Abstract
In vertebrates, seven signal transducer and activator of transcription (STAT) proteins bind to palindromic sites separated by spacers of two or three nucleotides (STAT1), four nucleotides (STAT6) or three nucleotides (STAT2 to STAT5a/b). This diversity of binding sites provides specificity to counter semiredundancy and was thought to be a recent evolutionary acquisition. Here, we examine the natural DNA-binding sites of the single Drosophila Stat and show that this is not the case. Rather, Drosophila Stat92E is able to bind to and activate target gene expression through both 3n and 4n spaced sites. Our experiments indicate that Stat92E has a higher binding affinity for 3n sites than for 4n sites and suggest that the levels of target gene expression can be modulated by insertion and/or deletion of single bases. Our results indicate that the ancestral STAT protein had the capacity to bind to 3n and 4n sites and that specific STAT binding preferences evolved with the radiation of the vertebrate STAT family.
Keywords: STAT, binding sites, Drosophila, evolution
Introduction
The Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signalling pathway was originally discovered in vertebrates on the basis of its transduction of activation of γ-interferon (Darnell, 1997; Levy & Darnell, 2002). Analysis of the pathway rapidly identified a family of seven closely related STATs as well as many pathway-activating cytokines, several receptors and four JAK kinases. The seven STAT transcription factors share several features including an SH2 domain, an invariant tyrosine residue phosphorylated as a result of the activation of STAT, and a characteristic DNA-binding domain. According to the established canonical model, activation of the JAK/STAT pathway by cytokine signalling brings about tyrosine phosphorylation of cytoplasmic STATs and leads to their dimerization. This complex translocates to the nucleus where it binds to DNA, thus activating target gene transcription (Kisseleva et al, 2002; Levy & Darnell, 2002). STAT DNA-binding sites, also known as Gamma interferon activation site (GAS) elements, consist of an essential core comprising the palindromic sequence TTC(n)GAA where n represents a spacer of 2–4 nucleotides. STAT6 shows a preference for 4n spacing, whereas other STATs preferentially bind to 3n, although they can also bind to 2n sites with low affinity (Ehret et al, 2001).
The Jak/Stat pathways identified in invertebrates seem to be much simpler. Of these, only Drosophila has a ‘complete' pathway comprising three unpaired-like cytokines (Upd, Upd2 and Upd3); one receptor (Domeless; Dome); one JAK kinase (Hop) and one Stat (Stat92E; Binari & Perrimon, 1994; Hou et al, 1996; Yan et al, 1996; Harrison et al, 1998; Brown et al, 2001; Chen et al, 2002; Agaisse et al, 2003; Hombría et al, 2005; reviewed in Hombría & Brown, 2002; Arbouzova et al, 2006).
In vitro site selection and electrophoretic mobility shift assays (EMSAs) showed that Drosophila Stat has a binding preference for sites with 3n spacing (Yan et al, 1996). In vivo, Stat92E binding of 3n sites was confirmed for the even skipped (eve) gene (Small et al, 1996). In cell culture, it was also shown that Drosophila raf (Draf) and Suppressor of cytokine signalling at 36E (Socs36E), enhancers containing 3n sites are activated by Jak/Stat (Kwon et al, 2000; Baeg et al, 2005; Müller et al, 2005). Furthermore, vertebrate 3n GAS elements act as reporters for the activation of Stat in Drosophila melanogaster and in Caenorhabditis elegans (Gilbert et al, 2005; Wang & Levy, 2006).
These data suggest that the ancestral STAT bound to 3n sites and that the preference for sites with other spacing evolved after the vertebrate STAT radiation. Here, we present evidence that the converse is true, with binding site plasticity of STAT transcription factors representing an ancestral state. We show that Drosophila Stat is able to activate transcription through 4n sites. We show that Stat binds to 3n sites with higher affinity in vitro, and that the transformation of 4n into 3n sites increases the activation of Stat targets both in vitro and in vivo. These observations clarify how the Stat binding preferences evolved and illustrate an unanticipated plasticity of DNA binding that will help in the definition of direct Stat targets outside the vertebrate lineage.
