SUMMARY
Two distinct partitiviruses, Penicillium stoloniferum viruses S and F, can be isolated from the fungus Penicillium stoloniferum. The bisegmented dsRNA genomes of these viruses are separately packaged in icosahedral capsids containing 120 coat-protein subunits. We used transmission electron cryomicroscopy and three-dimensional image reconstruction to determine the structure of Penicillium stoloniferum virus S at 7.3-Å resolution. The capsid, ~350 Å in outer diameter, contains 12 pentons, each of which is topped by five arched protrusions. Each of these protrusions is in turn formed by a quasisymmetric dimer of coat protein, for a total of 60 such dimers per particle. The density map shows numerous tubular features, characteristic of α-helices and consistent with secondary-structure predictions for the coat protein. This is the first three-dimensional structure of a virus from the family Partitiviridae and exhibits both similarities to and differences from the so-called “T=2” capsids of other dsRNA viruses.
INTRODUCTION
Viruses with dsRNA genomes infect a broad range of hosts (vertebrates, invertebrates, fungi, plants, protozoa, and bacteria), several of medical, veterinary, or agricultural importance. The International Committee on Taxonomy of Viruses recognizes six families of dsRNA viruses with icosahedral capsids: Totiviridae, Birnaviridae, Partitiviridae, Cystoviridae, Chrysoviridae, and Reoviridae (Fauquet et al., 2005). Except for totiviruses, all have segmented genomes, with the segments numbering from two to twelve depending on the family or genus.
Despite some similarities, the dsRNA viruses constitute a diverse group. Their icosahedral capsids range from simpler single-shelled structures (toti-, birna-, partiti-, and chrysoviruses) to concentric multi-shelled ones (cysto- and reoviruses). All, however, include one specialized capsid that directly surrounds the genome and is involved in both replicating and transcribing it. Except in birna- and chrysoviruses, this capsid encloses the genome in a mostly continuous icosahedral shell containing 120 chemically identical copies of coat protein (CP) in a so-called “T=2” lattice (form of T=1 in which a protein dimer constitutes the asymmetric unit) (Cheng et al., 1994; Grimes et al., 1998; Hill et al., 1999; Huiskonen et al., 2006; Lawton et al., 2000; Nakagawa et al., 2003; Reinisch et al., 2000). The 60-subunit T=1 capsid of chrysoviruses appears to be an exception, but because each of their subunits is thought to contain two genetically duplicated domains, these 120 putative domains might adopt a pseudo-“T=2” structure (Castón et al., 2003). Thus, although the 780-subunit, T=13 capsid of birnaviruses (Coulibaly et al., 2005) is a clear exception, the 120-subunit capsid appears to be a common attribute of the replication/transcription machinery of dsRNA viruses.
The genome-enclosing capsids of five dsRNA viruses have been examined to near atomic resolution by X-ray crystallography. These crystal structures represent one member of the family Totiviridae (Saccharomyces cerevisiae virus L-A, “T=2” virion with 60 asymmetric dimers of Gag (coat) protein (Naitow et al., 2002)), one member of the family Birnaviridae (infectious bursal disease virus, T=13 virion with 260 trimers of VP2 (Coulibaly et al., 2005)), and three members of the family Reoviridae (bluetongue virus, “T=2” core with 60 asymmetric dimers of VP3 (Grimes et al., 1998); mammalian orthoreovirus, “T=2” core with 60 asymmetric dimers of λ1 (Reinisch et al., 2000); and rice dwarf virus, “T=2” core with 60 asymmetric dimers of P3 (Nakagawa et al., 2003)). In addition, several other dsRNA viruses have been examined at low or moderate resolutions by transmission electron cryomicroscopy (cryoTEM) and three-dimensional (3D) image reconstruction. These include Ustilago maydis virus P4 and Helminthosporium victoriae 190S virus, family Totiviridae (Castón et al., 2006; Cheng et al., 1994) bacteriophages ϕ6 and ϕ8, family Cystoviridae (Huiskonen et al., 2006; Jäälinoja et al., 2007); Penicillium chrysogenum virus, family Chrysoviridae (Castón et al., 2003); and rhesus and simian rotaviruses, striped bass reovirus, and cytoplasmic polyhedrosis virus, family Reoviridae (Hill et al., 1999; Lawton et al., 2000; Shaw et al., 1996; Yeager et al., 1994; Zhou et al., 2003).
As inferred from the facts above, the partitiviruses are the only dsRNA viruses with icosahedral capsids for which a 3D structure has remained to be determined. Because the structural proteins of dsRNA viruses share so little primary-sequence similarity between families (Mertens, 2004), comparative sequence analyses cannot provide conclusive insights about capsid structure. It was therefore necessary to employ ab initio methods to determine the first partitivirus structure.
Viruses in the family Partitiviridae have isometric capsids with uniform diameters ranging from 28 to 38 nm in electron micrographs of negatively stained samples (Boccardo et al., 1985; Bozarth et al., 1971; Buck and Kempson-Jones, 1973; Crawford et al., 2006; Lim et al., 2005). The essential genome of partitiviruses comprises two linear dsRNA segments, each 1.4 to 2.3 kbp and packaged in separate particles. These two genome segments encode two proteins, one the viral CP and the other the viral RNA-dependent RNA polymerase (RdRp). In addition, satellite dsRNAs that are dependent on helper virus for replication and encapsidation are associated with infections by some partitiviruses (Ghabrial et al., 2005).
The family Partitiviridae includes three recognized genera: Partitivirus, Alphacryptovirus, and Betacryptovirus (Ghabrial et al., 2005). Members of the genus Partitivirus infect fungi (fungal partitiviruses) whereas those of the genera Alphacryptovirus and Betacryptovirus infect plants (plant partitiviruses; also called cryptoviruses). Among members of the genus Partitivirus, Penicillium stoloniferum viruses S (PsV-S) and F (PsV-F), each with an ~47-kDa CP and an ~62-kDa RdRp (Kim et al., 2003), are two of the simplest dsRNA viruses. PsV-S and -F can co-infect the filamentous fungus Penicillium stoloniferum. The respective names of these viruses reflect their particle mobilities upon electrophoresis: S, slow; F, fast (Bozarth et al., 1971). Both viruses form nonenveloped spherical particles, with uniform diameters of 30 to 34 nm (Bozarth et al., 1971; Buck and Kempson-Jones, 1973), but which are also distinguishable by serological properties (Bozarth et al., 1971). One estimate has indicated PsV-S CP to be present in 120 copies per particle (Buck and Kempson-Jones, 1974), leading one to expect another “T=2” structure. The genome of PsV-S specifically comprises a 1754-dsRNA1 (or S1), encoding the 539-aa RdRp, and a 1582-bp dsRNA2 (or S2), encoding the 434-aa CP (Kim et al., 2003).
Here we report a cryoTEM and image-reconstruction study of PsV-S, the first partitivirus for which a 3D structure has been determined. The icosahedral capsid of PsV-S (~350 Å in outer diameter) contains 12 pentons, each of which is topped by five arched protrusions. Each of these protrusions is in turn formed by a closely associating, quasisymmetric CP dimer. Sixty such dimers are arranged in a “T=2” lattice, which as noted above, is a common, though not universal, attribute of the genome-enclosing capsids of dsRNA viruses. The 3D map of PsV-S, at an estimated resolution of 7.3 Å, exhibits numerous tubular densities, characteristic of α-helices and consistent with secondary-structure predictions for the CP. Thus, though the complete fold of PsV-S CP is not yet resolved, it is clearly dominated by α-helices, as are the 120-subunit CPs of other dsRNA viruses.
RESULTS
Preparation and Purification of PsV-S
Large quantities of purified virions (~1 mg virions per 1 g mycelium), representing a mix of PsV-S and -F, were obtained from a co-infected Penicillium stoloniferum culture by gradient sedimentation. After ion-exchange chromatography, the gradient-purified virions separated into three distinct peaks, which were harvested as such (Figure 1, top). Peak 1 contained highly purified PsV-S with essentially no trace of PsV-F, whereas peaks 2 and 3 were greatly enriched in PsV-F but respectively contained small or trace amounts of PsV-S. The identities of the particles in each fraction were verified by SDS/polyacrylamide-gel electrophoresis (SDS/PAGE) (Figure 1, top).
