Abstract
Sgt1 is an adaptor protein implicated in a variety of processes, including formation of the kinetochore complex in yeast, and regulation of innate immunity systems in plants and animals. Sgt1 has been found to associate with SCF E3 ubiquitin ligases, the CBF3 kinetochore complex, plant R proteins and related animal Nod-like receptors, and with the Hsp90 molecular chaperone. We have determined the crystal structure of the core Hsp90–Sgt1 complex, revealing a distinct site of interaction on the Hsp90 N-terminal domain. Using the structure, we developed mutations in Sgt1 interfacial residues, which specifically abrogate interaction with Hsp90, and disrupt Sgt1-dependent functions in vivo, in plants and yeast. We show that Sgt1 bridges the Hsp90 molecular chaperone system to the substrate-specific arm of SCF ubiquitin ligase complexes, suggesting a role in SCF assembly and regulation, and providing multiple complementary routes for ubiquitination of Hsp90 client proteins.
Keywords: complex assembly, crystal structure, molecular chaperone, protein–protein interactions, ubiquitin ligase
Introduction
The Hsp90 molecular chaperone, in collaboration with a plethora of co-chaperones, is involved in the assembly and stabilization of key regulatory proteins in the eukaryotic cell (Pearl and Prodromou, 2006). Although the protein clientele of Hsp90 covers a broad range of structural classes, it is nonetheless selective and specific; the biochemical basis for this remains one of the least understood aspects of Hsp90 biology. Recruitment of one important class of Hsp90 clients, protein kinases, requires the adaptor co-chaperone Cdc37, which interacts simultaneously with the kinase and Hsp90, and regulates Hsp90s ATPase cycle in the process (Roe et al, 2004; Pearl, 2005; Caplan et al, 2007). Coupling of the Hsp90 chaperone system to the ubiquitin-regulated signalling and targeted destruction systems is mediated by a dimeric co-chaperone adaptor, CHIP (Connell et al, 2001), which binds to Hsp90 through its tetratricopeptide repeat (TPR) domain and E2 ubiquitin conjugating enzymes through its U-box domain (Zhang et al, 2005). Other co-chaperones, such as p23 and Aha1, have no apparent role in mediating interactions with clients or other systems, but exert an effect instead as regulators of Hsp90s inherent ATPase cycle (Sullivan et al, 1997; Panaretou et al, 2002; Meyer et al, 2004; Ali et al, 2006).
One of the most recently recognized co-chaperones of Hsp90, Sgt1 (Takahashi et al, 2003), was originally identified as a suppressor of a temperature-sensitive defect in the budding yeast Skp1 protein (Kitagawa et al, 1999). Sgt1p associates with Skp1p as an essential component of the CBF3 kinetochore complex, and is required for the assembly of yeast and human kinetochores (Bansal et al, 2004; Steensgaard et al, 2004). Sgt1p also associates with Skp1p in the context of the SCF (Skp1p–Cdc53p–F-box protein) class of E3 ubiquitin ligases (Kitagawa et al, 1999). Sgt1 is not exclusively associated with Skp1 and interacts independently of Skp1 with adenylate cyclases (Dubacq et al, 2002; Schadick et al, 2002), the CHORD-domain protein RAR1 (Azevedo et al, 2002), and members of a family of proteins including plant disease resistance R-gene products (Bieri et al, 2004; Leister et al, 2005) and animal Nod-like receptors (Mayor et al, 2007; da Silva Correia et al, 2007), that mediate innate immunity to parasites and infection. The ability of Sgt1 to bind to Hsp90 and simultaneously interact with other proteins (Takahashi et al, 2003; Lingelbach and Kaplan, 2004; Catlett and Kaplan, 2006; Mayor et al, 2007; da Silva Correia et al, 2007) implicates it as a client protein adaptor co-chaperone in a similar manner to Cdc37, albeit with a more structurally varied clientele.
Sgt1 consists of three identifiable domains: an N-terminal tetratricopeptide repeat (TPR) domain similar to those found in many other Hsp90 and Hsp70 co-chaperones; a central ‘CHORD and Sgt1' (CS) domain related to the β-sandwich domain of small heat-shock proteins, α-crystallin and the Hsp90 co-chaperone p23/Sba1; and a C-terminal Sgt-specific (SGS) domain. All three domains mediate protein–protein interactions. The SGS region has been implicated in interaction with leucine-rich repeat domains in the yeast adenylate cyclase Cdc35p (Dubacq et al, 2002), plant R proteins (Bieri et al, 2004; Leister et al, 2005) and the mammalian Nod1 receptor (da Silva Correia et al, 2007). Although initial studies implicated the TPR domain as essential for interaction with both Hsp90 and Skp1 (Bansal et al, 2004), further analysis suggests that the N-terminal TPR domain mediates interactions with Skp1, whereas the central CS domain is responsible for the recruitment of Sgt1 to Hsp90 (Lee et al, 2004; Lingelbach and Kaplan, 2004; Catlett and Kaplan, 2006). Immunoprecipitation and yeast two-hybrid (Y2H) assays identify the N-terminal half of Hsp90 as sufficient for interaction with Sgt1 (Takahashi et al, 2003).
To gain some insight into the coupling of the Hsp90 and Sgt1 protein interaction ‘hubs', we have determined the crystal structure of the Hsp90-N–Sgt1-CS core complex. The structure reveals a hitherto unknown locus of interaction on Hsp90, and facilitates a targeted mutational analysis that reveals the Hsp90 dependence of Sgt1 functions in vivo. Biochemical analysis of Sgt1 interactions suggests a role for Hsp90–Sgt1 in the assembly of SCF ubiquitin ligase complexes.
Results
We explored a range of Hsp90 and Sgt1 constructs from different species and obtained diffracting crystals of the core Hsp90–Sgt1 complex using an N-terminal domain construct of Hordeum vulgare (barley) Hsp90 and a CS domain construct from the Arabidopsis thaliana homologue, AtSgt1a. HvHsp90 is 64% identical to yeast Hsp82 and 70% identical to human Hsp90β, whereas AtSgt1a is 32 and 36% identical, respectively, to its yeast and human homologues. The structure of the complex was solved by molecular replacement with the crystal structure of yeast Hsp90-N domain and the NMR structure of human Sgt1-CS domain. The crystals contain three independent copies of the complex, each with a bound ADP molecule, and has been refined at 3.3-Å resolution (see Materials and methods and Table I).
Table 1.
