Abstract
Inter-specific hybridization leading to abrupt speciation is a well-known, common mechanism in angiosperm evolution; only recently, however, have similar hybridization and speciation mechanisms been documented to occur frequently among the closely related group of sensu stricto Saccharomyces yeasts. The economically important lager beer yeast Saccharomyces pastorianus is such a hybrid, formed by the union of Saccharomyces cerevisiae and Saccharomyces bayanus-related yeasts; efforts to understand its complex genome, searching for both biological and brewing-related insights, have been underway since its hybrid nature was first discovered. It had been generally thought that a single hybridization event resulted in a unique S. pastorianus species, but it has been recently postulated that there have been two or more hybridization events. Here, we show that there may have been two independent origins of S. pastorianus strains, and that each independent group—defined by characteristic genome rearrangements, copy number variations, ploidy differences, and DNA sequence polymorphisms—is correlated with specific breweries and/or geographic locations. Finally, by reconstructing common ancestral genomes via array-CGH data analysis and by comparing representative DNA sequences of the S. pastorianus strains with those of many different S. cerevisiae isolates, we have determined that the most likely S. cerevisiae ancestral parent for each of the independent S. pastorianus groups was an ale yeast, with different, but closely related ale strains contributing to each group’s parentage.
Ale-type beer, which is fermented using Saccharomyces cerevisiae strains, has been brewed since ancient times, possibly as early as 6000 BC (for review, see Hornsey 2003); in contrast, lager beer, with its hallmark low-temperature fermentations (5°C–14°C), is a more recently developed alcoholic beverage, arising in Bavaria (Corran 1975) near the end of the Middle Ages. Lager beer gained worldwide popularity starting in the late 1800s, concomitant with the advent of refrigeration, which allowed the necessary cool fermentation temperatures year-round. The lager yeast, Saccharomyces pastorianus, is distinct from S. cerevisiae in both physiological and genetic characteristics and is thought to have arisen in response to selective pressures from cold brewing temperatures. This selection may have taken place during successive rounds of cold-temperature fermentations resulting from a 16th-century Bavarian law that prohibited brewing during summer months because of the inferior quality of summer-brewed beers (Hornsey 2003). As described in more detail below, S. pastorianus has been shown to be a hybrid organism, and it is likely that lager yeast arose by “instantaneous speciation” due to an interspecific hybridization event (Martini and Kurztman 1985; Martini and Martini 1987) that occurred during these selective growth conditions. Not only is this mechanism known to be common in angiosperm speciation (Hegarty and Hiscock 2005), but interspecific hybridization (or “allopolyploidy”) among the closely related sensu stricto Saccharomyces yeast species is known to occur in both industrial and natural settings (Masneuf et al. 1998; Groth et al. 1999; de Barros Lopes et al. 2002; Liti et al. 2005; Gonzalez et al. 2007, 2008; Lopandic et al. 2007), and indeed may also have been the mechanism leading to the original whole-genome duplication in the ancestor of the Saccharomyces clade (Scannell et al. 2006).
Many isolates of S. pastorianus have been collected from breweries since Hansen pioneered pure culturing of yeast in the late 1800s (Hansen 1883). The fact that this organism is an interspecific hybrid has been known for more than two decades (e.g., Nilsson-Tillgren et al. 1981; Martini and Martini 1987; for reviews, see Kodama et al. 2005; Smart 2007), and work since then has shown conclusively that different brewing yeast isolates can contain different combinations of the genomes (or partial genomes) of the following closely related Saccharomyces sensu stricto species: S. cerevisiae, Saccharomyces bayanus, Saccharomyces bayanus var. uvarum, and possibly a fourth, unknown “lager-type” species (Casaregola et al. 2001; Naumova et al. 2005; Rainieri et al. 2006; for reviews, see Hornsey 2003; Kodama et al. 2005). Pulsed-field gel/Southern blot analysis of several S. pastorianus strains has shown distinct differences in the distribution of transposable elements (Pederson 1985; Liti et al. 2005), while array-CGH studies, using S. cerevisiae-only arrays, showed limited chromosomal changes among several strains (Bond et al. 2004; Kodama et al. 2005), also indicating complexity within this group. It is of note that two of the non-cerevisiae species whose genomes are present in S. pastorianus strains, that is, S. bayanus and S. bayanus var. uvarum, are known to be more cold-tolerant than S. cerevisiae (Giudici et al. 1998; Sato et al. 2002), and thus the presence of these genomes in the hybrid S. pastorianus may have led to its observed ability to carry out fermentation better at cold temperatures than S. cerevisiae alone (Sato et al. 2002). Of the component species listed above, only the S. cerevisiae (Goffeau et al. 1996) and S. bayanus var. uvarum (Cliften et al. 2003; Kellis et al. 2003) genomes have been fully sequenced; note that the S. bayanus var. uvarum isolate was called “Saccharomyces bayanus” in the latter references; however, we refer to it here as “Saccharomyces bayanus var. uvarum.” Most of the yeasts collected from actual lager fermentations have hybrid genomes consisting of S. cerevisiae, S. bayanus, and the “lager-type” sequences, and seem to lack S. bayanus var. uvarum sequences (Rainieri et al. 2006). Here, we report array-CGH and DNA sequence analysis of 17 S. pastorianus strains that are hybrids of S. cerevisiae and S. bayanus, and describe the detailed determination of their genomic structure, with the reconstruction of both the origin(s) and the subsequent evolutionary lineages of the strains.
