Abstract
Eukaryotic cells encode AMP-lysine hydrolases related to the rabbit histidine triad nucleotide-binding protein 1 (Hint1) sequence. Bacterial and archaeal cells have Hint homologs annotated in a variety of ways but the enzymes have not been characterized, nor have phenotypes been described due to loss of enzymatic activity. We developed a quantitative 31P NMR assay to determine whether Escherichia coli possesses an adenosine phosphoramidase activity. Indeed, soluble lysates prepared from wild-type laboratory Escherichia coli exhibited activity on the model substrate adenosine monophosphoramidate (AMP-NH2). The Escherichia coli Hint homolog, which had been comprehensively designated ycfF and is here named hinT, was cloned, over-expressed, purified and characterized with respect to purine nucleoside phosphoramidate substrates. Bacterial hinT was several times more active than mammalian Hint on three model substrates. In addition, bacterial and mammalian enzymes preferred guanosine versus adenosine phosphoramidates as substrates. Analysis of the lysates from a constructed hinT knockout strain of Escherichia coli demonstrated that all of the cellular purine nucleoside phosphoramidase activity is due to hinT. Physiological analysis of this mutant revealed that the loss of hinT enzymatic activity results in failure to grow in media containing 0.75 KCl, 0.9 M NaCl, 0.5 M NaOAc and 10 mM MnCl2. Thus, bacteria may possess nucleotidylylated phosphoramidate substrates that must be hydrolyzed to support growth under certain high salt conditions.
Keywords: Phosphoramidase, E. coli hinT, AMP-lysine hydrolase, ycfF
INTRODUCTION
Histidine triad enzymes are a superfamily of nucleoside monophosphate hydrolases and transferases containing an active site motif related to His-X-His-X-His-X-X, where X is a hydrophobic amino acid (1). The Hint branch, named for similarity with rabbit Hint1, is the most ancient branch, having representatives in all forms of life (1,2). The best biochemical substrates of these enzymes are model compounds containing a sidechain adenylylated-lysine residue from which Hint hydrolyzes AMP, leaving unmodified lysine (3,4). Loss of enzymatic activity in the S. cerevisiae homolog hnt1 produces cells that are temperature-sensitive on galactose media and hypersensitive to mutations in components of the general transcription apparatus, TFIIH (namely Kin28, Ccl1, Tfb3) and to mutations in the cyclin-dependent kinase activating kinase Cak1 (3). Birds encode a typical Hint gene on the Z chromosome, such that males have two doses, while avian Asw genes, which may function as dominant negative inhibitors of Hint via Gln127, are repeated 40 times on the female-specific W chromosome (5). The hint1 homolog of mouse is a nonessential gene (6) whose loss predisposes to an increased incidence of tumors (7). Two less typical members of the Hint branch of the superfamily are Aprataxin, which is mutated in ataxia-oculomotor apraxia1 (8,9), and the scavenger decapping enzyme, Dcps (10-12). The second and third branches of the HIT superfamily are homologs of the Fhit tumor suppressor protein and enzymes related to galactose-1-phosphate uridylyltransferase (1). Fhit and homologs are diadenosine polyphosphate hydrolases but Fhit functions as a tumor suppressor is not limited by substrate hydrolysis but rather substrate-binding (13). Enzymes in the GalT branch of the superfamily are nucleoside monophosphate transferases for a variety of unusual nucleotide substrates (1). Galactosemia in humans is usually caused by mutations that inactivate GalT activity (1), consistent with the idea that Hint branch enzymes and GalT branch enzymes have a conventional relationship between loss of enzymatic and biological function.
Although Hint hydrolases have been associated only recently with adenosine phosphoramidase activity, nucleoside phosphoramidase activity has been observed in partially purified extracts from rabbit liver (14,15), whole cells and extracts of the human T-leukemia, CEM cells (16), peripheral blood mononuclear cells (PBMCs) (17,18) and green monkey Vero cells (19). Part of the reason for continued interest in phosphoramidates has been their demonstrated utility as prodrugs of antiviral and anticancer nucleoside monophosphates, or pronucleotides (20-24). In general there is a requirement that therapeutic nucleosides be converted to at least the corresponding monophosphate before demonstrating biological activity. Nevertheless, many nucleosides are not substrates for the requisite nucleoside kinase. To overcome this hurdle, several pronucleotide approaches have been investigated, including the use of nucleoside monophosphoramidates (23,25). Although, the enzyme responsible for phosphoramidate hydrolysis has not been determined, direct evidence of intracellular P-N bond hydrolysis by a putative phosphoramidase has been demonstrated by studies of the intracellular metabolism of fluorodeoxyuridine phosphoramidates with permeablized cells (16) and O18 labeled AZT tryptophan methyl ester phosphoramidate with capillary LC-ESI-MS/MS (24). Recently, Bieganowski and coworkers reported that both S. cerevisiae Hnt1 and rabbit Hint1 hydrolyze AMP-NH2, AMP-lysine, AMP-Mor and AMP-alanine and suggested that Hint hydrolases might be the enzymes responsible for nucleoside monophosphoramidate prodrug activation in vivo (3). Consequently, in addition to gaining an understanding of their natural function, the study of Hint hydrolases may facilitate the design of tissue and species specific pronucleotides of potential therapeutic utility.
