Abstract
Hint, histidine triad nucleotide-binding protein, is a universally conserved enzyme that hydrolyzes AMP linked to lysine and, in yeast, functions as a positive regulator of the RNA polymerase II C-terminal domain kinase, Kin28. To explore the biochemical and structural bases for the adenosine phosphoramidate hydrolase activity of rabbit Hint, we synthesized novel substrates linking a p-nitroaniline group to adenylate (AMP-pNA) and inhibitors that consist of an adenosine group and 5′ sulfamoyl (AdoOSO2NH2) or N-ethylsulfamoyl (AdoOSO2NHCH2CH3) groups. AMP-pNA is a suitable substrate for Hint that allowed characterization of the inhibitors: titration of each inhibitor into AMP-pNA assays revealed their Ki values. The N-ethylsulfamoyl derivative has a 13-fold binding advantage over the sulfamoyl adenosine. The 1.8 Å co-crystal structure of rabbit Hint with N-ethylsulfamoyl adenosine revealed a binding site for the ethyl group against Trp123, a residue that reaches across the Hint dimer interface to interact with the alkyl portion of the inhibitor and, presumably, the alkyl portion of a lysyl substrate. Ser107 is positioned to donate a hydrogen bond to the leaving group nitrogen. Consistent with a role in acid-base catalysis, the Hint S107A mutant protein displayed depressed catalytic activity.
Histidine triad (HIT)1 proteins are a superfamily of enzymes that act as hydrolases and transferases on substrates that contain a nucleoside monophosphate group linked to an amino group, a nucleotide, or a phosphorylated sugar (1). The first branch of the HIT superfamily is the most ancient and contains enzymes related to rabbit Hint (2). The crystal structure of Hint demonstrated that the protein is a dimer with two identical AMP/GMP-binding sites per dimer formed by the most highly conserved amino acids in the superfamily (2). By screening a large number of compounds, the adenosine monophosphoramidates AMP-NH2, AMP-N-ε-(N-α-acetyl lysine methyl ester) and AMP-N-alanine methyl ester were identified as rabbit Hint and yeast Hnt1 substrates (3). Building on the observation that the RNA polymerase II C-terminal domain kinase Cdk7/Kin28 may be functionally linked to Hint/Hnt1 (4), it was demonstrated that, in yeast, Hnt1 enzymatic activity acts as a positive regulator of Kin28 function (3). Based on the observation that Hint and Hnt1 hydrolyze AMP linked to lysine (3), we developed the hypothesis that Hint homologs may hydrolyze adenylyl-modified proteins to regulate their functions in vivo (1).
The mouse homolog of rabbit Hint, termed hint1, has been knocked out and found to be encoded by a nonessential gene (5) that may protect the forestomach from carcinogenesis induced by N-nitrosomethylbenzamine (6). In birds, Hint-related genes are sex-linked. Male birds are homogametic (ZZ) while females are heterogametic (ZW). A typical HINT gene is located on the Z chromosome (7), while an extremely unusual HINT-related gene, termed ASW was found repeated approximately 40 times on the female-specific W chromosome (7,8). Though every Hint-related sequence examined to date conserved the 15 amino acids surrounding the AMP-binding site more than any other amino acids (2), these residues were specifically altered in Asw, apparently to eliminate or modify catalytic activity (9). Molecular modeling indicated that Asw protein has conserved the amino acids required to heterodimerize with avian Hint and, potentially, to insert across the dimer interface a nonconserved Gln126 in the vicinity of the Hint active site to depress or alter Hint specificity in a putative Hint-Asw heterodimer (9).
Additional interest in Hint-related hydrolases was generated by the observation that human ataxia-oculomotor apraxia 1, the second most common of the autosomal recessive ataxias, is caused by loss of a gene on 9p13 that encodes an apparent Hint-related hydrolase with an N-terminal FHA domain and a C-terminal sequence reminiscent of zinc fingers (10,11). The physical association of Aprataxin with DNA repair proteins Xrcc1 and Xrcc4 suggests that a repair deficiency may underlie the ataxia-telangiectasia-like neurological symptoms of ataxia-oculomotor apraxia.2
Though Hint genes are found in all organisms, reasonable Hint substrates were only identified (3) and a catalytic mechanism proposed (1) in 2002. To study the mechanism and specificity of Hint hydrolases, here we developed adenosine 5′-O-p-nitrophenylphosphoramidate (AMP-pNA) as a novel spectroscopic substrate. Using methods we established for analysis of Fhit with the fluorogenic substrate GpppBODIPY (12), we titrated nonlabeled inhibitors into assays of wild-type and mutant Hint enzymes to determine their equilibrium inhibitory binding constants. Biochemical and crystallographic analysis of wild-type Hint with newly synthesized adenosine sulfamoyl inhibitors indicated and located a binding site for an alkylamino leaving group. The inhibited Hint crystal structure also provides information about hydrogen-bonding that suggests that the carbonyl oxygen of Gly105 may assist the sidechain hydroxyl of Ser107 as the acid-base catalyst, in contrast to the earlier suggestion of His114 as the acid-base catalyst (1). Indeed, biochemical characterization of the S107A mutant indicates that the Ser hydroxyl plays a facilitative role in catalysis, though in its absence, the Gly105 carbonyl may assist a water molecule bound by the S107A enzyme to provide residual activity for protonation of the leaving group and activation of the hydrolytic water.