Results And Discussion
Stat92E activates dome through 4n sites
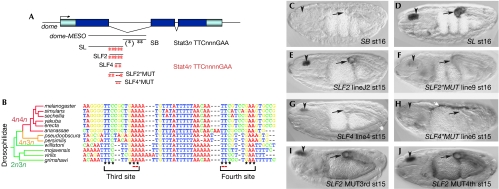
Upd expressed in the ectoderm of the Drosophila pharynx and hindgut signals to the adjacent mesoderm where it enhances dome transcription (Hombría et al, 2005). This effect is mediated by a mesoderm-specific enhancer (dome-MESO) present in the first intron of the dome gene (Hombría et al, 2005). To prove that this enhancer is regulated directly by Stat, we searched for potential Stat92E-binding sites [TTC(3n)GAA] in dome-MESO (Fig 1A; Yan et al, 1996) and identified three such sites at the 3′ end of the reporter. However, a 1.2-kb lacZ construct, dome-SB, containing these 3n sites, is unable to drive mesodermal expression (Fig 1C), whereas the complementary 1.6-kb proximal fragment, dome-SL, reproduces the dome-MESO pattern of expression (Fig 1D). The subdivision of this fragment locates the enhancer within a 746-bp fragment that we named dome-SLF2 (Fig 1A,E). Although no canonical 3n sites are present in SLF2, five 4n sites are present—a sequence bound in vertebrates by the STAT6 protein (Ehret et al, 2001). As previous in vitro binding site selection experiments using Stat92E isolated the 3n sites exclusively, we set out to test whether Drosophila Stat92E could bind to 4n sites in vivo.
Figure 1.
Stat regulates dome transcription through 4n sites. (A) Schematic representation of the dome gene showing the localization of the putative Stat-binding sites. Sites in the first intron are represented by black (3n) or red (4n) asterisks. (B) Comparison of the conserved SLF4 region in several Drosophilidae. The element has two putative Stat sites with varying spacer lengths as indicated by the tree branch colours (4n4n (red), 4n3n (orange) and 2n3n (green)). Asterisks under the sequence label the putative Stat-binding sites and red dashes indicate the mutated bases in the SLF2*MUT and SLF4*MUT. (C) The SB first intron fragment containing the 3n sites does not drive expression in the embryo. The SL (D) and the SLF2 (E) fragments drive expression in the pharynx and hindgut. (F) Simultaneous mutation of the conserved third and fourth Stat-binding sites in SLF2*MUT abolishes most expression from the pharynx and hindgut, although low levels remain at late stages. (G) The SLF4 fragment drives expression, albeit at low levels, in the pharynx and hindgut. Hindgut expression is only observed in inserts with higher levels of expression (compare G with Fig 2C). (H) Mutation of both Stat-binding sites abolishes pharynx and hindgut expression in SLF4*MUT. This particular line has been chosen as it has Jak/Stat-independent expression in the amnioserosa (white arrow) that acts as an internal control for staining. Mutation of only one conserved Stat site in SLF2, either the third site (I) or the fourth site (J), is not sufficient to abolish the expression (compare wild-type SLF2 in (E) with (I) and (J) with the double mutant in (F)). The tree in (B) is modified from the assembly, alignment and annotation of 12 species as published in FlyBase. Stat, signal transducer and activator of transcription.
To identify which of the five potential sites drive mesodermal regulation, we compared the first intron sequence of dome in several Drosophilidae (Drosophila 12 Genomes Consortium, 2007). The only conserved sequence is a 43 bp element containing the third and fourth D. melanogaster Stat 4n sites (Fig 1B). To test the possible function of these two sites, we made an SLF2 construct with the conserved third and fourth 4n sites mutated (SLF2*MUT), a 137-bp fragment containing only the two 4n sites present within the 43 bp conserved region (SLF4), and an SLF4 construct with these two 4n sites mutated (SLF4*MUT; Fig 1B).