Figure 1. Protein and dsRNA Gels of PsV-S and PsV-F.
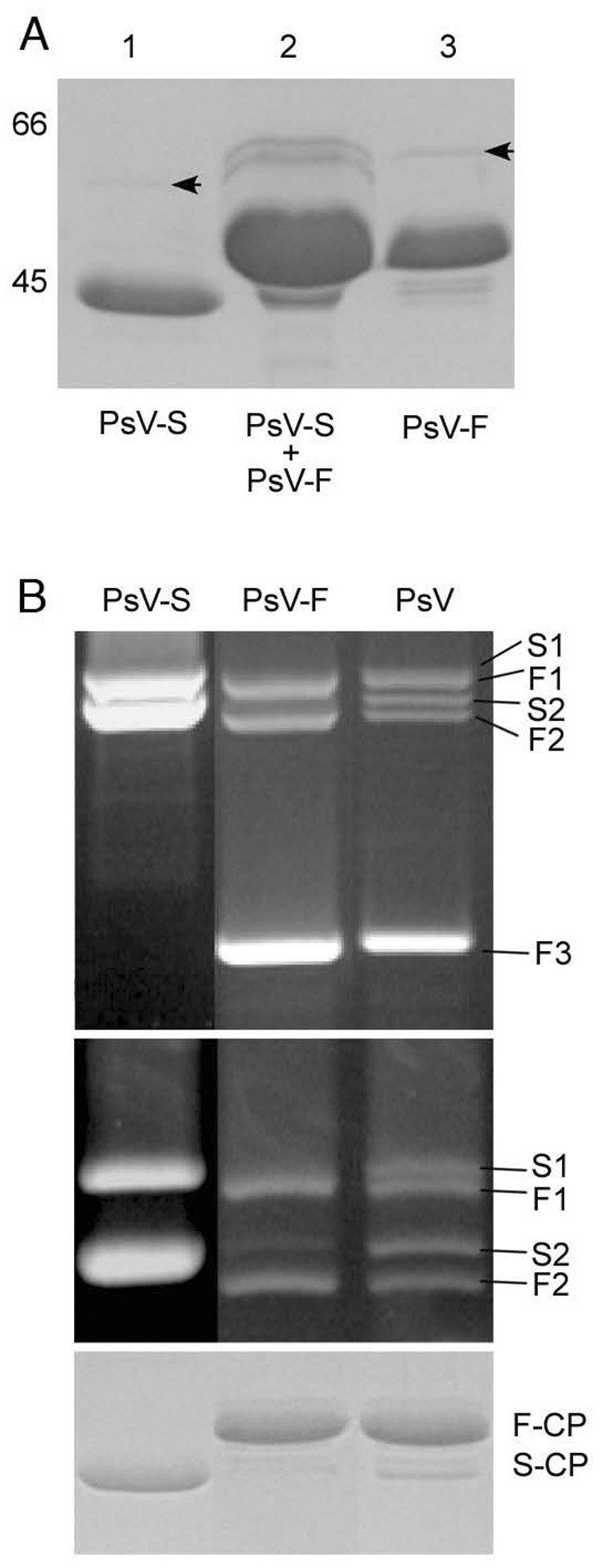
Top panel: SDS/PAGE analysis of ion-exchange chromatography fractions. Lane 1, peak 1 contained only PsV-S; lane 2, peak 2 contained a mixture of PsV-S and -F; lane 3, peak 3 was highly enriched in PsV-F and contained only traces of PsV-S. The relative positions of 66- and 45-kDa markers (data not shown) are indicated to the left of lane 1. The virus preparation analyzed in this panel was independent from that analyzed in the middle and bottom panels.
Middle panels: Agarose-gel electrophoresis of virion dsRNA isolated from ion-exchange-purified PsV-S and PsV-F as well as the unfractionated gradient-purified mixture of both viruses. The agarose gel was run for 1 h (upper) or 3 h (lower). S1 and S2 correspond to PsV-S dsRNA1 and dsRNA2, respectively; F1, F2, and F3 correspond to PsV-F dsRNA1, dsRNA2 and satellite dsRNA, respectively.
Bottom panel: SDS/PAGE analysis of the CPs of PsV-S and -F, purified by ion-exchange chromatography, as well as of the unfractionated gradient-purified PsV preparation. F-CP, PsV-F capsid protein; S-CP, PsV-S capsid protein.
The calculated molecular masses of the CPs of PsV-S (434 aa) and PsV-F (420 aa) are similar (47 kDa), but Mr values determined using SDS/PAGE were 43,000 and 47,000, respectively (Figure 1, top). It is unclear why the PsV-S CP exhibits a smaller Mr value, but this is consistent with previous results (Buck and Kempson-Jones, 1974). On the other hand, the Mr values of the more slowly migrating, minor bands resolved by SDS/PAGE (Figure 1, top, arrows) approximated the calculated molecular masses (62 and 63 kDa) of the putative RdRps of PsV-S and -F, respectively.
Agarose-gel electrophoresis of virion dsRNAs demonstrated the presence of three major species in the more highly enriched fraction of PsV-F (peak 3). These included a more slowly migrating pair of dsRNAs, consistent with genome segments F1, and F2, and a more rapidly migrating species attributable to the satellite dsRNA, F3 (Figure 1, middle upper, lane PsV-F). F1 and F2 were present in similar amounts, but much less than F3, consistent with previous results (Kim et al., 2003; Kim et al., 2005). In contrast, the PsV-S fraction (peak 1) showed only two species, present in similar amounts and consistent with genome segments S1 and S2 (Figure 1, middle upper and lower, lane PsV-S). The S1–S2 pair migrated more slowly than the F1–F2 pair (Figure 1, middle lower), also consistent with previous results (Kim et al., 2003; Kim et al., 2005). SDS/PAGE analysis of the CPs of the separated PsV-S and -F fractions versus the unfractionated preparation (Figure 1, bottom, lane PsV) correlated well with the dsRNA-gel analyses (Figure 1, middle upper and lower).
Electron Micrographs of PsV-S
Because of the greater purity achieved for PsV-S, we focused on it for the remainder of this study. Transmission electron micrographs were recorded from negatively stained samples (data not shown) or unstained, vitrified samples (Figure 2). In both cases, the virions exhibited circular profiles, consistent with a spherical morphology. The diameters of stained particles varied from 33 to 40 nm, with the variation explained in part because some particles were disrupted, were flattened on the carbon support, or had surface features masked by stain. The diameters of vitrified particles showed less variation (all ~35 nm), consistent with a conserved and well preserved structure. Close inspection of PsV-S images revealed the presence of coarse features, projecting ~5 nm away from the capsid surface.
Figure 2. Electron Micrograph of Unstained, Vitrified PsV-S.
