Abstract
Human telomeric DNA consists of tandem repeats of the sequence d(TTAGGG). Compounds that can stabilize the intramolecular DNA G-quadruplexes formed in the human telomeric sequence have been shown to inhibit the activity of telomerase and telomere maintenance, thus the telomeric DNA G-quadruplex has been considered as an attractive target for cancer therapeutic intervention. Knowledge of intramolecular human telomeric G-quadruplex structure(s) formed under physiological conditions is important for structure-based rational drug design and thus has been the subject of intense investigation. This review will give an overview of recent progress on the intramolecular human telomeric G-quadruplex structures formed in K+ solution. It will also give insight into the structure polymorphism of human telomeric sequences and its implications for drug targeting.
Keywords: human telomere, G-quadruplex, structure polymorphism, potassium structure, anticancer drug target, NMR
1. Introduction
Telomeres are specialized DNA protein complexes that cap the ends of linear chromosomes and provide protection against gene erosion at cell divisions, chromosomal non-homologous end-joinings and nuclease attacks [1-3]. Human and vertebrate telomeres consist of tandem repeats of the hexanucleotide d (TTAGGG)n 5-10 kb in length 5′ to 3′ toward the chromosome end [4-7], terminating in a single-stranded 3′-overhang of 100-200 bases that plays an important role in telomere structure and function [8-10]. The structure and stability of telomeres are closely related with cancer [11-14], aging [15, 16], and genetic stability [3, 17, 18]. Telomeric DNA is extensively associated with various proteins, such as Pot1 (protection-of-telomeres 1), TRF1 (telomeric-repeat-binding factor 1) and TRF2. In normal cells, each cell replication results in a 50- to 200- base loss of the telomere. After a critical shortening of the telomeric DNA is reached, the cell undergoes apoptosis [19]. However, telomeres of cancer cells do not shorten on replication, because of the activation of a reverse transcriptase telomerase that extends the telomeric sequence at the chromosome ends [20]. Telomerase is activated in 80-85% of human cancer cells [21], and has been suggested to play a key role in maintaining the malignant phenotype by stabilizing telomere length and integrity [22].
The G-rich telomeric sequence can fold into G-quadruplex (Figure 1), a DNA secondary structure consisting of stacked G-tetrad planes connected by a network of Hoogsteen hydrogen bonds and stabilized by monovalent cations, such as Na+ and K+. Vertebrate telomeric DNA repeats are highly conserved, which is suggested to be related with the G-quadruplex formation [14, 23]. The G-quadruplex formation by the single-stranded human telomeric DNA was shown to inhibit the activity of telomerase [24]. In addition, human Pot1 was shown to disrupt telomeric G-quadruplexes presumably by trapping the single-stranded form of telomeric DNA, thus allowing telomerase extension [25]. Pot1 is a highly conserved telomeric protein that binds to the 3′ end of single-stranded telomeric DNA and plays an important role in telomere end-capping and protection [26-28]. Compounds that stabilize the intramolecular DNA G-quadruplexes formed in the human telomeric sequence have been shown to inhibit the activity of telomerase and disrupt telomere capping and maintenance, making the intramolecular human telomeric DNA G-quadruplex an attractive target for cancer therapeutic intervention [11, 13, 14, 29-31].
Figure 1.
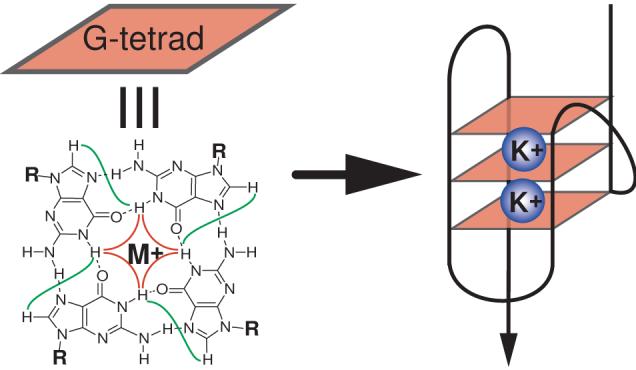
A schematic drawing of an intramolecular G-quadruplex (right) that is composed of three G-tetrads. A G-tetrad with H1-H1 and H1-H8 connectivity patterns detectable in NOESY experiments is also shown (left).
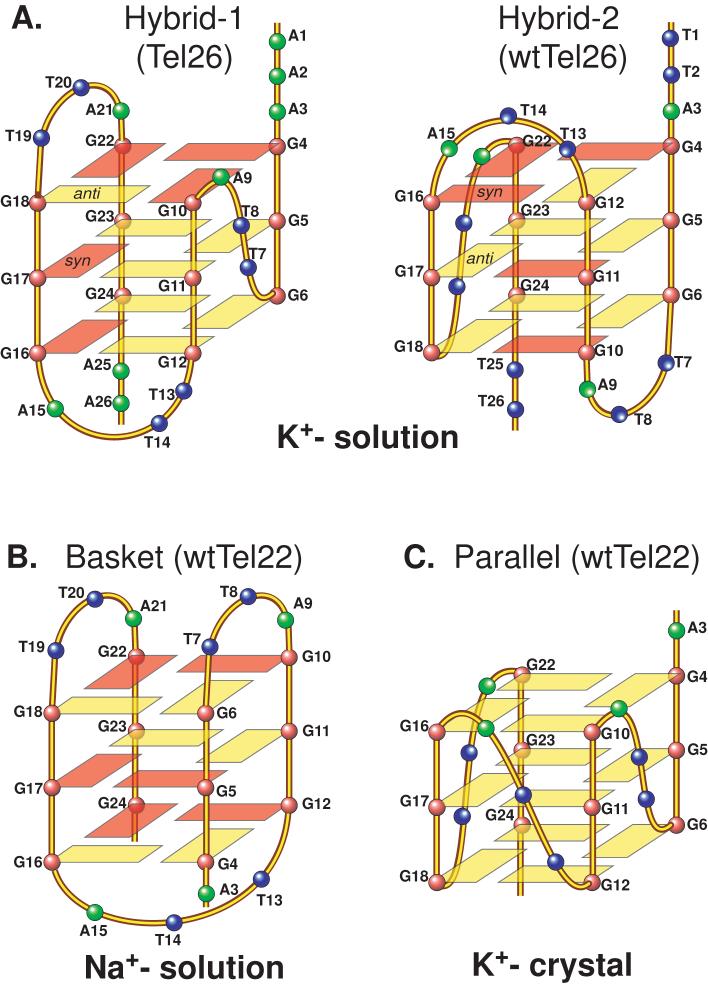
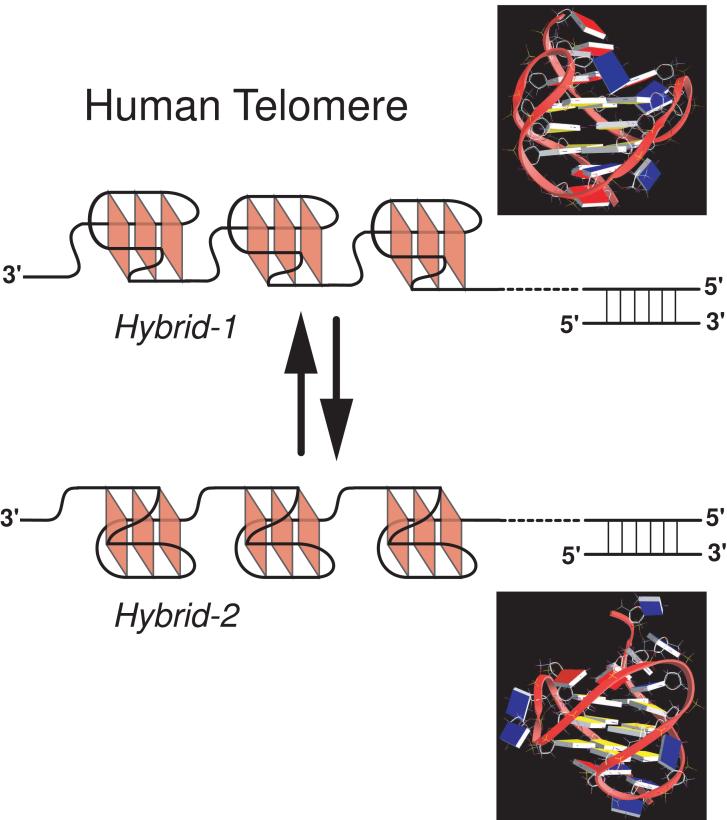
Structural information of the human telomeric G-quadruplex formed under physiologically relevant conditions is necessary for structure-based rational drug design. Because the K+ structure is considered to be biologically more relevant due to the higher intracellular concentration of K+, it has been the subject of intense investigation [32-45], although the Na+ structure was reported more than a decade ago [46]. In 2002, a crystal structure of the parallel-stranded G-quadruplex was reported using a 22-nt human telomeric sequence in the presence of K+ [47]. Recent results have shown that hybrid-type intramolecular G-quadruplexes (Figure 2A) appear to be the predominant conformation formed in human telomeric sequences in K+ solution [48-55]. The hybrid-type structures are distinct from the Na+-structure in solution (Figure 2B) and the crystal structure in the presence of K+ (Figure 2C). The hybrid-type conformation appear to be the major form for human telomeric sequences in solution