EXPERIMENTAL PROCEDURES
Synthesis of substrates
AMP-pNA was prepared in a three step procedure employing oxathiaphospholane ring opening condensation chemistry (13). First, to a solution of p-nitroaniline (0.276 g, 2 mmol) in dry pyridine (10 ml), elemental sulfur (0.128g, 4mmol) was added followed by dropwise addition of 2-chloro-1,3,2-oxathiaphospholane (0.314 g, 2.2 mmol). The reaction mixture was stirred at room temperature for 12 h. Solvent was then removed in vacuo and the residue was triturated with 15 ml acetonitrile. Excess sulfur was filtered off and the filtrate was concentrated in vacuo. The residue was dissolved in 2.5 ml of chloroform and applied to a silica gel column (2.5 × 18 cm). The column was eluted with methanol in chloroform (0→1%). Appropriate fractions were combined and evaporated to give N-(2-thiono-1,3,2-oxathiaphospholanyl)-p-nitroaniline (0.348g, 63%); 31P NMR (CD3OD) δ: 91.09 ppm; FAB-MS m/z (M-1) 275. Second, to a solution of N-(2-thiono-1,3,2-oxathiaphospholanyl)-p-nitroaniline (0.276 g, 1 mmol) in dry acetonitrile (10ml), N6,N6,O2′,O2′-tetrabenzoyladenosine (0.683 g, 1 mmol) was added followed by 1,8-diazabicyclo-(5.4.0)-undec-7-ene (DBU, 0.182 g, 1.2 mmol). The reaction mixture was stirred for 12 h at room temperature at which time solvent was evaporated and the residue was dissolved in a mixture of chloroform/methanol (50:1, v/v) and chromatographed on a silica gel column. The column was eluted with methanol in chloroform (2→10%). The appropriate fractions were combined and evaporated to give N6,N6,O2′,O2′-tetrabenzoyladenosine-5′-O-p-nitrophenylphosphoramidothioate (0.557g, 62%); 31P NMR (CD3CN) δ: 49.66 and 49.93ppm; FAB-MS m/z (M-1) 898. Third, to a solution of N6,N6,O2′,O2′-tetrabenzoyladenosine-5′-O-p-nitrophenylphosphoramidothioate (0.449g, 0.5mmol) in acetonitrile (3 ml) was added a solution of Oxone (Dupont Chemicals, 0.41g, 0.65mmol) in sodium citrate (pH 6.2, 7 ml). The reaction mixture was stirred for 16 h at room temperature. Aqueous Na2SO3 (1.0 ml) was then added and a solid precipitate was removed by filtration. The filtrate was extracted with three 2 ml volumes of chloroform and the solvent was removed from the filtrate. Residual solid was suspended in 10 ml of 20% aqueous ammonia and left for 24 h at room temperature in a tightly closed vial. Ammonia was then evaporated and the residue was dissolved in a mixture of methanol and water (6:1, v/v) and purified on a Sephadex A-25 column, eluted with a linear gradient of triethylammonium bicarbonate buffer (pH 7.5) from 0.05 to 0.5 M. The appropriate fractions were combined and evaporated to yield adenosine 5′-O-p-nitrophenylphosphoramidate (0.047g, 20%); 31P NMR (D2O) δ: 0.76 ppm; FAB-MS m/z (M-1) 466.
AMP-N-ε-(N-α-Boc-lysinamide) was obtained in a three step procedure. First, reaction of N-α-Boc-lysine methyl ester with one molar equivalent of 2-chloro-1,3,2-oxathiaphospholane in the presence of pyridine and elemental sulfur gave N-ε-(2-thiono-1,3,2-oxathiaphospholanyl)-N-α-Boc-lysine methyl ester in 51% yield [31P NMR (CD3OD) δ: 97.04 and 96.99 ppm; FAB-MS m/z (M-1) 397]. This compound in reaction with a molar equivalent of N6,N6,O2′,O2′-tetrabenzoyladenosine in acetonitrile solution, in the presence of DBU, provided N6,N6,O2′,O2′-tetrabenzoyladenosine-5′-O-[N-ε-(N-α-Boc-lysinamide)]phosphoramidothioate [31P NMR (CD3CN) δ: 59.62 and 59.45 ppm; FAB-MS m/z (M-1)1019], which subsequently under treatment with Oxone followed by aqueous ammonia was converted into AMP-N-ε-(N-α-Boc-lysinamide) [yield 26%, 31P NMR (D2O) δ: 9.94 ppm; FAB-MS m/z (M-1) 573].