Results
Array-CGH analysis of multiple S. pastorianus strains shows complex genome structures
We chose 17 strains of S. pastorianus to study; these strains represent a variety of collection dates, ranging from 1883 to 1976, as well as a variety of geographical locations and beer types (Table 1); many of them correspond to the strains studied by Rainieri et al. (2006) as described above. Using custom two-species 60-mer oligonucleotide microarrays, specifically designed to detect and distinguish between sequences from both the S. cerevisiae and S. bayanus var. uvarum genomes (Supplemental Fig. 1), we performed array-based comparative genomic hybridization, or aCGH (also called microarray karyotyping), on these strains. This technique allows us to visualize copy number changes specific to either subgenome within these hybrids that may have arisen from amplifications, deletions, and nonreciprocal translocations. However, we cannot detect copy-neutral rearrangements, such as inversions or reciprocal translocations within or between the two genome moieties. Note that while the lager yeasts we studied here contain only S. cerevisiae and S. bayanus genomic moieties, the S. bayanus sequences hybridize quite well to most S. bayanus var. uvarum probes on our microarrays and negligibly to the S. cerevisiae probes.
Table 1.
Strains used in this study, and their culture collection aliases
Boldface indicates the collection from which we obtained the isolate.
a(CBS) Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; (DBVPG) Dipartimento Biologia Vegetale Perugia, Yeast Industrial Collection, Perugia, Italy; (NCYC) National Collection of Yeast Cultures, Norwich, UK.
bSuperscript numbers 1 and 2 indicate strains in Liti et al. (2005) that were placed into two groups based on repeat sequences.
Note that most strains were most likely serially propagated at breweries until the time of collection, at which point they were “evolutionarily arrested” by being kept in a frozen state; this may not be true for the oldest cultures, however, due to the lack of freezers until the mid-century.
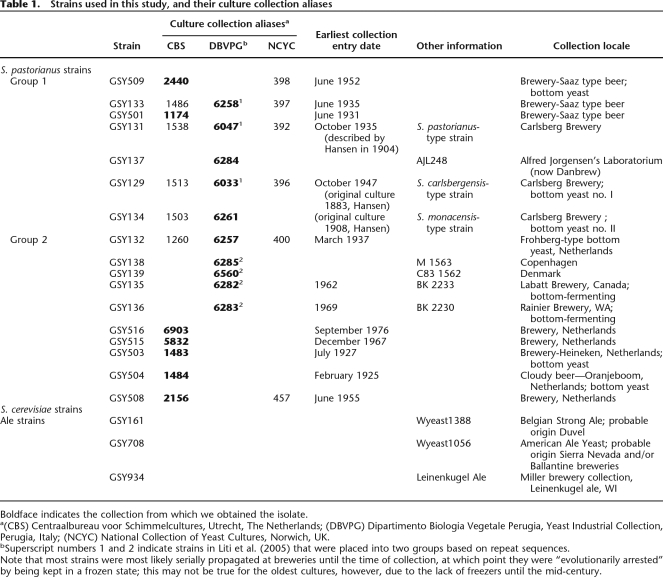
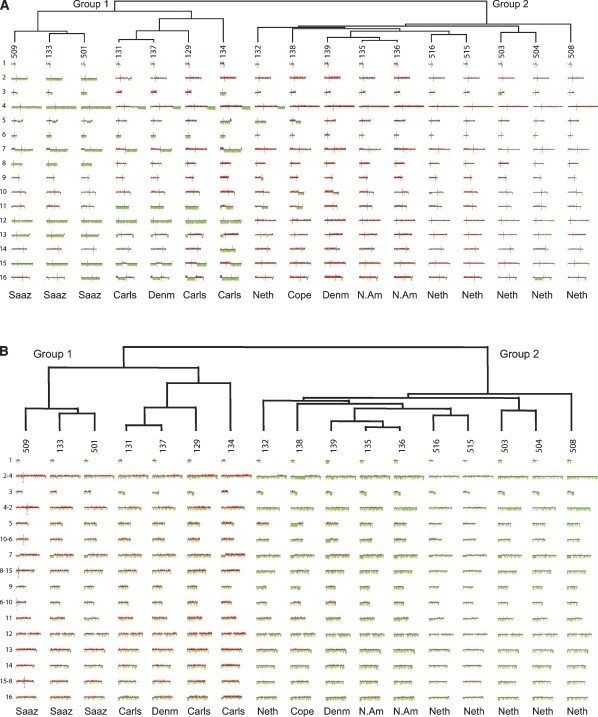
Genomic DNA from each of the lager strains was applied to the arrays, and hierarchical clustering of the data (using both the S. cerevisiae and S. bayanus genome portions) revealed that the strains fall into two obvious groups as shown in the “Caryoscope” view in Figure 1, A and B. Group 1 has lost a significant portion of the S. cerevisiae genome through whole-chromosome or segmental aneuploidy, but retains virtually all of the S. bayanus genome, while Group 2 retains nearly all genomic content of both genomes (Fig. 1A,B). In both groups, the S. bayanus genome is much more invariant than that of S. cerevisiae in terms of both breakpoints and ploidy changes.
Figure 1.