in the presence of K+, despite the co-presence of high concentration Na+ ion [48]. Furthermore, addition of K+ readily converts the Na+-form conformation to the K+-form hybrid-type structures. Depending on the flanking segments, the telomeric sequences can form two distinct but related hybrid-type structures, i.e., hybrid-1 or hybrid-2 telomeric G-quadruplexes [49, 53] (Figure 2A). The hybrid-2 type structure appears to be the major conformation on the extended human telomeric sequences [49]. Both hybrid-type structures contain three G-tetrads linked with mixed parallel/antiparallel-G-strands. The two hybrid-type G-quadruplexes differ in their loop arrangements, strand orientations, tetrad arrangements, and capping structures. Specifically, the three TTA loop segments adopt sequential side (double-chain-reversal, propeller)-lateral (edge-wise)-lateral conformations in the hybrid-1 structure and lateral-lateral-side conformations in the hybrid-2 structure, respectively. A naturally-occurring adenine triple platform was found to cap the 5′-end of the hybrid-1 telomeric G-quadruplex [50] while a T:A:T triple platform was found to cap the 3′-end of the hybrid-2 telomeric G-quadruplex [49]. The distinct capping structures appear to be crucial for the favored formation of the specific hybrid-type structure, and may provide specific drug binding sites. It has also been shown that human telomeric sequences are in equilibrium between hybrid-1 and hybrid-2 structures in K+ solution, which is largely determined by the 3′-flanking sequence [49]. Furthermore, both hybrid-type G-quadruplexes suggest a straightforward means for multimer formation with effective packing in the elongated human telomeric sequence and thus provide important implications for drug targeting of G-quadruplexes in human telomeres [48-50]. The structure polymorphism and dynamic equilibrium of the human telomeric sequence appear to be intrinsic to this highly conserved sequence. The low energy barrier between different forms of the human telomeric sequence may provide a means for different protein recognition to control the biology of human telomeres.
Figure 2.
(A) Schematic drawing of the folding topologies of the Hybrid-1 (major conformation in Tel26) and Hybrid-2 (major conformation in wtTel26) intramolecular telomeric G-quadruplexes in K+ solution. Yellow box = (anti) guanine, red box = (syn) guanine; red ball = guanine, green ball = adenine, blue ball = thymine. (B) Folding topology of the basket-type intramolecular G-quadruplex formed by wtTel22 in Na+ solution as determined by NMR. (C) Folding topology of the propeller-type parallel-stranded intramolecular G-quadruplex formed by wtTel22 in the presence of K+ in crystalline state. The numbering system is based on wtTel26.
In addition to human telomeres, DNA G-quadruplex structures have also been found to form in the promoter regions of oncogenes (see review by L. Hurley in this issue) and other biologically relevant regions of the genome [56]. Interestingly, the structural studies have shown a great conformational diversity in different G-quadruplexes [49, 50, 57-59], such as folding topologies, loop conformations, and capping structures, indicating that these structural motifs may be differentially regulated and targeted.
2. Intramolecular human telomere G-quadruplex structures formed in K+ solution
Distinct structures are formed by a 22-nt human telomeric sequence wtTel22 in Na+ solution and crystalline state in the presence of K+
The minimal requirement for an intramolecular telomeric G-quadruplex is a four G-tract human telomeric sequence 5′-GGG(TTAGGG)3 (wtTel21, Figure 3A). More than a decade ago, the intramolecular human telomeric G-quadruplex structure in Na+ solution by NMR was reported using the 22-nt human telomeric sequence 5′-AGGG(TTAGGG)3 (wtTel22, Figure 3B) [46]. wtTel22 is a four G-tract human telomeric sequence with a 5′-flanking-A and no 3′-flanking sequence. The structure in Na+ solution is a basket-type, mixed antiparallel/parallel stranded intramolecular G-quadruplex, consisting of three G-tetrads connected with one diagonal and two lateral TTA loops (Figure 2B). In 2002, using the same 22-nt human telomeric sequence, a crystal structure of the intramolecular human telomeric G-quadruplex in the presence of K+ was reported [47], which was a parallel-stranded structure (Figure 2C) distinctly different from the Na+ solution structure. The crystal structure in the presence of K+ consists of three G-tetrads connected with three symmetrical side, propeller (double-chain-reversal) TTA loops.
Figure 3.
(A) Four-G-tract native human telomeric sequences with different flanking sequences. The numbering system is shown above wtTel27. (B) Four-G-tract human telomeric DNA sequences that have been used for structure determination. The numbering system is shown above wtTel26.