Synthesis of inhibitors
5′-O-Sulfamoyladenosine was synthesized by modification of the methods of Shuman (14). 2′,3′-O-Isopropylidene-5′-O-sulfamoyladenosine was first purified as follows: A mixture of 2′,3′-O-isopropylidene adenosine (3.00 g, 9.76 mmol) and hexabutyldistannoxide (11.50 g, 20 mmol) in anhydrous benzene was heated to reflux under argon for 2 h. The resulting clear solution was cooled to 5 °C and a solution of sulfamoyl chloride (4.50g, 39 mmol), synthesized as described (15), in dioxane (80 ml) was added dropwise. After stirring at room temperature for 30 min, solvent was evaporated and the residue was extracted with two 50 ml volumes of hexane. The insoluble residue was treated with a dilute solution of methanolic ammonia (50 ml), evaporated and purified by SiO2 column chromatography using 9 volumes chloroform to 1 volume methanol as the mobile phase. The appropriate fractions were evaporated to give pure 2′,3′-O-isopropylidene-5′-O-sulfamoyl adenosine (2.27 g, 60 %) as a white foam. 1H-NMR (DMSO-d6) δ 1.31 (s, 3H, CH3-isopropylidene), 1.50 (s, 3H, CH3-isopropylidene), 4.20 (m, 2H, H-5′,5”), 4.36 (m, 1H, H-4′), 5.05 (dd, 1H, H-3′), 5.45 (dd, 1H, H-2′), 6.20 (d, 1H, H-1′, J1-2 = 2 Hz), 7.35 (br s, 2H, NH2, D2O-exchangeable), 7.60 (br s, 2H, SO2-NH2, D2O exchangeable), 8.15 (s, 1H, H-2), 8.30 (s, 1H, H-8). 2′,3′-O-Isopropylidene-5′-O-sulfamoyl adenosine (386 mg, 0.10 mmol) was dissolved in a solution of aqueous TFA (90%, 20 ml) and stirred at room temperature for 30 min. The solution was then evaporated to dryness and the residue dissolved in 15 ml aqueous methanolic ammonia and evaporated to dryness leaving a white residue, crystallized from water yielding pure 5′-O-sulfamoyl adenosine (246 mg, 72 %) as white needles. 1H-NMR (DMSO-d6) δ 4.25 (m, 4H, H-5′,5”, H-4′, and H-3′), 5.51 (m, 1H, H-2′), 5.45 (d, 1H, OH, D2O-exchangeable), 5.65 (d, 1H, OH, D2O-exchangeable) 5.90 (d, 1H, H-1′, J1-2 = 4 Hz), 7.30 (s, 2H, SO2-NH2, D2O-exchangeable), 7.65 (s, 2H, NH2-6, D2O-exchangeable, H 8.15 (s, 1H, H-2), 8.30 (s, 1H, H-8). ESMS (m/z) 347 (M+1, 100%), 338 (10%), 308, 280, 250.
5′-O-Ethylsulfamoyl adenosine was synthesized as follows. Adenosine (3.26 g, 12.2 mmol) was dissolved in dry dimethylformamide (40 ml), heated to 50°C, and the resulting solution treated with excess imidazole (1.66 g, 24.4 mmol). Ethylsulfamoyl chloride (1.75 g, 12.2 mmol), purified by modification of the method of Weiss (16), was added at 0°C and the solution stirred 30 min at room temperature. The resulting oil was dissolved in dichloromethane and methanol (9:1), then solvent evaporated and the oily product decanted from a solid residue, which was twice extracted with dichloromethane. The crude oily product crystallized on standing. The dichloromethane-extracts and the solid product were dissolved in methanol, solvent was evaporated and the residue was taken up in dichloromethane and methanol (9:1) for silica chromatography, eluting with a gradient of methanol (10 - 20%) in dichloromethane. Appropriate fractions were identified by TLC, pooled, and evaporated to give 5′-O-ethylsulfamoyladenosine as a white solid, further purified by reverse phase HPLC. 1H-NMR (DMSO) δ 0.95 (t, 3H, Me), 2.85 (q, 2H, CH2), 4.10-4.25 (m, 4H, H-3′,4′,5′,5”), 4.65 (dd, 1H, H-2′), 5.45 (d, 1H, OH, D2O-exchangeable), 5.65 (d, 1H, OH, D2O-exchangeable), 5.90 (d, 1H, H-1′, J1-2 = 4 Hz), 7.30 (bs, 2H, NH2-6, D2O-exchangeable), 7.85 (bs, 1H, SO2NH, D2O-exchangeable)8.15 (s, 1H, H-2), 8.35 (s, 1H, H-8). HRMS (FAB +ve) found 375.1072; calc for C12H19N6O6S 375.1087.
Mutagenesis and enzymology
Wild-type rabbit Hint was expressed and purified in bacteria using pSGA02-HINT as described (3). The S107A mutant of rabbit Hint was generated by site-directed mutagenesis of the wild-type expression vector using primer 7129 (5′ ATGAACGTGATAGACGGCCTGTCCACCATCGGA) to generate plasmid pB415, which was used to produce homogeneous mutant Hint enzyme as above. AMP-pNA substrate at concentration 1mM was incubated with homogeneous rabbit Hint enzymes in reactions at 30°C containing 20 mM Na HEPES, pH 7.2 and 0.5 mM MgCl2. Reaction samples were spotted on silica TLC plates (E. Merck). Plates were developed in 2-propanol:NH4OH:1,4-dioxane:H 2O (50:35:8:7). Developed plates were imaged by epi-UV illumination on a Bio-Rad Fluor S instrument. Initial rate assays for AMP-pNA were performed in spectrophotometric cuvettes. Pre-mixes containing AMP-pNA (50, 100, 300, 400 or 700 μM), 20 mM Na HEPES, pH 7.2 and 0.5 mM MgCl2 were equilibrated at 30°C and then reactions were initiated with addition of 97.5 to 239.3 pmol of rabbit Hint. To determine kinetic parameters for AMP-N-ε-(N-α-Boc-lysinamide), initial rate reactions were completed in the same buffer at 0.15, 0.3, 0.6, 1.25, 2.5, 5, 10, 20 μM concentration of substrate and the products were analyzed by HPLC on a HQ column (Applied Biosystems) with NH4HCO3, pH 8 as the mobile phase. The pH-dependence of kcat/Km were determined at 6 pH values (5, 5.5, 6, 6.5, 7, 7.25) with 4 or 5 concentrations of AMP-pNA (from 12 to 400 μM) for the wild-type and mutant enzyme. These reactions were performed in 66 mM Na, K phosphate buffers with 0.5 mM MgCl2 using 964 pmol of wild-type Hint or 1776 pmol of Ser107Ala mutant Hint.