Array CGH of the S. cerevisiae and S. bayanus genome portions of 17 S. pastorianus strains. (A) Array CGH of the S. cerevisiae genomes. Array CGH data for the S. cerevisiae chromosomes of the S. pastorianus strains are shown in numerical order with chromosome I at the top and chromosome XVI at the bottom. Regions that are strongly green represent loss of that portion of the S. cerevisiae genome; regions that are strongly red represent regions of the S. cerevisiae genome that are amplified; and regions that are neither strongly green nor strongly red have a normal complement of the S. cerevisiae genomic content for that region. (B) Array CGH of the S. bayanus genomes. Array CGH data for the S. bayanus chromosomes of the examined strains are shown in a manner identical to that described in A; note that the three pairs of S. bayanus chromosomes that have experienced reciprocal translocations relative to the S. cerevisiae genome are shown as they exist in S. bayanus, that is, in their translocated form. For these six chromosomes, the translocation breakpoint is shown (for GSY509 only, but are the same for all strains) as a blue vertical line. In every case for the translocated chromosomes, the number of the chromosome is such that the chromosome to the left of the breakpoint is listed first in the name (e.g., chromosome 2-4 has the chromosome homologous to the S. cerevisiae chromosome II to the left side of the blue vertical line, and has the homologous chromosome IV to the right side of the vertical line). For the translocated chromosomes, centromeres are shown as shorter black vertical lines; for all nontranslocated chromosomes, the centromeres are in the same location as for the S. cerevisiae chromosomes in A.
It is notable that the two groups correlate with geographical/brewery groupings, with Group 1 representing both the Saaz-type beers (typically brewed in regions now in the Czech Republic) as well as Carlsberg brewery strains (Denmark), while Group 2 contains strains from the Netherlands (Heineken, Oranjeboom and other breweries) and from non-Carlsberg Danish breweries and two North American breweries. Note that it is most likely that all lager yeasts originally arose in Europe; nevertheless, transport of the yeasts to the various brewery locations would essentially have led to their “geographic isolation” for, in most of the strains we studied, a minimum of decades or possibly centuries. In addition, the two groupings correlate with two types of lager yeast identified by some brewers as Saaz type (Group 1) and Frohberg type (Group 2) (Noonan 1996). The separation of S. pastorianus into two genomically distinct groups has been previously suggested based on the analysis of transposon sequence distribution among a subset of seven of the strains we have studied (Liti et al. 2005); note that the transposon-based groupings show complete agreement with our groupings (Table 1), providing independent confirmation of our results.
Further genome differences exist between Group 1 and Group 2 strains
In Group 1, many apparent rearrangement breakpoints within the S. cerevisiae genome have resulted in changes in the ploidy of a chromosomal segment (Fig. 1A). Most changes within this group have resulted in the loss of S. cerevisiae sequences. There is a distinct subgroup within Group 1, consisting exclusively of the Saaz-type strains and distinguished by showing the greatest degree of loss of the S. cerevisiae genome. For all of the Group 1 strains, the reduced copy number (strongly green) regions in Figure 1A indicate the complete absence of that region of the S. cerevisiae genome, as we are unable to amplify fragments in those regions we tested by PCR using known S. cerevisiae-specific primers. In Group 2, very few of the S. cerevisiae chromosomes show rearrangements or other changes that result in either whole-chromosome or large segmental changes in ploidy (Fig. 1A). Breakpoints and segmental ploidy changes do exist in these Group 2 strains, but they tend to occur closer to the ends of the chromosomes than those in Group 1, and often do not result in complete loss of that portion of the S. cerevisiae genome.
Within the S. bayanus portion of the genomes, there are no whole-chromosome losses seen in either group except for the loss of chromosome III in GSY137 (Group 1) and of chromosome I in GSY139 (Group 2). Instead, relatively small telomeric segmental losses are seen in many of the strains: for example, at the left ends of chromosomes VII, IX, XIV, and XVI, and the right ends of chromosomes III, V, X–VI, and XIII. Interestingly, no amplifications of the S. bayanus portion of the genome are observed. Note that because our probes are designed using the sequenced S. bayanus var. uvarum strain, while the lager strains contain S. bayanus sequences (which are ∼10% diverged from the S. bayanus var. uvarum sequence, our observations; see below), there is less consistent hybridization to the microarrays, giving a noisier and greener “Caryoscope” than for the S. cerevisiae portion of the genome. Additionally, the probable presence of an additional S. cerevisiae genome in the Group 2 strains (see below) reduces the relative S. bayanus signal, causing even more green bias for this group. Nevertheless, these telomeric losses can be seen as areas of strong green intensity with little or no red signal within the region; for example, see the left end of chromosome VII or the right end of chromosome III in most of the strains (Fig. 1B). Interestingly, much of the variation in the S. bayanus portion does not separate the Group 1 and Group 2 designations, but instead, with the exception of GSY134, distinguishes the Saaz subgroup of Group 1 from the remaining Group 1 members (e.g., loss of the left end of chromosomes VII and XVI). Intriguingly, Group 2 is like the Saaz group with respect to chromosome VII, but like the non-Saaz group for chromosome XVI.
With regard to the mitochondrial genomes found in these hybrid organisms, our arrays contained probes only for the S. cerevisiae mitochondrial genome as the S. bayanus mitochondrial sequence was not available. However, in all 17 lager strains that we studied, we observed a lack of S. cerevisiae mitochondrial sequences (data not shown), implying, since these strains are respiration-capable, that all lager strains contain mitochondria derived from the S. bayanus ancestor. Our results agree with those of Rainieri et al. (2008), who have similarly described the absence of S. cerevisiae mitochondrial sequences and the presence of S. bayanus–like mitochondrial sequences in 22 lager yeast strains (which include 10 of the strains used in this study).