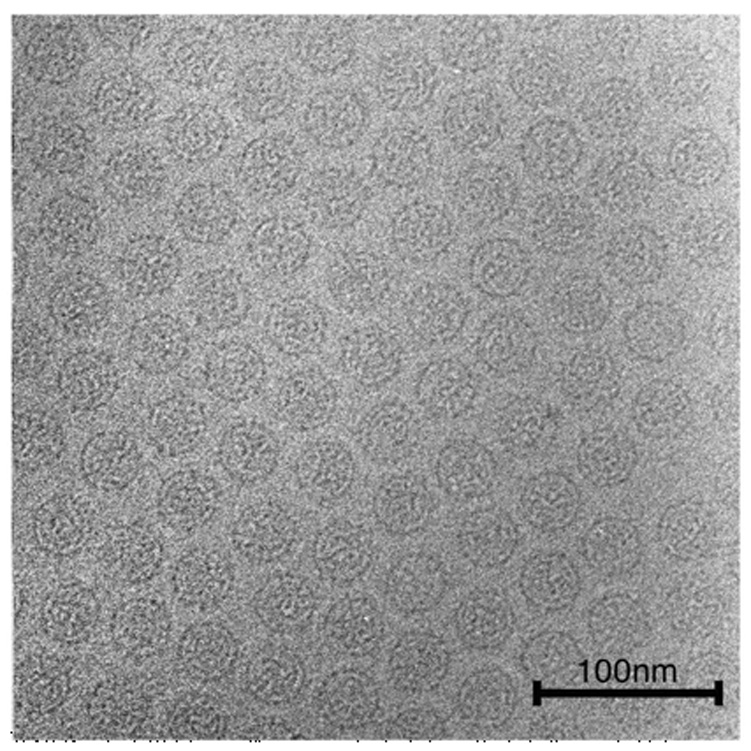
Micrographs of PsV-S particles suspended in a layer of vitreous ice over holes in a carbon support film were recorded at a nominal magnification of 50,000 in an FEI Sphera microscope at 200 keV. Particles show consistently circular profiles and coarse surface features.
3D Structure of PsV-S
A 3D reconstruction of PsV-S was computed from 14,246 particle images recorded at a nominal magnification of 50,000 in an FEI Sphera microscope at 200 keV and with underfocus settings ranging between 1.1 and 2.3 µm (see Experimental Procedures). The resolution of the reconstruction was estimated conservatively to be 7.3 Å, using a Fourier-shell correlation of 0.5 (Figure 3).
Figure 3. Fourier-shell Correlation Plot.
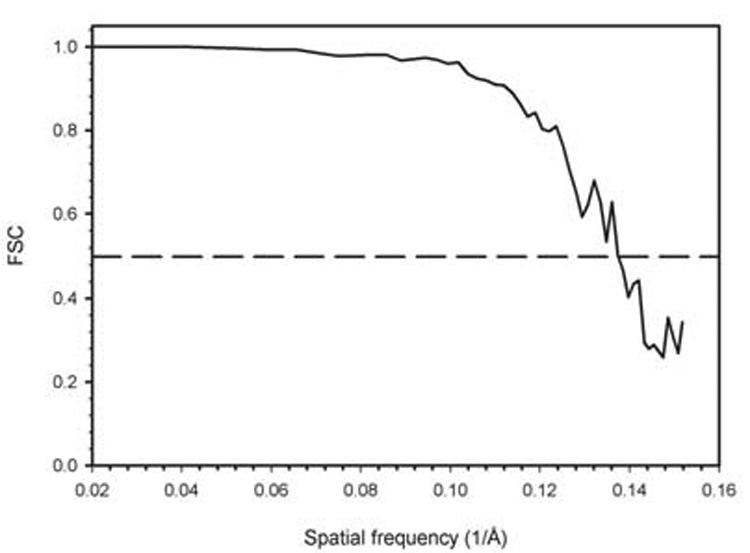
Fourier-shell correlation (FSC) plot (Harauz and van Heel, 1986) obtained from comparison of 3D reconstructions from two halves of the 14,246-image PsV-S data set and calculated for spatial frequencies ranging from 1/60 to 1/6.6 Å−1. The resolution of the PsV-S density map was estimated to be 7.3 Å based on the conservative FSC=0.5 threshold test (first spatial frequency at which the FSC value drops below 0.5).
The outer surface of the PsV-S capsid is ~350 Å in diameter and is markedly uneven (Figure 4A–C). Sixty arched protrusions (“arches”), each topped by two pointed spikes, give the virus particle an eerie resemblance to a medieval weapon, the morning star or martial flail. Each arch has a dimeric morphology (35 Å long, 21 Å wide, and 28 Å high), with the dyad axis in an approximately radial orientation, but not coincident with any of the 30 icosahedral twofold axes. Indeed, all of the icosahedral symmetry axes (two-, three-, and fivefold) are located within low-lying regions in the surface topography.
Figure 4. 3D Reconstruction of PsV-S.
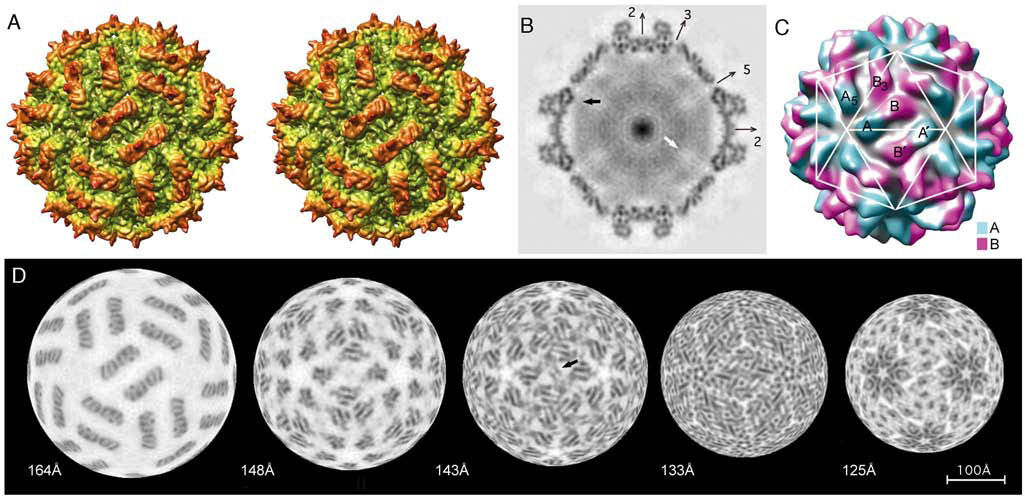
(A) Stereo image of a radially color-cued, surface-shaded representation of the PsV-S reconstruction, viewed along an icosahedral twofold axis and showing the corrugated nature of the outer surface.
(B) Density-projection image of a planar, one-pixel-thick equatorial section through the reconstruction; densest regions are darkest. Positions of several icosahedral symmetry axes that lie in the plane of the section are indicated. Black arrow points to a region where putative interactions between capsid and genomic RNA occur. White arrow points to a lobe-shaped region of reduced density along the fivefold axis, where genome appears to be at least partially excluded. Punctate dots, especially apparent in the capsid shell near the vertical twofold axis, are interpreted as density features belonging to α-helices seen end-on in this plane.
(C) Monoscopic view of the reconstruction rendered at 21-Å resolution and colored to highlight the approximate boundaries of the two different positions of CP subunits (cyan, CP-A; purple, CP-B) as well as to highlight the organization of these subunits in the “T=2” lattice. The white, T=1 lattice overlay helps to clarify the arrangement of CP subunits within the icosahedron.
(D) Radial density projections (each one pixel or 1.27 Å thick), excised from the front hemisphere of the reconstruction. The series progresses left to right from high to low radii as indicated beneath. Arrowhead in projection at radius 143-Å indicates the plate-like density described in the text. The scale bar in (D) is equally representative of all panels.
Orientation of the PsV-S surface arches could potentially exhibit one or the other handedness, for example, tilting to the right (as shown in Figure 4A) or the left when viewed down a particle twofold axis. To be able to display this aspect in the proper fashion, we performed tilt analyses on vitrified samples as described previously (Belnap et al., 1997). The results confirmed the handedness to be that shown in Figure 4A.
Density projection images, representing one-pixel- (1.27-Å-) thick, planar cross-sections through the 3D map of PsV-S (e.g., Figure 4B, which is an equatorial section), provide edge-on views of the capsid structure and reveal an inner surface that is markedly smooth. In such views, the arches appear to extend from an underlying thin, mostly continuous shell. The arches and shell are each composed of numerous punctate or linear density features (see further analysis in next section). Two small pores, visible in the shaded-surface representation (Figure 4A) as well as in the equatorial section (Figure 4B), penetrate the shell. These occur at the icosahedral three- and fivefold axes, where three CP-B and five CP-A monomers surround the respective axes. In addition, at higher contour thresholds, a laterally bilobed pore appears at each A–B–B3 junction (data not shown; see next section for nomenclature). Aside from these pores, the PsV-S capsid forms a rather impenetrable vessel that houses and protects the single dsRNA genome segment, S1 or S2, within each particle.