The 22-nt human telomeric sequence wtTel22 does not form a single G-quadruplex in K+ solution
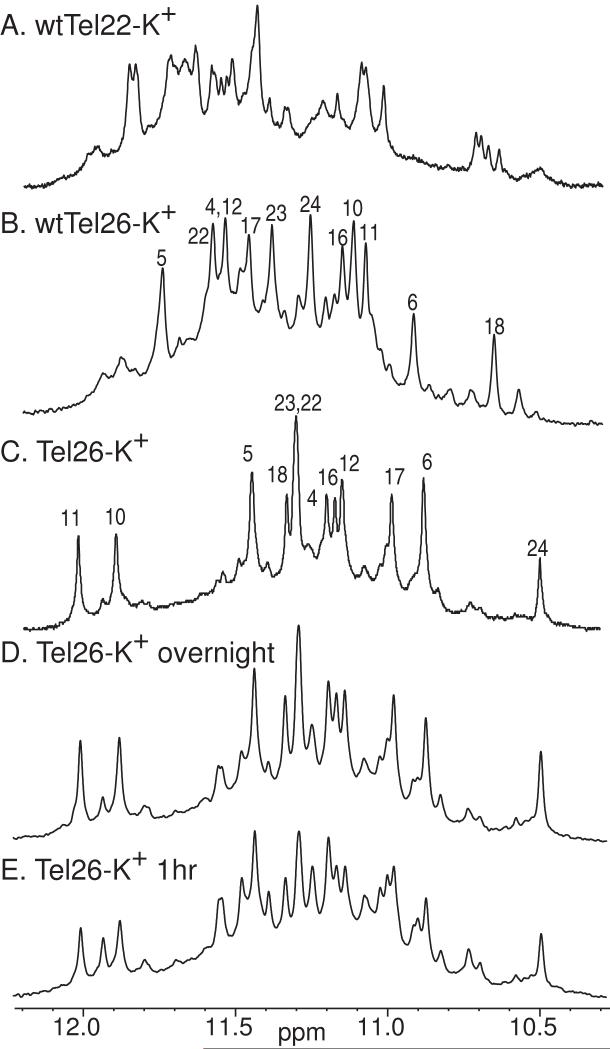
However, the wtTel22 sequence does not form a single G-quadruplex structure in K+ solution, as indicated by the imino proton peaks in the 1D NMR spectra [48] (Figure 4A). The NMR data of the wtTel22 sequence in K+ solution indicate the presence of multiple G-quadruplex conformations with relatively sharp peaks. A large number of variant four G-tract sequences containing the same core sequence of wtTel22 with different flanking segments has been screened, and the 3′-flanking segment has been found to be critical for the formation of a stable G-quadruplex structure in K+ solution [48]. 1H NMR has identified that a few sequences form one stable major G-quadruplex in K+ solution, including the 26-nt sequence Tel26 (Figure 4C).
Figure 4.
The imino proton region of the 1D 1H NMR of wtTel22 (A), wtTel26 with assignment (B), and Tel26 with assignment (C) in K+ solution. The imino proton region of the 1D 1H NMR of the 1hr (D) and overnight (E) K+ solution samples of Tel26 is also shown.
Tel26 forms a hybrid-type G-quadruplex structure in K+ solution: the hybrid-1 type human telomeric G-quadruplex
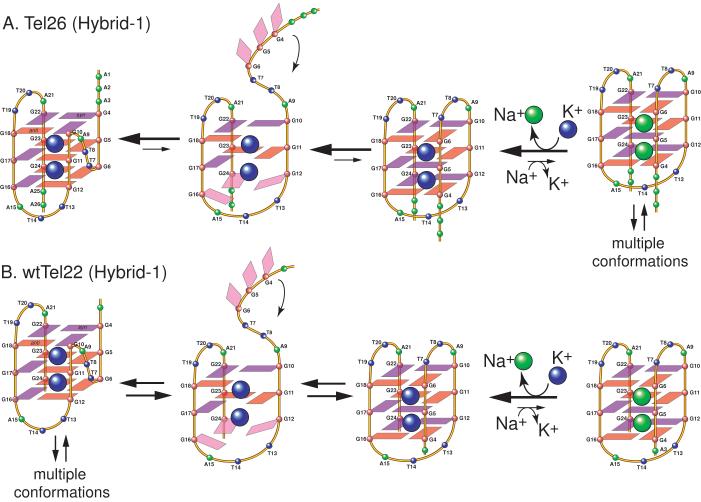
The Tel26 sequence 5′-AAAGGG(TTAGGG)3AA was used for full NMR structure determination in K+ solution [48, 50]. Tel26 contains the core 22-nt human telomeric sequence wtTel22 (5′-AGGG(TTAGGG)3) with modified flanking segments (Figure 3B). By using site-specific labeling of 15N, 13C-enriched guanine in Tel26, the H1 and H8 protons of the twelve guanines involved in the three G-tetrads were assigned. The thymine H6 protons were assigned by using deoxyuridine (dU) substitution for each dT of the Tel26 sequence. Complete assignment of the proton and 31P resonances of Tel26 was accomplished by using multiple 2D homonuclear and heteronuclear NMR experiments. Based on these NMR experiments, the folding topology (Figure 2A) [48] and the molecular structure (Figure 5A) (PDB ID 2HY9) [50] of the intramolecular G-quadruplex formed by Tel26 in K+ solution have been determined. Tel26 adopts a hybrid-type (hybrid-1) intramolecular G-quadruplex consisting of three G-tetrads linked with mixed parallel/antiparallel G-strands in K+ solution (Figure 2A). The three TTA loop segments sequentially adopt double-chain-reversal (side), lateral, and lateral loop conformations. Using NOE-restrained distance geometry (DGSA) and molecular dynamics (RMD) methods, the NMR structure determination was performed from a starting model of an arbitrary extended single-stranded DNA. A total of 552 NOE distance restraints, of which 279 are from inter-residue NOEs, were incorporated into the NOE-restrained structure calculation. Dihedral angle restraints were used for the glycosidic torsion angles as well as for a number of unusual torsion angles ε of loop residues based on the 3JH3′P3′-couplings. The NMR structures are very well defined, including the three TTA loops and the two flanking sequences at the 5′- and 3′- ends (Figure 5A). The RMSD of the 10 lowest energy structures is 1.22 Å for all residues; and is only 0.91 Å for residues 3-25, including the 5′-flanking A3, three TTA loops, and the 3′-flanking A25. Remarkably, global interaction information is well observed in the G-quadruplex structures because of extensive DNA folding. Thus very well defined molecular structures of a G-quadruplex can be derived from an arbitrary extended single-stranded DNA by the NMR-restrained structure calculation, analogous to that of a globular protein.