Ploidy differences between Groups 1 and 2
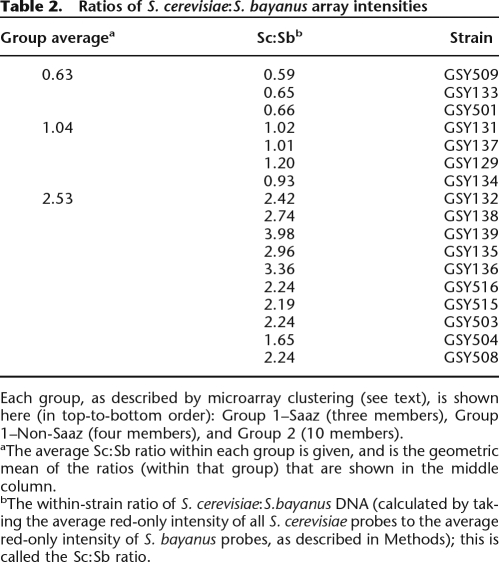
We also roughly measured the relative ploidies of the S. cerevisiae and S. bayanus genomes within the genomes of each of the 17 strains. With the same microarray data used to generate the Caryoscopes shown in Figure 1, A and B, we first separately calculated the mean (average) of the net red intensities for all S. cerevisiae probes, and likewise for all S. bayanus probes, and then calculated the ratio of the S. cerevisiae:S. bayanus mean net red intensities. This ratio represents the average amount of total S. cerevisiae DNA relative to the average total amount of S. bayanus DNA within a given hybrid genome; note that this ratio is internally controlled because only the one lager strain’s genomic DNA is labeled with the red dye, and the relative proportions of S. cerevisiae and S. bayanus genomes are being determined within this single sample. The calculated ratios showed distinct differences between Group 1 and Group 2 strains. As seen in Table 2, all Group 1 strains have ratios that are much lower than those for the Group 2 strains; we observe ratios of 0.6–0.65 (average value of 0.63) for the Group 1 Saaz subgroup and ratios of 1.0–1.2 (average 1.04) for the remaining Group 1 strains, compared to ratios ranging from 1.65 to almost 4.0 in Group 2 (average 2.53). These results indicate that in Group 2 there is an average of approximately twofold to threefold more S. cerevisiae DNA sequences relative to S. bayanus DNA within the genome, while among the non-Saaz Group 1 strains, there is roughly an equal amount of DNA sequences contributed by the two genome moieties. It appears therefore that there may be two or more relatively complete S. cerevisiae genomes present in the Group 2 strains, but roughly only one genome each of S. cerevisiae and S. bayanus in the non-Saaz Group 1 strains. Finally, in the Saaz subgroup, there are actually proportionately more S. bayanus DNA sequences relative to S. cerevisiae sequences; this seems reasonable because of the large number of S. cerevisiae chromosomes and chromosome portions that are lost from these strains as seen on the S. cerevisiae Caryoscopes (Fig. 1A).
Table 2.
Ratios of S. cerevisiae:S. bayanus array intensities
Each group, as described by microarray clustering (see text), is shown here (in top-to-bottom order): Group 1–Saaz (three members), Group 1–Non-Saaz (four members), and Group 2 (10 members).
aThe average Sc:Sb ratio within each group is given, and is the geometric mean of the ratios (within that group) that are shown in the middle column.
bThe within-strain ratio of S. cerevisiae:S.bayanus DNA (calculated by taking the average red-only intensity of all S. cerevisiae probes to the average red-only intensity of S. bayanus probes, as described in Methods); this is called the Sc:Sb ratio.
We have also performed FACS analysis on a subset of the lager strains (GSYs 131, 133–139; representing both Group 1 subgroups and Group 2) to determine absolute DNA content and have found a strict correlation that strains from Group 2 have a higher absolute DNA content than those from Group 1. Group 1 strains (from both subgroups) appear to have DNA content similar to a diploid S. cerevisiae strain that was derived from the S288C laboratory strain, while Group 2 strains appear to have triploid DNA content (data not shown). Taken with the S. cerevisiae:S. bayanus genomic ratios calculated above, we believe that Group 1 strains contain approximately one genome each of S. cerevisiae and S. bayanus (with slight differences due to aneuploidies), while Group 2 strains most likely contain two genomes of S. cerevisiae and only one genome of S. bayanus. These results show further that Group 1 and Group 2 strains differ genomically from each other beyond the differences seen in their aCGH profiles.
Rearrangement breakpoints are shared within S. pastorianus Groups 1 and 2 and often occur clustered within the genome
Upon examination of the Caryoscopes shown in Figure 1, A and B, we noticed that there often appeared to be similarities among the Group 1 and 2 strains in the regions where S. cerevisiae and S. bayanus chromosomal “breakpoints” occurred, where “breakpoint” indicates the junction between adjacent differences in segmental copy number. We examined each Caryoscope manually for such junctions as described in Methods. In this way, we obtained a list of those breakpoints that occurred in each of the Group 1 and Group 2 strains for the S. cerevisiae genome, and the combined Group 1 and 2 S. bayanus genomes (see Supplemental Table S2 for a detailed list of breakpoints).