In this study, we developed a simple 31P NMR assay capable of detecting total phosphoramidase activity in cell lysates. We scanned the E. coli genome (26) for a Hint homolog and discovered an ORF named ycfF at the 16min position (161090-1161467) on the E. coli genetic map with 47% amino acid sequence identity to rabbit and human Hint1. Testing the hypothesis that ycfF encodes a E. coli Hint homolog (here refer to hinT) that is responsible for observed cellular phosphoramidase activity, we cloned, purified and characterized the activity of hinT in vitro and also constructed a mutant strain in which the hinT gene was deleted. Supporting the view that hinT is entirely responsible for nucleoside monophosphoramidate hydrolysis, such substrates were completely stable in E. coli hinT mutant extracts. Further characterization demonstrated that bacterial hinT is homodimeric and capable of hydrolyzing adenosine and guanosine 5′-phosphoramidate monoesters significantly faster than mammalian Hint (3). In addition, evaluation of E. coli hinT mutants established a requirement for protein expression when grown on NaCl, KCl and MnCl2, and phosphoramidase activity when grown on NaOAc.
EXPERIMENTAL PROCEDURES
Evaluation of Phosphoramidase Activity in E. coli Cell Lysates
Adenosine 5′-monophosphoramidate (AMP-NH2) was used to investigate the phosphoramidase activity in E. coli cell lysates since it was earlier shown to be a specific substrate for rabbit and yeast Hint hydrolases (3). Phosphoramidase activity in cell lysates was measured by the 1H-decoupling mode of 31P NMR spectroscopy (Varian VAC-300 spectrometer), which can clearly distinguish the substrate, AMP-NH2, and the product, adenosine 5′-monophosphate (AMP). NMR spectrometry is suitable for quantitation by integration of the area under the peak, which is proportional to the amount of nuclei present. The relative ratio of peak areas can be used to calculate the amount of turnover corrected based on a standard curve (Supplementary, Fig. 1). The information about chemical shifts is described in Fig. 1 of supplementary information. Cell lysates from E. coli Tuner(DE3)pLacI cells (Novagen) were obtained by treatment with lysozyme (1mg/ml) and the DNA was precipitated by protamine sulfate followed by centrifugation to remove cell debris. The supernatant was dialyzed extensively against Buffer A (20mM Tris pH 7.0; 1mM EDTA; 1mM DTT, 0.01mM PMSF) to remove cellular phosphates and nucleotides. Phosphate free cell lysates (0.75mg total protein) were incubated with 5mM AMP-NH2 in 0.5 ml reaction buffer (0.5mM MgCl2, 20mM HEPES pH 7.2) at 37°C for intervals of 15min, 30 min, and 12h. The reactions were terminated by snap freezing in a dry ice/acetone slush bath and lyophilized for 5 h. The samples were then dissolved in NMR buffer (600μL, 50mM EDTA, 20mM HEPES pH 7.2, and 1mM TMP (trimethylphosphate, internal standard)) and the insoluble portion was spun down by centrifugation at 12,500g for 5min. A 31P NMR spectrum was then collected for the supernatant. For the low pH control experiment, 10mM AMP-NH2 in reaction buffer (500μL, pH 2.0) was incubated at 37°C for 12h and the 31P signal was detected under the same conditions.
Genetic Deletion of E. coli hinT
The E. coli hinT gene in strain BW25113 was disrupted as described (27). Plasmid pKD3 and primers 7024 and 7025 (primer sequences in Supplemental Table 1) were used for amplification of the chloramphenicol resistance marker (Cmr). Stable chloramphenicol resistant transformants of BW25113 were tested by PCR with primers 7026 and 7027 to confirm correct recombination of the Cmr marker into the hinT locus. A strain that tested positive by PCR was named BB1. Strain BB3 was constructed by removing the antibiotic resistance marker using Red recombinase as previously described (27). The hinT gene was amplified from the chromosomal DNA of E. coli strain BW25113 with primers 7328 and 7329. The product of this reaction was cloned into Bluescript SK+ using EcoRI and BamHI sites included in the primers’ sequences. The resulting plasmid was named pB429. Mutagenesis (28) of phagemid DNA with primer 7330 resulted in plasmid pB431 containing hinT with an H101A substitution. Plasmids pB432 and pB433 were obtained by cloning HindIII - EcoRI fragments containing wild type and H101A mutated alleles of hinT from plasmids pB429 and pB431 into plasmid pACYC184. All E. coli transformations were performed by electroporation
Plasmid Constructions
The expression construct pPH70D was described previously (29). In which the E. coli DHFR (dihydrofolate reductase) gene followed by a thrombin cleavage site were incorporated in the expression vector. The N-terminus of desired proteins were fused to DHFR with a 15 a.a. thrombin linker (29). The fusion proteins can be first purified using MTX (methotrexate) affinity chromatography followed by cleavage with human thrombin to release the native proteins. Both plasmids were constructed by replacing the NAT2 (hamster polymorphic N-acetyltransferase 2) in pPH70D with the desired proteins.