Crystallographic statistics
| Data collection | HvHsp90-N–AtSgt1a-CS |
| Space group | P212121 |
| a, b, c (Å) | 100.268, 129.654, 135.998 |
| Wavelength (Å) | 0.9537 |
| Resolution (Å) | 38.1–3.3 (3.48–3.3) |
| Rmerge | 0.161 (0.561) |
| I/σI | 8.5 (2.3) |
| Completeness (%) | 99.5 (99.9) |
| Redundancy | 3.6 (3.6) |
| Refinement | |
| Number of reflections | 50 605 |
| Rwork/Rfree | 0.20/0.24 |
| Number of atoms | |
| Protein+ADP | 7404 |
| B-factors | |
| Protein+ADP | 59.7 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.012 |
| Bond angles (deg) | 1.483 |
| Ramachandran statistics | |
| Preferred | 88.8% |
| Allowed | 8.7% |
| Outliers | 2.4% |
The structure of yeast and human Hsp90-N domains has been described earlier (Prodromou et al, 1997; Stebbins et al, 1997) and the barley Hsp90-N domain is essentially identical in structure (r.m.s.d. ∼0.75 and 0.67 Å, respectively for 198 and 172 common Cα positions). The mobile ‘lid' segment, which closes over the nucleotide-binding pocket in the ATP-bound closed state of the Hsp90 (Ali et al, 2006), is in a fully open conformation in the Hsp90-N–Sgt1-CS complex, and an ADP molecule is present in the nucleotide-binding pocket. The AtSgt1a CS domain has the same β3–β4 ‘sandwich' structure seen in the NMR structure of HsSgt1-CS domain (Lee et al, 2004), with small differences in the conformation of the loops connecting the β-strands.
Hsp90-N–Sgt1-CS interface
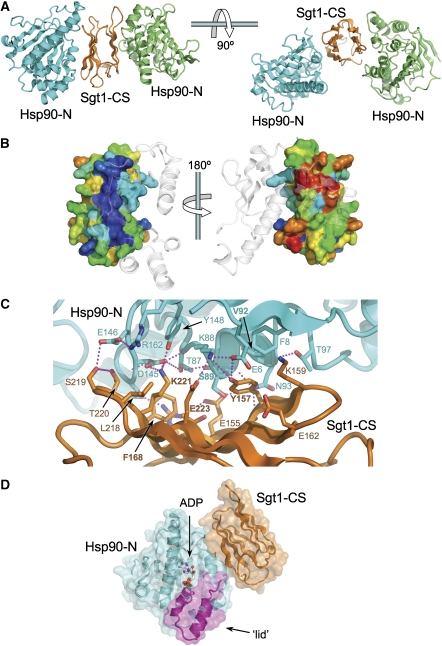
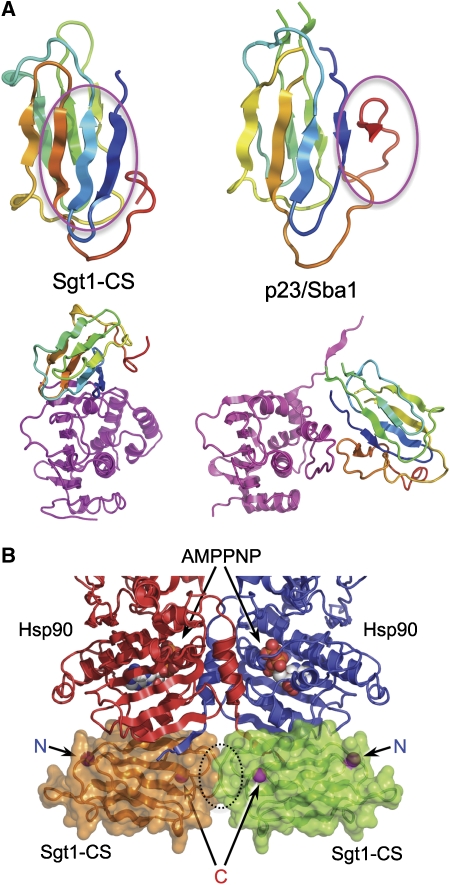
The crystal lattice is such that each of the β-sheet faces of the Sgt1-CS domain structure contacts a different Hsp90-N domain through distinct regions (Figure 1A). Although a single Sgt1 molecule might interact simultaneously with both-N domains in an Hsp90 dimer, this is inconsistent with the ∼1:1 binding stoichiometry suggested by ITC (Supplementary Figure 1). It is most likely that one of the Hsp90–Sgt1 interfaces is the biologically significant interaction, whereas the other is a lattice contact necessary for crystal formation. To identify the biologically significant interface, we analysed the conservation of the residues involved. In the case of Hsp90, which is highly conserved throughout, there was no clear difference between the two points of contact with Sgt1-CS in the crystal. However, Sgt1 showed a marked difference, with the residues involved in one interaction being very much more conserved overall, and with some contributing residues (Tyr157, Lys221 and Glu223) identical in all species (Figure 1B; Supplementary Figure 2).
Figure 1.
Crystal structure of Hsp90-N-Sgt1-CS complex. (A) In the crystals, opposite faces of the Sgt1-CS domain β-sandwich structure interact with different parts of adjacent Hsp90-N domains. (B) Conservation colouring of the Sgt1-CS surface (blue → red; most → least conserved) based on the alignment in Supplementary Figure 2. The surface shown on the left is highly conserved with a ridge of essentially invariant residues that form the interface with Hsp90. The very high degree of conservation suggests that this interface is biologically authentic, whereas that on the right is a crystal lattice contact. (C) Detail of the conserved Hsp90-N–Sgt1-CS interface. The interface is built around two hydrophobic patches, centred on Sgt1-Tyr157 and Phe168, reinforced by a network of hydrogen bonding interactions (dotted lines). Formation of the complex buries ∼1100 Å2 of molecular surface, which is consistent with a reversible interface. (D) Overview of the Hsp90-N–Sgt1-CS interaction based on the conserved interface. The ADP bound in the pocket of the Hsp90-N domain and the mobile ‘lid' segment that closes in the ATP-bound state of Hsp90 are indicated.
The conserved interface is formed by residues from strands one, two and three from the four-stranded face of Sgt1-CS and the first strand and third helix of Hsp90-N domain (Figure 1C). The core interaction is provided by Tyr157 from AtSgt1a, the hydroxyl group of which hydrogen bonds to the side chains of Glu6 and Lys88 of HvHsp90, whereas its aromatic face packs into a hydrophobic recess on HvHsp90 formed by the side chains of Phe8, Val92 and Lys88, and the main chain of Ser89. Sgt1 Tyr157 is absolutely conserved, as are all the interacting residues in Hsp90, apart from Ser89, which is alanine in non-plant Hsp90s. Phe168 provides a second hydrophobic interaction, packing between the side chains of Thr87, Asp144 and Asp145 on Hsp90. Phe168 is also highly conserved, being phenylalanine in plants and yeast, and methionine in metazoa. These hydrophobic interactions are supported by an extensive network of polar interactions involving Arg153, Glu155, Thr166, Lys170, Lys221 and Glu223 from Sgt1, and Ser89, Asn93, Asp144, Asp145 and Glu146 from Hsp90 (Figure 1C). All the Sgt1 residues involved are at least very highly conserved, whereas the Hsp90 residues, with the exception of Ser89, are invariant. The Sgt1-CS interaction site on Hsp90 does not involve the mobile lid segment nor does it impinge upon the ATP-binding pocket at the heart of the N domain (Figure 1D).