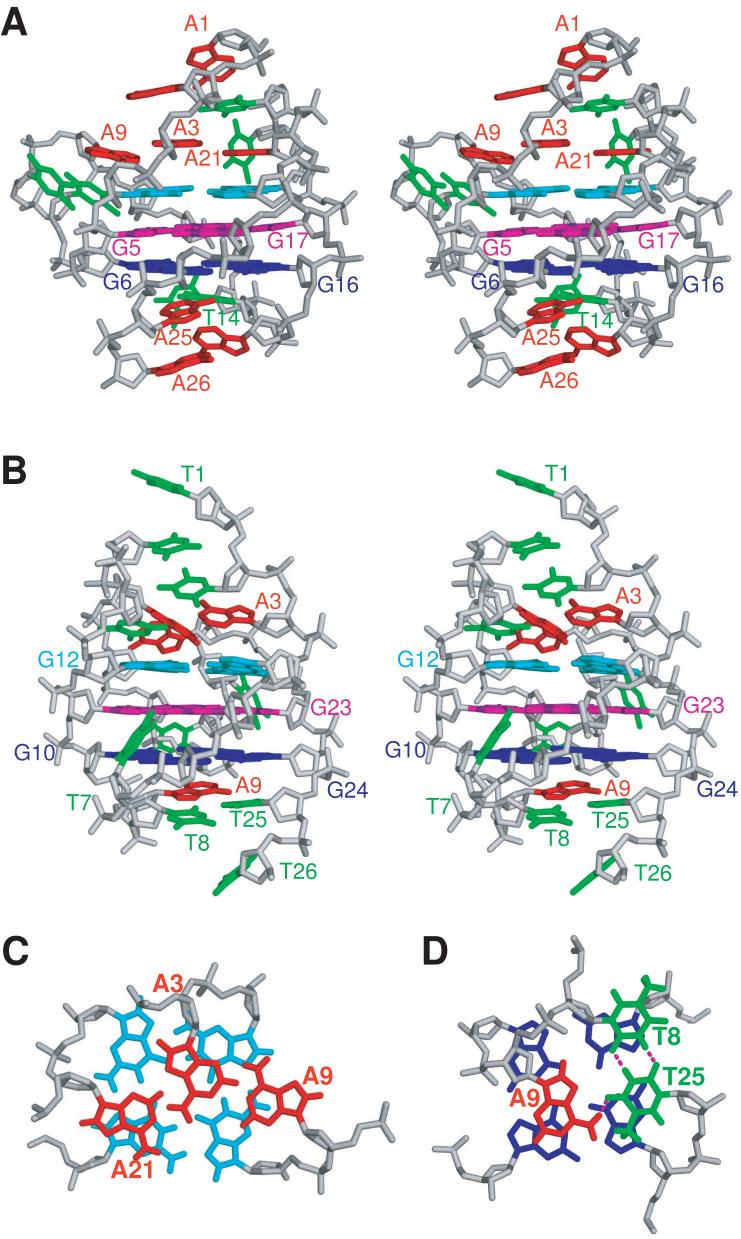
Figure 5.
(A) Stereo view of the representative model of the NMR-refined structure of the hybrid-1 type telomeric G-quadruplex formed by Tel26 in K+ solution. (B) Stereo view of the representative model of the NMR-refined structure of the hybrid-2 type telomeric G-quadruplex formed by wtTel26 in K+ solution. (C) The top view of the adenine triple (red) capping the top G-tetrad (cyan). (D) The bottom view of the T:A:T triple capping the bottom G-tetrad (blue), with the potential hydrogen-bonds shown in dashed lines. The loop adenines are in red, and the loop thymines are in green. The top G-tetrad (as in Figure 2A) is in cyan, the middle G-tetrad is in magenta, and the bottom G-tetrad is in blue.
An adenine triple capping structure specific to the hybrid-1 type human telomeric G-quadruplex is observed for the hybrid-1 structure
In the molecular structure of the hybrid-1 G-quadruplex formed by Tel26 in K+, a well-defined adenine triple platform is found to form with three naturally occurring adenine residues, A21, A3, and A9, capping the top end of the hybrid-1 type human telomeric G-quadruplex (Figure 5C) [50]. A potential hydrogen bond may be formed between A3 and A21. The naturally occurring adenine triple capping structure appears to be specific to the hybrid-1 type structure of the human telomeric DNA sequence. An A:T base pair (A25:T14) capping at the bottom end was also observed in the hybrid-1 telomeric G-quadruplex formed by Tel26. However, A25 is a mutated residue from the native thymine in the human telomeric sequence (Figure 3B), which may explain our observation that the 3′-flanking T-to-A mutation selectively stabilizes the hybrid-1 type telomeric G-quadruplex in K+ solution [48].
The wild-type 26-nt human telomeric sequence wtTel26 forms a second hybrid-type G-quadruplex structure in K+ solution: the hybrid-2 type human telomeric G-quadruplex
Tel26 forms two well-defined G-quadruplex conformations when freshly dissolved in K+ solution, as indicated by two separate sets of relatively sharp guanine imino peaks. The powder of oligonucleotide Tel26 was prepared by lyophilization after it was thoroughly dialyzed in water. One conformation (∼ 40%) slowly converts to the other (∼ 60%) overnight, and the complete conversion takes about a day (Figure 4D and 4E). This observation led to the careful examination of the native 26-nt human telomeric sequence wtTel26, (TTAGGG)4TT (Figure 3B)[49]. The 1D 1H NMR spectrum of the wtTel26 sequence in K+ solution shows a major intramolecular G-quadruplex structure with twelve resolved imino proton resonances between 10.5-12 ppm (Figure 4B); however, this major conformation only accounts for ∼ 75% of the total population, with about 25% population of minor conformations shown as weak and broader resonances. Thus it is a more challenging process to get complete resonance assignment for wtTel26. The complete spectral assignment for all proton resonances of wtTel26 was accomplished by using the 2D NMR methods and a large number of different conditions [49]. Based on the NMR experimental data, the folding topology (Figure 2A) and molecular structure (Figure 5B, PDB ID 2JPZ) of the major G-quadruplex formed by wtTel26 in K+ solution have also been determined [49]. WtTel26 adopts a second hybrid-type (hybrid-2) intramolecular G-quadruplex consisting of three G-tetrads linked with mixed parallel/antiparallel G-strands in K+ solution; however, this hybrid-type structure is different from the hybrid-1 structure in loop arrangement (Figure 2A). The three TTA loop segments sequentially adopt lateral, lateral, and double-chain-reversal (side) loop conformations. The solution structure of the hybrid-2 type human telomeric G-quadruplex formed by wtTel26 in K+ solution was determined by an NOE-restrained distance geometry (DGSA) and molecular dynamics (RMD) approach, starting from an arbitrary extended single-stranded DNA. A total of 727 NOE distance restraints, of which 324 are from inter-residue NOE interactions, were used in the NOE-restrained structure calculation. Dihedral angle restraints were used for the glycosidic torsion angle (χ) based on the experimentally determined syn and anti conformations. Like the hybrid-1 human telomeric G-quadruplex structure [50], the hybrid-2 telomeric G-quadruplex structure is very well defined. For the 10 best NMR refined structures, the RMSD is 1.38 Å for all residues except the 5′-terminal T1.
A T:A:T triple capping structure specific to the hybrid-2 type human telomeric G-quadruplex is observed for the hybrid-2 structure
In the molecular structure of the hybrid-2 G-quadruplex formed by wtTel26 in K+, a very well-defined T:A:T triple platform is found to form with T8 and A9 of the first TTA lateral loop and T25 of the 3′-flanking segment, capping the bottom end of the hybrid-2 human telomeric G-quadruplex (Figure 5D) [49]. Potential H-bonds may be formed between T8 and T25, and between T25 and A9. The three residues of the T:A:T triple platform, T8, A9 and T25, all stack very well with the bottom tetrad. This T:A:T triple capping structure appears to be specific to the hybrid-2 type structure (Figure 2A) of the human telomeric DNA sequence. The important role of the T:A:T capping structure in stabilizing the hybrid-2 human telomeric G-quadruplex structure was demonstrated by mutational analysis of wtTel26 with single A-to-T and single T-to-U substitutions [49].