To create plasmid pTFCF15DmY, E. coli genomic DNA was purified from K12 strain EMG2 (ATCC) according to a standard protocol (30). Using isolated genomic DNA as the template, a 385bp fragment was amplified by PCR with primers 101 and 102, which contain the restriction sites XhoI and XbaI, respectively. The PCR product was ligated into the T/A cloning vector, pSTBlue 1 (Novagen). Novablue supercompetent cells were transformed with the ligation mixture, and insert-containing clones were selected by blue/white screening on X-gal/IPTG indicator plates. The resulting plasmid pTFCY-TA was digested with XhoI and XbaI and subcloned into previously XhoI and XbaI digested and purified pPH70D. The ligation mixture was transformed into chemically competent E. coli DH5α cells (Novagen). The plasmid pTFCF15DmY was purified with QIAprep Spin Miniprep kit (Qiagen) and the desired sequence was confirmed by DNA sequencing (Advanced Genetic Analysis Center, University of Minnesota). The over-expression E. coli strain Tuner(DE3)pLacI cells (Novagen) were made competent by the Hanahan method (31) and transformed with pTFCF15DmY. To express the E. coli hinT-H101A mutant, the E. coli hinT-pSAG02 plasmid was subjected to site-directed mutagenesis using primer 103 and 104 (3).
To express human Hint1, the pJLCF15DmH plasmid was created following a similar procedure described above. Total mRNA was isolated using Straight A’s mRNA isolation System (Novagen) from CEM cell line, a human T-lymphoblast leukemia cell line, and the cDNA library was synthesized by reverse transcriptase and DNA polymerase I/ RNase H according to the manufacturer’s protocol (Novagen). Using this cDNA library as template, the human Hint1 cDNA was amplified by PCR with primers 105 and 106.
Expression and Purification of Recombinant Proteins
The cell growth and cell lysates extraction were described previously except that 0.5 mM IPTG was used for induction (29). Bacterial or human Hint-DHFR fusion proteins were purified by methotrexate (MTX)-agarose (Sigma) using a 12.5-ml column, washed with 40 column volumes of wash buffer A (20mM Tris pH 7.0; 1mM EDTA; 1mM DTT, 0.01mM PMSF), 60 column volumes of wash buffer A with 1M NaCl, and followed by folate elution (5mM folate; 32 mM Tris pH 9.0; 1mM EDTA; 1mM DTT, 0.01mM PMSF). Fractions (8ml) were collected at a flow rate 3ml/min and an aliquot (10μL) of each fraction was assayed for protein concentration with the Bradford dye reagent (Biorad). Fractions containing more than 0.1mg/ml of protein were analyzed by 12% SDS-PAGE, and DHFR activities were determined (32,33). The standard DHFR assay mixture contained 50μM DHF, 100μM NADPH and 1mM DTT in MTEN buffer (50 mM 2-morpholinoethanesulfonic acid, 25 mM tris(hydroxymethyl)aminomethane, 25 mM ethanolamine, and 100 mM NaCl, pH 7.0), and the enzyme in a final volume of 1.0 ml. The reaction was started by the addition of DHF (33). Fractions containing pure Hint-DHFR fusion proteins were pooled and concentrated to 2mg/ml. A 2.5-ml protein solution was further purified and exchanged with buffer A without PMSF by Sephadex G-25 PD-10 desalting column (Amersham Pharmacia).
Purified Hint-DHFR fusion proteins were dialyzed against thrombin-cleavage buffer (50 mM Tris, pH 8.0; 0.1 M NaCl; 2.5mM CaCl2) and digested with human thrombin (Sigma) at 4° C for 19h, with 8 units of human thrombin per milligram of protein. To separate Hint hydrolases from DHFR domains, reaction mixtures were applied onto a 10ml AMP-agarose affinity column (Sigma) and washed with 16 column volumes of buffer A with 1M NaCl, 4 column volumes of buffer A, and eluted with adenosine buffer (2mM adenosine, 20mM Tris pH 7.0; 1mM EDTA; 1mM DTT). Fractions (8ml) were collected at a flow rate of 3ml/min and aliquots (10μL) from each fraction were used to determine the protein concentration. Fractions containing more than 0.1mg/ml of protein were analyzed by 18% SDS-PAGE. Fractions containing pure Hint protein were pooled and concentrated to 2.6mg/ml.
Rabbit Hint1 and E. coli hinT-H101A were purified by AMP-agarose affinity chromatography as previously described (3,34) except that expression was performed in the E. coli hinT deletion mutant (BB1) described in this study.