Ki values for sulfamoyl and N-ethylsulfamoyl adenosine were obtained by titrating the inhibitors (zero, 5, 10, 40, 60, and 100 μM) into complete digests of 100 μM AMP-pNA. Using known amount of enzyme (96 pmol), first order decay curves for remaining substrate were used to calculate kcat/Km and the inhibitor concentration-dependence of reduction of kcat/Km (apparent) was calculated as described (12).
X-ray crystallography
Rabbit Hint crystals were grown as described (2). Crystals were soaked in a well solution saturated with either sulfamoyl or N-ethylsulfamoyl adenosine for two hours. Cryoprotectant solutions were prepared from the well solution containing 13% glycerol and saturated with the inhibitor. Crystals were scooped from the soak solution and briefly submerged in cryoprotectant solution just prior to freezing in liquid nitrogen. Hint-sulfamoyl adenosine data were collected in the cryostream at -180° C at the X-25 beamline of the National Sychrotron Light Source at Brookhaven National Laboratory. Hint-ethylsulfamoyl adenosine data were collected on the Kimmel Cancer Center home source. We employed CNS (17) for model refinement and calculation of electron density maps. Model building was carried out using O (18). Collection and refinement statistics are provided in Table II. Coordinates and structure factors have been deposited with the Research Collaboratory for Structural Bioinformatics Protein Databank.3
Table II.
Crystallographic data collection and refinement statistics for Hint-ethylsulfamoyl adenosine
| space group | P43212 |
| unit cell dimensions (Å) | a=b=39.88, c=142.3 |
| reflections (measured / unique) | 95,355 / 10,698 |
| resolution limits (Å) | 23-1.8 |
| completeness (%) | 93.9 (88.3)1 |
| Rsym (%)2 | 6.5 (14.3)1 |
| I/σ | 9.2 (5.1)1 |
| multiplicity | 8.4 (3.4)1 |
| Rwork (%)3 | 21.6 (29.9)1 |
| Rfree (%)4 | 24.2 (28.1)1 |
| protein and nucleotide nonhydrogen atoms | 961 |
| water molecules | 72 |
| root mean square difference, bond lengths (Å) | 0.013 |
| root mean square difference, bond angles (°) | 1.4 |
| average B factor (Å2) | 16.9 |
Numbers in parentheses refer to data in the 1.86 to 1.80Å shell.
Rsym = Σ|I-<I>|/Σ<I> in which I is a measured intensity and <I> is the average intensity from multiple measurements of symmetry-related reflections.
Rwork = Σ|Fo- Fc|/ΣFo.
Rfree was calculated as for Rwork using a test set of reflections (7%) not used for refinement.
RESULTS AND DISCUSSION
Biochemical evidence for an alkylamine-binding site in rabbit Hint
To study the enzymatic activity of the Hint AMP-lysine hydrolase, we synthesized AMP-pNA. To test whether Hint would tolerate pNA as a leaving group, we incubated Hint with AMP-pNA and analyzed the progress of the reaction by TLC and HPLC. Both assays made it clear that Hint cleaves this molecule generating one equivalent of AMP and pNA (data not shown). Release of the chromophore allowed the new substrate to be followed spectroscopically at 410 nm. To compare AMP-pNA with a more biologically authentic substrate, AMP-N-ε-(N-α-Boc-lysinamide) was prepared and assayed by HPLC. As shown in Table 1, consistent with rabbit Hint as an AMP-lysine hydrolase, the enzyme liberated AMP from AMP-N-ε-(N-Boc-lysinamide) with a kcat of 0.23 s-1 and a Km of 470 nM. In contrast, the convenience of the continuously and spectroscopically monitored substrate came at the cost of a 280-fold higher Km and about 100-fold lower kcat, presumably because the enzyme tolerates the bulky leaving group very poorly. As the hydrolytic step for both of these substrates is expected to proceed identically through adenylylated His112 (1), the pNA leaving group is expected to inhibit adenylylation, i.e., productive covalent attack of the α-phosphate by His112.
Table I.
Steady state characterization of Hint substrates and inhibitors
| compound | kcat (s-1) | Km (μM) | kcat/Km(M-1s-1) | KI (μM) |
|---|---|---|---|---|
| AMP-pNA | 0.00187 ±0.00006 | 134. ± 11. | 14.0 | - |
| AMP-N-ε- (N-Boc-Lys-NH2) | 0.234 ±0.011 | 0.472±0.057 | 496000. | - |
| Ado-SO2-NH2 | - | - | - | 16.1± 0.2 |
| Ado-SO2-NH-C2H5 | - | - | - | 1.26± 0.28 |
Despite the relatively poor kinetic values, the AMP-pNA substrate is reasonably suited for assays of competitive inhibitors and determination of their Ki values with methods we established for GpppBODIPY and Fhit (12). We synthesized new Hint inhibitors consisting of adenosine and 5′-sulfamoyl or N-ethylsulfamoyl groups. Both were titrated into AMP-pNA assays and competitive Ki values were obtained by calculating the inhibitor concentration-dependence in reduction of kcat /Km (apparent) for substrate hydrolysis (12). As shown in Table I, the sulfamoyl adenosine inhibited Hint with a Ki value of 16.1 μM while addition of the ethyl group to sulfamoyl adenosine resulted in an inhibitor with a Ki value of 1.25 μM. The 13-fold advantage in equilibrium binding conferred by addition of the ethyl group suggests a favorable interaction with an alkylamine leaving group such as a lysine or protein-lysine.
