Abstract
The mechanism of proton transport in the D-pathway of cytochrome c oxidase (CcO) is further elucidated through examining a protonated water/hydroxyl cluster inside the channel. The second generation multi-state empirical valence bond (MS-EVB2) model was employed in a molecular dynamics study based on a high resolution X-ray structure, to simulate the interaction of the excess proton with the channel environment. Our results indicate that a hydrogen-bonded network consisting of about 5 water molecules surrounded by three side chains and two backbone groups (S197, S200, S201, F108) is involved in storage and translocation of an excess proton to the extracellular side of CcO.
INTRODUCTION
The D-pathway of cytochrome c oxidase (CcO) has been the focus of extensive experimental and theoretical research in the past few years due to its high capacity for proton transport and the ability to transport protons destined for both chemical catalysis (substrate protons) and the pumping process (pumped protons). Both simulation and experiment suggest that a number of confined water molecules, together with a number of hydrophilic residues from several transmembrane helices, provide a hydrogen-bonded (H-bonded) chain that is able to transport protons from the inner surface of CcO to a carboxylic residue E286 (R. sphaeroides, Rs, notation) buried 25 Å away in the interior of the enzyme.1–3 This has lead to the suggestion that water molecules may act as protonatable donor/acceptors in the mechanism of CcO.4–6
It has been recently demonstrated through multi-state empirical valence bond5 (MS-EVB) molecular dynamics (MD) simulations of the bovine CcO6 (PDB ID: 1V54) that a “proton trap” region may exist in the channel where an excess proton resides “waiting” for the deprotonation of E242 (bovine notation, or E286 in Rs notation). These free energy pathway, i.e., potential of mean force (PMF), calculations indicate that the free energy barrier for this proton to escape from the trap region and approach the protonated E242 in bovine CcO is greater than 18 kcal/mol.7 Given the high degree of structural conservation of the D-pathway region among different CcOs, one would expect similar results from other species. This expectation is indeed confirmed and reported here for the first time through a careful examination of a recently solved crystal structure of the R. sphaeroides CcO (PDB ID: 2GSM)8 in comparison with MS-EVB simulation data. This analysis reveals a protonated water/alcohol/carbonyl oxygen cluster near the top of the D-pathway. In the high resolution crystal structure, two water molecules that make up the base of the protonated cluster are only 2.53 Å apart, thus they are more closely related to a Zundel cation, H5O2+, or part of a (distorted) Eigen cation, H9O4+, than a neutral water-water H-bonded pair.
Since x-ray studies present a static picture of a protein in a crystal unit cell, averaged over all elementary cells during data collection, only limited information about fluctuations within an H-bonded network is provided. It is therefore particularly informative to further characterize the excess channel proton in the Rs CcO by using the MS-EVB MD method. In this paper, we employ the coordinates obtained from the 2.0 Å resolution x-ray diffraction studies of the Rs CcO in such calculations using the second generation MS-EVB model (MS-EVB2).5, 9 The important distinction in this approach is that the proton can fully delocalize and hop along multiple water molecules, consistent with the Grotthuss mechanism of proton transfer.10, 11 The ultimate aim here is to further our understanding of the mechanisms behind the operation of the D-channel in CcO.
COMPUTER SIMULATIONS
The MS-EVB2 model has been successfully used to treat proton transport in bulk solution and several biomolecular systems.5 Briefly, in an EVB model, the state of a chemical reaction can be described as a linear combination of a number of independent localized valence bond states |i〉:
| (1) |
The MS-EVB potential energy surface is defined as the electronic ground-state wave function that is obtained by solving the eigenvalue problem:
| (2) |
Here, c is the eigenvector with elements ci[i=1,N] for the lowest eigenvalue E0; N is the number of EVB states. Since the MS-EVB amplitude is proportional to the contribution of the corresponding EVB state to the overall MS-EVB potential energy function, this quantity can be used as a measure of the hydronium character of the hydrated excess proton structure. In bulk aqueous systems, when the population of the highest occupied state, , is ~0.5, the excess proton assumes a Zundel cation structure. An Eigen cation, having an amplitude , is more hydronium-like and slightly more stable than Zundel cation in bulk water.5, 12
In eq (2), H is the EVB Hamiltonian with matrix elements hij = 〈i|Ĥ|j〉. Each matrix element is a function of the nuclear degrees of freedom of the system under consideration. The diagonal matrix elements depend on the nuclear degrees of freedom of the system and can be described by molecular mechanics force fields. The off-diagonal elements are then empirically chosen to reproduce as best as possible the actual potential energy surface, typically as determined from ab initio calculations or experimental results. After one determines the MS-EVB potential surface, the forces on the system nuclei can then be generated from the ground state in eq (2) using the Hellmann-Feynman theorem. For a complete description of this model, see the available literature.5, 9, 12, 13 The interactions between hydronium and the surrounding water molecules were modeled here using the MS-EVB2 parameter set.9 The proton transfer between water and amino acids, such as E286, was not explicitly allowed in the present study.