PsV-S dsRNA and RdRp in Core
Diffuse density inside the capsid (Figure 4B) is attributed to the dsRNA but must also include contributions from the ~1 RdRp molecule that is thought to be present inside each PsV-S particle (Buck and Kempson-Jones, 1974). This density is not as well resolved as that in the capsid, for several likely reasons. First, since the dsRNA and RdRp components are present in only single copies per particle, they must occupy either asymmetric positions or a subset of the symmetric positions available to them. In either case, this arrangement leads to a “smearing out” of their contributions to the 3D map when icosahedral symmetry is imposed during reconstruction. Second, it has not been possible to compute separate reconstructions for particles that contain either of the two different genome segments, contributing further heterogeneity to the internal density. Despite these limitations, it is intriguing that the “RNA density”, with some exceptions (e.g., Figure 4B, black arrow), avoids contact with the capsid inner surface. In addition, this density appears to avoid large, lobe-shaped regions along the fivefold axes (Figure 4B, white arrow), suggesting that some portions of the genome may loosely follow the capsid symmetry.
Organization of PsV-S Capsid Subunits
The dimeric structure of each arch and the “T=2” organization of the CP subunits (Figure 4C) are clearly revealed in radial sections of the 3D density map, five of which are illustrated (Figure 4D). Careful inspections, progressing from high to low radius, show that two nearly identical monomers (roughly resembling two symmetrically oriented “E”s) comprise each arch (Figure 4D, 164Å). Beneath the base of each arch, at progressively lower radii, the two monomers splay apart, with one forming a pentameric cluster with its four nearest neighbors and the other forming a trimeric cluster with its two nearest neighbors (Figure 4D, 148Å and 143Å). At even lower radii within the shell, the number of intersubunit contacts increases substantially and makes it more difficult to discern monomer boundaries (Figure 4D, 133Å and 125Å). Nevertheless, at the higher radii, it is clear that the PsV-S capsid comprises 60 copies each of two very similar subunits, hereafter designated “CP-A” (Figure 4C, cyan) and “CP-B” (Figure 4C, purple). These clusters of CP-A and -B subunits are what respectively surround the pores at the five- and threefold axes as noted above. Further scrutiny of the radial sections shows that the CP-A and -B subunits are staggered radially by ~4 Å, with CP-B at higher radius. Also, the portions of the CP-A and -B subunits that comprise adjacent arches lie in approximately the same orientation, except for slight differences to accommodate curvature of the shell.
Defining the Asymmetric Unit of PsV-S
The asymmetric unit of the 120-subunit PsV-S capsid consists of one CP-A and one CP-B subunit (Figure 4C). These subunits are chemically identical but occupy nonquasiequivalent environments in the icosahedron. From geometrical considerations alone, any adjacent pair of CP-A and -B subunits could be chosen to represent the icosahedral asymmetric unit. Knowledge about the role of subunit oligomers in virion assembly (dimers, trimers, etc.) can sometimes identify a functional asymmetric unit (Rossmann et al., 1985). However, because little is known about the pathway of partitivirus assembly, the choice for PsV-S is largely arbitrary and based on dimer compactness and extent of contacts. Two obvious alternatives involve (i) the quasisymmetric dimer of CP-A and -B subunits that form one arch or (ii) the asymmetric dimer of approximately parallel CP-A and -B subunits in neighboring, antiparallel arches. Alternative ii has been favored to describe other 120-subunit capsids of dsRNA viruses (Castón et al., 1997; Castón et al., 2006; Grimes et al., 1998; Hill et al., 1999; Huiskonen et al., 2006; Lawton et al., 2000; Nakagawa et al., 2003; Reinisch et al., 2000), but as suggested below based on structural features, we favor alternative i to describe PsV-S.
In any virion structure, including that of PsV-S, a variety of distinct intersubunit interactions are involved in forming a stable capsid. Neglecting the possible presence of extended regions of each CP that might be forming additional contacts with adjacent or more distant subunits, simple interpretation of the arrangement depicted in Figure 4C suggests the presence of at least five classes of intersubunit contacts. Using CP-A and -B monomers from a single quasisymmetric dimer (one arch and shell density most closely connected to the arch) to define the asymmetric unit (alternative i above), the intersubunit contacts would include the following: A–B (intradimer), A–A5 (intrapentamer), B–B3 (intratrimer), and A–B′ and B–B′ (interdimer, i.e., between neighboring, antiparallel dimers represented by each pair of arches). Except for the A–B interactions within each arch, all others occur in the shell. The B–B′ interactions include ones between neighboring subunits from adjacent CP-B trimers across the icosahedral twofold axes.
Consideration of these different interactions, as discerned by inspection of the radial sections, makes it feasible to assign densities to each of the different CP subunits. For example, B–B3 contacts produce a clustering of subunits around the threefold axes in comparison to A–A5 contacts around the fivefold axes. A particularly striking interaction (B–B′) occurs between the CP-B subunits in adjacent, antiparallel dimers. A flat, plate-like density feature, oriented perpendicular to the shell, extends away from each CP-B subunit, generating a pair of parallel plates that pass close to the particle twofold axis and can be seen edge-on in both the shaded-surface representation (Figure 4A) and some of the radial sections (e.g., Figure 4D, 143Å, arrowhead). B–B′ interactions are also visible through lower radii within the shell (Figure 4D, 133Å and 125Å). The B–B′ interactions thus appear to “seal off” the icosahedral twofold axis from the neighboring subunits in adjacent CP-A pentamers so that a sixth class of intersubunit contacts, A–A′, appears not to be found in the PsV-S structure. A–B interactions occur at three levels. Two sets of well-defined contacts are located at different radii in the arch (e.g., Figure 4D, 164Å). A third, more extensive but less obvious set of A–B contacts spans the radial thickness of the shell domain (Figure 4D, 143Å–125Å). As noted above, all A–B′ and B–B′ contacts are confined to the shell, and they are thus similarly less well defined than the intra-arch contacts.
Detecting and Modeling the PsV-S CP Secondary Structures
Inspection of the PsV-S 3D map revealed numerous tubular features of ~6-Å diameter, suggestive of α-helices, and a few planar features, ~6-Å thick, suggestive of β-sheets. Such features are recognizable in both radial (Figure 4D) and planar (Figure 4B and Figure 5B–D) sections. Helices parallel to the viewing direction appear as dense, circular dots, whereas helices perpendicular (i.e., lying “in plane”) appear as dark, thin lines.
Figure 5. Substructure in PsV-S Capsid.