The hybrid-1 and hybrid-2 human telomeric G-quadruplexes are closely related yet distinct in their folding and structures
Both hybrid-type structures contain three G-tetrads linked with mixed parallel/antiparallel G-strands, but they differ in loop arrangements, strand orientations, G-tetrad arrangements and capping structures (Figure 2A). The hybrid-1 structure has sequential side-lateral-lateral loops with the first TTA loop adopting the double-chain-reversal conformation, whereas the hybrid-2 structure has lateral-lateral-side loops with the last TTA loop adopting the double-chain-reversal conformation. Both structures contain three parallel G-strands and one antiparallel G-strand: in hybrid-1 the third G-strand is antiparallel whereas in hybrid-2 it is the second G-strand that is antiparallel. Both hybrid-type structures contain five syn guanines and mixed asymmetrical G-arrangements. The first and second G-tetrads (from the 5′ end) have reversed G-arrangements and the second and third G-tetrads have the same G-arrangements: For the hybrid-1 structure, the first G-tetrad is (syn:syn:anti:syn) (starting from the first G-strand and around the G-tetrad) and the bottom two are (anti:anti:syn:anti), whereas for the hybrid-2 structure, the first G-tetrad is (syn:anti:syn:syn) and the bottom two are (anti:syn:anti:anti).
The two hybrid-type human telomeric G-quadruplexes show distinct molecular structures with four grooves of different widths (Figure 5A and 5B). For the hybrid-1 structure, the widths of the four grooves are 13.06 Å (groove I), 11.89 Å (groove II), 8.04 Å (groove III), and 9.20 Å (groove IV), with groove I occupied by a double-chain-reversal loop [50]. For the hybrid-2 structure, the widths of the four grooves are 11.74 Å (groove I), 8.62 Å (groove II), 13.56 Å (groove III), and 9.88 Å (groove IV), with groove III occupied by a double-chain-reversal loop [49]. A groove width is measured by the closest P-P distance across groove. Both the hybrid-type human telomeric G-quadruplexes have the backbone δ torsion angles in the range of 110° to 150°, consistent with the C2′-sugar pucker conformations.
Capping structures determine the selective formation of hybrid-1 or hybrid-2 telomeric G-quadruplex structure
The native 26-nt human telomeric sequence wtTel26 and the modified 26-nt sequence Tel26 contain the same core four G-tract 22-nt human telomeric sequence 5′-AGGG(TTAGGG)3 (wtTel22), with Tel26 containing the modified 5′- and 3′- flanking-AA instead of the native TT (Figure 3B). However, in K+ solution, wtTel26 forms a major hybrid-2 structure whereas Tel26 forms a predominant hybrid-1 structure (Figure 2A) [49, 50]. The molecular structures of the two hybrid-type human telomeric G-quadruplexes indicate that specific capping structures selectively stabilize the specific hybrid-type telomeric G-quadruplex.
In the hybrid-2 telomeric G-quadruplex, the top end is covered by the 5′-flanking segment and the second TTA lateral loop, with the bottom end covered by the first TTA lateral loop and the 3′-flanking segment (Figure 2A). A well-defined T:A:T triple capping structure is formed at the bottom end of the hybrid-2 G-quadruplex and appears to selectively stabilize the hybrid-2 structure formed by wtTel26 [49] (Figure 5D). This T:A:T triple capping structure formed by T8, A9, and T25 is specific to the hybrid-2 folding and is not possible to form in the hybrid-1 folding (Figure 2A). In the modified Tel26 sequence (Figure 3B), T25 is mutated to A25, and as such the T8:A9:T25 triple can no longer form (Figure 2A) hence the hybrid-2 structure is no longer favored. Rather, Tel26 forms a hybrid-1 telomeric structure, in which the top end is covered by the 5′-flanking segment, the first TTA side loop, and the third TTA lateral loop, and the bottom end covered by the second TTA lateral loop and the 3′-flanking segment (Figure 2A); thus two capping platforms specific to hybrid-1 can form: a naturally occurring adenine triple capping structure at the top end of the G-quadruplex (Figure 5C), and an A25:T14 base pair capping structure involving the mutated A25 at the bottom end (Figure 5A) [50].
Insights into the G-quadruplex loop conformations
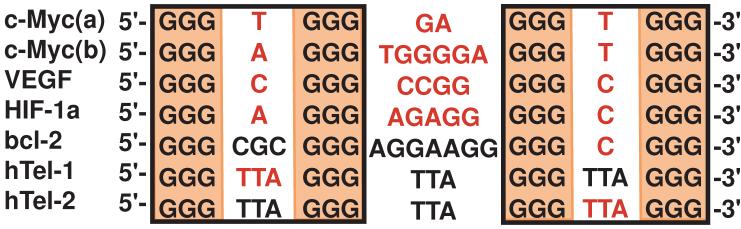
The hybrid-type human telomeric G-quadruplexes contain a 3-nt double-chain-reversal side loop, which is the first TTA segment and the third TTA segment in the hybrid-1 and hybrid-2 structures, respectively [48, 50] (Figure 2A). Such double-chain-reversal loop conformations are favored by short loop sizes of 1 and 2 nt, as shown in other G-quadruplex structures [48, 57, 58, 60-63] (Figure 6). A loop longer than 2 nt is in general not as favored for the double-chain-reversal loop conformation, likely due to unfavored base-solvent interactions of groove-positioned loop residues whose stacking interactions are lacking. Interestingly, however, in the molecular structures of the two hybrid-type human telomeric G-quadruplexes, the adenines of the TTA double-chain-reversal loops adopt a similar conformation, which is positioned above and partially stacked with the top G-tetrad. In particular, the A9 residue of the first TTA double-chain-reversal loop of the hybrid-1 structure is positioned partially above the top tetrad and participates in the adenine triple capping structure (Figure 5A), while A21 of the third TTA double-chain-reversal loop of the hybrid-2 structure is also positioned partially above the top G-tetrad (Figure 5B). This specific conformation essentially makes the 3-nt TTA double-chain-reversal loop equivalent to a 2-nt sized loop in the groove, as only the two thymine residues are positioned in the groove region. This may explain why a 3-nt double-chain-reversal loop can be present in the hybrid-type human telomeric G-quadruplexes. The presence of a 3-nt double-chain-reversal loop may also contribute to the structural flexibility and polymorphism of the hybrid-type telomeric G-quadruplexes.
Figure 6.
Comparison of G-quadruplex-forming sequences. Loops colored in red have been shown to adopt parallel-stranded double-chain-reversal side loops (see text for more details).
Human telomeric sequences form a mixture of hybrid-1 and hybrid-2 structures, the population of which largely depends on the 3′-flanking sequences
As human telomeric sequences can form two different intramolecular hybrid-type G-quadruplex conformations in K+ solution, various extended four-G-tract human telomeric sequences were systematically examined using site-specific incorporation of 15N-labeled guanines for unambiguous assignment of tetrad guanine imino protons for each sequence [49]. While human telomeric sequences form predominantly hybrid-type G-quadruplex structures in K+ solution, we found they are in equilibrium between hybrid-1 and hybrid-2 conformations, and that the equilibrium of the two conformations appears to be largely determined by the 3′-flanking sequence (Figure 3A).
The hybrid-2 structure appears to be the major conformation formed in K+ solution by extended four-G-tract human telomeric sequences with a 3′-flanking-TT (wtTel26, wtTel25b, wtTel24b) and a 3′-flanking-T (wtTel25a) (Figure 3A). Furthermore, the hybrid-2 structure appears to be the major conformation formed in K+ solution by the 27-nt human telomeric sequence wtTel27 with a 3′-flanking TTA segment [49]. In contrast, the hybrid-1 structure appears to be the major conformation in human telomeric sequences with no 3′-flanking segment (wtTel23 and wtTel24, Figure 3A) [53]. These results are in good agreement with the structural data, that the 3′-flanking-T (T25) is necessary for the formation of the T:A:T triple capping structure, which selectively stabilizes the hybrid-2 type telomeric G-quadruplex (Figure 5D) [49]. The telomeric sequences lacking this 3′-flanking-T are unable to form the T:A:T capping and thus unable to form a stable hybrid-2 structure.