Crystallographic identification of the alkylamine binding site in rabbit Hint
Previously, the most informative crystal structures of Hint have been bound to GMP, 8-Br-AMP (2), and adenosine tungstate (19). The GMP and 8-Br-AMP crystal structures can be considered product complexes that represent the form of the enzyme bound to a nucleoside monophosphate following hydrolysis of a putative nucleoside monophosphoramidate substrate while the adenosine tungstate structure can be considered to be an analog of the transition state. An additional crystal structure of human Hint bound to adenosine α,β-methylene diphosphonate was described as an analog of a substrate complex (19) but, as ADP is an extremely poor substrate of Hint (3), it is not clear that this complex can serve as a model of a good enzyme-substrate complex.
To examine active site geometry with amine-containing nucleotides, we soaked rabbit Hint crystals (2) with the two inhibitors synthesized in this study. Both soaks resulted in crystals that diffracted to 2.0 Å or higher resolution. When the difference electron density of the sulfamoyl adenosine-soaked crystal was examined, we could easily identify the adenosine density and place the sulfur atom in the position of the 5′ phosphorus but we could not interpret which two of the sulfur ligands were oxygen and which one was the amino group (not shown). In contrast, the ethyl group of N-ethylsulfamoyl adenosine allowed unequivocal assignment of every non-hydrogen atom at 1.8 Å resolution. Because this inhibitor was shown to possess a 13-fold binding advantage over the sulfamoyl adenosine, we suggest that the alkyl portion of this inhibitor is bound in a substrate-analogous fashion. Data collection and refinement statistics for this crystal structure are provided in Table II. Representative electron density and the overall structure of the Hint dimer bound to N-ethylsulfamoyl adenosine are shown in Figure 2. Difference electron density for the inhibitor is shown in Figure 3.
Figure 2.
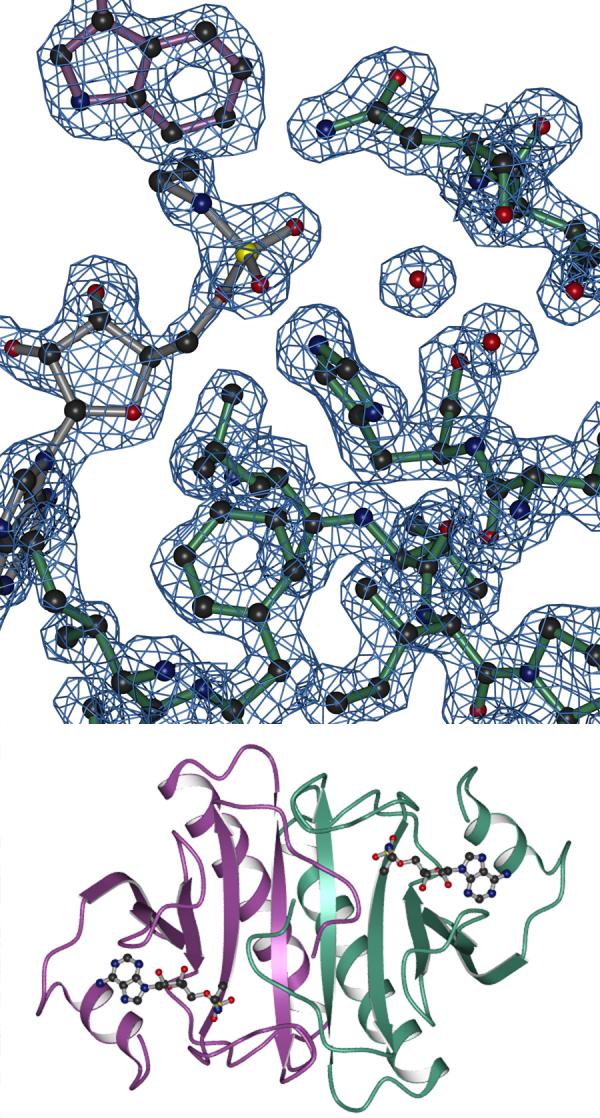
Hint-ethylsulfamoyl adenosine complex: electron density and overall structure. One Hint monomer is depicted in green, the second Hint monomer in purple and the N-ethylsulfamoyl adenosine in gray. A) Representative 2Fo - Fc electron density, contoured at 1 σ used to fit the coordinates. Note the position of Trp123 in purple as proximal to the nucleotide-binding site of the green monomer. B) The dimeric Hint-ethylsulfamoyl adenosine crystal structure. Ethylamino groups are visible protruding from the α-sulfates of the bound inhibitors in the Hint homodimer. Molecular graphics were generated as described (2).
Figure 3.
Active site geometry of Hint. Stereo view depicting the refined N-ethylsulfamoyl adenosine inhibitor inside 1.5 σ Fo-Fc difference electron density calculated from protein and water coordinates alone. As in Figure 2, the Hint monomer binding the inhibitor is in green and Trp123 from the other monomer is in purple. Atomic distances (Å) are shown for the carbonyl oxygen of Gly105 and the γ oxygen of Ser107 to the nitrogen of N-ethylsulfamoyl adenosine, the hydrogen bond network connecting His51, His114 and the 5′ oxygen of the inhibitor, and for the two interactions of the adenylylating His112.