We then visualized all of the independent breakpoints on the S. cerevisiae genome using GBrowse at SGD (Nash et al. 2007); note that for the S. bayanus chromosomes, we used the equivalent region of the S. cerevisiae genome in GBrowse. Out of a total of 107 breakpoints (90 seen in the S. cerevisiae genome, and only 17 seen in the S. bayanus genome), many appeared to occur fairly close to each other in clusters (Supplemental Fig. 2a–p). To determine whether any of these groups of breakpoints are closer together than expected by chance, we first determined that the breakpoints were not randomly dispersed across the genome, both using a simple Poisson test, and a K-means clustering (Everitt 1974) to determine statistically significant cluster groups and their locations (see Methods). We found that 10 of the chromosomes had significant clustering of breakpoints (see Supplemental Fig. 2a–p), suggesting that these breakpoints may have been driven by particular genomic features or contexts close to those clusters.
When looking at the genomic features that occur near these breakpoints, it appeared that both the solo and clustered breakpoints tend to occur near tRNAs, Ty retrotransposon elements (or their solo long terminal repeats [LTRs]), and origins of replication, or, more often, where a combination of these features occur in fairly close proximity to each other (Supplemental Fig. 2a–p), although these types of chromosomal features occur too frequently in the genome to be able to statistically prove their direct association with the breakpoints we see. Additionally, since these features are described for the S288C genome but the S. cerevisiae genomes in our S. pastorianus strains are presumably not derived from S288C, the features may not reside at the same locations in S. pastorianus. However, given the fact that breakpoints of gross chromosomal rearrangements such as translocations have been previously noted as occurring near these types of chromosomal features, especially tRNAs and Ty and LTR elements (Dunham et al. 2002), it seems likely that at least some of the breakpoints we observe may also be colocalized with (and possibly caused by) such elements.
Breakpoints and regions of amplification or deletion of either genome often include genes of possible selective significance in brewing environments
In the lager strains, we also see breakpoints occurring at other distinct genomic sites of interest, leading to either the amplification or elimination of S. cerevisiae genes possibly involved in brewing and/or general growth or mating performance. For example, on chromosome III, we see a cluster of two independent S. cerevisiae genome breakpoints (i.e., seen in Group 1 and Group 2 strains) occurring exactly at the HML silent mating locus, and even more interesting, another cluster of three independent breakpoints (i.e., seen in the S. cerevisiae and the S. bayanus genomes of both Groups 1 and 2) occurring exactly at the MAT locus; these breakpoints occur in most of the lager strains, in both Groups 1 and 2. The S. cerevisiae copy of chromosome III appears to be particularly susceptible to variable genomic rearrangements; the S. cerevisiae chromosome III distal region extending from the MAT locus breakpoint to the right end can either be lost (in two of the Saaz strains) or amplified (all Group 2 strains), or the entire chromosome may be lost (GSYs 501 and 134 of Group 1) or duplicated (GSYs 131, 137, and 129 of Group 1); in one case (GSY503), the region extending from the MAT breakpoint to the left end of the chromosome is deleted. Chro-mosomes V, VII, X, and XVI also appear to contain more genomic rearrangements than the remaining chromosomes.
To visualize better the changes in copy number of genes that occur across all strains as a result of breakpoints or whole-chromosome aneuploidies, we used the program CGH-Miner (see Supplemental Fig. 3; Supplemental Table 3) (Wang et al. 2005). Using this analysis, we have identified regions of amplification or deletion shared across most or all of the 17 lager strains, and thus presumably contain genes that are of selective advantage in brewing environments (Supplemental Fig. 3). For example, a breakpoint in most of the strains is seen immediately adjacent to the MAL3 gene cluster on chromosome II, resulting in an increased copy number of these genes, which are known to be involved in maltose utilization (see Fig. 1A; Supplemental Fig. 3); we have observed that all of these strains are able to use maltose as a sole carbon source, as would be expected for brewing strains. Several breakpoints lead to changes in flocculation (FLO) gene copy numbers: on chromosome I are a cluster of breakpoints on the left end that bracket the FLO9 gene, apparently eliminating it; on the right end of this chromosome, the FLO1 and PHO11 genes are eliminated in most strains. Additionally, the region of chromosome VIII in the S. cerevisiae genome immediately proximal to the FLO5 gene is missing in all isolates in all groups, either due to loss of the entire chromosome (Saaz subgroup) or by segmental aneuploidy adjacent to a breakpoint; this results in the absence of the S. cerevisiae copy of the FLO5 gene but apparent retention of the S. bayanus version of the gene (known as Lg-FLO1). These genes have been implicated in normal flocculation behavior of S. pastorianus and the absence of FLO5 and its replacement by Lg-FLO1 have recently been noted in S. pastorianus (Ogata et al. 2008), correlating with our observations.
Larger regions of amplification of the S. cerevisiae genome are seen in most of the strains on the left ends of chromosomes VII and XVI, and a complicated pattern of amplification and/or deletion occurs on the right arm of chromosome III as described above. Additionally, a large region of deletion is seen in most strains at the right end of chromosome XVI (Supplemental Fig. 3). These regions involve so many genes that it is difficult to determine whether any single gene is responsible to drive the copy number change of these regions, but they do often contain groups of genes involved in metabolism and transport of nitrogen and metals (Supplemental Fig. 3). Interestingly, the amplifications of the S. cerevisiae genome at the left ends of chromosomes VII and XVI are accompanied by deletions of the homologous regions of the S. bayanus genome (Fig. 1B), implying that in these two cases, there has most likely been a gene-conversion-type event between homologs, “converting” the S. bayanus distal portion of the chromosome into the S. cerevisiae portion, yielding a chimeric chromosome. However, repeated attempts to clone and characterize the junction regions of the chimeric chromosomes have been unsuccessful. We suspect that this may be due to an inability to design good primers because of the abovementioned sequence differences between the sequenced strain S. bayanus var. uvarum and the S. bayanus genome in the lager strain.