Structure polymorphism of G-quadruplexes formed in extended human telomeric sequences in K+ solution
Importantly, in all four-G-tract human telomeric sequences we examined, both hybrid-1 and hybrid-2 forms appear to form and coexist in K+ solution [49]. For extended human telomeric sequences (such as wtTel26 and wtTel27), the hybrid-2 form appears to be the major conformation in K+ solution, accounting for approximately 75% in wtTel26 and 65% in wtTel27, but the hybrid-1 form can also be detected in both sequences (Figure 3A). However, the kinetics of the interconversion between the two conformations appears to be slow on the NMR time scale, as very few exchange peaks were observed in NOESY experiments. Actually, even for the truncated wtTel22 sequence which shows the coexistence of at least two major conformations with comparable populations, the exchange rate between the multiple conformations is in the slow-exchange regime on the NMR time scale as demonstrated by separate sharp NMR peaks [48] (Figure 4A).
The energy difference between the two hybrid-type telomeric G-quadruplex conformations appears to be rather small, as the equilibrium of hybrid-1 and hybrid-2 structures can be readily shifted by minor changes, such as the lengths or minor modifications of the flanking sequences. For example, while wtTel26 and Tel26 contain the same 22-nt four-G-tract core sequence and differ only in the 2-nt flanking segments (Figure 3B), they respectively form hybrid-2 and hybrid-1 as the major conformation in K+ solution (Figure 2A)[49]. Another example can be found for the wtTel24a and wtTel25a sequences, whose only difference is the 5′-flanking segment, i.e., TA for wtTel24a and TTA for wtTel25a (Figure 3A). While both sequences contain the 3′-flanking-T that can form the T:A:T triple capping that specifically stabilizes the hybrid-2 structure, the population of the hybrid-2 form is 75% in wtTel25a (with 25% hybrid-1), but only about 55% in wtTel24a. This is likely due to the fact that the 5′-terminal-T of the truncated sequence wtTel24a can form another 5′-end fold-back capping structure which stabilizes the hybrid-1 form [52]. The low energy barrier may also suggest that the in vivo equilibrium of the two forms can be affected and readily shifted by different factors, such as body temperature, ion concentration, and protein binding.
Structure polymorphism of human telomeric G-quadruplexes may be related with the highly conserved asymmetric human telomeric DNA sequence
The human telomeric sequence is highly conserved. Both hybrid-type human telomeric G-quadruplexes are asymmetric structures (Figure 2A), which are related with the intrinsic asymmetric residue distribution of the tandem TTA loops in the human telomeric DNA sequence. This structural asymmetry determines the possibility of the formation of the two very closely related but distinct hybrid-1 and hybrid-2 structures. Unlike the G-rich sequences in gene promoter regions [57, 58, 60-67], the telomeric DNA sequence is unique in that it contains the same tandem repeats with the same linker segments. Thus, in a telomeric sequence, any four-G-tract region has the same potential of forming an intramolecular G-quadruplex structure. The linker segment in the human (and vertebrate) telomeric sequence is TTA, whereas linker segments in the telomeric sequences of most lower organisms contain thymines only. The presence of adenine in the TTA loops adds an asymmetry in human telomeric sequences, thereby providing a more selective basis for different capping structures and thus different G-quadruplex conformations. For example, the T:A:T triple capping with T8, A9 and T25 can be formed in the hybrid-2 folding only, whereas the adenine triple capping with A3, A9, and A21 can be formed in the hybrid-1 folding only (Figure 2A).
Potential of the hybrid-type human telomeric G-quadruplexes to form higher-order multimers
Human telomeric DNA terminates with a 3′ single-stranded overhang of 100-200 nt. Significantly, both the hybrid-type human telomeric G-quadruplexes structures provide an efficient scaffold for a compact-stacking structure of multimers in human telomeric DNA (Figure 7). The 5′- and 3′- ends of the hybrid-type G-quadruplex structure point in opposite directions, allowing the hybrid-type G-quadruplex to be readily folded and stacked end to end in the elongated linear telomeric DNA strand (Figure 2A). The energy barrier between the two hybrid type structures is low, and as such it can be easily surpassed by binding of a specific ligand, thus changing the G-quadruplex conformation. Consequently, the changed G-quadruplex structure may disrupt existing protein interactions and introduce new protein recognitions. Intriguingly, the capping structures observed, such as the adenine triple in the hybrid-1 structure and the T:A:T triple in hybrid-2 structure, can provide not only stacking interactions between the two adjacent telomeric G-quadruplexes but also specific binding sites for small molecule ligands to target G-quadruplexes in the elongated human telomeres (Figure 7).
Figure 7.
A model of DNA secondary structure composed of compact-stacking multimers of hybrid-type G-quadruplexes in human telomeres, with an equilibrium between hybrid-1 and hybrid-2 forms in K+ solution. The hybrid-1 and hybrid-2 telomeric G-quadruplexes have distinct molecular structures, as shown by the representative NMR structures (guanine = yellow, adenine = red, thymine = blue).
Review of multiple biophysical studies
A large number of biophysical and crosslinking studies have been reported on the K+-structure of human telomeric G-quadruplexes [32-34, 36-40]. Structure polymorphism of the human telomeric G-quadruplexes, particularly for the 22-nt wtTel22 sequence, has been reported by various methods. The different CD spectra have been observed for human telomeric sequences in Na+ and K+ solution [32, 37, 39, 40]. Multiple structures formed by wtTel22 in Na+ and K+ solution have also been reported in a 125I-radioprobing study [36], a FRET study [33], a platinum cross-linking study [34], a chemical ligation study [40], and a photon correlation spectroscopic study [39]. The hybrid-type G-quadruplex structure can explain the results and predictions made in previous biophysical studies where physiologically relevant conditions were used with the wild-type human telomeric sequence. In a recent biophysical study [38] sedimentation coefficients and 2-aminopurine (2-AP) fluorescence were used to probe the G-quadruplex structure formed by wtTel22 in K+ solution. The order of the accessibility of the single substitution of 2-AP as probed by fluorescence quenching was consistent with the hybrid-1 type structure. The sedimentation coefficient of wtTel22 in Na+ solution was in good agreement with the calculated value from the Na+-solution structure of wtTel22, while the sedimentation coefficient of wtTel22 in K+ solution was found to be different from the calculated value of the parallel-stranded structure as observed in the crystalline state [47]. Actually, a calculation on the hybrid-1 structure [50] gave a sedimentation coefficient (S) value of 2.09 with two K+ ions bound, which is in good agreement (1.5% difference) with the experimentally observed value of 2.11 (unpublished data from Prof. John Trent at the University of Louisville). It was also shown in the same report that the addition of PEG200, which may mimic the crystal condition, can shift the conformation of wtTel22 to a parallel-stranded structure [38]. A more recent study has elaborated on this observation, that the presence of a high concentration PEG200 as a crowding agent can completely shift the conformation of wtTel21 (Figure 3A) to the parallel-stranded structure [68]. This result again indicates a small energy barrier of different telomeric G-quadruplexes. Molecular crowding agents are volume exclusion macromolecules intended to increase the true concentration of the test molecule to mimic the crowded biological fluids. It is of note that the concentration used for NMR structure determination is in the 2-3 mM range, more than 40 fold higher than the crowding condition used in CD and fluorescence studies with PEG200 [68]. In addition, molecular crowding agents are not supposed to have specific interactions with the test molecular structures [69, 70], whereas PEG has been shown to induce enhanced hydrophobic surface area and hydrophobic interactions with the test molecules [71-74]. However, it is important to note that while the hybrid-type G-quadruplexes are the predominant structures formed by the human telomeric DNA sequence under aqueous conditions, the small energy barrier of different forms, including the parallel form, can make them readily shifted by different protein and ligand bindings.