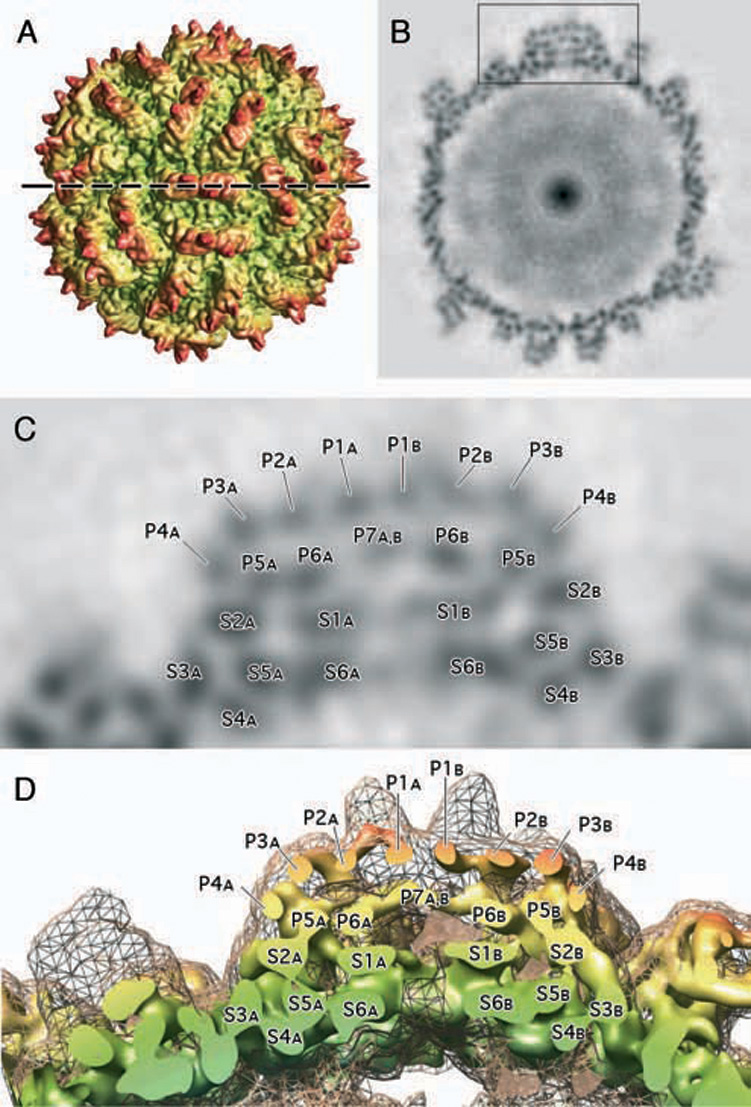
(A) Radially color-cued, surface-shaded representation of the PsV-S reconstruction, similar to that shown in Figure 4A, but viewed along the local quasi-dyad axis of one arch. The dashed horizontal line marks the location of the thin, density section shown in (B).
(B) Density-projection image of the reconstruction, similar to that shown in Figure 4B, but for an equatorial cross-section that passes through the middle of a single arch along its long axis (density highlighted inside the boxed region). This section reveals numerous, punctate dots inside the arch and also distributed throughout the rest of the capsid.
(C) Enlarged view of the region marked by the box in (B). The punctate features in a single arch and the underlying capsid shell are labeled to distinguish their locations within the CP-A or -B monomers. Each arch consists of 13 distinct features, with P1A to P7A located within the CP-A monomer and P1B to P7B in CP-B. One feature, P7A,B, is not subdivided into two as might be expected because it occurs where CP-A and -B monomers form a close A–B contact. Six distinct features, S1A to S6A and S1B to S6B, lie within the shell and beneath the arch in each subunit. Though no A–B contacts occur in the plane of this cross-section, several were seen on either side of this plane and within the shell.
(D) Same as (C), but for a 3D view of the density map sliced along the plane marked by the dashed line in (A). The outer envelope of the map, depicted in grey mesh, encompasses a solid model view like that in (A), but rendered at a higher contour threshold to highlight the locations of the punctate features identified in (C).
Given the preponderance of tubular density features in the PsV-S map, along with our tentative assignment of densities to the CP-A and -B subunits, we manually modeled all such “helix” densities within the CP-A subunit as 6-Å-diameter cylinders of appropriate lengths (see Figure 6). The results of this modeling suggested that the CP-A helices are distributed in four layers, two of which form the arch and the other two a part of the capsid shell. Six helices are clearly evident within the arch domain (Figure 5C, P1A–P6A), including four in the outermost layer (Figure 5C, P1A–P4A) and two below this (Figure 5C, P5A and P6A). All of the helices in the top layer are parallel to each other and to the shell, and are oriented approximately perpendicular to the long axis of the arch. Helix P6A is similarly oriented, but P5A, though parallel to the shell, lies almost perpendicular to the other five CP-A helices in the arch. In addition to these six helices, another tubular density feature (Figure 5C, P7A,B) lies at the interface between the CP-A and -B subunits and is not sufficiently resolved to permit an unambiguous assignment.
Figure 6. Modeling the Secondary Structures of PsV-S Capsid Subunits.
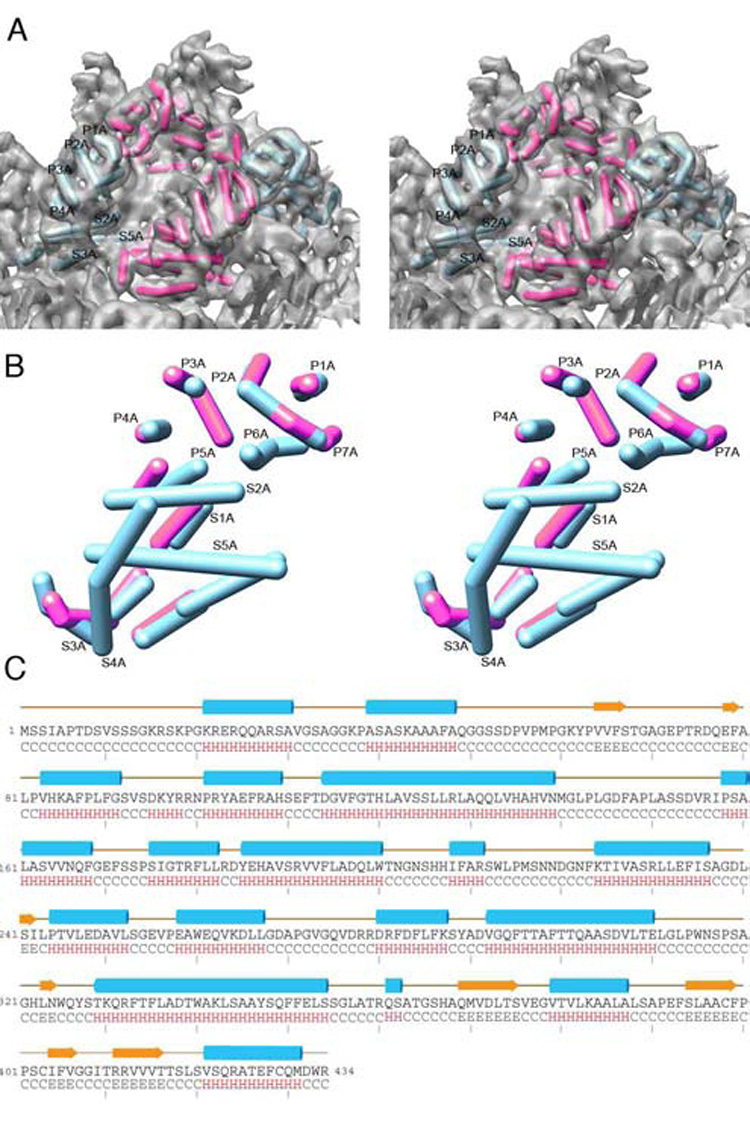
(A) Close-up, stereo view of the PsV-S 3D density map (in grey; contoured at a higher contour threshold to emphasize the most prominent features), encompassing two neighboring arches or one asymmetric unit. The tubular density features in both arches were modeled as cylinders, with those deemed to reside within each separate CP monomer rendered in either cyan (CP-A) or magenta (CP-B). Several of the tubular features are labeled according to the scheme adopted in Figure 5. The modeling was initially performed manually and then verified computationally (Yu and Bajaj, 2007).
(B) Further enlarged, stereo view of one CP-A and one CP-B model extracted from the PsV-S density map and superimposed to maximize their overlap. Greatest similarities are identified where only the CP-A model appears, because it obscures CP-B; differences in the subunit tertiary structures are identified where corresponding portions of the models diverge. The number of presumed α-helices identified in each subunit is identical, and these secondary-structure elements are distributed quite similarly, especially in the shell domain of each subunit.
(C) Consensus secondary-structure prediction for PsV-S CP, showing high α-helix content. The few predicted β-strands are located mainly near the C-terminal end of the sequence.