Targeted disruption of Hsp90–Sgt1 interaction
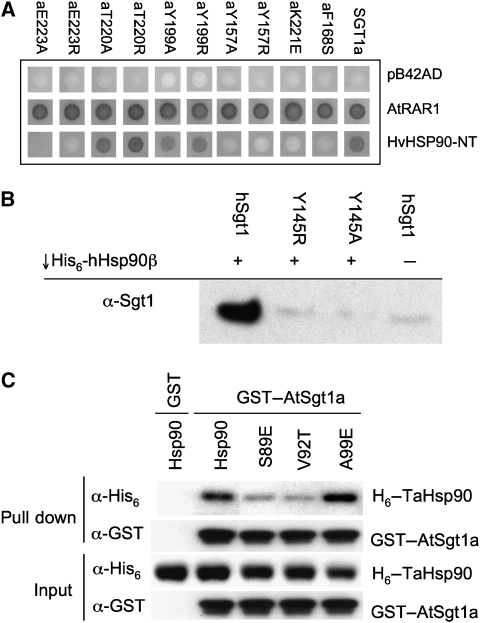
Although the conservation pattern clearly identifies the biologically significant Hsp90–Sgt1 interface, we were concerned to verify this interaction outside the context of a crystal lattice. We examined the effect of mutations in residues on the surface of AtSgt1a-CS, on its ability to interact with the HvHsp90-N domain, using the Y2H system (Boter et al, 2007). Mutations of AtSgt1a-Tyr199, at the heart of the non-conserved crystal lattice contact, had no effect on the Y2H interaction with Hsp90, nor did mutation of AtSgt1a-Thr220 on the periphery of the conserved interface. However mutations of Tyr157, Phe168, Lys221 and Glu223, in the conserved interface, all significantly diminished the interaction, confirming its functional significance (Figure 2A). None of these mutations affected the Y2H interaction with AtRAR1, the interaction of which is mediated by a different region of the AtSgt1a-CS domain (Boter et al, 2007), confirming that these mutations do not disrupt CS domain folding.
Figure 2.
Targeted disruption of Hsp90-N–Sgt1-CS interaction. (A) Yeast two-hybrid analysis of AtSgt1a-CS mutants. AtSGT1a or its derivatives were assayed for interaction with AtRAR1 and HvHSP90-N using LexA Yeast Two-Hybrid system. Mutation of residues Tyr157, Phe168, Lys221 or Glu223, in the conserved interface with Hsp90-N, abrogated reporter activation. (B) Co-precipitation assay with His6-tagged human Hsp90β, and human Sgt1. Hsp90 efficiently co-precipitates wild-type Sgt1, but not mutants of the core interface residue, Tyr145 (equivalent to Tyr157 in the plant protein). (C) Co-precipitation assay with GST-tagged Arabidopsis Sgt1a, and plant Hsp90. AtSgt1a co-precipitates wild-type TaHsp90 or TaHsp90 with mutation of a residue not in the observed interface, but does not efficiently co-precipitate TaHsp90 with mutations in interfacial residues Ser89 or Val92.
To confirm that this interaction was conserved across species, we used an in vitro co-precipitation assay with human homologues to follow Hsp90–Sgt1 interactions in solution (see Materials and methods), and compared the interaction of wild-type Sgt1 with mutants intended to disrupt the conserved Sgt1–Hsp90 interface. GST-tagged full-length wild-type human Sgt1 efficiently co-precipitated hHsp90β (Figure 2B). In contrast, Arg or Ala mutations of Tyr145 (equivalent to AtSgt1a-Tyr157), significantly decreased the ability of hSgt1 to co-precipitate hHsp90β.
Using the plant system, we mutated several residues in the wheat (Triticum aestivum) Hsp90-N domain that are involved in the interface with Sgt1, which also involves the equivalent of Tyr157. Mutation of TaHsp90 Ser89 and Val92, both of which directly contact AtSgt1a Tyr157 to Glu and Thr, respectively, significantly decreased their ability to be co-precipitated by GST–AtSgt1a when compared with wild type. By contrast an Ala99Glu mutant, which is not directly involved in this interface, was efficiently co-precipitated (Figure 2C).
Taken together, the strong conservation of the residues on the four-stranded face involved in interaction with Hsp90 and the disruption of that interaction, in vitro and in the Y2H system, by mutations on either side of that interface argue very strongly that this is the biologically authentic site of interaction between the two proteins and that it is both necessary and sufficient for that interaction.
Sgt1 couples Hsp90 to SCF complexes
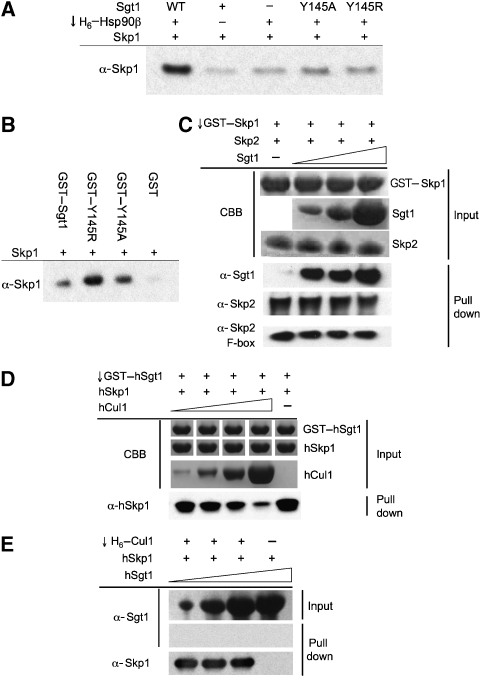
Yeast Sgt1p binds simultaneously to yHsp90 (Hsp82p) and ySkp1p, suggesting that it functions as an adaptor linking the two proteins (Catlett and Kaplan, 2006). We verified this interaction using the human proteins and observed co-precipitation of hSkp1 with His6-tagged hHsp90α only in the presence of hSgt1 (Figure 3A). With the Tyr145Arg or Tyr145Ala hSgt1 mutants, no hSkp1 was co-precipitated. However, GST fusions of both hSgt1 mutants were fully competent in co-precipitating hSkp1, confirming that the Hsp90- and Skp1-interacting regions of Sgt1 are functionally independent (Figure 3B).
Figure 3.
Sgt1 interactions with SCF complex. (A) Bridged co-precipitation assay with His6-tagged human Hsp90β, Sgt1 and Skp1. Hsp90 efficiently co-precipitates Skp1 (visualized by western blot) only when wild-type Sgt1 is present to bridge the interaction. Mutations in the Sgt1 Hsp90-binding residue Tyr145 prevent Skp1 co-precipitation. (B) Direct co-precipitation of Skp1 by Sgt1 (visualized by western blot) is not affected by Sgt1 Tyr145 mutations. (C) Sgt1 is co-precipitated by GST–Skp1–Skp2 or GST–Skp1–Skp2(F-Box) complexes, with no competitive displacement of Skp2 by increasing concentrations of Sgt1. Input protein loadings (10%) are visualized with Coomassie Brilliant Blue (CBB) and co-precipitated proteins are visualized by western blot. (D) Co-precipitation of Skp1 by GST-tagged Sgt1 is diminished by increasing concentrations of Cul1, showing competition between Sgt1 and Cul1 for binding to Skp1. Protein is visualized as in (C). (E) His6-tagged Cul1 efficiently co-precipitates Skp1, but not Sgt1, showing that there is no direct interaction between Cul1 and Sgt1, and that Skp1 cannot bind Cul1 and Sgt1 simultaneously. Increasing concentrations of Sgt1 fail to displace Cul1, which binds Skp1 ∼70-fold tighter than Sgt1.