We have adopted a truncated model for CcO represented by subunit I, crystal water within 8 Å of this subunit, an excess proton and a counter ion Cl− for total charge neutrality. The approximation associated with the truncated model is justified because the immediate D-channel environment is the most relevant for the transport properties of the excess proton.
Initial configurations for the MS-EVB2 proton transport simulations were prepared by replacing a water molecule at the bottom of the channel above the N121/139 with a hydronium. The coordinates of atoms were taken from a snapshot saved after 2 ns of an equilibrium NVT (constant number of atoms, volume and temperature) trajectory. For a detailed description of the molecular dynamics (MD) simulation protocol, see ref.6
MS-EVB2 simulations were carried out with the DL_POLY14 package modified to include the MS-EVB2 algorithm. E286 was protonated, as suggested on the basis of electrostatic calculations,15 and the rest of the residues remained in their default protonation state. The system was simulated at a constant temperature of 300K in the constant NVT ensemble maintained by a Nose-Hoover thermostat with a relaxation constant of 0.2 ps. The electrostatic interactions were calculated with the Ewald method, and the cutoff radii for both Lennard-Jones and real-space Coulombic interactions were 10 Å. The MD time step was 1 fs, and the protein interactions were modeled by the AMBER9916 force field with water represented by the flexible TIP3P model.
RESULTS AND DISCUSSION
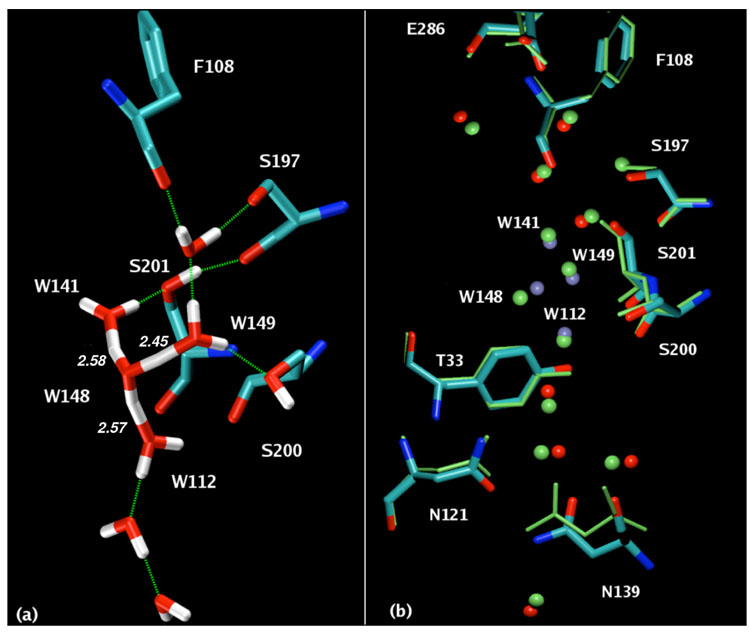
A representative snapshot taken from the simulation trajectory is shown in Figure 1(a). After traveling half the length of the channel within the first 50 ps of the MS-EVB simulation, the excess proton assumes residence in a wide pocket, previously referred to as a proton trap,6, 7 near the top of the channel and 6–7 Å away from residue E286. The excess proton is seen to be delocalized across four water molecules, W148, W141, W149 and W112, but preferentially residing on a central hydronium-like structure, W148, having the dominant MS-EVB amplitude (~ 0.65), therefore indicating an Eigen-like species. It should be noted that an Eigen cation resonance structure is intrinsically a multi-state (i.e., non-classical and non-two-state EVB) species in the MS-EVB model. The solvation structure of the excess proton is also stabilized by the hydrogen bond interactions mediated by the OH groups of two serine residues, S200/201. These H-bonds are largely present throughout the simulation, leading to more stability in the trapped proton structure. The mean water and hydroxyl oxygen distances are 2.74 Å and 2.85 Å, values closely comparable with 2.78 for the O(W141)-OH(S201) and 2.82 for the O(W149)-OH(S200) distance, respectively, in the crystal structure.
