lassa virus infection [34]. How natural mutations in GPC, NP, and/or L proteins contribute to the altered production of proinflammatory cytokines and the suppressed immune signaling mechanism are not understood. Several mechanisms to explain the phenomenon are proposed: 1) mutation of the GPC protein may change the receptor specificity and thus allows for a more efficient spreading of the virus within an infected host that can lead to an increased proinflammatory cytokine production; 2) mutations of GPC protein may enable the virus to more efficiently infect host immune cells and impair their functions, leading to the observed suppressed immune response; 3) mutations in NP and/or L proteins may enhance viral replication in vivo, and thus overwhelm the normal host immune responses to viral infection; and 4) mutations of any of the viral proteins identified in this study may modulate its interaction with host cell signaling proteins, leading to the differential responses to viral infection. Development of a reverse genetics system for PICV will help to unravel the molecular mechanisms leading to the acquired virulence of viral infection in guinea pigs, and the roles of the individual viral proteins in viral replication and pathogenesis. These efforts will undoubtedly provide important insights into the pathogenesis of hemorrhagic fever caused by the different arenaviruses.
Acknowledgments
This research was supported by the Emory University Research Committee (URC) and the Southeast Regional Center of Excellence for Emerging Infections and Biodefense (SERCEB) to HL and YL, the Emory Digestive Diseases Research and Development Center P/F fund (DK64399) and the Center for AIDS Research (CFAR P30 AI050409) to HL, and the pilot project component of Dr. Ahmed’s U19 grant (RFA-AI-02-042) to YL.
Footnotes
The original publication is available at www.springerlink.com
References
- 1.Aronson JF, Herzog NK, Jerrells TR. Pathological and virological features of arenavirus disease in guinea pigs. Comparison of two Pichinde virus strains. Am J Pathol. 1994;145:228–35. [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson JF, Herzog NK, Jerrells TR. Tumor necrosis factor and the pathogenesis of Pichinde virus infection in guinea pigs. Am J Trop Med Hyg. 1995;52:262–9. doi: 10.4269/ajtmh.52-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot MC, Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J Immunol. 2004;172:2861–9. doi: 10.4049/jimmunol.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 4.Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. 2003;77:2866–72. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowick GC, Fennewald SM, Elsom BL, Aronson JF, Luxon BA, Gorenstein DG, Herzog NK. Differential signaling networks induced by mild and lethal hemorrhagic fever virus infections. J Virol. 2006;80:10248–52. doi: 10.1128/JVI.01384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowick GC, Fennewald SM, Scott EP, Zhang L, Elsom BL, Aronson JF, Spratt HM, Luxon BA, Gorenstein DG, Herzog NK. Identification of differentially activated cell-signaling networks associated with pichinde virus pathogenesis by using systems kinomics. J Virol. 2007;81:1923–33. doi: 10.1128/JVI.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchmeier MJ, Bowen MD, Peters CJ. Arenaviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Lippincott-Raven Publisher; Philadelphia, PA: 2001. pp. 1635–1668. [Google Scholar]
- 8.Connolly BM, Jenson AB, Peters CJ, Geyer SJ, Barth JF, McPherson RA. Pathogenesis of Pichinde virus infection in strain 13 guinea pigs: an immunocytochemical, virologic, and clinical chemistry study. Am J Trop Med Hyg. 1993;49:10–24. doi: 10.4269/ajtmh.1993.49.10. [DOI] [PubMed] [Google Scholar]
- 9.Cornu TI, de la Torre JC. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol. 2001;75:9415–26. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornu TI, de la Torre JC. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J Virol. 2002;76:6678–88. doi: 10.1128/JVI.76.13.6678-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichler R, Strecker T, Kolesnikova L, ter Meulen J, Weissenhorn W, Becker S, Klenk HD, Garten W, Lenz O. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP) Virus Res. 2004;100:249–55. doi: 10.1016/j.virusres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Feldmann H, Volchkov VE, Volchkova VA, Klenk HD. The glycoproteins of Marburg and Ebola virus and their potential roles in pathogenesis. Arch Virol Suppl. 1999;15:159–69. doi: 10.1007/978-3-7091-6425-9_11. [DOI] [PubMed] [Google Scholar]
- 13.Feldmann H, Volchkov VE, Volchkova VA, Stroher U, Klenk HD. Biosynthesis and role of filoviral glycoproteins. J Gen Virol. 2001;82:2839–48. doi: 10.1099/0022-1317-82-12-2839. [DOI] [PubMed] [Google Scholar]
- 14.Fennewald SM, Aronson JF, Zhang L, Herzog NK. Alterations in NF-kappaB and RBP-Jkappa by arenavirus infection of macrophages in vitro and in vivo. J Virol. 2002;76:1154–62. doi: 10.1128/JVI.76.3.1154-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franze-Fernandez MT, Iapalucci S, Lopez N, Rossi C. Subgenomic RNAs of Tacaribe virus. In: Salvato MS, editor. The Arenaviridae. Plenum Press; New York: 1993. pp. 113–132. [Google Scholar]
- 16.Freed EO. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 2003;11:56–9. doi: 10.1016/s0966-842x(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Sastre A. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology. 2001;279:375–84. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- 18.Garcin D, Rochat S, Kolakofsky D. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J Virol. 1993;67:807–12. doi: 10.1128/jvi.67.2.807-812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–6. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunther S, Lenz O. Lassa virus. Crit Rev Clin Lab Sci. 2004;41:339–90. doi: 10.1080/10408360490497456. [DOI] [PubMed] [Google Scholar]