Analysis of SLF2*MUT shows that mutation of the third and fourth 4n sites results in the almost complete loss of mesoderm expression (Fig 1F), although low levels of expression were still observed at late embryogenesis.
The SLF4 fragment alone is able to drive expression in the mesoderm of the pharynx and hindgut. The expression of SLF4 is more variable than that of SLF2 with some insertions showing expression exclusively in the pharynx (Fig 1G), whereas others showing low levels of general mesoderm expression in addition to expression in the pharynx and hindgut (Fig 2C). Mutation of the 4n sites in SLF4*MUT is sufficient to ablate all expression in both the pharynx and the hindgut (Fig 1H).
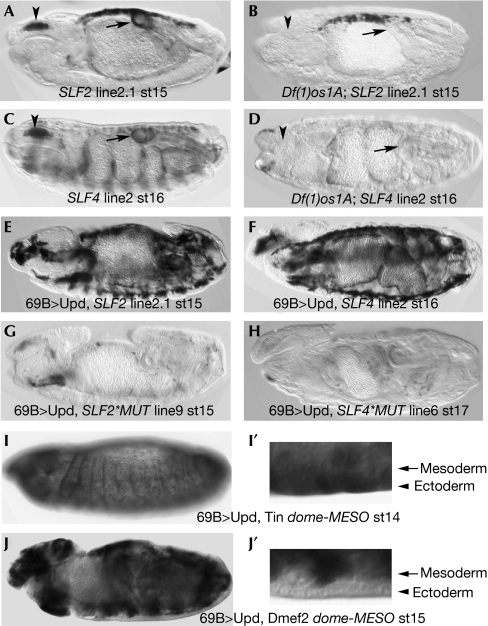
Figure 2.
Expression of SLF2 and SLF4 depends on Jak/Stat pathway activity. (A) An SLF2 line in wild type. This insertion has expression on the amnioserosa unrelated to the enhancer. (B) The same insertion as in (A) in a Df(1)os1A embryo lacking all upd genes. The expression in the pharynx and hindgut disappears, whereas the unrelated amnioserosa staining persists, acting as an internal control for staining levels. (C) A wild-type SLF4 strong insertion. (D) The same insertion in a Df(1)os1A background. (E,F) Ectopic upd in the ectoderm driven by the 69B-Gal4 line induces ectopic SLF2 (E) or SLF4 (F) expression in the mesoderm. (G,H) Ectopic upd does not induce ectopic expression when the two conserved Stat-binding sites are mutated as in SLF2*MUT (G) or in SLF4*MUT (H). (I) Simultaneous expression in the ectoderm of upd and tin can activate dome-MESO in the ectoderm. (J) Simultaneous expression of upd and Dmef2 does not result in the activation of dome-MESO in the ectoderm. (I′,J′) Close-up views of the ventral side of the embryos in (I,J) showing the position of the ectoderm and mesoderm cells. Only Upd and Tin co-expression can activate the enhancer in the ectoderm. In both cases, because of the presence of the endogenous mesodermal cofactors, there is mesodermal enhancer expression. dome-MESO, domeless-mesoderm-specific enhancer; Jak, Janus kinase; Stat, signal transducer and activator of transcription.
The lower levels of expression of SLF4 relative to those of SLF2 in the pharynx and hindgut and the slight remnant expression observed in the pharynx of SLF2*MUT embryos suggest that some of the non-conserved 4n sites might contribute to dome-MESO expression. We tested whether both of the conserved 4n sites in SLF2 are necessary by independently mutating them (Fig 1I,J). Mutation of a single site is not sufficient to abolish mesoderm expression, indicating that these sites are redundant in the context of the SLF2 enhancer.
To confirm that SLF2 and SLF4 are responsive to Jak/Stat signalling, we studied their expression in Df(1)os1A mutants that lack all Upd ligands. As expected, SLF2 expression disappeared from the pharynx and hindgut (Fig 2A,B) and the same is true for SLF4 (Fig 2C,D). Conversely, ectopic activation of the pathway by expression of Upd or Upd2 using the ectoderm-specific 69B-Gal4 line activates SLF2 (Fig 2E; data not shown) and SLF4 (Fig 2F) in the mesoderm. This ectopic activation requires the conserved 4n sites as it is not observed in SLF2*MUT (Fig 2G) or SLF4*MUT (Fig 2H).