Figure 1.
(a). A snapshot of the H-bonded network of the protonated water cluster. The structure is colored by atom name (cyan, C; blue, N; red, O; white, H). The hydrogen bonds are shown as green lines. The O-O distances between W148 and three solvating water molecules are 2.58 Å, 2.57 Å, 2.45 Å, respectively. (b). Superposition of the time-averaged structure and the crystal structure. The crystal structure residues and water oxygen atoms are colored green. The simulation structure is colored by atom name, with the central hydronium and the first solvation shell water oxygen atoms labeled and highlighted in ice blue.
In Figure 1(b), the time-averaged MS-EVB simulation structure based on a 2 ns trajectory has been overlaid with the Rs CcO D-pathway x-ray structure in order to compare the positions of amino acid side chains and internal water molecules. As far as the heavy-atom positions are concerned, the MD-averaged structure is in very good alignment with the high resolution crystallographic x-ray data. The hydrogen atoms of the water molecules in the simulation are omitted in this picture because of their considerable fluctuations and they are also invisible in the experimental x-ray data. The three solvating waters undergo minor oscillations (root-mean-square-fluctuation ~ 0.7 Å) and hardly deviate from their initial crystal positions. The hydronium and water oxygen coordination distances range from 2.3 to 2.8 Å, with a mean value of 2.5 Å. Note that in bulk water the radial distribution function of water oxygen atoms is essentially zero at 2.5 Å, supporting our assertion that the observed oxygen-oxygen distances in the x-ray results reflect a protonated water cluster. In the crystal structure, W141 and W149 are apparently the base of the protonated cluster as the O-O bond distance of 2.53 Å is shorter than the O-O H-bond distance of liquid water (> 2.9 Å), indicating a much stronger hydrogen bond. The assignment of these two water molecules is validated by the relatively low temperature factors (see Supporting Information). We note the subtle difference here between the MD-averaged structure and the x-ray structure: In the latter the short distance between W141 and W149 favors a more Zundel-like structure for the excess proton. However, in the MD-averaged structure the protonated water cluster was investigated at 300K under non-crystal conditions within the fluctuating protein. Both the internal water molecules and side chain groups will behave somewhat differently in such a dynamic environment. In fact, the solvating structure of the excess proton is sometimes found to be Zundel-like in the MS-EVB simulations, but this is not the prevailing structural arrangement.
For simplicity the MS-EVB2 methodology used in the current study is not capable of completely delocalizing the proton on amino acids side chains, such as E286, through dynamical protonation/deprotonation events. Nevertheless, given the 7 Å distance between the protonated E286 and the proton trap region, its contribution to the Hamiltonian can be viewed as being insignificant. The main conclusions of this study concerning the storage of an excess proton in a protonated water cluster at top of the D-pathway and the proton solvation structure are expected to hold qualitatively.