Substrate Specificity
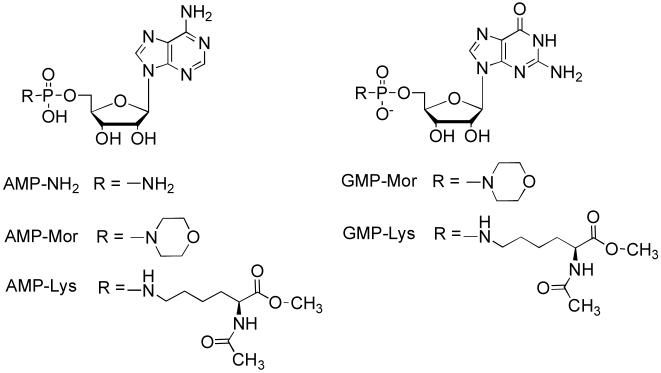
AMP-NH2, adenosine 5′-monophosphomorpholidate (AMP-Mor) and guanosine 5′-monophosphomorpholidate (GMP-Mor) (10mM) were incubated with hHint1-DHFR, E. coli hinT-DHFR, hHint1, rabbit Hint1, and E. coli hinT (2.5 or10μg) in the reaction buffer (500μL, 0.5mM MgCl2, 20mM HEPES pH 7.2) at 22°C for intervals of 36 and 66min. To determine the residual phosphoramidase activity in E. coli hinT-H101A, AMP-NH2(10mM) was incubated with E. coli hinT-H101A (1200 and 2700μg) in the reaction buffer at 22°C for 14h. The reactions were quenched by adding 5M NaOH (20μL), followed by snap freezing in a dry ice/acetone slush bath, and lyophilized. The synthetic substrates, AMP-Lysine and GMP-Lysine (Fig. 2), (2.5mM) were incubated with E. coli hinT (0.25μg) and hHint1 (0.625μg) for 20 and 40min. The reactions were stopped by snap freezing in a dry ice/acetone slush bath and lyophilized. The sample preparation for 31P NMR measurement was as described previously for the cell-lysate experiment except that the pH was adjusted to 7.0 by the addition of 12μL 6M HCl before NMR spectra were collected.
Figure 2.
Structures of Substrates.
RESULTS
E. coli Cell Lysates Possess Adenosine Phosphoramidase Activity
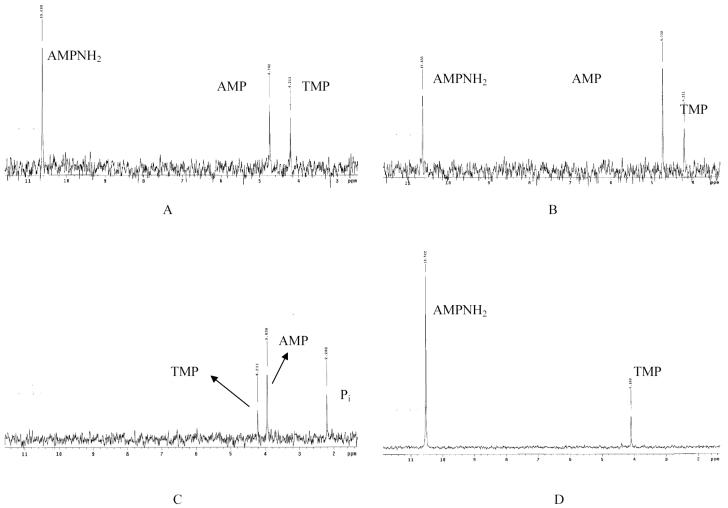
Tuner(DE3)pLacI, a modified E. coli BL21 strain, was used to determine phosphoramidase activity in cell lysates. At pH 7.2, the AMP-NH2 hydrolysis rate by E. coli cell lysates was 76.3 (± 20.9) nmol mg-1 min-1; data were averaged for incubation time intervals of 15min, and 30min (Fig. 1A, B). The 12h-incubation reaction showed complete hydrolysis of AMP-NH2 to AMP, as well as limited further cleavage of AMP into inorganic phosphate (Pi) and adenosine. The production of Pi may be the result of E. coli phosphatases or chemical hydrolysis (Fig. 1C). To verify that the formation of AMP was due to an enzymatic process and not chemical hydrolysis, a control experiment was performed by incubating 10 mM AMP-NH2 in pH 2.0 buffer at 37°C for 12h (Fig. 1D). The pH 2.0 test conditions were chosen due to the acid lability of phosphorous-nitrogen bond (P-N bond) (35). Clear AMP-NH2 and TMP signals were visible by 31PNMR, with no detectable AMP or Pi, confirming that AMP-NH2 is stable at pH 2.0 for at least 12h.
Figure 1.
Adenosine monophosphoramidase activity in E. coli cell lysates.
(A) AMP-NH2 (5mM) incubated with E. coli cell lysates (0.75mg total protein) at 37°C for 15min; (B) for 30min; (C) for 12h. (D) Control: AMP-NH2 (10mM) incubated with pH 2.0 buffer at 37°C for 12h.
hinT is the Only Bacterial Source of Adenosine Monophosphoramidase Activity
To determine if E. coli hinT is solely responsible of the observed lysates phosphoramidase activity, a mutant strain, BB1, in which the hinT gene was replaced with the chloramphenicol resistance marker, was constructed. The phosphoramidase activities of the wild-type E. coli BL21 Star strain and the mutant strain lysates were determined (Supplementary Fig. 3). When using AMP-NH2 as a substrate, the specific activity of BL21 Star WT cell lysates was found to be 20.1 (± 7.7) nmol mg-1 min-1. However, no AMP formation could be observed with lysates derived from the hinT deletion strain, BB1. (Supplementary Information; Fig. 3 and table 2). Therefore, we conclude that the adenosine phosphoramidase activity observed for E. coli cell lysates is totally dependent on hinT.