The dome-MESO enhancer and its derivatives are expressed specifically in the mesoderm, suggesting that Stat is interacting with tissue-specific cofactors. We tested whether dome-MESO could also be activated in the ectoderm if Upd was co-expressed with various mesoderm-specific proteins. We observed ectopic ectoderm expression after co-expression of Upd with Tinman (Tin; Fig 2I,I′), but not with Dmef2, Bagpipe or Biniou (Fig 2J,J′; data not shown). This suggests that Tin or one of its downstream targets is a STAT cofactor necessary for dome activation in the mesoderm. The requirement for this interaction explains why only the 4n sites in dome-MESO are functional.
Stat92E binds to 4n and 3n sites with different affinity
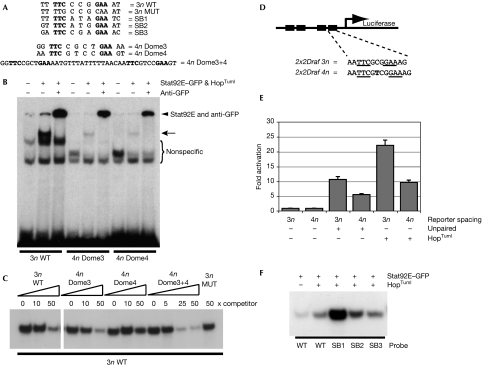
To test whether Stat92E can bind to the conserved 4n sites in vitro, oligonucleotides containing these sites (known as 4n Dome3 and 4n Dome4; Fig 3A) were used in EMSAs together with a Stat92E–GFP (green fluorescent protein) fusion protein activated by co-expression of the constitutively active Jak allele HopTuml (Luo et al, 1995; Karsten et al, 2006). By using the established 3n wild-type consensus (Yan et al, 1996) as a positive control, a strong band is detectable (Fig 3B, arrow) that is supershifted by the addition of the GFP antibody (Fig 3B, arrowhead). The same binding conditions with the 4n Dome3 and 4n Dome4 sites also give clear band shifts that can be supershifted (and possibly stabilized) by anti-GFP (Fig 3B). However, 4n band shifts are considerably weaker than those produced by the 3n control under these in vitro conditions. We next tested whether 4n Dome sites could compete with wild-type radiolabelled 3n sites (3n wild type) for binding to activated Stat92E–GFP. Although unlabelled 3n wild-type-binding sites are strong competitors (Fig 3C, lanes 1–3), a 50-fold excess of mutant 3n-binding sites is not able to compete with labelled 3n wild type (Fig 3C, lane 14). By contrast, unlabelled 4n Dome3 and, to a lesser extent, 4n Dome4 sites can compete with the labelled 3n wild-type probe (Fig 3C, lanes 6 and 9). A Dome3+4 probe containing both 4n sites separated by their D. melanogaster spacer (Fig 1B) at 25-fold excess (Fig 3C, lane 12—a concentration that provides a 50-fold excess of 4n sites) produces stronger competition than the equivalent concentration of either 4n site alone, suggesting that two adjacent sites are able to bind Stat92E better than a single binding site in isolation.
Figure 3.