As observed earlier in crystal structures of rabbit Hint bound to GMP and 8-Br-AMP (2), the most highly conserved amino acids in Hint homologs and, indeed, among all HIT enzymes, surround the purine base, the ribose and the 5′ phosphate groups. Recently, we observed that an additional amino acid is highly conserved in Hint hydrolases—the amino acid was missed in our initial analysis because Trp123 of one monomer is not in contact with the nucleotide bound in that monomer but rather makes a close approach to the nucleotide across the dimer interface (9). This amino acid is one of a remarkable 15 residues that are sexually dimorphic in avian Hint-related genes: it is Trp in the typical Hint polypeptide encoded on the Z chromosome and it is Gln on the unusual and potentially inhibitory Asw polypeptide that is encoded in a tandem array of ∼40 repeats on the female-specific W chromosome (9). The present biochemical studies establish a binding site for the alkyl portion of the alkylamine leaving group of Hint substrates. As shown in Figures 2A and 3, that site is located across the Hint dimer interface against Trp123.
Evidence that Ser107 is the acid-base catalyst for Hint
Earlier, we argued that for Hint hydrolases, whose phosphoramidate substrates necessarily have an amine leaving group, protonation of the phosphoramidate nitrogen may be an important kinetic component of the adenylylation step in catalysis (1). The crystal structure of Hint bound to N-ethylsulfamoyl adenosine allowed us to analyze how conserved features of Hint hydrolases interact with an analog of the 5′-phosphoramidate nitrogen, equatorial oxygens, and the alkylamine leaving group.
Though we considered His114 to be the candidate hydrogen bond donor (1) extrapolating from product complexes (2), this structure suggests that the Ser107 hydroxyl is positioned for proton donation with alignment that is assisted by the carbonyl oxygen of Gly105. In the present structure the ε-nitrogen of His114 is 3.1 Å from the ribose 5′ oxygen and 3.3 Å from one of the α sulfur’s equatorial oxygen atoms and not in hydrogen-bonding distance or orientation with the sulfamoyl nitrogen. In contrast, the sulfamoyl nitrogen is 2.7 Å from the carbonyl oxygen of Gly105 and 3.2 Å from the Ser107 γ-oxygen. Thus, we propose that Ser107 donates a hydrogen bond to the phosphoramidate nitrogen of a Hint substrate.
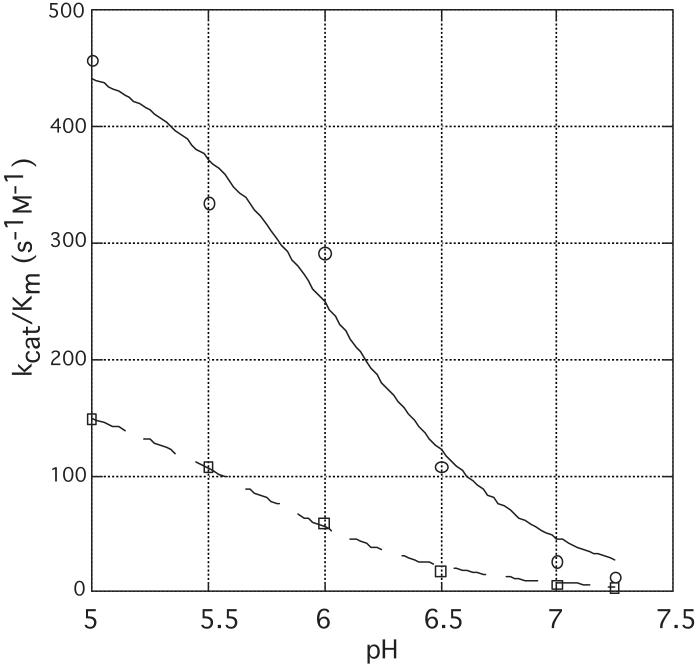
To test the hypothesis that Ser107 plays a role in catalysis, we performed a site-directed alteration of this residue to Ala. Rabbit Hint-S107A was expressed and purified in E. coli and characterized with respect to AMP-pNA hydrolysis. The pH-dependence of kcat/Km was determined at 6 pH values for the wild-type and mutant enzymes. When, as shown in Figure 4, the specificity constant, kcat/Km, was plotted as a function of pH, we were able to model Hint enzymatic activity on AMP-pNA as being controlled by a single titratable group with a pKa of 6.0 ± 0.1 and maximal activity in the protonated form. The S107A mutant, in the same assays, exhibited approximately 3-fold reduced activity at all pH values, with little alteration (a 0.4 unit drop) in pKa. Given the positon of the Ser107 hydroxyl, the reduced activity of this mutant was expected, though one might have expected the magnitude of the defect to be greater than 3-fold. To test whether a larger catalytic defect of the S107A mutant was masked by the AMP-pNA substrate, we determined kcat and Km for AMP-N-ε-(N-Boc-lysinamide). While the mutant’s Km was unaffected, the kcat was depressed by 4-fold.
Figure 4.
pH-dependence of wild-type (o) and S107A (”) forms of Hint. Enzyme specificity constants (kcat / Km) with AMP-pNA were plotted as a function of pH and fitted to kcat / Km (observed) = (kcat / Km (protonated))10^(pKa -pH)/(1+10^(pKa - pH)).