CD has become a useful technique for the characterization of G-quadruplex-forming oligonucleotides. CD spectra are sensitive to DNA base stacking, e.g., guanine base stacking in G-quadruplexes, which is related to the orientations of G-strands [48, 51, 75, 76]. Guanines having the same anti glycosidic conformation along a G-strand give rise to a positive peak at 260 nm and a small negative peak at 240 nm in CD, as observed in the parallel-stranded G-quadruplex formed in the promoter region of c-Myc [60]. Guanines with alternating anti and syn glycosidic conformations give rise to a positive peak at 295 nm and a negative peak at 265 nm in CD spectra [48, 75], as observed in the antiparallel-stranded G-quadruplex motifs such as the basket-type G-quadruplex formed by wtTel22 in Na+ (Figure 2B and 8A) and the chair-type G-quadruplex formed by a thrombin-binding aptamer [77]. Interestingly, the two hybrid-type telomeric G-quadruplexes formed by wtTel26 and Tel26 in K+ solution exhibit very similar CD signature profiles (Figure 8A). The hybrid-type telomeric G-quadruplexes contain both the alternating and non-alternating glycosidic conformations for adjacent guanines on a G-strand, and thus exhibit a hybrid CD profile, namely, a positive peak around 290 nm and a smaller peak around 265 nm, and a smaller negative peak at 240 nm. The positive peak around 290 nm is likely due to the alternating guanine glycosidic conformations along G-strands between the top and middle G-tetrads in the hybrid-type telomeric G-quadruplexes, while the positive peak around 265 nm and the negative peak around 240 nm are likely due to the non-alternating guanine glycosidic conformations between the middle and bottom G-tetrads (Figure 2A).
Figure 8.
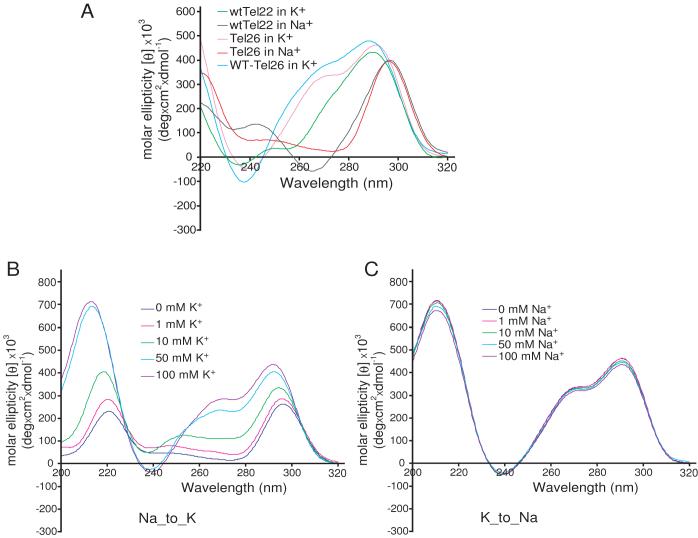
(A) CD spectra of wtTel26, Tel26, and wtTel22 in 100 mM Na+ or K+ solution at 25 °C. Similar CD signatures are observed for wtTel26 and Tel26 in K+ solution, while similarly distinct CD signatures are observed for wtTel26, Tel26, and wtTel22 in Na+ solution. (B) Titration experiments of K+ in the presence of 150 mM Na+ for Tel26, and (C) titration experiments of Na+ in the presence of 100 mM K+ for Tel26, monitored by CD spectroscopy.
Hybrid-type structures are the predominant conformations for wtTel26 and Tel26 in K+ solution, even in the co-presence of a high concentration of Na+
The wtTel22 sequence AGGG(TTAGGG)3, which forms a well-defined basket-type G-quadruplex in Na+ solution [46] (Figure 2B). In contrast, the extended sequences wtTel26 (TTAGGG)4TT and Tel26 AAAGGG(TTAGGG)3AA do not form a well-defined G-quadruplex conformation in Na+ solution, as indicated by broad envelopes in 1D 1H NMR, although the CD spectra still indicate a basket-type conformation [48, 49]. This is likely due to the steric interference between the diagonal loop and the two extended flanking segments of wtTel26 and Tel26 in the basket-type structure, as they are all positioned on the same end of the basket-type G-quadruplex (Figure 2B).
In K+ solution, the hybrid-type structures (Figure 2A) appear to be the more stable, and thus the predominant, conformation for wtTel26 and Tel26, even in the co-presence of a high concentration of Na+. Addition of K+ to Na+ solution readily converts the preformed basket-type Na+ structure to the hybrid-type K+ structure [48] (Figure 8B). In contrast, addition of Na+ to 50 mM K+ solution, even to a concentration of 200 mM, does not change the hybrid-type K+ structure (Figure 8C). The same results are also observed for the truncated 22-nt wtTel22 sequence, that the addition of K+ can convert Na+-form to K+-form in the presence of Na+, while the addition of Na+ does not have any effect on the K+-form structure. Full conversion of the Na+-form to the K+-form for the extended 26-nt sequences takes several hours to an overnight incubation depending on the K+ concentration, whereas such conversion for the truncated 22-nt wtTel22 sequence is faster than can be detected by either CD or NMR [48].
Close examination of the hybrid-type and basket-type telomeric G-quadruplex structures reveals a possible mechanism of the interconversion of the two forms through a strand-reorientation mechanism [48] (Figure 9). The 5′-G-strand of the basket-type G-quadruplex may dissociate from the structure and swing back to the other side of the second G-stand to form a parallel-stranded structural motif with a double-chain-reversal loop. The first two G-tetrad planes do not need to be completely melted for the new conformation as the other six guanines still keep the same relative positions and glycosidic sugar conformations, while the bottom G-tetrad is likely to be melted and dissociated to rearrange the guanine conformation. This suggested partial melting is consistent with recent reports showing a multi-step melting transition of telomeric G-quadruplexes [78, 79].
Figure 9.
Schematic diagram of a possible mechanism of the interconversions between the basket type (Na+) and the hybrid-type (K+) forms of telomeric G-quadruplexes. The hybrid-1 structure is used for illustration. The hybrid-type G-quadruplex structure is the most stable and thus the predominant form in the presence of K+, regardless of the presence or absence of Na+. The interconversion rate is much slower for the extended four repeat telomeric sequence Tel26 (A) than for the truncated four repeat telomeric sequence wtTel22 (B).