Drosophila Stat binds to 3n and 4n sites with various affinities. (A) Sequences of the oligonucleotides used in (B,C,F). SB1, SB2 and SB3 correspond to the potential Stat-binding sites present in the SB fragment (Fig 1A), and Dome3, Dome4 and Dome3+4 correspond to the D. melanogaster Stat-binding sites in SLF4 (Fig 1A,B). 3n wild type (WT) corresponds to the Stat92E-binding site consensus (Yan et al, 1996). (B) EMSA assay using the radiolabelled binding sites indicated and showing binding activities that are detectable after co-transfection of plasmids expressing Stat92E–GFP and the constitutively active HopTuml proteins. Specific shifted bands corresponding to DNA:Stat92E–GFP (arrow) and DNA:Stat92E–GFP complexes supershifted with anti-GFP (arrowhead) are indicated. (C) EMSA assay using radiolabelled 3n WT to detect activated Stat92E–GFP-binding activity. Each lane contains the same quantity of cell extract and labelled 3n WT-binding site and was co-incubated with the indicated fold excess of unlabelled competitor sites. (D) Schematic representation of the reporters used in (E) with black boxes representing Stat-binding sites. Underlined bases represent the core Stat92E-binding sequence. Another thymine residue (bold) was inserted into each binding site of 2x2Draf 3n to create the 4n reporter. (E) Firefly luciferase activity in cells transfected with the reporters shown in (D) and co-transfected with plasmids expressing either the pathway ligand unpaired or the constitutively active Jak HopTuml. Levels were normalized to a co-transfected constitutively expressed Renilla luciferase plasmid and are expressed as fold change over unstimulated state. (F) EMSA assay of unactivated and activated Stat92E–GFP binding to radiolabelled WT, SB1, SB2 and SB3 oligonucleotides. EMSA, electrophoretic mobility shift assay; GFP, green fluorescent protein; MUT, mutated; STAT, signal transducer and activator of transcription.
To measure in vivo the relative transcriptional activation potential of activated Stat92E–GFP at these sites, we devised a luciferase reporter plasmid containing either four 3n- or 4n-binding sites (Fig 3D; Methods). By using these reporters in an established Kc167 cell-based model (Müller et al, 2005), we stimulated cells by either co-expressing the Upd ligand or the activated Jak HopTuml (Luo et al, 1995). Under these conditions, both 3n and 4n reporters show significantly increased levels of activity over the unstimulated state, with the 3n reporter around twice as active as the 4n reporter (Fig 3E). Finally, we found that the SB 3n sites (Figs 1A, 3A) can bind to Stat92E in vitro (Fig 3F), underscoring the importance of cofactors for the activity of Stat in vivo.
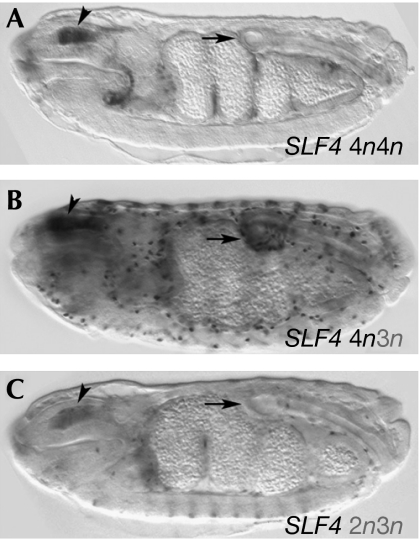
Comparison of the conserved SLF4 region in various Drosophilidae (Fig 1B) shows that species closely related to D. melanogaster share the 4n spacing of the Stat fourth site, with 3n sites present in more distantly related drosophilids. In some species in which the fourth site is 3n, the third site has a 2n spacer. The above experiments suggest that changes in spacer length during evolution might modulate the levels of transcription of target genes. To test this possibility, we mutated the fourth 4n spacer in SLF4 to a 3n spacer as observed in the obscura group. The resulting enhancer drove higher levels of expression in D. melanogaster than did SLF4 (Fig 4A,B), indicated by the consistent appearance of hindgut expression in all insertions. Further mutation in this enhancer of the third-site to a 2n spacer, as in D. virilis, restored the levels of expression similar to the original SLF4 enhancer (Fig 4C). These results indicate that varying the relative number of 3n compared with 4n sites might control the level of expression of Stat targets during evolution.
Figure 4.
Various Stat spacer lengths modify in vivo transcriptional activation. (A) Levels of expression of the SLF4 enhancer containing the normal 4n4n D. melanogaster Stat-binding sites. (B) Enhanced expression of an SLF4 enhancer in which the conserved fourth Stat 4n site is transformed to 3n. (C) An SLF4 enhancer in which the third site has been transformed to 2n and there is a 3n site transformation on the fourth site. Note that the levels of expression in (C) are lower than those in (B), and more similar to those of (A). Stat, signal transducer and activator of transcription.