Our modeling further indicates the presence of eight helices (five long and three short) within the shell domain of CP-A, six identifiable within a single planar section (Figure 5C, S1A–S6A). Unlike in the arch domain, where the helices are aligned and stacked compactly, the shell helices lie in different orientations. For example, a 15-Å-long tube of density lies adjacent and nearly parallel to the fivefold axis, and with the homologous densities from four symmetry-related CP-A partners, forms a funnel-shaped pore at the shell surface. In total, including the assignment of at least one clear, sheet-like feature involved in forming the B–B′ contact at the twofold axis, our model accounts for ~70% of the density we tentatively assigned to the CP-A subunit (Figure 6A). The remaining, unascribed density either appears as discontinuous “islands” or bears no clear resemblance to helix or sheet. At least some of this density likely represents loops that connect secondary-structure elements. For example, two ends of the medium-length helices, P1A and P2A, lie at the base of the pointed spike, so that the spike may simply be a large and exposed, though compact, loop that connects these helices.
Having modeled the CP-A subunit and recognized the similar density signatures of CP-A and -B, we duplicated the CP-A model and fitted it as a rigid body to the density we had tentatively assigned to CP-B (red cylinders in Figure 6A, B). Though the lengths and the positions of corresponding tubular density features in the two subunits varied slightly, to a first approximation the CP-A model fitted nicely to the CP-B density. For CP-A and -B subunits within one arch, the CP-A model was rotated by ~180° to fit the CP-B density, consistent with the description of this A–B pair as quasisymmetric (approximating twofold symmetry). The largest deviations between CP-A and -B, which become apparent when the models are superimposed (Figure 6B), occur at the two radial extremes: at the top of the arch and in the inner wall of the shell. The large, central core of each subunit appears to adopt a more stable, invariant structure.
PsV-S CP Secondary-Structure Predictions and Correlations with Structure Model
The deduced amino acid sequence of PsV-S CP permitted us to employ various secondary-structure prediction algorithms to test our model of the subunit structure. We used three different methods, SSPRO, PSIPRED, and PHD (Bryson et al., 2005; Cheng et al., 2005; Rost et al., 2004), all of which assigned essentially the same sequence segments to α-helices or β-sheets, though there were occasional, slight variances in the lengths of each element from the different methods. The consensus predicted structure for PsV-S CP (Figure 6C) classifies it in a mixed category, comprising 42% α-helix and 10% β-sheet, with the latter mostly found near the C-terminus. One interpretation of the predicted structure identifies 17 helices (Figure 6C), which we have subclassified as follows: two short (2–4 aa), eleven medium (8–13 aa), and four long (19–26 aa; here the stretch of residues from 175 to 200 is treated as one contiguous helix rather than two medium helices as implied by Figure 6C). Neglecting the short helices, which would likely not be identifiable in our 7.3-Å map, 14 of the 15 other predicted helices correlate nicely with the 14 tubular features that we modeled as helices for both subunits. The “missing” medium-sized helix either is not clearly resolved or constitutes unassigned density, which may appear nontubular if it is flexible or forms close contacts with a loop or β-strand. Though this close correlation with the number of predicted helices provides some confidence in our modeling, we were unable to assign sequences to appropriate-length tubular densities as a means to identify the complete fold of PsV-S CP. Except for the loops that we ascribe to the pointed spikes, lack of resolvable connectivity between the various secondary-structure elements necessitates future analysis at higher resolution.
DISCUSSION
Among the six recognized families of dsRNA viruses with icosahedral capsids (Fauquet et al., 2005), only the family Partitiviridae, comprising bisegmented viruses of fungi and plants, has previously not been represented by a 3D structure. This report resolves that situation and reveals the basic architectural design that many or all partitiviruses might be expected to share. Complete genome sequences are available for many fungal partitiviruses, currently classified in the genus Partitivirus, but only a few are reported for plant partitiviruses in the genus Alphacryptovirus and none for ones in the genus Betacryptovirus. Considering differences in host affinity and means of transmission among these viruses (Ghabrial et al., 2005), it will be especially interesting to determine if plant partitiviruses exhibit the same structural hallmarks as PsV-S. We have recently found that the capsid structures of two other fungal partitiviruses, PsV-F and Fusarium poae virus 1, resemble that of PsV-S (manuscript in preparation).
New structures may also be complementary to sequences in determining whether yet-to-be-classified virus isolates should be placed in the family Partitiviridae. For example, picobirnaviruses are a group of bisegmented dsRNA viruses from humans and other vertebrates, which have small virus particles and genome sizes similar to partitiviruses (Wakuda et al., 2005). Sequence comparisons, however, suggest that picobirnaviruses are distinct from the other bisegmented dsRNA viruses (Rosen et al., 2000), and analysis of their virion structure may support their assignment to a separate taxonomic family.
The 3D structure of PsV-S confirms “T=2” as a common, though not universal, capsid form among dsRNA viruses. Partitiviridae is the fourth dsRNA-virus family (in addition to Totiviridae, Cystoviridae, and Reoviridae) to exhibit this general design, and a fifth family (Chrysoviridae) may have a pseudo-“T=2” capsid as described in Introduction. The significance of this structure is not known, but it remains notable that these 120-subunit capsids have so far been seen in nature only among dsRNA viruses. This may simply reflect a common ancestor, but might also reflect some yet-to-be-understood aspect of RNA packaging or synthesis by these viruses. See below for discussion of “T=2” capsids formed by recombinant CPs of some T=3 viruses.
The PsV-S structure also reveals a previously unobserved feature of the 120-subunit capsid of a dsRNA virus: prominent surface arches, external to the shell, each formed by additional contacts between a pair of CP-A and -B subunits. These arches might be involved in host interactions, although the existence and/or nature of such interactions remain poorly defined for these viruses. Partitiviruses have generally smaller CPs than those of the other “T=2” viruses: ~39 to 76 kDa for partitiviruses (e.g., 47 kDa for PsV-S) vs. ~76 to 147 kDa for others (e.g., 76 kDa for Saccharomyces cerevisiae virus L-A; 85 kDa for bacteriophage ϕ6; and 142 kDa for mammalian orthoreovirus). Thus, since the smaller partitivirus CPs can mediate fewer intersubunit contacts within the shell, this may have favored the presence of arches, i.e., layers of contacts above the shell, to stabilize the capsid. The arch domain of PsV-S is, in a basic architectural sense, analogous to the protruding (P) domain of tomato bushy stunt virus, Norwalk virus, and others (Harrison, 2001).
The structure of PsV-S leads us to a particular view of how its “T=2” lattice is organized, suggesting a quasisymmetric A–B dimer as the icosahedral asymmetric unit instead of the asymmetric, approximately parallel A–B dimer (designated A–B′ in this report; Figure 4C) favored by previous investigators of other “T=2” dsRNA-virus structures. This view is most strongly promoted by the surface arches, which not only are compelling features but also contribute to making the A–B contacts within each quasisymmetric dimer more extensive than those within each asymmetric (A–B′) dimer. This view can also be applied to the other 120-subunit capsids of dsRNA viruses (Castón et al., 1997; Harrison, 2001), although in toti- and reoviruses the contacts within each quasisymmetric dimer are much less extensive than those within each asymmetric dimer (Grimes et al., 1998; Huiskonen et al., 2006; Naitow et al., 2002; Nakagawa et al., 2003; Reinisch et al., 2000; Zhou et al., 2003). In cystoviruses, the contacts within each quasisymmetric dimer appear similar in extent to those within each asymmetric dimer (Huiskonen et al., 2006; Jäälinoja et al., 2007).
The β-sheet-rich CPs of certain T=3, plus-strand RNA viruses, including brome mosaic virus and cowpea chlorotic mottle virus, have been shown to form strict (60-subunit) T=1 and/or “T=2”, in addition to T=3, capsids when expressed in recombinant form (Krol et al., 1999; Tang et al., 2006). For these viruses, the usual T=3 capsid is assembled from symmetric CP dimers, as a result of which particles with a strict T=1, “T=2”, or T=3 arrangement of CP subunits might be variably assembled from these same basic building blocks (i.e., from 30, 60, or 90 CP dimers, respectively) (Krol et al., 1999; Tang et al., 2006). Our view of the PsV-S capsid as being formed from 60 quasisymmetric CP dimers is consistent with this assembly pathway and suggests how the α-helix-rich “T=2” capsid of dsRNA viruses might have evolved from either a strict T=1 or a T=3 capsid ancestor by either addition or loss of 30 CP dimers per particle. Alternatively, the “T=2” capsid might have been ancestral. Direct evolution between the α-helix-rich and β-sheet-rich CPs seems unlikely, but the 120-subunit design was apparently accessible for selection with either type of subunit and favored by the helix-rich CPs of dsRNA viruses.