Skp1 is functionally associated with a variety of proteins containing an ‘F-box'—a small helical-coil domain that mediates interaction with the C-terminal part of Skp1 in the context of SCF E3 ubiquitin ligases (Hao et al, 2005). Previous studies have shown that interaction with Skp1 is mediated by the TPR domain of Sgt1 (Catlett and Kaplan, 2006). As TPR domains are also helical-coil structures, we considered the possibility that Sgt1 might be a cryptic ‘F-box' protein and interact with the same site on Skp1. To test this, we co-expressed human Skp1 with Skp2, an F-box protein that mediates the recruitment of p27kip1 to SCF complexes (Hao et al, 2005), which would therefore compete with Sgt1 if it interacted with the F-box-binding site on Skp1. We found that the Skp1–Skp2 complex or a Skp1–Skp2(F-box) complex was fully able to co-precipitate Sgt1, without competitive displacement of Skp2 with increasing concentrations of Sgt1 (Figure 3C). This suggests that Sgt1 interacts primarily with the N-terminal lobe of Skp1 and can coexist in Skp1 complexes with Skp2 (and probably other F-box proteins), the binding site of which is in the C-terminal lobe.
As well as binding F-box proteins, Skp1 binds to Cullin scaffold proteins such as Cul1, through its N-terminal lobe. Cullins couple Skp1 to the Rbx1 ring-finger protein that catalyses transfer of ubiquitin from an E2 ubiquitin conjugate, to a lysine on the target protein bound by the F-box protein, or to a ubiquitin molecule already attached to the target. Again, we considered the possibility that the TPR domain of Sgt1 might bind to Skp1 in competition with Cul1. Although GST–Sgt1 was able to co-precipitate Skp1 in isolation, the presence of Cul1 in the reaction decreased the co-precipitation of Skp1 in a dose-dependent manner (Figure 3D). In the inverse experiment, Cul1 co-precipitated Skp1, but not Sgt1 present in the same reaction (Figure 3E), confirming that Cul1 and Sgt1 do not interact directly, and that they bind to Skp1 with mutual exclusion. However, we were not able to decrease the amount of Skp1 co-precipitated by Cul1 with increasing amounts of Sgt1. To analyse this further, we directly determined the affinities of Sgt1 and of Cul1, for Skp1, using isothermal titration calorimetry (Supplementary Figure 3). Although Sgt1 bound in the low micromolar range (KD=1.64 μM), Cul1 bound Skp1 ∼70-fold more tightly, with a dissociation constant of ∼24 nM, so that Cul1 could not be competed off a Cul1–Skp1 complex by comparable levels of Sgt1. Taken together, these data are fully consistent with Sgt1 and Cul1 binding in mutual exclusion at a common or at least overlapping binding site on the N-terminal half of Skp1.
In vivo coupling of Hsp90- and Sgt1-centred complexes
Hsp90 is the hub of a nexus of physical and functional interactions with complexes involved in signalling and transcriptional regulation, the component proteins of which form the dependent clientele of the Hsp90 chaperone system (Pearl and Prodromou, 2006; Zhao and Houry, 2007). Sgt1 has also been implicated in the assembly and function of diverse protein complexes, including CBF3 kinetochore, SCF E3 ubiquitin ligase and adenyl cyclase complexes in yeast (Kitagawa et al, 1999; Dubacq et al, 2002) and R-proteins and Nod-like receptors in plants and animals (Peart et al, 2002; Bieri et al, 2004; Leister et al, 2005; da Silva Correia et al, 2007; Mayor et al, 2007). Sgt1 therefore also has the characteristics of an interaction hub. However, it is not clear whether Sgt1 has an independent function in any of these systems, or whether it functions purely as a scaffold protein, coupling them to Hsp90.
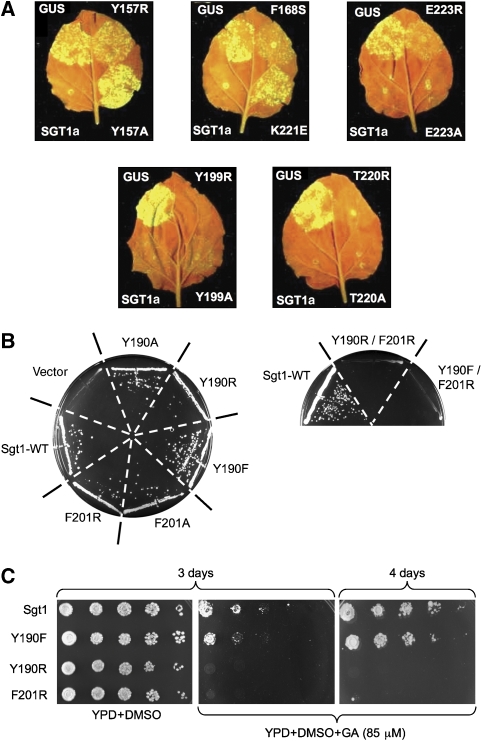
To gain some insight into this, we utilized our structural and biochemical data to engineer mutations in plant Sgt1, in vivo, which would be expected to specifically disrupt its interaction with Hsp90, but not with other known ligand proteins. Using a GFP-tagged fluorescent virus propagation assay, as described earlier (Azevedo et al, 2006; Boter et al, 2007), we determined the Rx-protein-mediated resistance to infection by potato virus X (PVX), of tobacco leaves (Nicotiana benthamiana) silenced for endogenous Sgt1, and heterologously complemented with wild-type or mutant AtSgt1a (see Materials and methods). Alanine mutations of Tyr199 on the three-stranded face of AtSgt1a-CS domain or of Thr220 on the four-stranded face, but not involved in direct contact with Hsp90, had no effect on the ability of the mutant protein to confer resistance to infection. Alanine mutations of Glu223 or Thr166, which are part of the Hsp90-interacting surface, also had little effect on resistance. However, alanine mutation of Tyr157, at the heart of the interface with Hsp90, or a charge reversal mutation of Lys221 close by, caused a substantial loss of resistance, comparable to an Sgt1a null (Figure 4A). The ability of a single missense mutation in Sgt1 that specifically disrupts its interaction with Hsp90 in vitro, to disrupt R-gene-mediated viral resistance in vivo, provides definitive evidence for the direct dependence of this class of innate immunity system on the Hsp90 chaperone system, with Sgt1 acting as a molecular scaffold coupling the two.
Figure 4.