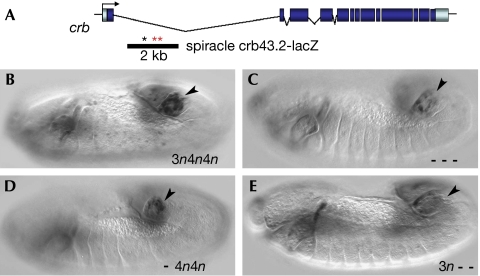
The direct Stat target crb is regulated through 4n sites
To determine whether other Drosophila Stat target genes are controlled through 4n sites, we analysed the Stat-dependent crb spiracle-specific enhancer (Fig 5A,B; Lovegrove et al, 2006). Simultaneous mutation of the three putative Stat-binding sites (one 3n and two 4n sites) reduces crb spiracle expression (Fig 5C). Mutation of the 3n site has little effect on the expression of enhancer (Fig 5D), whereas mutation of both 4n sites (Fig 5E) results in levels of expression similar to those obtained after mutation of both the 3n and 4n sites.
Figure 5.
The crb gene is regulated by Stat through 4n sites. (A) Schematic representation of the crb gene showing the localization in the spiracle enhancer of the putative Stat-binding sites analysed (red asterisks (4n sites), black asterisk (3n site)). (B) Expression of the spiracle enhancer in the wild type. (C) Expression in the triple 3n4n4n mutant (- - -). (D) Expression in the 3n mutant site (-4n4n). (E) Expression in the double 4n4n mutant sites (3n- -). Stat, signal transducer and activator of transcription.
Our results highlight the limitations of bioinformatic and in vitro DNA-binding analyses as sole methods for defining transcription-factor-binding sites if they overlook low-affinity binding sites that might be functional in vivo because of tissue-specific cofactors.
The capacity of Drosophila Stat to activate through both 3n and 4n sites suggests that the ancestral Stat protein had the ability to bind to both sites. This capacity has been retained in Drosophila and possibly in other invertebrates. Intriguingly, this suggests that the Drosophila Stat protein dimer has some flexibility in its ability to bind to DNA sites with different spacing. The loss of this flexibility after the radiation of the vertebrate STATs might have resulted in the differential 3n compared with 4n binding preferences observed in mammalian STAT proteins. In vertebrates, the specialization of various STAT proteins for different binding sites might have been advantageous for acquiring target specificity (Seidel et al, 1995), although it is possible that vertebrate STATs can also activate targets through low binding affinity sites. In Drosophila, in which it has been shown that Upd acts as morphogen (Xi et al, 2003), maintaining Stat binding flexibility might be advantageous, as the evolution of the number of Stat-binding sites and their spacer length could provide a flexible system to modulate the distance from the source at which a given Stat target could be activated.
Methods
Constructs and directed mutagenesis. All reporter constructs were generated in phs43lacZ. To create the SB and SL reporters, the dome-MESO 2.8-kb enhancer fragment (Hombría et al, 2005) was subdivided into a distal (SB) EcoRV-BamH1 and a proximal NotI-EcoRV (SL) fragment. PCR amplification of the SL fragment with the primers TAGGAGGGGAACTGGGATGG and ATGTTTGGCCTCGAAATTGC generated 746 bp SLF2. Amplification with CGAATACGTTAGGGCGAGCC and GTACATCGGCACTTCGGACG created 138 bp SLF4. Amplified fragments were subcloned into pGEMT and from there into phs43lacZ.
The conserved Stat sites in SLF4 and SLF2 were in vitro mutagenized into TTCCGCTGTT, the third site, and AACGTCCGAA, the fourth site (Fig 1B), using QuikChange (Stratagene; www.stratagene.com) and appropriate PAGE-purified primers to create SLF2*MUT and SLF4*MUT. These sites were mutated independently in SLF2 to create SLF2MUT3rd and SLF2MUT4th. The same sites were also mutated to create various spacer variants of SLF4. In SLF4 4n3n, the fourth site was mutated to TTC.TCCGAA where (.) indicates a deleted G. In SLF4 2n3n, apart from this deletion, the third site was mutated to TTCC..TGAA where (..) indicates deleted GC.