The PsV-S capsid appears to include contacts between CP-B, and not CP-A, subunits, across the icosahedral twofold axes. Such B–B′ contacts also appear to occur in cystoviruses (Huiskonen et al., 2006; Jäälinoja et al., 2007), but not in toti- or reoviruses; in the latter two virus families, A–A′ contacts occur across the twofold axes (Grimes et al., 1998; Lawton et al., 2000; Naitow et al., 2002; Nakagawa et al., 2003; Reinisch et al., 2000; Zhou et al., 2003). This difference among families does not strictly correlate with CP size, since the CPs of some partiti- cysto-, and totiviruses can all be near 80 kDa. An interesting correlation, however, is that partiti- and cystoviruses (with twofold B–B′ contacts) mediate semiconservative transcription (template strand released from the genomic duplex) whereas toti- and reoviruses (with twofold A–A′ contacts) mediate conservative transcription (template strand retained in the genomic duplex). The significance, if any, of this correlation, whether it be evolutionary and/or functional, remains to be ascertained.
The PsV-S RdRp is not identifiable in the current, icosahedrally averaged structure of the virion, as would be expected if it is present in only one copy per particle (Buck and Kempson-Jones, 1974). Whether—and if so, where—this RdRp molecule is noncovalently anchored to the inner surface of the CP shell remains unknown. Pores through the shell (at the five- and threefold axes as well as at the A–B–B3 junctions, as noted in Results) are logical candidates to be the sites for plus-strand RNA exit during viral transcription, and thus the RdRp seems likely to be anchored beneath one of these pores. Anchoring of the RdRp molecules near the transcript exit sites within other dsRNA viruses has been previously demonstrated or discussed (Castón et al., 1997; Dryden et al., 1998; Prasad et al., 1996; Zhang et al., 2003), and this hypothesis is now extended to partitiviruses.
The 120-subunit CPs of dsRNA viruses for which there are structure data at subnanometer resolution, including now the 47-kDa CP of PsV-S, are dominated by α-helices. This contrasts with the CPs of a large number of plus-strand RNA viruses, which contain the canonical BIDG-CHEF β-motif and little helix (Harrison, 2001; Rossmann and Johnson, 1989). Interestingly, modeled helices in the PsV-S completely differ in arrangement from those in Saccharomyces cerevisiae virus L-A (Naitow et al., 2002). Also, with the possible exception of three “core” helices (unpublished data), they differ substantially from those in the reoviruses (Grimes et al., 1998; Nakagawa et al., 2003; Reinisch et al., 2000; Zhou et al., 2003). Hence, though the fold of PsV-S CP is yet to be determined, it appears to be distinct from those of other dsRNA viruses.
EXPERIMENTAL PROCEDURES
Fungal Culture and Virion Purification
Penicillium stoloniferum ATCC 14586 was used as the source of PsV-S and -F. Mycelium was harvested from 7-day stationary cultures grown at room temperature (~22°C) on potato dextrose broth containing 0.5% yeast extract and was homogenized in a cold blender with buffer A (50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 5 mM EDTA, and 1 mM DTT), stored at 4°C. The homogenate was blended with an equal volume of chloroform, and the emulsion was broken by centrifugation at 8000 × g for 20 min. The aqueous layer was subjected to two cycles of differential centrifugation (27,000 rpm for 150 min in a Beckman type 30 rotor and 10,000 rpm for 10 min in a Beckman JA-20 rotor). Pellets were resuspended in buffer A. The final purification employed rate-zonal centrifugation in sucrose gradients (100–500 mg/ml). The gradients were made in buffer A and centrifuged at 24,000 rpm in a Beckman SW28 rotor for 4 h. The major band was withdrawn with a syringe from the side of the tube and diluted with buffer A. These virions were then concentrated by overnight centrifugation at 40,000 rpm in a Beckman 50Ti rotor. All centrifugation steps were performed at 4°C. The pellets were resuspended in 0.03 M sodium phosphate buffer (pH 7.6). PsV-S and -F were separated using DEAE–cellulose chromatography essentially as described by Buck and Kempson-Jones (1973) except that the column was eluted with a step gradient of 0.0 to 0.3 M NaCl solutions at intervals of 0.05 M NaCl. Three UV-absorbing peaks were resolved. Peak 1 eluted with 0.15 M NaCl and contained only PsV-S; peak 2 eluted with 0.2 M NaCl and contained both PsV-S and -F, with the latter predominating; and peak 3 eluted with 0.25 M NaCl and contained PsV-F virions with traces (>1%) of PsV-S, as determined by RNA- and protein-gel analyses described in the text.
CryoTEM
Small (3-µl) aliquots of purified PsV-S virions (~5 mg/ml) were vitrified for cryoTEM via standard, rapid freeze-plunging procedures. Samples were applied to Quantifoil holey grids that had been plasma-cleaned for 25 sec in an E. A. Fischione 1020 plasma cleaner. Grids were then loaded into an FEI Vitrobot maintained at 80% relative humidity, blotted for 7.5 sec, plunged into liquid ethane, and transferred into a precooled Gatan 626 cryo-transfer holder, which maintained the specimen at liquid-nitrogen temperature in the microscope. Micrographs were recorded on Kodak SO-163 electron-image film at 200 keV in an FEI Sphera microscope under low-dose conditions (~15 e/Å2) and at a nominal magnification of 50,000.
3D Image Reconstruction
Eighteen micrographs, exhibiting minimal specimen drift and image astigmatism and recorded at underfocus settings of between 1.1 and 2.3 µm, were digitized at 6.35-µm intervals (representing 0.79-Å pixels at the specimen) on a Nikon Coolscan 800 densitometer. RobEM (http://cryoEM.ucsd.edu/programs.shtml) was used to extract 14,246 individual particle images, each 511-x-511 pixels in size and to preprocess them as described (Baker et al., 1999). We next used the RMC (random-model computation) procedure (Yan et al., 2007a) to generate an initial reconstructed model at 23-Å resolution from 82 particle images. This map was then used to initiate full orientation and origin determinations and refinement of the entire set of images using the current version of AUTO3DEM (Yan et al., 2007b). Corrections to compensate in part for the effects of the microscope contrast-transfer function were performed as previously described (Bowman et al., 2002; Zhang et al., 2003). A final 3D map, reconstructed from the 14,246 particles extracted, was estimated to be reliable to at least 7.3-Å resolution, using a Fourier-shell correlation of 0.5 (see Figure 3). To aid our analysis and interpretation of the structure, we computed the 3D reconstruction to a resolution limit of 6.1 Å, using an inverse temperature factor of 1/200 Å2 to enhance the high-resolution features out to 7.3 Å (Havelka et al., 1995), after which the inverse factor was held constant and a Gaussian function was applied to attenuate the Fourier data smoothly to zero from 6.5 to 6.1 Å. Graphical representations were generated with the RobEM and Chimera visualization software packages (Goddard et al., 2007).
Handedness
To determine the handedness of the PsV-S structure, we recorded pairs of 0°- and 10°-tilt images from the same specimen areas. The handedness of the capsid was then determined by comparing the particle images in each tilt pair, using a procedure described previously (Belnap et al., 1997). Results were applied to the final 3D map.
Secondary-structure Predictions and Modeling
The amino acid sequence of PsV-S CP was retrieved from GenBank (accession number YP_052857). Secondary-structure predictions were generated with the SSPRO, PSIPRED, and PHD programs (Bryson et al., 2005; Cheng et al., 2005; Rost et al., 2004). Manual fitting of cylinders designed to represent α-helices at low resolution was performed with the CHIMERA software. Coordinates of L-A virus Gag protein (PDB-ID.1M1C), BTV VP3 protein (PDB-ID.2BTV), Reovirus λ1 (PDB-ID.1EJ6), and RDV P3 (PDB-ID.1UF2), all obtained from VIPERdb (Shepherd et al., 2006), were used to compare the folds of these viral subunits.