Functional dependence of Sgt1 on Hsp90 interaction in vivo. (A) Functional assay of SGT1a mutants in Rx-mediated resistance against potato virus X (PVX). Rx-containing N. benthamiana plants silenced for NbSGT1 were co-infiltrated with Agrobacterium expressing wild-type AtSGT1a (lower left, positive control), or AtSGT1a mutants as indicated (right half of the leaf) or GUS (upper left, negative control) together with PVX-GFP. Virus accumulation was monitored by GFP fluorescence under UV illumination 5 days after inoculation. Mutations in residues not involved in the core interface (Tyr199 and Thr220) with Hsp90 had little effect on the ability of AtSgt1a to facilitate viral resistance. Mutations of the hydrophobic Hsp90 interface residues Tyr157 or Phe168, and charge-reversal mutations in polar interface residues Lys221 or Glu223, severely impaired the biological function of Sgt1a. (B) Single-point mutations in Hsp90-interacting residues did not significantly impair essential Sgt1 functions in yeast (left), but double mutants abolished viability (right). (C) Single-point mutations in Hsp90-interacting residues in yeast Sgt1 sensitize yeast to killing by the Hsp90 inhibitor geldanamycin (GA).
Although yeast lacks homologues of plant R-proteins or animal Nod-like receptors, Sgt1 is essential in yeast, not least due to its involvement in the CBF3 kinetochore complex, and the SCF E3 ubiquitin ligases that regulate cell cycle transitions (Kitagawa et al, 1999). To determine whether essential Sgt1 functions in yeast are also dependent on its interaction with Hsp90, we engineered similar interface-disrupting point mutations, singly and in combination, into yeast Sgt1 and determined their ability to confer viability in the absence of a wild-type Sgt1 allele (see Materials and methods). Unlike the plant system, single-point mutations in either of the main hydrophobic Sgt1 interface residues (Tyr190, Phe201≡Tyr157, Phe168 in plants) had little effect (Figure 4B). However, double mutants abrogated viability. This is consistent with the observation of multiple missense mutations in previously identified ts SGT1 alleles (Kitagawa et al, 1999). Furthermore, although viable under normal conditions, yeast harbouring Sgt1 single-point mutants were significantly more sensitive to the Hsp90 inhibitor, geldanamycin, than wild type (Figure 4C). Taken together, these data confirm the dependence of essential Sgt1 functions in yeast on its ability to form a stable complex with Hsp90.
TPR co-chaperone compatibility
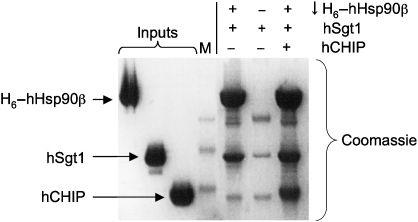
The extreme C terminus of Hsp90 consists of an MEEVD motif that provides the binding site for a range of co-chaperones containing TPR domains. Although Sgt1 possesses a TPR domain, this is not essential for Hsp90 binding (Lee et al, 2004; Lingelbach and Kaplan, 2004). Furthermore, a yeast TPR domain co-chaperone, Sti1 known to interact with the C terminus of Hsp90, bound simultaneously with Sgt1 (Catlett and Kaplan, 2006). We found that the human TPR domain co-chaperone CHIP was also able to bind to Hsp90 at the same time as Sgt1 (Figure 5), showing that Sgt1 can most likely coexist in Hsp90 complexes with a range of TPR domain co-chaperones. As Sgt1 is able to couple Hsp90 to the Skp1 and F-box components of SCF E3 ligase systems, its ability to coexist in Hsp90 complexes with CHIP, an active E3 ubiquitin ligase, is particularly interesting.
Figure 5.
Co-binding of TPR domain E3 ligase CHIP and Sgt1. His6-tagged human Hsp90β efficiently co-precipitates Sgt1 and CHIP, confirming that the TPR domain of Sgt1 does not interact with the C-terminal MEEVD sequence of Hsp90, which provides the binding site for CHIP, and other TPR domain co-chaperones.
Discussion
Hsp90 interaction and nucleotide dependence
The Hsp90–Sgt1 core complex structure reveals a previously unknown site of co-chaperone interaction on the chaperone. Binding sites have been characterized in the middle domain for the ATPase activator Aha1 (Meyer et al, 2004), and at the C-terminal MEEVD motif for a variety of TPR domain co-chaperones (reviewed in Pearl and Prodromou, 2006). Sgt1 also possesses a TPR domain, but this does not bind to Hsp90 in isolation, and Hsp90 constructs lacking the MEEVD motif are still able to bind Sgt1 (Catlett and Kaplan, 2006). Cdc37 interacts with the open conformation of the lid segment in the N-terminal nucleotide-binding domain and arrests Hsp90's ATPase cycle (Roe et al, 2004). p23/Sba1 also interacts with the lid segment in the N domain, but only in its closed conformation within the closed ATP-bound state of the chaperone, and in doing so reinforces the interaction of the N domains within the closed dimer to each other and to the middle domains (Ali et al, 2006). Although the CS domain of Sgt1 and the globular core domain of p23/Sba1 have similar three-dimensional structures, they bind to different regions of the Hsp90 surface, using different parts of their own structures. Sgt1 utilizes residues on the surface of the four-stranded β-sheet, whereas a C-terminal strand, not present in Sgt1-CS, provides the majority of p23/Sba1a interaction with Hsp90-N (Figure 6A).
Figure 6.
CS domain interactions. (A) Comparison of the binding surfaces used by the structurally homologous CS domains of the Hsp90 co-chaperones Sgt1 (left) and p23/Sba1 (right). Sgt1-CS uses residues on the face of its four-stranded β-sheet, whereas p23/Sba1 interacts with Hsp90-N through a C-terminal extension not present in Sgt1. The interaction site on Hsp90-N (bottom, magenta) is also completely different, with Sgt1-CS binding the side of the domain, whereas p23/Sba1 binds directly to the lid segment in its closed ATP-bound conformation. (B) Hypothetical model of Sgt1-CS binding to Hsp90 in the closed ATP-bound conformation, made by superimposing the Hsp90-N domain from the present structure on to the crystal structure of the full-length Hsp90-AMPPNP-p23/Sba1 structure (PDB code 2CG9). Consistent with the preference of Sgt1 for binding to the open ADP-bound state of Hsp90, the docked Sgt1-CS domains clash sterically, and this is likely to be exacerbated in the context of the full-length protein by the TPR and SGS domains, which extend from the N- and C terminus, respectively.