The wild-type crb43.2 enhancer and the triple mutant Stat-binding-site spiracle enhancer have been described earlier (Lovegrove et al, 2006). The mutations in the triple mutant (- - -) were TTCCATGCC (for 3n), TTCGTTTGTT (for 5′ 4n) and TTCAGGGGTT (for 3′ 4n). In the 3n mutated construct (-4n4n), the 3n site was TTCCATGTT. In the double 4n mutant construct (3n- -), the sites were transformed to TTCGTTTGTT (5′ 4n) and TTCAGGGGTT (3′ 4n). For each construct, several independent inserts were analysed using anti-β-galactosidase. For SLF4, SLF2*MUT and SLF4*MUT, 10 inserts were studied in each case to confirm that the expression was consistent. All fly strains have been described by Hombría et al (2005). Df(1)os1A is a deletion for all three Upd ligands (Upd, Upd2 and Upd3).
Electrophoretic mobility shift assays. EMSAs to detect DNA binding of Stat92E were undertaken as described by Karsten et al (2006). Double-stranded DNA probes were generated by annealing GGAGGGTTCCGCTGAAAAT and GACATTTTCAGCGGAACCC (4n Dome3), GGACAATTCGTCCGAAGTG and GACCACTTCGGACGAATTG (4n Dome4), GGAGGGTTCCGCTGAAAATGTTTATTTTTAACAATTCGTCCGAAGTG and GACCACTTCGGACGAATTGTTAAAAATAAACATTTTCAGCGGAACCC (4n Dome3+4), GGAATTTTCATAGAATCA and GACTGATTCTATGAAAAT (3n SB1), GGAGGTTTCATGGAAATC and GACGATTTCCATGAAACC (3n SB2), GGACGATTCCGAGAACTG and GACCAGTTCTCGGAATCG (3n SB3), GGATTTTTCCCGGAAATG and GACCATTTCCGGGAAAAA (3n wild type) or GGATTTTTGCCGCAAATG and GACCATTTGCGGCAAAAA (3n MUT). The recessed ends of annealed oligonucleotide pairs were filled in using Klenow polymerase and dNTPs containing either 32P-γ-dCTP (to generate radioactive probes) or unlabelled dNTPs (to generate cold competitors). EMSAs shown in Fig 3B,F used 0.15 pmol of radiolabelled probe per lane and 50 μg ml−1 poly dI-dC, whereas supershifts included 0.3 μl α-GFP antibody (Abcam; www.abcam.com). For competition assays (Fig 3C), binding to 0.06 pmol of radiolabelled 3n wild-type probe was assayed in the presence of either 0, 0.6 or 3 pmol of unlabelled Dome3, Dome4, 3 pmol 3n MUT or 0.3, 1.5 or 3 pmol of Dome3+4 added as competitor with 83 μg ml−1 poly dI-dC.
Luciferase assays. The 2x2Draf 3n luciferase reporter is based on the p5′-663Drafwt-luc originally containing two 3n Stat-binding sites (Kwon et al, 2000) that we duplicated to generate a reporter containing four 3n sites. We also generated another version of p5′-663Drafwt-luc in vitro which was mutated to include another spacer nucleotide that we duplicated to generate a reporter containing four 4n sites (known as 2x2Draf 4n).
Transformation of the 3n to 4n sites was generated in vitro using the QuikChange method (Stratagene) and the oligonucleotides GGGGATCCTAAAATTCGTCGGAAAGTAATAAAATTCGTCGGAAAGTAAAGATCCCCCG and CGGGGGATCTTTACTTTCCGACGAATTTTATTACTTTCCGACGAATTTTAGGATCCCC (underlined bases represent the core Stat92E-binding sequence and bold indicates the added base). Both the original 3n and the newly generated 4n vectors were then multimerized by cutting out the binding sites using BamHI and XbaI, filling in and re-ligating into the parental vector cut SmaI to generate 2x2Draf 3n and 2x2Draf 4n, respectively. Constructs were sequence verified. Activity assays were undertaken in Kc167 cells as described by Müller et al (2005). Equal quantities of both 3n and 4n reporters were transfected for each experiment repeated in quadruplicate and normalized to unstimulated background activity.