In the absence of the Ser107 hydroxyl, Hint clearly has the ability to adenylylate and hydrolyze substrates at a reduced rate. We hypothesize that the carbonyl oxygen of Gly105 assists the function of Ser107 as the acid-base catalyst for Hint. In the absence of Ser107, we propose that a water molecule bound to the S107A enzyme functions as the acid-base catalyst and that the cost of loss of the protein-positioned hydroxyl group is the observed 3 to 4-fold reduced activity of the mutant enzyme. In GalT, a member of the distinctive branch 3 HIT nucleoside monophosphate transferases (1), the corresponding Ser161 also functions on the leaving group side (20). However, rather than donating a hydrogen bond to the α-β bridging oxygen in a UDP-hexose substrate, which would be analogous to donating a hydrogen bond to the phosphoramidate nitrogen in a Hint substrate, Ser161 donates to a β phosphate equatorial oxygen (20).
CONCLUSIONS
We have used organic synthesis, enzymology, X-ray crystallography and mutagenesis to develop novel substrates and inhibitors of the Hint adenosine monophosphoramidate hydrolase. Consistent with our view that adenylylated lysine residues are natural substrates, this work establishes that Hint kinetics are superior with an alkylamine substrate such as AMP-N-ε-(N-α-Boc-lysinamide versus AMP-pNA and that the ethyl group of an ethylsulfamoyl adenosine inhibitor confers a 13-fold binding advantage over the non-derivatized sulfamoyl adenosine. The crystal structure of the N-ethylsulfamoyl adenosine inhibitor located the alkylamine binding site against conserved residue Trp123, which interacts with the inhibitor across the dimer interface and also identified Ser107 as the candidate acid-base catalyst for the enzyme. pH-titration of wild-type and mutant enzymes establishes that Hint functions with a protonated titratable group and that Ser107 may donate and receive back a proton in a manner that can be substituted by bound water in adenosine monophosphoramidate hydrolysis. Our proposed catalytic mechanism for Hint hydrolases is provided in Figure 5.
Figure 5.
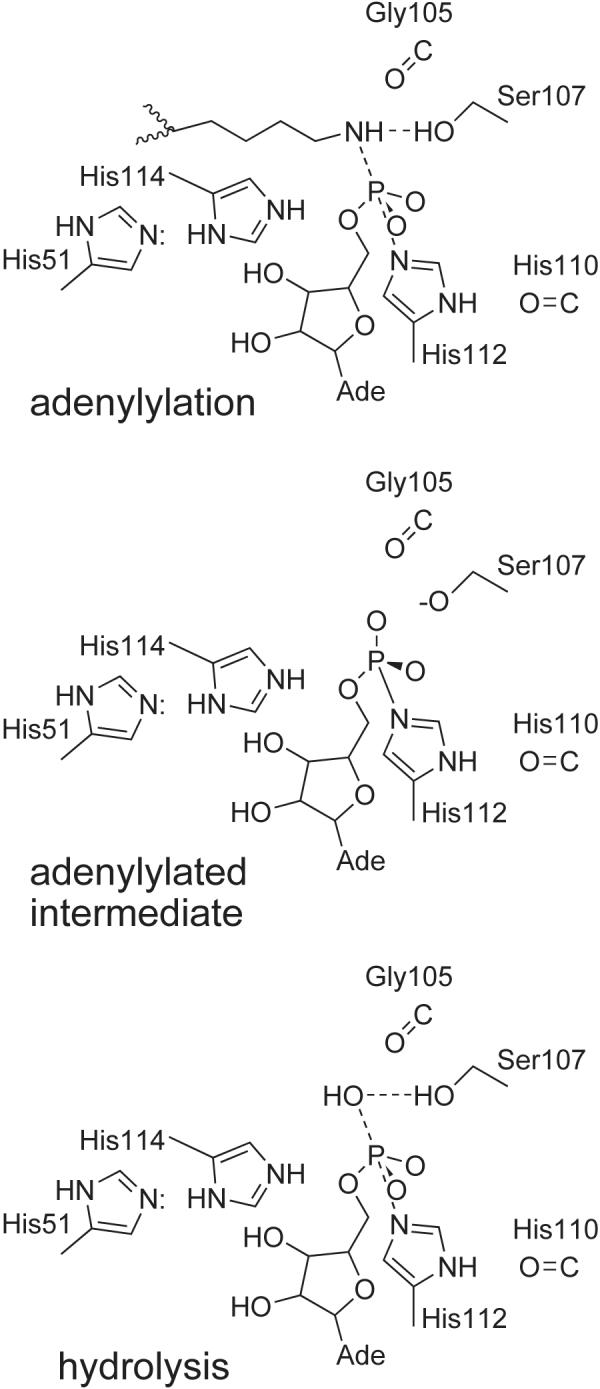
Proposed catalytic mechanism for Hint hydrolases. In the adenylylation step, we propose that Ser107 donates a proton to the leaving group such that the adenylylated intermediate, with inverted configuration at phosphorus, contains a serine alkoxide ion. The serine alkoxide would then abstract a proton from the attacking water to hydrolyze the adenylylated intermediate, returning the phosphorus to its original configuration. As shown in Figure 3, the reaction geometry is supported by additional hydrogen bonds. In particular, the leaving group and hydrolytic geometry is thought to be enforced by the carbonyl group of Gly105. Note that this reaction would proceed in an identical manner with a Ser107Ala enzyme containing a water molecule bound in place of the γ hydroxyl group.
Figure 1.