Most significant mechanistically are the different sites of interaction of p23/Sba1 and Sgt1-CS on Hsp90-N, and their effect on the conformational changes associated with Hsp90s ATPase cycle. p23/Sba1 interacts with residues on the lid segment of Hsp90 that are only available in the ATP-bound conformation of the chaperone (Ali et al, 2006), explaining the strongly ATP-dependent binding of p23/Sba1 (Sullivan et al, 1997; Siligardi et al, 2004). Sgt1-CS binds to Hsp90-N away from the lid and ATP-binding pocket, so that its preferential binding in the absence of ATP (Lee et al, 2004; Catlett and Kaplan, 2006) is difficult to explain at first sight. However, part of the interaction is provided by Glu6 and Phe8 from the N-terminal strand of Hsp90, which undergoes a large movement during the ATPase cycle, swapping from its own N domain in the open ATP-free state (Prodromou et al, 1997) to bind to the other N domain in the closed ATP-bound conformation (Ali et al, 2006), recapitulating its previous interactions. Although Sgt1-CS could interact with the N-terminal strand in the ATP-free or ATP-bound states, it would certainly be displaced during the ATP-driven conformational change, so that in the presence of ATP the equilibrium concentration of Hsp90–Sgt1 complexes would be lower than in its absence. Furthermore, when the Hsp90-N–Sgt1-CS complex is used to model binding of Sgt1-CS domains to both Hsp90-N domains in the closed ATP-bound structure of the dimer (Figure 6B), there is a steric clash between the two Sgt1-CS domains. This may be exacerbated by the presence of the N-terminal TPR and C-terminal SGS domains, the disposition of which within the overall Hsp90–Sgt1 complex is not currently known.
Function of the Hsp90–Sgt1 couple
The range of functions in which Hsp90 is an essential participant continues to increase (Pearl et al, 2008). Rather than clarifying the biochemical basis of Hsp90 involvement, the diversity of the expanding protein clientele makes it increasingly difficult to understand selective interaction with Hsp90 in terms of any common structural motif. At least part of this selectivity comes from co-chaperones such as Cdc37 that function as adaptors, recruiting specific classes of clients to Hsp90.
Sgt1 fulfils the requirements of an Hsp90 adaptor co-chaperone, interacting with Hsp90 through its CS domain, and with a range of putative client proteins through its TPR and SGS domains. Unlike Cdc37 however, it does not appear to regulate the ATPase activity of Hsp90 (Catlett and Kaplan, 2006). In plant innate immunity systems, Sgt1 is essential not only for the activity of R-proteins but also for their stable accumulation (Bieri et al, 2004; Azevedo et al, 2006; Boter et al, 2007). However, the related mammalian Nod-like receptors lose activity, but not stability, when Sgt1 is knocked down (da Silva Correia et al, 2007). They are nonetheless depleted by pharmacological inhibition of Hsp90, suggesting that this biologically widespread family of proteins are bone fide Hsp90 clients.
Chaperone–client relationships are less clear in the Sgt1-mediated coupling of Hsp90 and SCF complexes. A variety of different SCF subassemblies can be expressed in a functional state in bacteria, which argues against a stringent client dependence on Hsp90, and no depletion of SCF components on Hsp90 inhibition has been reported. Sgt1 physically couples Hsp90 and Skp1 (Catlett and Kaplan, 2006) and we have extended that observation to show that Sgt1 binds Skp1 in the presence of an F-box protein. Thus, Sgt1 links Hsp90 to the target-specific ‘arm' of an SCF complex. The ability of Sgt1 to bind Skp1 alongside an F-box protein explains the association of Hsp90–Sgt1 with the yeast CBF3 kinetochore complex (Kitagawa et al, 1999; Stemmann et al, 2002; Bansal et al, 2004; Lingelbach and Kaplan, 2004; Rodrigo-Brenni et al, 2004; Steensgaard et al, 2004), as the Ctf13 component of CBF3 is an F-box protein, that is itself the target of ubiquitin-dependent degradation (Kaplan et al, 1997).
In contrast to F-box proteins, we found that Sgt1 binding to Skp1 was mutually exclusive with binding of Cul1, which provides the generic arm of the SCF complex, recruiting the ring-finger E3 ubiquitin ligase enzyme Rbx1. At first sight, this appears to be in contradiction to a reported co-precipitation from yeast lysates of the cullin Cdc53p, as well as Skp1p, by Sgt1p (Kitagawa et al, 1999). However, when SCF components were expressed in insect cells in the same study, the amount of Sgt1p co-precipitated by the F-box protein Cdc4p was decreased when Cdc53p was also present, consistent with our clear observation using purified human proteins that Cul1 and Sgt1 compete for Skp1 binding.
Association of the cullin arm with the specific arm of SCF complexes, through binding to Skp1, is regulated by a complex cycle of reversible modification involving covalent attachment of NEDD8, a ubiquitin-like protein, to Cul1. Neddylated Cul1 in complex with Rbx1 associates with Skp1 and its attendant F-box protein and is fully competent to recruit E2 ubiquitin conjugates and ubiquitinate the substrate protein bound to the F-box protein (Wu et al, 2006). This active SCF complex is inactivated when NEDD8 is removed by the COP9 signalosome complex (Lyapina et al, 2001). De-neddylated Cul1 is then bound by Cand1 with concomitant displacement of the Skp1–F-Box subcomplex and blockade of the NEDD8 attachment site on the cullin (Goldenberg et al, 2004). Regeneration of an active SCF complex thus requires displacement of Cand1, neddylation and association with a new Skp1–F-Box substrate complex (Wu et al, 2006). The ability of Sgt1 to couple Hsp90 to the specific arm of SCF complexes, along with its competitive displacement by the cullin arm, suggests a clear chaperone role for Hsp90–Sgt1 in binding and stabilizing a Skp1–F-Box substrate complex in the absence of Cul1–Rbx1. This is fully consistent with the observation of the ∼70-fold higher affinity we observe with Cul1, as compared with Sgt1, for binding to Skp1, which would ensure efficient displacement of Hsp90–Sgt1 from a new Skp1–F-Box substrate complex by Cul1, once freed of its inhibitory association with Cand1. The ATPase cycle of Hsp90 could have a function in facilitating the release of Cul1 from Cand1, and allowing its re-association with Skp1 to regenerate an active SCF complex (Figure 7A and B); however, further work will be required to test this possibility. A role for Hsp90–Sgt1 in facilitating the assembly of SCF complexes is fully consistent with observations that defective SGT1 alleles in yeast impair SCF-dependent turnover of Sic1p in vivo, and extracts from SGT1-impaired strains are defective in cyclin ubiquitination in vitro (Kitagawa et al, 1999).
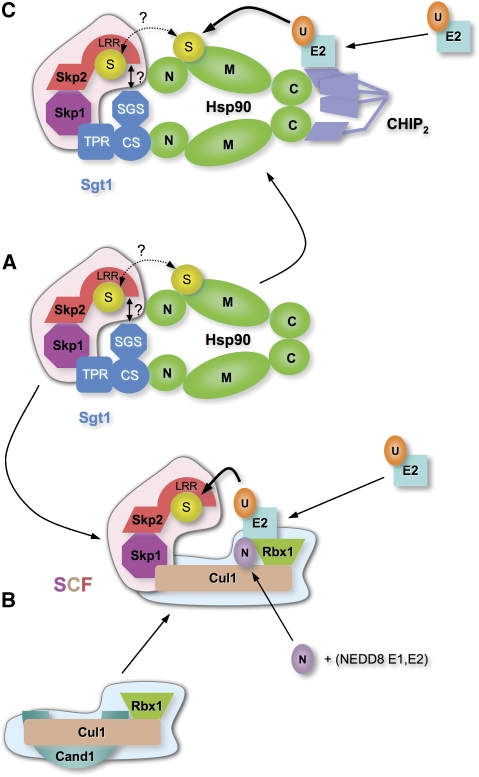
Figure 7.