3. Concluding remarks
Recent results show that the hybrid-type G-quadruplex structures appear to be the major conformations formed in four-G-tract human telomeric sequences in K+ solution, with a dynamic equilibrium between hybrid-1 and hybrid-2 conformations that appears to be largely determined by the 3′-flanking sequence. The two hybrid-type telomeric G-quadruplexes are distinct and yet closely related. Different capping structures appear to determine the favored formation of a specific hybrid-type telomeric G-quadruplex structure, and may provide specific binding sites for drug interaction. Both hybrid-type G-quadruplex structures suggest a straightforward means for multimer formation with effective packing in human telomeres and provide important implications for drug targeting of telomeric G-quadruplexes. The structure polymorphism and dynamic equilibrium of telomeric G-quadruplexes, which appear to be intrinsic to the highly conserved telomeric sequence, may be important for the biology of human telomeres. Nature may have chosen this specific sequence with its asymmetry and the low energy barrier between different forms, which may provide a means to affect the protein recognition and to control the biology of telomeres. It is important to note that while the hybrid-type G-quadruplexes are the predominant forms under aqueous conditions, other conformations may still be selectively recognized by different protein binding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- [2].van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- [3].Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- [4].Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- [5].Allshire RC, Dempster M, Hastie ND. Human Telomeres Contain at Least 3 Types of G-Rich Repeat Distributed Non-Randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, J.R. W. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- [9].Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Colgin LM, Reddel RR. Telomere maintenance mechanisms and cellular immortalization. Current Opinion in Genetics & Development. 1999;9:97–103. doi: 10.1016/s0959-437x(99)80014-8. [DOI] [PubMed] [Google Scholar]
- [11].Mergny JL, Helene C. G-quadruplex DNA: A target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- [12].Sun DY, Hurley LH. Targeting telomeres and telomerase. In: Chaires JB, Waring MJ, editors. METHODS IN ENZYMOLOGY, Drug-Nucleic Acid Interactions. Vol. 340. Academic Press Inc; San Diego: 2001. pp. 573–592. [DOI] [PubMed] [Google Scholar]
- [13].Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- [14].Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- [15].Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- [16].Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutation Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- [17].Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s Syndrome Helicase Unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- [18].Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- [20].Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- [21].Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- [22].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- [23].Salazar M, Thompson BD, Kerwin SM, Hurley LH. Thermally induced DNA:RNA hybrid to G-quadruplex transitions: possible implications for telomere synthesis by telomerase. Biochemistry. 1996;35:16110–16115. doi: 10.1021/bi961442j. [DOI] [PubMed] [Google Scholar]
- [24].Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- [25].Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- [27].Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- [28].Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- [29].Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochemical Society Transactions. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- [30].Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han HY, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- [31].Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- [32].Balagurumoorthy P, Brahmachari SK. Structure and Stability of Human Telomeric Sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- [33].Ying LM, Green JJ, Li HT, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Platinum cross-linking of adenines and guanines on the quadruplex structures of the AG(3)(T(2)AG(3))(3) and (T(2)AG(3))(4) human telomere sequences in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].He YJ, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with I-125-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- [38].Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wlodarczyk A, Grzybowski P, Patkowski A, Dobek A. Effect of ions on the polymorphism, effective charge, and stability of human telomeric DNA. Photon correlation spectroscopy and circular dichroism studies. J. Phys. Chem. B. 2005;109:3594–3605. doi: 10.1021/jp045274d. [DOI] [PubMed] [Google Scholar]
- [40].Qi JY, Shafer RH. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Risitano A, Fox KR. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005;15:2047–2050. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]
- [44].Ourliac-Garnier I, Elizondo-Riojas MA, Redon S, Farrell NP, Bombard S. Cross-links of quadruplex structures from human telomeric DNA by dinuclear platinum complexes show the flexibility of both structures. Biochemistry. 2005;44:10620–10634. doi: 10.1021/bi050144w. [DOI] [PubMed] [Google Scholar]
- [45].Lee JY, Okumus B, Kim DS, Ha TJ. Extreme conformational diversity in human telomeric DNA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang Y, Patel DJ. Solution Structure of the Human Telomeric Repeat D[Ag(3)(T(2)Ag(3))3] G-Tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- [47].Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- [48].Ambrus A, Chen D, Dai JX, Bialis T, Jones RA, Yang DZ. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dai JX, Carver M, Punchihewa C, Jones RA, Yang DZ. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dai JX, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang DZ. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)(3)] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- [52].Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: An intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions. as determined by NMR in a K+ solution, FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- [55].Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- [57].Ambrus A, Chen D, Dai JX, Jones RA, Yang DZ. Solution structure of the biologically relevant g-quadruplex element in the human c-MYC promoter. implications for g-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- [58].Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel G-strands Formed in the Human BCL-2 Promoter Region in Solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dai JX, Chen D, Jones RA, Hurley LH, Yang DZ. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- [61].Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sun DY, Guo KX, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. Epub 2007 Mar 4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].De Armond R, Wood S, Sun DY, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1 alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- [65].Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hurley LH, Von Hoff DD, Siddiqui-Jain A, Yang DZ. Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Semin. Oncol. 2006;33:498–512. doi: 10.1053/j.seminoncol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- [67].Yang DZ, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides & Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- [68].Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- [69].Charlton LM, Pielak GJ. Peeking into living eukaryotic cells with high-resolution NMR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11817–11818. doi: 10.1073/pnas.0605297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Minton AP. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: Macromolecular crowding and protein stability revisited. Biophys. J. 2005;88:971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chebotareva NA, Kurganov BI, Livanova NB. Biochemical effects of molecular crowding. Biochem.-Moscow. 2004;69:1239. doi: 10.1007/s10541-005-0070-y. + [DOI] [PubMed] [Google Scholar]
- [72].Naeem A, Khan A, Khan RH. Partially folded intermediate state of concanavalin A retains its carbohydrate specificity. Biochemical and Biophysical Research Communications. 2005;331:1284–1294. doi: 10.1016/j.bbrc.2005.04.041. [DOI] [PubMed] [Google Scholar]
- [73].Farruggia B, Nerli B, Pico G. Study of the serum albumin-polyethyleneglycol interaction to predict the protein partitioning in aqueous two-phase systems. J. Chromatogr. B. 2003;798:25–33. doi: 10.1016/j.jchromb.2003.08.044. [DOI] [PubMed] [Google Scholar]
- [74].Farruggia B, Garcia G, Dangelo C, Pico G. Destabilization of human serum albumin by polyethylene glycols studied by thermodynamical equilibrium and kinetic approaches. Int. J. Biol. Macromol. 1997;20:43–51. doi: 10.1016/s0141-8130(96)01150-6. [DOI] [PubMed] [Google Scholar]
- [75].Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucl. Acids. Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kypr J, Fialova M, Chladkova J, Tumova M, Vorlickova M. Conserved guanine-guanine stacking in tetraplex and duplex DNA. Eur. Biophys. J. Biophys. Lett. 2001;30:555–558. doi: 10.1007/s002490100174. [DOI] [PubMed] [Google Scholar]
- [77].Schultze P, Macaya RF, Feigon J. Three-dimensional Solution Structure of the Thrombin-binding DNA Aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- [78].Antonacci C, Chaires JB, Sheardy RD. Biophysical characterization of the human telomeric (TTAGGG)(4) repeat in a potassium solution. Biochemistry. 2007;46:4654–4660. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- [79].Li J, Trent JO, Bishop GR, Chaires JB. Uncovering the Energetic Basis of G-Quadruplex Stability; First Internationl Meeting on Quadruplex DNA; Louiville, KY, USA. 2007. [Google Scholar]