Reconstruction of last common ancestor genome structures for S. pastorianus Groups 1 and 2
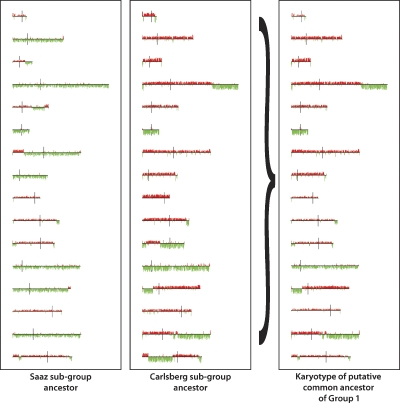
Using the microarray karyotypes shown in Figure 1A, we were able to infer an ancestral karyotypic lineage of the S. cerevisiae genomic portion of the S. pastorianus strains, by sequentially generating an ancestor for each node in the tree as described in Methods. As shown in Figure 2, the reconstructed last common ancestor (LCA) of Group 1 is inferred to have been missing two S. cerevisiae-derived chromosomes, as well as having several additional segmental aneuploidies, while it is apparent that the Group 2 LCA would have had two copies (based on above ploidy evidence) of an essentially complete S. cerevisiae genome, given that for each chromosome, at least one member of this group has a complete copy of that chromosome with no segmental ploidy change, except for the loss of a region near the right end of chromosome XVI that is seen in all Group 2 strains (Fig. 1A).
Figure 2.
Reconstruction of the ancestral karyotypic lineages of the S. cerevisiae portion of the S. pastorianus genome. By examining the karyotypes of the two strains that share the furthest divergence point within Group 1, and assuming that S. cerevisiae DNA sequences can only be lost, not gained de novo, each S. cerevisiae chromosome of the putative shared ancestor can be reconstructed as the merging of all S. cerevisiae segments present in both strains, using the iterative process described in Methods. This figure shows the reconstructed ancestors for the two subgroups within the Group 1 S. pastorianus strains on the left, and the final reconstructed ancestor for the entire Group 1 on the right. Applying this logic to Group 2, the reconstructed ancestor for Group 2 is an organism with a complete S. cerevisiae genome (data not shown).
DNA sequencing suggests separate origins of S. pastorianus groups
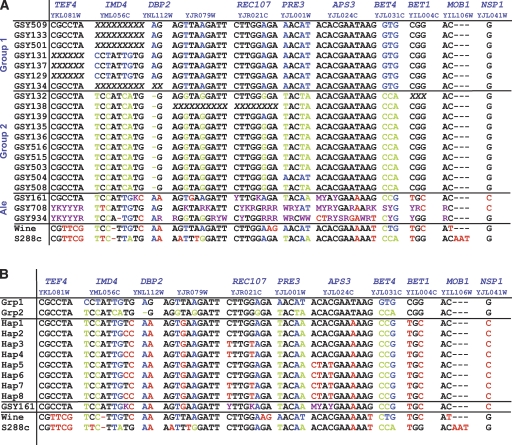
The clear separation of two groups based on our array-CGH data suggests that they derived from distinct and separate hybrid ancestors rather than a single common hybrid ancestor. However, the aCGH data alone are not enough to establish this as fact; we therefore performed DNA sequencing to help make this determination. We sequenced the intronic and adjacent regions of 11 intron-containing genes (representing five different chromosomes); some of these loci have been previously used to establish phylogenetic relationships among wild Saccharomyces paradoxus strains (Kuehne et al. 2007). We performed PCR-amplification from genomic DNA, using S. cerevisiae-specific primers, which yielded fragments for sequencing from only the S. cerevisiae regions of the hybrid S. pastorianus genomes. Alignment of the resulting sequences (Fig. 3A) showed that 14 nucleotides out of the combined 4710 bp (0.3%) differ between the two main groups of S. pastorianus strains; these changes correlate almost exactly with the groupings that were derived by clustering of the microarray karyotyping data (Fig. 1A,B). Only two exceptions are seen, both within Group 2: GSY139 has one polymorphic nucleotide (in REC107) in common with the Group 1 strains, but all its other nucleotides are identical to those of Group 2, while GSY504 has the three polymorphisms of PRE3 that are associated with Group 1 strains, but all other nucleotides are identical to those of Group 2. Note that despite the fact that we sequenced PCR products in these experiments, we did not observe any heterozygous nucleotides for any of our strains, implying that if multiple genomes of S. cerevisiae are present within some of the strains, they are almost certainly homozygous copies.
Figure 3.
(A) Sequence differences between the Group 1 and Group 2 S. pastorianus strains (S. cerevisiae genomic portion only). Only nucleotides showing differences among the sequenced strains are shown. The five different gene regions sequenced are shown along the top; the primers used for both PCR and sequencing are given in Supplemental Table S1. Nucleotides in blue and green are those that distinguish Group 1 from Group 2 strains; nucleotides in red are those that are not shared with either Group 1 or Group 2 strains, and nucleotides in purple are those that are heterozygous in the ale strains, as determined from manual inspection of the sequence traces. Where a gene is not present in the genome because of deletion of that region of the S. cerevisiae genomic portion, the sequences are shown as a series of “X”s. Note that all of the heterozygous nucleotides or regions seen in the ale strains can yield (in at least one configuration) nucleotides consistent with the corresponding residues in the lager strains (either Group 1, Group 2, or both). (B) Sequences of the ale haplotypes inferred from spores of GYS161. As described, the ale strain GSY161 was sporulated and seven spores were sequenced across the 11 loci shown in the figure. For each unlinked locus with heterozygosities in the parent strain, unambiguous haplotypes were derived. Because there are three heterozygous loci, eight haplotypes can be inferred; all eight are shown in this figure with the one most similar to the Group 1 and Group 2 lager strains shown at the top of the list (“Hap1”).