A role for Hsp90–Sgt1 in SCF assembly. (A) Previous observations and data presented here that Hsp90–Sgt1 can interact with Skp1 in complex with an F-box protein such as Skp2. In such a complex, a client/substrate protein (S) could be bound to either the chaperone or to the F-box protein, and in principle could be transferred from one to the other. (B) Competitive displacement of Hsp90–Sgt1 from Skp1 by Cul1–Rbx1, concomitant with the release of Cul1–Rbx1 from Cand1, would permit formation of an active SCF complex, able to be neddylated and bind E2. Disruption of the Cand1–Cul1 interaction might require the ATPase activity of Hsp90. (C) Alternatively, as a parallel pathway during stress conditions or when the SCF route is overloaded, an Hsp90–Sgt1–Skp1–F-box protein complex could allow ubiquitination of a client/substrate protein by the recruitment of CHIP to the C-terminal binding site on Hsp90.
Hsp90 might also have a function in facilitating or regulating access of the substrate protein to the SCF system. A number of proteins known to be Hsp90 clients are degraded through SCF complexes, most significant of which is probably the cell cycle regulatory tyrosine kinase Wee1 (Aligue et al, 1994; Watanabe et al, 2004). However, the interplay between these two Wee1-interacting pathways has not so far been characterized, although both are required for proper mitotic entry. It has long been known that inhibition of Hsp90 promotes ubiquitin-dependent degradation of its client proteins (Mimnaugh et al, 1996), but the E3 systems responsible, particularly for protein kinases, have not been unambiguously identified. The TPR domain U-box protein CHIP is an E3 ubiquitin ligase able to associate with Hsp90 or Hsp70 through the C-terminal EEVD motif (Cyr et al, 2002; Zhang et al, 2005), and is a clear candidate for that role, although there are only a few clear demonstrations of CHIP's involvement in the degradation of major Hsp90 clients such as protein kinases (Xu et al, 2002; Zhang et al, 2007). The ability of CHIP to bind to an Hsp90–Sgt1 complex raises the possibility that it could have a function in ubiquitination of Skp1–F-Box-bound substrate proteins, possibly as a backup under stress conditions, or in some circumstances in direct collaboration with the SCF system (Nelson et al, 2006).
Materials and methods
Y2H analysis
AtSGT1a point mutations were obtained by site-directed mutagenesis (BIO S&T) using AtSGT1a-pGEX-6P-1 construct. AtSGT1a WT and mutant derivates were cloned in pLexA (Clontech) as EcoRI/NotI. AtRAR1 and HvHSP90-NTD clones in pB42AD were described earlier (Takahashi et al, 2003). Interaction analyses were carried out as described in the manufacturer's protocol (MATCHMAKER; Clontech).
Rx-mediated resistance assay in N. benthamiana
AtSGT1a and mutant derivates were PCR amplified, cloned into pBIN61 as XbaI/BamHI and transformed into Agrobacterium tumefaciens C58C1 carrying plasmid pCH32. Transgenic N. benthamiana plants expressing Rx:HA under the control of its own promoter were silenced for NbSGT1 as described earlier (Azevedo et al, 2006) and co-infiltrated with Agrobacterium expressing AtSGT1a or mutant derivates (OD=0.3) and PVX-GFP (OD=0.001). PVX accumulation was monitored by GFP fluorescence under UV illumination 5 days after inoculation.
Strains and plasmids
The Saccharomyces cerevisiae YKK39 (matα ura3-52, lys2-801, ade2-101, trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1Δ1∷HIS31) was used as the host strain for the expression of SGT1 mutant alleles (Kitagawa et al, 1999). Its viability was maintained by the SGT1 gene of the centromeric (single copy) URA3 plasmid pRS316-SGT1 (Kitagawa et al, 1999).
Mutagenesis and plasmid construction
Amino-acid changes were generated in the SGT1 gene of the centromeric (single copy) LEU2 plasmid pRS315-SGT1 (Lingelbach and Kaplan, 2004), using the QuickChange site-directed mutagenesis kit (Stratagene). The following mutations were introduced and confirmed by sequencing: Y190A, Y190R, Y90F (TAT to GCT, AGA and TTT, respectively); F201A, F201R (TTT to GCT and AGA, respectively) and the double mutants Y190R/F201R and Y190F/F201R.
Media and genetic techniques
The S. cerevisiae strain YKK39 was transformed with pRS315-SGT1 bearing wild-type or mutant SGT1, as indicated in figure legends (Rose et al, 1990). Transformants were selected on drop-out media without uracil and leucine. The ability of SGT1 mutants to maintain cell viability was assessed by curing for pRS316-SGT1 by incubating the transformants on drop-out media without leucine, but containing uracil (25 mg/l) and 5-FOA (0.1%).
DNA constructs
cDNAs for H. vulgare Hsp90 and A. thaliana Sgt1a were as described (Boter et al, 2007). Human Hsp90β was as described (Panaretou et al, 2002). Human Skp1, Skp2, Sgt1 and Cul1 were amplified from IMAGE ESTs (4 243 711, 2 962 938, 2 985 858 and 40 118 923, respectively). Different constructs of Skp1 (residues 1–163, 1–70, 81–163 and Δ71–80) and Sgt1 were cloned into pGEX6P-1, Skp2 constructs (residues 1–411 and 89–141) were cloned into pET28a. Cul1 coding region was split into two halves (residues 1–410 and 411–776) as described earlier (Zheng et al, 2002) and cloned into pST38 (Tan, 2001) for co-expression.
Protein expression and purification
Plant and mammalian Hsp90 and Sgt1 constructs were all expressed in the Escherichia coli strain Rosetta (DE3) pLysS. Hsp90 constructs, were purified as described earlier (Prodromou et al, 1997). GST-tagged Sgt1 constructs were purified by ion exchange on Q-Sepharose and immobilized on glutathione Sepharose beads. Sgt1 was then released by PreScission cleavage and further purified on a Superdex 75 PG gel-filtration column. Human Skp1, Skp2 and Cul1 constructs were all purified as described earlier (Zheng et al, 2002).
In vitro protein co-precipitation assay
In total, 50 μg of each indicated protein (except protein in gradient titrations) was incubated in 50 μl glutathione Sepharose resin or TalonTM metal affinity resin in 200 μl of binding buffer consisting of 50 mM Tris pH 7.5, 150 mM NaCl, 0.1% NP-40 and 1 mg/ml BSA (pH 7.5). For gradient titration experiments, 5 μg of gradient protein was used in the first reaction, with three-fold increase in subsequent reactions (i.e. 15, 45 μg and so on). Reactions were performed overnight at 4°C. Resin was washed three times with 1 ml binding buffer later. One-fiftieth of the washed resin was resolved by SDS–PAGE, electroblotted and probed with antibody to the particular protein construct (‘α-') as indicated. Quantities of protein consumed were shown by SDS–PAGE using 10% of each input proteins and visualized with Coomassie Brilliant Blue.