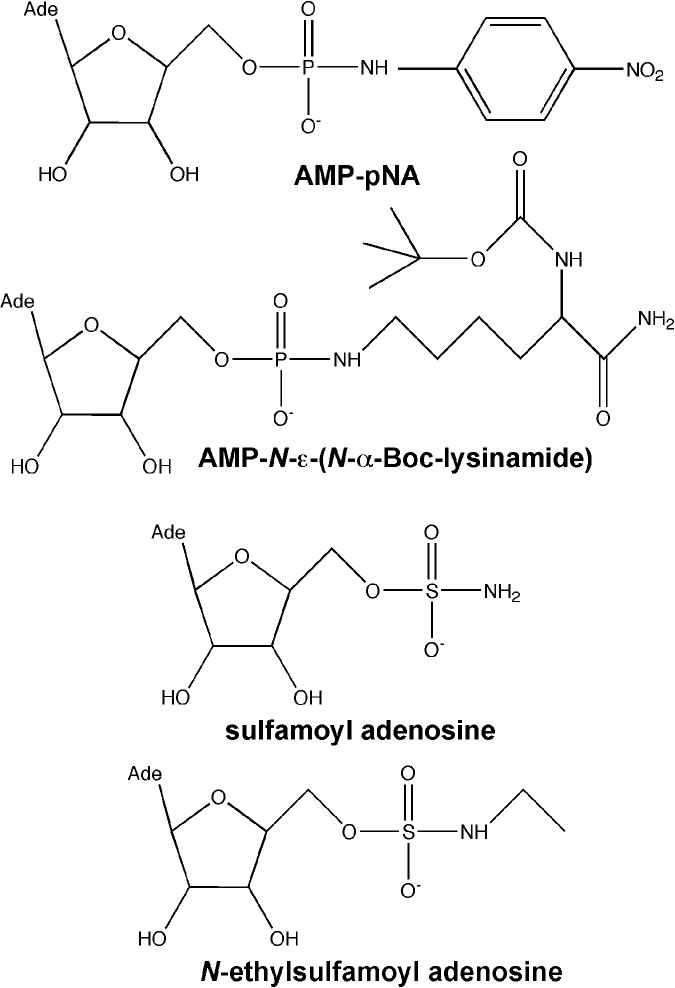
Novel Hint substrates and inhibitors. Chemical structures of Hint substrates AMP-pNA and AMP-N-ε-(N-α-Boc-lysinamide), and Hint inhibitors, sulfamoyl adenosine and N-ethylsulfamoyl adenosine
Footnotes
This work was supported by research grant CA75954 from the National Cancer Institute to CB and by the State Committee for Scientific Research (KBN), grant PBZ-KBN 059/T09/06 to JB. X-ray crystallographic data for this study were measured at beamline X-25 of the National Synchrotron Light Source, supported by the NIH and the Department of Energy.
- HIT
- histidine triad
- AMP-pNA
- adenosine 5′-O-p-nitrophenylphosphoramidate
- DBU
- 1,8-diazabicyclo-(5.4.0)-undec-7-ene
Clements, P.M., Breslin, C., Deeks, E.D., Byrd, P.J., Ju, L., Bieganowski, P., Brenner, C., Moreira, M.-C., Taylor, A.M.R., Caldecott, K.W., submitted.
Research Collaboratory for Structural Bioinformatics Protein Databank = PDB # 1RZY
References
- 1.Brenner C. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner C, Garrison P, Gilmour J, Peisach D, Ringe D, Petsko GA, Lowenstein JM. Nat. Struct. Biol. 1997;4:231–238. doi: 10.1038/nsb0397-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieganowski P, Garrison PN, Hodawadekar SC, Faye G, Barnes LD, Brenner C. J. Biol. Chem. 2002;277:10852–10860. doi: 10.1074/jbc.M111480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korsisaari N, Makela TP. J. Biol. Chem. 2000;275:34837–34840. doi: 10.1074/jbc.C000505200. [DOI] [PubMed] [Google Scholar]
- 5.Korsisaari N, Rossi DJ, Luukko K, Huebner K, Henkemeyer M, Makela TP. Mol. Cell. Biol. 2003;23:3929–3935. doi: 10.1128/MCB.23.11.3929-3935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su T, Suzui M, Wang L, Lin CS, Xing WQ, Weinstein IB. Proc. Natl. Acad. Sci. U S A. 2003;100:7824–7829. doi: 10.1073/pnas.1332160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hori T, Asakawa S, Itoh Y, Shimizu N, Mizuno S. Mol. Biol. Cell. 2000;11:3645–3660. doi: 10.1091/mbc.11.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 9.Pace HC, Brenner C. Genome Biol. 2003;4:R18. doi: 10.1186/gb-2003-4-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Nature Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 11.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. Nature Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 12.Draganescu A, Hodawadekar SC, Gee KR, Brenner C. J. Biol. Chem. 2000;275:4555–4560. doi: 10.1074/jbc.275.7.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guga P, Okruszek A, Stec WJ. Topics Curr. Chem. 2002;220:169–200. [Google Scholar]
- 14.Shuman DA, Robins RK, Robins MJ. J. Amer. Chem. Soc. 1969;91:3391–3392. doi: 10.1021/ja01040a062. [DOI] [PubMed] [Google Scholar]
- 15.Graf R. Chem. Ber. 1959;92:509–555. [Google Scholar]
- 16.Weiss G, Schultz G. Liebigs Ann. Chem. 1969;729:40–51. [Google Scholar]
- 17.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kuntsleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Jones TA, Zou IY, Cowan S,W, Kjeldgaard M. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 19.Lima CD, Klein MG, Hendrickson WA. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 20.Geeganage S, Ling VWK, Frey PA. Biochemistry. 2000;39:5397–5404. doi: 10.1021/bi992594s. [DOI] [PubMed] [Google Scholar]