Earlier site-directed mutagenesis experiments17 have shown that the D132N mutant of CcO, in which proton uptake into the D-pathway is blocked, is still able to form the F (Ferryl) intermediate from the fully reduced enzyme. The formation of the F intermediate requires that the binuclear center becomes protonated, but in the D132N mutant the inner aqueous phase cannot be the immediate source of this proton. This suggests that the proton required to form the F state is taken internally from the D-pathway, presumably from E286 or groups in the D-pathway located further away from the binuclear center than E286. The results presented in this work establish the plausibility on both structural and computational grounds for the storage of an excess proton in the H-bonded network inside the D-pathway. Our results suggest that E286 may play a role as a proton shuttle through which the proton is transferred to electrostatically compensate for the electron that is transferred to the binuclear center from heme a. The proton is stored in an internal water network inside the D-pathway, not on any particular protein side chain. It is suggested here that although the proton uptake from the bulk solution is impaired in the D132N mutant, the protonated network formed by residues S200, S201 and water molecules resident in the channel could be the primary source for the reprotonation of E286.
The present results help to refine the picture of the proton transport mechanism in CcO, which appears to occur in several discrete steps. In each of these steps, proton movement is governed by the electrostatic potential or the protonation state of key groups. A proton is first taken up from bulk solution by D132 and temporarily stored in the proton trap. The rate-limiting step of proton transfer from E286 to the binuclear center is followed by a facile reprotonation,17 extracting the proton internally from the proton trap through a transiently formed water chain. The facile reprotonation of E286 by the trapped proton could contribute to its high preference for the protonation state, consistent with its high pKa.18 Such step-by-step transfer ensures efficient and fast proton conduction, which is important because decreased proton transport through the D-pathway will increase the lifetime of deprotonated E286, and as has been suggested19 anionic E286 may affect the ligation of CuB in the binuclear center and cause enzyme inactivation. The delayed reprotonation of E286 may also expect to have influence on the pKa value of the pumping site.
It should be noted that the present results are based on the structure determined on a two-subunit form of the enzyme. It is known from the functional studies by Hosler and colleagues that in the subunit III-depleted form of the enzyme the rate of proton uptake through the D-pathway is slowed by at least an order of magnitude, perhaps due to an increased exposure of D132 and/or partial loss of proton-collecting groups.20 However, the present studies were restricted to the interior of the channel, and did not concern the rate of proton uptake into the channel. The essence of the functioning of CcO as a vectorial proton pump is that proton movements within the enzyme are strongly coupled to the electron transfer. Since the D-pathway is used for the transfer of both pumped and substrate protons, to prevent the transfer of all protons to the binuclear center for O2 reduction, proton transfer processes in the D-pathway must be stringently regulated so the pace is determined by electron transfer, either by controlling the proton conduction rates or the magnitudes of the driving forces. A proton trap inside the D-pathway may manage to tune the proton transport to a certain rate by storing an excess proton so that E286 is rapidly protonated from the inner region. According to our simulations of the decoupled N98D mutant,7 in which all the protons through the D-pathway are delivered to the catalytic site,21 this mutation could in fact abolish the proton pumping by perturbing the proton trap region and consequently accelerating proton conductivity. This result supports the hypothesis that the proton trap region plays a key role in the CcO proton pumping mechanism.
CONCLUSIONS
MS-EVB2 simulations have been carried out using the recently available 2 Å resolution x-ray crystallographic structure of the R. sphaeroides CcO, and the MD-averaged structure is found to be in reasonable agreement with the crystallographic data. In contrast to our earlier studies using the bovine structural data, this agreement is achieved without manipulations of the crystallographically determined heavy atom positions. We conclude that a protonated water cluster in the D-pathway, mostly stabilized by S200/S201, is a key feature of the proton translocation to the extracellular side.
The present results also reveal that, in addition to protonatable amino acid side chains such as D132 and E286, water networks in the D-pathway of CcO can also constitute proton-binding sites. Such features have also been found in other biomolecular systems. For example, in bacteriorhodopsin a protonated water cluster at the proton release site close to the surface was first proposed through pKa calculations22 and then verified by IR measurements;23, 24 a released proton is suggested to be stored in an H5O2+ complex rather than on one of the nearby glutamic acids. In green fluorescent protein (GFP), an analogous pathway to the CcO D-pathway has been identified,25 and transient fluorescence experiments suggest that the proton is delocalized on the proton-wire connecting D82 and E222.26 The computer simulation method used here may also be applicable to these bimolecular systems and to others that may store a proton in a similar fashion. Further research on such systems in addition to CcO will be the target of future research.