Nucleoside and Leaving Group Specificity of Bacterial and Mammalian Hint Hydrolases
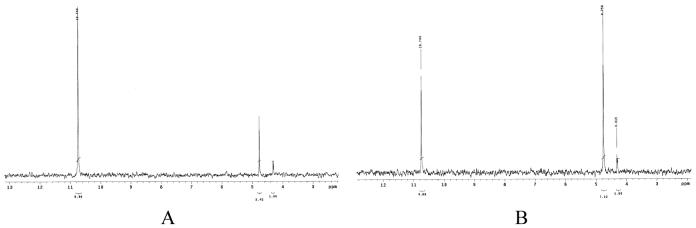
Both E. coli hinT and human Hint1 were purified to homogeneity as DHFR fusion proteins by MTX affinity chromatography (see Supplementary Fig. 4, Lanes 1, 2 and 6). Removal of the DHFR affinity handle by thrombin digestion (Supplementary Fig. 4, Lane 7) and AMP affinity chromatography afforded the purified recombinant enzymes (Supplementary Fig.4, Lanes 3-5). The apparent molecular weights of recombinant purified E. coli hinT and E. coli hinT-DHFR fusion protein were analyzed by analytical gel filtration chromatography on a Superdex G-75 size exclusion column strongly indicating, as has been observed for rabbit and human Hint1, the formation of a stable homodimer in solution (Supplementary Fig. 5). Based on our 31P NMR assay, we found that E. coli hinT and human Hint1 are adenosine phosphoramidases with specific activities of 526 (± 27) and 196 (± 33) nmol nmol-1 min-1, respectively. Phosphoramidase activity could not be observed in cell extracts expressing the E. coli hinT-H101A mutant. (Data not shown) However, when the protein was purified and sufficiently concentrated, enzyme concentration dependent activity for the E. coli hinT-H101A mutant (0.017 (± 0.002) nmol nmol-1 min-1) was determined. (Fig. 3)
Figure 3.
Phosphoramidase activity of the E. coli hinT-H101A by 31 P NMR Assay. (A) E. coli hinT-H101A, 1200μg; (B) 2700μg were incubated with AMPNH2 (10mM) at 22°C for 14h.
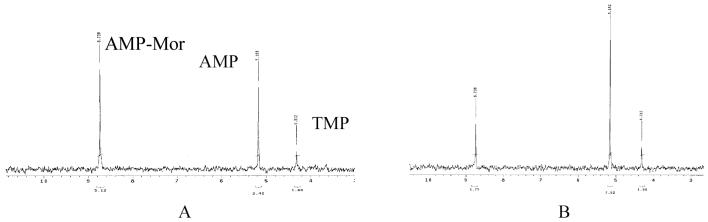
The co-crystal structure of rabbit Hint1 with GMP (2) and the fact that GMP can elute the rabbit Hint1 bound to the adenosine affinity column (34) imply that guanosine phosphoramidates may also be substrates for Hint hydrolases. To evaluate the substrate specificity with respect to the different purine nucleoside phosphoramidates, the purified Hint hydrolases were evaluated with AMP-Mor and GMP-Mor (Fig. 2). AMP-Mor has previously been shown to be a reasonable substrate for rabbit Hint1 and yeast Hnt1 (3). All three Hint hydrolases were fully capable of utilizing both compounds as substrates with a 2-fold preference for the GMP-Mor. Since AMP-lysine was shown to be a substrate for rabbit Hint1, both AMP-lysine and GMP-lysine were also evaluated as substrates for E.coli hinT and human Hint1. As observed for the morpholino compounds, GMP-lysine was approximately a 2-fold better substrate than AMP-lysine for both enzymes. (4) Hydrolysis rates for the three Hint hydrolases with each substrate are summarized in table 1. Two representative NMR spectra of E. coli hinT incubation with AMP-Mor are shown in figure 4.
Table 1.
Summary of the specific activity (nmol nmol-1 min-1) of the recombinant purified Hint hydrolases
| Substrate | E. coli hinT-DHFR | Human Hint1-DHFR | E. coli hinT | Human Hint1 | Rabbit Hint1 |
|---|---|---|---|---|---|
| AMP-NH2 | 753±2 | 388±45 | 526±27 | 196±33 | 70.0±1.3 |
| AMP-Mor | 411±4 | 43.4±4.0 | 360±1 | 45.0±0.1 | 26.6±0.6 |
| GMP-Mor | 675±17 | 87.2±11.8 | 669±1 | 78.7±1.3 | 48.6±0.4 |
| AMP-lysine | ND | ND | 529±7 | 102±19 | ND |
| GMP-lysine | ND | ND | 836±262 | 232±68 | ND |
Figure 4.
Phosphoramidase activity of the E. coli hinT by 31 P NMR Assay. (A) E. coli hinT protein (2.5μg) incubated with AMP-Mor (10mM) at 22°C for 36min. (B) for 66min.