We additionally performed preliminary sequencing on the S. bayanus portion of the genome, using a subset of the lager strains (again including representatives from all groupings: GSYs 129, 131–135, 137–139, 503, 504). Because of the divergence between the sequenced S. bayanus var. uvarum genome and the S. bayanus-derived genome in the lager strains, we were able to design successful PCR primers for only one of the genes used in our S. cerevisiae sequencing study, YRA1. Of the ∼700 bp of sequence obtained, 9% (63 bp) were divergent between the sequenced S. bayanus var. uvarum genome and the lager S. bayanus genome. Within just the lager strains, however, only one difference was seen, but strikingly, this single nucleotide difference segregated strictly with Group 1 versus Group 2 strains (data not shown), showing that although the two groups share 99.9% (699/700 bp) identical S. bayanus genomes, there may have been separate origins of the S. bayanus genome between the two groups as well.
To shed light on what type of S. cerevisiae yeast may have been the ancestral S. cerevisiae parent of S. pastorianus strains, we additionally sequenced the same gene regions from three different ale strains (which are diploid), using the rationale that an ale strain, being present in the beers of middle Europe at the inferred time of S. pastorianus’s origin, might be a likely candidate (Fig. 3A). These three ale strains were obtained from geographically different sources (Table 1) and therefore represent some amount of diversity; note, however, that it is believed that all ale and lager strains originally arose in Europe. Our sequence trace data revealed striking heterozygosity in the ale strains, with two of the strains, GSY708 and GSY934, being particularly heterozygous across most of the loci sequenced. Examination of the ale strain sequences showed that in all cases of heterozygous nucleotides, at least one of the two possible nucleotides is identical to the corresponding nucleotide in the lager strains (either Group 1, Group 2, or both). As a group, the ale strains show the least number of nucleotides that are incompatible with the lager strain sequences (i.e., nucleotides shown in red in Fig. 3A) when compared to the wine strain RM11 and the laboratory strain S288C. However, with the amount of heterozygosity seen in the ale strains, we wished to determine haplotype information to ascertain the actual level of similarity of the ale strains to the lager strains. We therefore sporulated the ale strains; only GSY161 gave enough tetrads with four viable spores to allow us to determine its component haplotypes. Note that in this strain, six heterozygous nucleotides were seen (Fig. 3A): one nucleotide within the IMD4 locus, two nucleotides within the REC107 locus, and three nucleotides within the APS3 locus (all three loci are located on different chromosome arms and therefore unlinked). We sequenced these meiotic segregants and found the haplotypes shown in Figure 3B. Indeed, one of the haplotypes (Fig. 3B, “Hap1”) contains sequences identical to the Group 1 sequences for all six heterozygous loci. This result shows that there are segregants of GSY161 that, aside from the seven “pre-existing” non-heterozygous divergent nucleotides (shown in red for “Hap1” in Fig. 3B), are identical in sequence to the Group 1 and/or Group 2 strains, with an overall nucleotide variation of <0.15% (7/4710 bp).
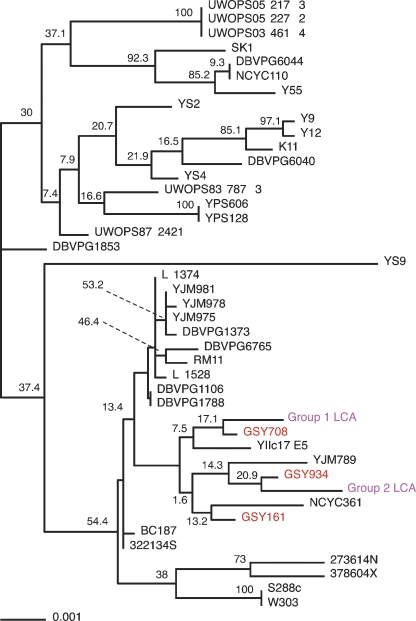
To further determine whether ale strains in particular are more similar to the S. cerevisiae genome portion of S. pastorianus strains than are other types of S. cerevisiae strains, we also needed sequence data for these loci from a set of diverse S. cerevisiae strains. Thus, we retrieved data for the 37 strains of S. cerevisiae that have recently been sequenced (between 1× and 3× coverage) and publicly released by the Sanger Center (The Saccharomyces Genome Resequencing Project). These strains were originally collected from wine, sake, other fermented beverages, fruits, oak tree exudates, nectar, bakeries, clinical isolates, soil, and so on; however, none were ale strains. Using the sequence data that we had generated from the Group 1 and Group 2 lager strains and from our sequenced ale strains, in conjunction with sequence data from the Sanger Center strain set, as well as that of the S. cerevisiae laboratory strain S288C (Nash et al. 2007) and the S. cerevisiae wine strain RM11, we constructed a phylogenetic tree (Fig. 4). Strikingly, among all of the S. cerevisiae sequences examined, the inferred sequences of the LCAs of the two S. pastorianus groups are both found closest in the tree to ale strains.