Supplementary Material
The temperature factors of the atoms at the top of the D-pathway in the crystal structure of I-II subunit Rs CcO (2GSM)
Acknowledgments
The research was supported by the National Institutes of Health (Grants GM053148 to GAV and GM26916 to SFM). The authors gratefully acknowledge the National Center for Supercomputing Applications for computational resources.
References
- 1.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 2.Namslauer A, Brzezinski P. FEBS Lett. 2004;567:103–110. doi: 10.1016/j.febslet.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 3.Riistama S, Hummer G, Puustinen A, Dyer RB, Woodruff WH, Wikstrom M. FEBS Lett. 1997;414:275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- 4.Sharpe MA, Qin L, Ferguson-Miller SJ. In: Proton entry, exit and pathways in cytochrome oxidase: Insight from 'Conserved' water" in 'Biophysical and Structural Aspects of Bioenergetics'. Wikstrom M, editor. Cambridge, UK: Royal Society of Chemistry; 2005. [Google Scholar]
- 5.Voth GA. Acc. Chem. Res. 2006;39:143–150. doi: 10.1021/ar0402098. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Voth GA. Proc. Natl. Acad. Sci. 2005;102:6795–6800. doi: 10.1073/pnas.0408117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Voth GA. Biochim Biophys Acta. 2006;1757:852–859. doi: 10.1016/j.bbabio.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 8.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Proc Natl Acad Sci U S A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day TFJ, Soudackov AV, Cuma M, Schmitt UW, Voth GA. J. Chem. Phys. 2002;117:5839–5849. [Google Scholar]
- 10.Agmon N. Chem. Phys. Lett. 1995;244:456–462. [Google Scholar]
- 11.de Grotthuss CJT. Ann. Chim. (Paris) 1806;58:54–74. [Google Scholar]
- 12.Schmitt UW, Voth GA. J. Chem. Phys. 1999;111:9361–9381. [Google Scholar]
- 13.Schmitt UW, Voth GA. J. Phys. Chem. B. 1998;102:5547–5551. [Google Scholar]
- 14.Smith WL, Forester RT. DL-Poly Tutorial. Vol. 2. Warrington: Central Laboratory of the Research Councils; [Google Scholar]
- 15.Kannt A, Lancaster CR, Michel H. Biophys J. 1998;74:708–721. doi: 10.1016/S0006-3495(98)73996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 17.Smirnova IA, Adelroth P, Gennis RB, Brzezinski P. Biochemistry. 1999;38:6826–6833. doi: 10.1021/bi982865j. [DOI] [PubMed] [Google Scholar]
- 18.Namslauer A, Pawate AS, Gennis RB, Brzezinski P. Proc. Natl. Acad. Sci. 2003;100:15543–15547. doi: 10.1073/pnas.2432106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills DA, Hosler JP. Biochemistry. 2005;44:4656–4666. doi: 10.1021/bi0475774. [DOI] [PubMed] [Google Scholar]
- 20.Gilderson G, Salomonsson L, Aagaard A, Gray J, Brzezinski P, Hosler J. Biochemistry. 2003;42:7400–7409. doi: 10.1021/bi0341298. [DOI] [PubMed] [Google Scholar]
- 21.Pawate AS, Morgan JE, Namslauer A, Mills DA, Brzezinski P, Ferguson-Miller S, Gennis RB. Biochemistry. 2002;41:13417–13423. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- 22.Spassov VZ, Luecke H, Gerwert K, Bashford D. J Mol Biol. 2001;312:203–219. doi: 10.1006/jmbi.2001.4902. [DOI] [PubMed] [Google Scholar]
- 23.Garczarek F, Brown LS, Lanyi JK, Gerwert K. Proc Natl Acad Sci U S A. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garczarek F, Gerwert K. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 25.Agmon N. Biophys J. 2005;88:2452–2461. doi: 10.1529/biophysj.104.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leiderman P, Huppert D, Agmon N. Biophys J. 2006;90:1009–1018. doi: 10.1529/biophysj.105.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The temperature factors of the atoms at the top of the D-pathway in the crystal structure of I-II subunit Rs CcO (2GSM)