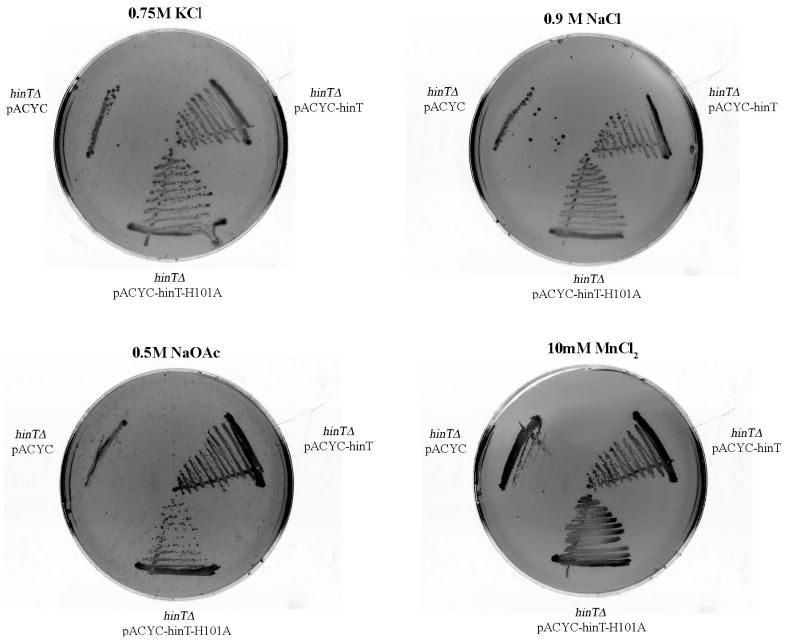
Hint is required for growth on the LB-agar media containing 0.75, KCl, 0.9 M NaCl, 0.5 M NaOAc, and 10mM MnCl2. The E. coli hinT knockout strain BB3 (hinTΔ) was transformed with either an empty vector (hinTΔ pACYC), a plasmid expressing E.coli hinT (hinTΔ pACYC-hinT), or a plasmid expressing the His-101 to Ala mutant (hinTΔ pACYC-hinT-H101A). Colonies were observed for BB3 cells transformed with pACYC-hinT (hinTΔ pACYC-hinT) and pACYC-hinT-H101A (hinTΔ, pACYC-hinT-H101A) when grown with LB-agar media containing 0.75M KCl, 0.9 M NaCl, and 10mM MnCl2. When cultures were grown in the presence of 0.5 M NaOAc, colonies were observed for BB3 cells transformed with pACYC-hinT, while a marked reduction in the number of colonies for cells transformed with pACYC-hinT-H101A could be seen.
hinT is Required for Growth Under High Salt Conditions
The E. coli hinT knockout strain BB3 was transformed with either a control plasmid, pACYC, a plasmid expressing E.coli hinT, pACYC-hinT, or a plasmid expressing the catalytically inefficient HINT His-101 to Ala mutant, pACYC-hinT-H101A. When cultures were grown under high chloride salt conditions (LB-agar media containing 0.75M KCl, 0.9 M NaCl, and 10mM MnCl2), colonies were observed for BB3 cells transformed with pACYC-hinT (hinTΔ pACYC-hinT, Fig. 4) and pACYC-hinT-H101A (hinTΔ, pACYC-hinT-H101A Fig. 4). However, when cultures were grown in the presence of 0.5 M NaOAc, colonies were observed for BB3 cells transformed with pACYC-hinT, while a marked reduction in the number of colonies for cells transformed with pACYC-hinT-H101A could be seen. Consequently, the expression of catalytically active hinT appears to be required by E. coli for growth under high salt conditions.
DISCUSSION
Nucleoside phosphoramidates have been exploited as a general strategy for the delivery of nucleoside monophosphate antiviral and anticancer agents. Extensive analysis of their metabolism by cell extracts and whole cells has provided supporting evidence that this transformation is enzyme mediated. Recently, both yeast Hnt1 and rabbit Hint1 have been shown to be AMP-lysine hydrolases, thus implying that this class of enzymes are at least partially responsible for earlier observations of this activity (3,4). A test of this hypothesis required development of an assay that could measure total cellular nucleoside monophosphoramidate hydrolase activity and a genetic system to knock out or knock down candidate genes. Here we accomplished this in E. coli.
The 31P NMR assay requires little or no sample preparation and is capable of accurately determining the concentration of substrates and products because the chemical shifts of the phosphoramidate and the monophosphate are generally separated by 4-6 ppm. In addition, we have demonstrated that this assay is independent of the structure of either the nucleoside or amine that compose the nucleoside phosphoramidate.
Applying our assay to the investigation of the phosphoramidate hydrolase potential of bacteria cells, we found that AMP-NH2 was rapidly hydrolyzed by E. coli cell extracts. When cloned and expressed, E. coli hinT and human Hint1 were shown to be adenosine/guanosine phosphoramidases. In addition, rabbit Hint1, known to be an adenosine phosphoramidase, was shown to be a guanosine phosphoramidase as well. We can also conclude, based on gene replacement in the BB1 cell lysates studies, that hinT is the only purine phosphoramidase expressed by E. coli. Evaluation of the substrate specificity of the E.coli hinT and hHint1 revealed that GMP-lysine was a 3-fold better substrate for hHint1 than GMP-Mor, but only modestly preferred by E. coli hinT. A similar conclusion was observed AMP-NH2 and AMP-Mor. These results suggest that the two enzymes differ in their capacity to accommodate variability in substrate steric bulk. The preference of all three Hint proteins for the guanosine may result from hydrogen bonding between the N2 amino group of guanine and the backbone carbonyl oxygen of an active site residue, as observed in the GMP-bound rabbit Hint1 crystal structure (2). However, although E. coli hinT has almost 50% sequence identity with human and rabbit Hint1, the overall greater catalytic activity of the bacterial enzyme suggests that key differences do exist between these enzymes. On going structural and enzymatic analysis of these enzymes should provide the insights necessary to develop a rational for this discrepancy.