Crystallization, data collection and structure determination
HvHsp90-N and AtSgt1a-CS were combined in a 1:1 molar ratio in the presence of 5 mM ADP, incubated for 30 min and concentrated to 10 mg/ml by ultrafiltration. Initial multiple crystals were grown by vapour diffusion at 4°C against 26% w/v PEG4000, 100 mM Tris (pH 8.5) and 200 mM magnesium sulphate. Subsequent streak seeding into solutions of 16% w/v PEG4000, 100 mM Tris (pH 8.5) and 200 mM magnesium sulphate produced single thin plates. Crystals were harvested into reservoir solution with addition of glycerol (20% v/v) before flash cooling to 100 K. X-ray data were collected on beamline ID23 at the ESRF, Grenoble, from single crystals, and processed by using MOSFLM (Leslie, 1995) and the CCP4 package (CCP4, 1994). Crystals had space group P212121, with three copies of the complex in the asymmetric unit. The thin plate crystals have a high solvent content (59% v/v) and only gave useful diffraction to 3.3 Å. The structure was solved by molecular replacement with Phaser (McCoy, 2007) using ADP-bound yeast Hsp90-N (PDB code: 1AMW) and the solution structure of human Sgt1-CS domain (PDB code: 1RL1) as search models, built using COOT (Emsley and Cowtan, 2004), and refined with CNS in PHENIX (Adams et al, 2002), using NCS restraints. Data collection and refinement parameters are given in Table I. Crystallographic data have been deposited in the Protein Databank with code: 2JKI
Supplementary Material
Supplementary Figures 1
Supplementary Figures 2
Supplementary Figures 3
Supplementary Material
Acknowledgments
We are grateful to Kenneth Kaplan for the kind gift of yeast strain YKK39 and the pRS315-SGT1 vector, to Mark Roe for help with data collection and processing and to Cara Vaughan for useful discussion. This study was supported by The Wellcome Trust (LHP), The Gatsby Foundation and KAKENHI 19678001 (KS), Marie Curie Fellowship Program (MB) and the Japan Society for the Promotion of Science (YK).
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH (2006) Crystal structure of an Hsp90—nucleotide–p23/Sba1 closed chaperone complex. Nature 440: 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aligue R, Akhavan-Niak H, Russell P (1994) A role for Hsp90 in cell cycle control: Wee1 tyrosine kinase activity requires interaction with Hsp90. EMBO J 13: 6099–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Bansal PK, Abdulle R, Kitagawa K (2004) Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol Cell Biol 24: 8069–8079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH, Shirasu K, Schulze-Lefert P (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16: 3480–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Amigues B, Peart J, Breuer C, Kadota Y, Casais C, Moore G, Kleanthous C, Ochsenbein F, Shirasu K, Guerois R (2007) Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 19: 3791–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Mandal AK, Theodoraki MA (2007) Molecular chaperones and protein kinase quality control. Trends Cell Biol 17: 87–92 [DOI] [PubMed] [Google Scholar]
- Catlett MG, Kaplan KB (2006) Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J Biol Chem 281: 33739–33748 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol 3: 93–96 [DOI] [PubMed] [Google Scholar]
- Cyr DM, Hohfeld J, Patterson C (2002) Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends BiochemSci 27: 368–375 [DOI] [PubMed] [Google Scholar]
- da Silva Correia J, Miranda Y, Leonard N, Ulevitch R (2007) SGT1 is essential for Nod1 activation. Proc Natl Acad Sci USA 104: 6764–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C (2002) Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell 1: 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N (2004) Structure of the Cand1–Cul1–Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119: 517–528 [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP (2005) Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell 20: 9–19 [DOI] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger PK (1997) Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91: 491–500 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Lee YT, Jacob J, Michowski W, Nowotny M, Kuznicki J, Chazin WJ (2004) Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J Biol Chem 279: 16511–16517 [DOI] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW (1995) MOSFLM Users Guide. Cambridge, UK: MRC Laboratory of Molecular Biology [Google Scholar]
- Lingelbach LB, Kaplan KB (2004) The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol Cell Biol 24: 8938–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J (2007) A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol 8: 497–503 [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Liao C, Hu B, Mark Roe S, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH (2004) Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J 23: 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimnaugh EG, Chavany C, Neckers L (1996) Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem 271: 22796–22801 [DOI] [PubMed] [Google Scholar]
- Nelson RF, Glenn KA, Miller VM, Wen H, Paulson HL (2006) A novel route for F-box protein-mediated ubiquitination links CHIP to glycoprotein quality control. J Biol Chem 281: 20242–20251 [DOI] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C (2002) Activation of the ATPase activity of Hsp90 by the stress-regulated co-chaperone Aha1. Mol Cell 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Pearl LH (2005) Hsp90 and Cdc37—a chaperone cancer conspiracy. Curr Opin Genet Dev 15: 55–61 [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem 75: 271–294 [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C, Workman P (2008) The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J 410: 439–453 [DOI] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DA, Xiao S, Coleman MJ, Dow M, Jones JD, Shirasu K, Baulcombe DC (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH (1997) Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90: 65–75 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Thomas S, Bouck DC, Kaplan KB (2004) Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol Biol Cell 15: 3366–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe SM, Ali MM, Meyer P, Vaughan CK, Panaretou B, Piper PW, Prodromou C, Pearl LH (2004) The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37). Cell 116: 87–98 [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schadick K, Fourcade HM, Boumenot P, Seitz JJ, Morrell JL, Chang L, Gould KL, Partridge JF, Allshire RC, Kitagawa K, Hieter P, Hoffman CS (2002) Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgtlp family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot Cell 1: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligardi G, Hu B, Panaretou B, Piper PW, Pearl LH, Prodromou C (2004) Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J Biol Chem 279: 51989–51998 [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP (1997) Crystal structure of an Hsp90–geldanamycin complex: targetting of a protein chaperone by an antitumor agent. Cell 89: 239–250 [DOI] [PubMed] [Google Scholar]
- Steensgaard P, Garre M, Muradore I, Transidico P, Nigg EA, Kitagawa K, Earnshaw WC, Faretta M, Musacchio A (2004) Sgt1 is required for human kinetochore assembly. EMBO Rep 5: 626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Neidig A, Kocher T, Wilm M, Lechner J (2002) Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc Natl Acad Sci USA 99: 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri ES, Litwack G, Toft D (1997) Nucleotides and two functional states of hsp90. J Biol Chem 272: 8007–8012 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc Natl Acad Sci USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S (2001) A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif 21: 224–234 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H (2004) M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci USA 101: 4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JT, Chan YR, Chien CT (2006) Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol 16: 362–369 [DOI] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L (2002) Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA 99: 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Nephew KP, Gallagher PJ (2007) Regulation of death-associated protein kinase. Stabilization by HSP90 heterocomplexes. J Biol Chem 282: 11795–11804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH (2005) Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zhao R, Houry WA (2007) Molecular interaction network of the Hsp90 chaperone system. Adv Exp Med Biol 594: 27–36 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1
Supplementary Figures 2
Supplementary Figures 3
Supplementary Material