Acknowledgments
We thank Stephen Brown, Fernando Casares and Sol Sotillos for comments on the paper; Manfred Frasch and Michael Taylor for stocks; Nan Hu, Kirsty Johnstone and Sabine Häder for technical assistance. J.C.-G.H. was supported by Junta de Andalucía, Ministerio de Educación y Ciencia, Consolider and the European Regional Development Fund (ERDF); M.P.Z. by the Deutsche Forschungsgemeinschaft (DFG), the Max Planck Society and The Royal Society. M.P.Z. is a Cancer Research-UK Senior Cancer Research Fellow.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N (2003) Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell 5: 441–450 [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Bach EA, Zeidler MP (2006) Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol 16: 80–88 [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N (2005) Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev 19: 1861–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binari R, Perrimon N (1994) Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev 8: 300–312 [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombría JC (2001) Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol 11: 1700–1705 [DOI] [PubMed] [Google Scholar]
- Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX (2002) mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev 16: 388–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE Jr (1997) STATs and gene regulation. Science 277: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P (2001) DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem 276: 6675–6688 [DOI] [PubMed] [Google Scholar]
- Gilbert MM, Weaver BK, Gergen JP, Reich NC (2005) A novel functional activator of the Drosophila JAK/STAT pathway, unpaired2, is revealed by an in vivo reporter of pathway activation. Mech Dev 122: 939–948 [DOI] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N (1998) Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev 12: 3252–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombría JC, Brown S (2002) The fertile field of Drosophila Jak/STAT signalling. Curr Biol 12: R569–R575 [DOI] [PubMed] [Google Scholar]
- Hombría JC, Brown S, Hader S, Zeidler MP (2005) Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol 288: 420–433 [DOI] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N (1996) Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 84: 411–419 [DOI] [PubMed] [Google Scholar]
- Karsten P, Plischke I, Perrimon N, Zeidler MP (2006) Mutational analysis reveals separable DNA binding and trans-activation of Drosophila STAT92E. Cell Signal 18: 819–829 [DOI] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW (2002) Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285: 1–24 [DOI] [PubMed] [Google Scholar]
- Kwon EJ, Park HS, Kim YS, Oh EJ, Nishida Y, Matsukage A, Yoo MA, Yamaguchi M (2000) Transcriptional regulation of the Drosophila raf proto-oncogene by Drosophila STAT during development and in immune response. J Biol Chem 275: 19824–19830 [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE Jr (2002) Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3: 651–662 [DOI] [PubMed] [Google Scholar]
- Lovegrove B, Simoes S, Rivas ML, Sotillos S, Johnson K, Knust E, Jacinto A, Hombría JC (2006) Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Curr Biol 16: 2206–2216 [DOI] [PubMed] [Google Scholar]
- Luo H, Hanratty WP, Dearolf CR (1995) An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J 14: 1412–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M (2005) Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436: 871–875 [DOI] [PubMed] [Google Scholar]
- Seidel HM, Milocco LH, Lamb P, Darnell JE Jr, Stein RB, Rosen J (1995) Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA 92: 3041–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M (1996) Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol 175: 314–324 [DOI] [PubMed] [Google Scholar]
- Wang Y, Levy DE (2006) C. elegans STAT: evolution of a regulatory switch. FASEB J 20: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Xi R, McGregor JR, Harrison DA (2003) A gradient of JAK pathway activity patterns the anterior–posterior axis of the follicular epithelium. Dev Cell 4: 167–177 [DOI] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE Jr (1996) Identification of a Stat gene that functions in Drosophila development. Cell 84: 421–430 [DOI] [PubMed] [Google Scholar]