The physiological role of mammalian Hint proteins remains controversial, with a mixture of conflicting evidence and isolated experiments. An interaction between a Hint1 homolog and Cdk7 was observed in a yeast-two hybrid assay (36). However, studies with transgenic mice, in which the Hint1 was deleted, failed to reveal a phenotype consistent with the regulation of Cdk7 (6). Our observation of the association of E. coli hinT with growth under high salt conditions is the first suggestion of the possible physiological role of these enzymes in bacteria. Since the sensitivity of BB3 to high concentrations of KCl, NaCl, NaOAc and MnCl2 could be remedied by complementation with a plasmid capable of expressing catalytically active E. coli hinT, it can be concluded that lose of hinT phosphoramidase activity is responsible for this phenotype and not other ORFs residing in the operon containing hinT. Optimal growth on NaOAc appears, however, appears to require a significantly more active enzyme than on chloride salts, which may be reflective of discrepancies in the molecular events responsible for this phenotype. Given the wide distribution of highly sequence similar genes in a wide variety of over 126 different gram positive and gram-negative bacteria (Data not shown), the expression of these proteins may be a general requirement for bacterial cell growth in an environment of high salt.
A possible unique role for Hint proteins has been proposed in regulating the nucleotidylation of protein substrates (3). The “nucleotidylation” post-translational modification has been observed for the regulation of the glutamine synthetase activity by two enzymes: uridylyl transferase and adenylyl transferase (37). AMP-N-ε-(N-α-acetyl lysine methyl ester) can be easily hydrolyzed by rabbit HINT1, yeast HNT1 (3). Our data also indicate that both AMP-lysine and GMP-lysine are excellent substrates for E. coli hinT and human Hint1 (Table 1). Based on these observations, it is likely that Hint proteins maybe involved in regulating either protein adenylylation and/or guanylylation. Ongoing studies should define the natural substrates and physiological role of Hint proteins and relate them to the observed phenotype. In addition, the potential differences in substrate specificity observed between the bacterial and mammalian enzymes suggest that the design of bacteria-specific pronucleotides may be achievable.
Supplementary Material
Glossary
Abbreviations
- AMP
adenosine 5′-monophosphate
- AMP-NH2
adenosine 5′-monophosphoramidate
- AMP-lysine
AMP-N-ε-(N-α-acetyl lysine methyl ester) 5′-phosphoramidate
- AMP-Mor
adenosine 5′-monophosphomorpholidate
- AZT
3′-azido-3′-deoxythymidine
- Cdk7
cyclin dependent kinase 7
- DHF
dihydrofolate
- DHFR
dihydrofolate reductase
- DTT
dithiothreitol
- EDTA
ethylenediaminetetracetic acid
- Fhit
fragile histidine triad
- FLT
3′-fluoro-3′-deoxythymidine
- GalT
galactose-1-phosphate uridylyltransferase
- GMP-lysine
GMP-N-ε-(N-α-acetyl lysine methyl ester) 5′-phosphoramidate
- GMP-Mor
guanosine 5′-monophosphomorpholidate
- HEPES
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- Hint
histidine triad nucleotide-binding protein
- IPTG
isopropyl-β-D-thiogalactopyranoside
- LC-ESI-MS/MS
liquid chromatograph negative mode electrospray ionization tandem mass spectrometry
- MTX
methotrexate
- NADPH
β-nicotinamide adenine dinucleotide 2′-phosphate reduced
- PAGE
polyacrylamide gel electrophoresis
- 31P NMR
phosphorus nuclear magnetic resonance
- Pi
inorganic phosphate
- PMSF
phenylmethanesulfonyl fluoride
- P-N bond
phosphorous-nitrogen bond
- X-gal
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
- RNase H
ribonuclease H
- Tris
tris(hydroxymethyl)aminomthane
- SDS
sodium dodecyl sulfate
- TFIIH
transcription factor II H
- TMP
trimethylphosphate
Reference
- 1.Brenner C. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner C, Garrison P, Gilmour J, Peisach D, Ringe D, Petsko GA, Lowenstein JM. Nat. Struct. Biol. 1997;4:231–238. doi: 10.1038/nsb0397-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieganowski P, Garrison PN, Hodawadekar SC, Faye G, Barnes LD, Brenner C. J. Biol. Chem. 2002;277:10852–10860. doi: 10.1074/jbc.M111480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krakowiak A, Pace HC, Blackburn GM, Adams M, Mekhalfia A, Kaczmarek R, Baraniak J, Stec WJ, Brenner C. J. Biol. Chem. 2004;279:18711–18716. doi: 10.1074/jbc.M314271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pace HC, Brenner C. Genome Biol. 2003;4:R18. doi: 10.1186/gb-2003-4-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korsisaari N, Rossi DJ, Luukko K, Huebner K, Henkemeyer M, Makela TP. Mol. Cell. Biol. 2003;23:3929–3935. doi: 10.1128/MCB.23.11.3929-3935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su T, Suzui M, Wang L, Lin CS, Xing WQ, Weinstein IB. Proc. Natl. Acad. Sci. USA. 2003;100:7824–7829. doi: 10.1073/pnas.1332160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Nat. Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 9.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. Nat. Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Rodgers ND, Jiao X, Kiledjian M. The EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salehi Z, Geffers L, Vilela C, Birkenhager R, Ptushkina M, Berthelot K, Ferro M, Gaskell S, Hagan I, Stapley B, McCarthy JE. Mol. Microbiol. 2002;46:49–62. doi: 10.1046/j.1365-2958.2002.03151.x. [DOI] [PubMed] [Google Scholar]
- 12.Kwasnicka DA, Krakowiak A, Thacker C, Brenner C, Vincent SR. J. Biol. Chem. 2003;278:39051–39058. doi: 10.1074/jbc.M306355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapasso F, Krakowiak A, Cesari R, Arkles J, Yendamuri S, Ishii H, Vecchione A, Kuroki T, Bieganowski P, Pace HC, Huebner K, Croce CM, Brenner C. Proc. Natl. Acad. Sci. USA. 2003;100:1592–1597. doi: 10.1073/pnas.0437915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledneva RK, Preobrazhenskaya NN, Shinskii NG, Shabarova ZA, Prokof’ev MA. Dokl. Akad. Nauk Sssr. 1970;193:1308–1310. [Google Scholar]
- 15.Dudkin SM, Ledneva RK, Shabarova ZA, Prokofiev MA. FEBS Lett. 1971;16:48–50. doi: 10.1016/0014-5793(71)80682-8. [DOI] [PubMed] [Google Scholar]
- 16.Abraham TW, Kalman TI, McIntee EJ, Wagner CR. J. Med. Chem. 1996;39:4569–4575. doi: 10.1021/jm9603680. [DOI] [PubMed] [Google Scholar]
- 17.McIntee EJ, Remmel RP, Schinazi RF, Abraham TW, Wagner CR. J. Med. Chem. 1997;40:3323–3331. doi: 10.1021/jm960694f. [DOI] [PubMed] [Google Scholar]
- 18.Chang SL, Griesgraber GW, Southern PJ, Wagner CR. J. Med. Chem. 2001;44:223–231. doi: 10.1021/jm000260r. [DOI] [PubMed] [Google Scholar]
- 19.Abraham TW, McIntee EJ, Vidhya VI, Schinazi RF, Wagner CR. Nucleosides, Nucleotides and Nucleic Acids. 1997;16:2079–2092. [Google Scholar]
- 20.McGuigan C, Kinchington D, Nicholls SR, Nickson C, O’Connor TJ. Bioorg. Med. Chem. Lett. 1993;3:1207–1210. [Google Scholar]
- 21.Balzarini J, Karlsson A, Aquaro S, Perno CF, Cahard D, Naesens L, De Clercq E, McGuigan C. Proc. Natl. Acad. Sci. USA. 1996;93:7295–7299. doi: 10.1073/pnas.93.14.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGuigan C, Cahard D, Sheeka HM, De Clercq E, Balzarini J. Bioorg. Med. Chem. Lett. 1996;6:1183–1186. [Google Scholar]
- 23.Wagner CR, Iyer VV, McIntee EJ. Med. Res. Rev. 2000;20:417–451. doi: 10.1002/1098-1128(200011)20:6<417::aid-med1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Chou T.-f., Griesgraber GW, Wagner CR. Molecular Pharmaceutics. 2004:102–111. doi: 10.1021/mp0340338. [DOI] [PubMed] [Google Scholar]
- 25.Drontle DPW, C R. Mini-reviews in Medicinal Chemistry. 2004;4:409–419. doi: 10.2174/1389557043403945. [DOI] [PubMed] [Google Scholar]
- 26.Brenner C, Bieganowski P, Pace HC, Huebner K. J. Cell. Physiol. 1999;181:179–187. doi: 10.1002/(SICI)1097-4652(199911)181:2<179::AID-JCP1>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunkel TA, Roberts JD, Zakour RA. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 29.Sticha KR, Sieg CA, Bergstrom CP, Hanna PE, Wagner CR. Protein Expr. Purif. 1997;10:141–153. doi: 10.1006/prep.1997.0734. [DOI] [PubMed] [Google Scholar]
- 30.Current protocols in molecular biology supplement 56, Unit 2.4
- 31.Hanahan D. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 32.Stone SR, Morrison JF. Biochemistry. 1982;21:3757–3765. doi: 10.1021/bi00259a006. [DOI] [PubMed] [Google Scholar]
- 33.Morrison JF, Stone SR. Biochemistry. 1988;27:5499–5506. doi: 10.1021/bi00415a017. [DOI] [PubMed] [Google Scholar]
- 34.Gilmour J, Liang N, Lowenstein JM. Biochem. J. 1997;326:471–477. doi: 10.1042/bj3260471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song H, Johns R, Griesgraber GW, Wagner CR, Zimmerman CL. Pharm. Res. 2003;20:448–451. doi: 10.1023/a:1022616523678. [DOI] [PubMed] [Google Scholar]
- 36.Korsisaari N, Makela TP. J. Biol. Chem. 2000;275:34837–34840. doi: 10.1074/jbc.C000505200. [DOI] [PubMed] [Google Scholar]
- 37.Jaggi R, van Heeswijk WC, Westerhoff HV, Ollis DL, Vasudevan SG. The EMBO J. 1997;16:5562–5571. doi: 10.1093/emboj/16.18.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The value includes the Gly-Thr-Leu-Glu tetrapeptide in N-terminal after thrombin cleavage.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.