ACKNOWLEDGMENTS
We thank Norm Olson for discussions and management of the cryoTEM facilities and for help in preparing Figure 2 and Figure 5. We also thank Zeyun Yu and Chandrajit Bajaj for advice in using undistributed versions of their segmentation and secondary-structure analysis routines. This work was supported in part by grants R01 AI47904 (to M.L.N.) and R37 GM033050 (to T.S.B.) from the U.S. National Institutes of Health, as well as by grant 2001-35319-10010 from the U.S. Department of Agriculture National Research Initiative Competitive Research Program (to S.A.G.). In addition, the San Diego Supercomputer Center provided partial support for R.S.S. and for access to TeraGrid computing. Shared instrumentation grant 1S10 RR020016-01 from the U.S. National Institutes of Health as well as support from the University of California-San Diego and the Agouron Foundation (all to T.S.B.) were used to establish new cryoTEM facilities at the University of California-San Diego, where all PsV-S samples were vitrified and imaged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The reconstruction has been deposited in the Electron Microscopy Database at the European Bioinformatics Institute with the accession number EMD-1459.
REFERENCES
- Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Molec. Biol. Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Baker TS. A method for establishing the handedness of biological macromolecules. J. Struct. Biol. 1997;120:44–51. doi: 10.1006/jsbi.1997.3896. [DOI] [PubMed] [Google Scholar]
- Boccardo G, Milne RG, Disthaporn S, Chettanachit D, Putta M. Morphology and nucleic acid of rice gall dwarf virus. Intervirology. 1985;23:167–171. doi: 10.1159/000149600. [DOI] [PubMed] [Google Scholar]
- Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 2002;76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozarth RF, Wood HA, Mandelbrot A. The Penicillium stoloniferum virus complex: two similar double-stranded RNA virus-like particles in a single cell. Virology. 1971;45:516–523. doi: 10.1016/0042-6822(71)90352-7. [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck KW, Kempson-Jones GF. Biophysical properties of Penicillium stoloniferum virus S. J. Gen. Virol. 1973;18:223–235. doi: 10.1099/0022-1317-18-3-223. [DOI] [PubMed] [Google Scholar]
- Buck KW, Kempson-Jones GF. Capsid polypeptides of two viruses isolated from Penicillium stoloniferum. J. Gen. Virol. 1974;22:441–445. [Google Scholar]
- Castón JR, Trus BL, Booy FP, Wickner RB, Wall JS, Steven AC. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J. Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castón JR, Ghabrial SA, Jiang D, Rivas G, Alfonso C, Roca R, Luque D, Carrascosa JL. Three-dimensional structure of penicillium chrysogenum virus: a double-stranded RNA virus with a genuine T=1 capsid. J. Mol. Biol. 2003;331:417–431. doi: 10.1016/s0022-2836(03)00695-8. [DOI] [PubMed] [Google Scholar]
- Castón JR, Luque D, Trus BL, Rivas G, Alfonso C, Gonzalez JM, Carrascosa JL, Annamalai P, Ghabrial SA. Three-dimensional structure and stoichiometry of Helmintosporium victoriae 190S totivirus. Virology. 2006;347:323–332. doi: 10.1016/j.virol.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RH, Caston JR, Wang G-J, Gu F, Smith TJ, Baker TS, Bozarth RF, Trus BL, Cheng N, Wickner RB, et al. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric gag dimers. J. Mol. Biol. 1994;244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Crawford LJ, Osman TA, Booy FP, Coutts RH, Brasier CM, Buck KW. Molecular characterization of a partitivirus from Ophiostoma himal-ulmi. Virus Genes. 2006;33:33–39. doi: 10.1007/s11262-005-0028-6. [DOI] [PubMed] [Google Scholar]
- Dryden KA, Farsetta DL, Wang G, Keegan JM, Fields BN, Baker TS, Nibert ML. Internal structures containing transcriptase-related proteins in top component of mammalian orthoreovirus. Virology. 1998;245:33–46. doi: 10.1006/viro.1998.9146. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus Taxonomy: Eighth Report of the International Committee for the Taxonomy of Viruses. San Diego: Academic Press; 2005. [Google Scholar]
- Ghabrial SA, Buck KW, Hillman BI, Milne RG. In: Partitiviridae, In Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. London: Elsevier/Academic Press; 2005. pp. 581–590. [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Ziéntara S, Mertens PPC, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Harrison SC. The familiar and the unexpected in structures of icosahedral viruses. Curr. Opin. Struct. Biol. 2001;11:195–199. doi: 10.1016/s0959-440x(00)00189-5. [DOI] [PubMed] [Google Scholar]
- Havelka WA, Henderson R, Oesterhelt D. Three-dimensional structure of halorhodopsin at 7 Å resolution. J. Mol. Biol. 1995;247:726–738. doi: 10.1006/jmbi.1995.0176. [DOI] [PubMed] [Google Scholar]
- Hill CL, Booth TF, Prasad BVV, Grimes JM, Mertens PP, Sutton GC, Stuart DI. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat. Struct. Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ. Structure of the bacteriophage □6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Jäälinoja HT, Huiskonen JT, Butcher SJ. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages ϕ6 and ϕ8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kim JW, Kim SY, Kim KM. Genome organization and expression of the Penicillium stoloniferum virus S. Virus Genes. 2003;27:249–256. doi: 10.1023/a:1026343831909. [DOI] [PubMed] [Google Scholar]
- Kim JW, Choi EY, Lee JI. Genome organization and expression of the Penicillium stoloniferum virus F. Virus Genes. 2005;31:175–183. doi: 10.1007/s11262-005-1793-y. [DOI] [PubMed] [Google Scholar]
- Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton JA, Estes MK, Prasad BVV. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 2000;55:185–229. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WS, Jeong JH, Jeong RD, Yoo YB, Yie SW, Kim KH. Complete nucleotide sequence and genome organization of a dsRNA partitivirus infecting Pleurotus ostreatus. Virus Res. 2005;108:111–119. doi: 10.1016/j.virusres.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 2002;9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Prasad BVV, Rothnagel R, Zeng CQ-Y, Jakana J, Lawton JA, Chiu W, Estes MK. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Rosen BI, Fang ZY, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology. 2000;277:316–329. doi: 10.1006/viro.2000.0594. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht H-J, Johnson JE, Kamer G, Luo M, Mosser AG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Ann. Rev. Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AL, Samal SK, Subramanian K, Prasad BVV. The structure of aquareovirus shows how the different geometries of the two layers of the capsid are reconciled to provide symmetrical interactions and stabilization. Structure. 1996;4:957–967. doi: 10.1016/s0969-2126(96)00102-5. [DOI] [PubMed] [Google Scholar]
- Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, Johnson JE, Brooks CL, 3rd, Reddy VS. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 2006;34:D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Johnson JM, Dryden KA, Young MJ, Zlotnick A, Johnson JE. The role of subunit hinges and molecular "switches" in the control of viral capsid polymorphism. J. Struct. Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J. Virol. Meth. 2005;126:165–169. doi: 10.1016/j.jviromet.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J. Struct. Biol. 2007a;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM-an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007b;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Berriman JA, Baker TS, Bellamy AR. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Bajaj C. Computational approaches for automatic structural analysis of large bio-molecular complexes. IEEE/ACM Trans. Comp. Biol. Bioinformatics. 2007 doi: 10.1109/TCBB.2007.70226. in press. [DOI] [PubMed] [Google Scholar]
- Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Zhang H, Jakana J, Lu XY, Zhang JQ. Cytoplasmic polyhedrosis virus structure at 8 Å by electron cryomicroscopy. Structural basis of capsid stability and mRNA processing regulation. Structure. 2003;11:651–663. doi: 10.1016/s0969-2126(03)00091-1. [DOI] [PubMed] [Google Scholar]