- 21.Hass M, Westerkofsky M, Muller S, Becker-Ziaja B, Busch C, Gunther S. Mutational analysis of the lassa virus promoter. J Virol. 2006;80:12414–9. doi: 10.1128/JVI.01374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–2. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 23.Iapalucci S, Lopez N, Franze-Fernandez MT. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology. 1991;182:269–78. doi: 10.1016/0042-6822(91)90670-7. [DOI] [PubMed] [Google Scholar]
- 24.Jacamo R, Lopez N, Wilda M, Franze-Fernandez MT. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. J Virol. 2003;77:10383–93. doi: 10.1128/JVI.77.19.10383-10393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahrling PB, Hesse RA, Rhoderick JB, Elwell MA, Moe JB. Pathogenesis of a pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect Immun. 1981;32:872–80. doi: 10.1128/iai.32.2.872-880.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson KM, McCormick JB, Webb PA, Smith ES, Elliott LH, King IJ. Clinical virology of Lassa fever in hospitalized patients. J Infect Dis. 1987;155:456–64. doi: 10.1093/infdis/155.3.456. [DOI] [PubMed] [Google Scholar]
- 27.Katz MA, Starr JF. Pichinde virus infection in strain 13 guniea pigs reduces intestinal protein reflection coefficient with compensation. J Infect Dis. 1990;162:1304–8. doi: 10.1093/infdis/162.6.1304. [DOI] [PubMed] [Google Scholar]
- 28.Kirk WE, Cash P, Peters CJ, Bishop DH. Formation and characterization of an intertypic lymphocytic choriomeningitis recombinant virus. J Gen Virol. 1980;51:213–8. doi: 10.1099/0022-1317-51-1-213. [DOI] [PubMed] [Google Scholar]
- 29.Lee KJ, Perez M, Pinschewer DD, de la Torre JC. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J Virol. 2002;76:6393–7. doi: 10.1128/JVI.76.12.6393-6397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A. 2001;98:12701–5. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez N, Jacamo R, Franze-Fernandez MT. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol. 2001;75:12241–51. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez N, Franze-Fernandez MT. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res. 2007;124:237–44. doi: 10.1016/j.virusres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Lukashevich IS, Maryankova R, Vladyko AS, Nashkevich N, Koleda S, Djavani M, Horejsh D, Voitenok NN, Salvato MS. Lassa and Mopeia virus replication in human monocytes/macrophages and in endothelial cells: different effects on IL-8 and TNF-alpha gene expression. J Med Virol. 1999;59:552–60. [PMC free article] [PubMed] [Google Scholar]
- 34.Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170:2797–801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2006;80:9192–9. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Sobrido L, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matloubian M, Kolhekar SR, Somasundaram T, Ahmed R. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J Virol. 1993;67:7340–9. doi: 10.1128/jvi.67.12.7340-7349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCormick JB, Webb PA, Krebs JW, Johnson KM, Smith ES. A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis. 1987;155:437–44. doi: 10.1093/infdis/155.3.437. [DOI] [PubMed] [Google Scholar]
- 39.Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A. 2003;100:12978–83. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez M, de la Torre JC. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2003;77:1184–94. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinschewer DD, Perez M, de la Torre JC. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol. 2003;77:3882–7. doi: 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinschewer DD, Perez M, de la Torre JC. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol. 2005;79:4519–26. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. Embo J. 1989;8:3867–74. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Oldstone MB. Genetic mapping of lymphocytic choriomeningitis virus pathogenicity: virulence in guinea pigs is associated with the L RNA segment. J Virol. 1985;55:704–9. doi: 10.1128/jvi.55.3.704-709.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–8. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 46.Salvato M, Borrow P, Shimomaye E, Oldstone MB. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J Virol. 1991;65:1863–9. doi: 10.1128/jvi.65.4.1863-1869.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–10. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 48.Strecker T, Eichler R, Meulen J, Weissenhorn W, Dieter Klenk H, Garten W, Lenz O. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected] J Virol. 2003;77:10700–5. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahl-Jensen VM, Afanasieva TA, Seebach J, Stroher U, Feldmann H, Schnittler HJ. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J Virol. 2005;79:10442–50. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson SM, Clegg JC. Sequence analysis of the S RNA of the African arenavirus Mopeia: an unusual secondary structure feature in the intergenic region. Virology. 1991;180:543–52. doi: 10.1016/0042-6822(91)90068-m. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Marriott K, Aronson JF. Sequence analysis of the small RNA segment of guinea pig-passaged Pichinde virus variants. Am J Trop Med Hyg. 1999;61:220–5. doi: 10.4269/ajtmh.1999.61.220. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Marriott KA, Harnish DG, Aronson JF. Reassortant analysis of guinea pig virulence of pichinde virus variants. Virology. 2001;290:30–8. doi: 10.1006/viro.2001.1127. [DOI] [PubMed] [Google Scholar]