Figure 4.
Phylogenetic tree, estimated using parsimony, of sequenced loci across a large number of S. cerevisiae strains. Sequence data from the deduced last common ancestors of Groups 1 and 2 (magenta) were aligned with sequence data from three ale strains (red) and 37 other sequenced S. cerevisiae strains. Bootstrap values are shown. An unrooted version of this tree is available in Supplemental Figure 4.
Discussion
Using two-species array-CGH with 17 different S. pastorianus lager yeast strains, we have shown that these strains have undergone multiple gross chromosomal rearrangements, mostly segmental and whole-chromosome aneuploidies, after the presumed original interspecific hybridization event that gave rise to this hybrid organism. The chimeric nature of the genome of S. pastorianus has been known for quite some time (e.g., Nilsson-Tillgren et al. 1981; Martini and Martini 1987; for reviews, see Kodama et al. 2005; Smart 2007), as has the existence of rearrangements and aneuploidies within the hybrid genome. However, previous work on such rearrangements and aneuploidies was performed only for single genes or chromosomes (e.g., Nillson-Tillgren et al. 1981; Holmberg 1982; Pederson 1985; Casey 1986; Petersen et al. 1987; Kielland-Brandt et al. 1995; Tamai et al. 1998), for multiple small regions across the genome (Rainieri et al. 2006), for only one of the two coexisting genomes (Bond et al. 2004), or for just one strain (Kodama et al. 2005).
Comparison with previous genomic studies of lager yeast
We compared our two-species aCGH results for the three lager “type strains” (Saccharomyces carlsbergensis CBS1513 [GSY129], Saccharomyces monacensis CBS1503 [GSY134], and S. pastorianus CBS1538 [GSY131]) (see Table 1) with previously reported genome composition studies that used these same strains. Most of these previous studies used single-locus techniques such as Southern blots (Yamagishi and Ogata 1999), or PCR-RFLP and single-locus DNA sequencing (Casaregola et al. 2001; Rainieri et al. 2006); however, limited S. cerevisiae-only aCGH results have also been reported (with data shown for chromosome XVI only) for these same three strains (Kodama et al. 2005). For strains CBS1503 and CBS1513, our results match almost perfectly with all of the above published results in terms of which chromosomal regions are represented within the genome by S. cerevisiae and/or S. bayanus sequences; that is, the whole-chromosome aneuploidies as well as the breakpoint locations (and associated regional copy number changes) for the segmental aneuploidies that we see are consistent with the data presented in the above papers. However, for strain CBS1538 (GSY131), the results from both Rainieri et al. (2006) and Kodama et al. (2005) do not agree with our findings. Our results show that this strain has lost three complete S. cerevisiae chromosomes (III, XI, and XII) from the hybrid genome, whereas both Rainieri et al. (2006) and Kodama et al. (2005) indicate that CBS1538 has lost eight complete S. cerevisiae chromosomes (the three we find missing plus II, IV, VIII, XV, and XVI); their findings are instead consistent with the hybridization pattern that we see for CBS1174 (GSY501) (see Fig. 1A). It is possible that because we obtained the CBS1538 strain from a different source from these authors, there has been either a change in the strain’s genome during propagation in the different culture collections, or, more likely, there may have been contamination or mislabeling of the cultures; this is a point that should be investigated further. Note that for all other strains that we studied in common with those used in Rainieri et al. (2006), our results on the presence/absence of S. cerevisiae versus S. bayanus portions are consistent with their data.
With regard to currently used production lager strains, aCGH studies have been reported for the commonly used Weihenstephan Nr. 34 (34/70) strain (Kodama et al. 2005), using two-species microarrays, and on strains CMBS-33 (Belgium) and 6701 (Ireland) (Bond et al. 2004), using S. cerevisiae-only microarrays. The set of lager strains we investigated in this study did not include these strains, nor any currently used production lager strains, with the most recent collection date being 1976 (Table 1). However, the aCGH results reported for strains Weihenstephan 34/70 and 6701 indicate that they are virtually identical in the S. cerevisiae genome structure to the two very similar strains GSY508 (CBS2156) and GSY515 (CBS5832) that we did investigate (Fig. 1A). In fact, for the Weihenstephan 34/70 strain, on which two-species aCGH was performed by Kodama et al. (2005), the same breakpoints and copy numbers of the S. bayanus genome portion are seen in GSY508 and GSY515 (Fig. 1B). The CMBS-33 strain is also similar to GSY508 and GSY515 except that it has a novel breakpoint on S. cerevisiae chromosome VIII that we do not see among any of the strains we investigated. These results indicate that at least some, and possibly all, current production lager strains belong to the Group 2 type lager yeast class. Note that both GSY515 and 508 are among the strains that have experienced the least loss of S. cerevisiae sequences from the hybrid genome of all the 17 strains we have investigated, which may be of importance in obtaining successful lager fermentations under current brewery practices.
Allopolyploidy as a window onto genome duplication and subsequent gene loss
In addition to their industrial importance, lager yeasts provide a unique view of the types of genomic changes that can occur after a relatively recent allopolyploidy event. Our work here, using a large number of S. pastorianus isolates to survey the global architecture of both genomes that coexist in these hybrids, shows a surprising amount of variation among these strains, despite their recent origins and the fact that they have all undergone selection for similar traits (i.e., adequate growth, as well as the production of desirable flavor profiles, during lager beer fermentations). These allopolyploid strains probably experienced their respective interspecific hybridization events on the order of mere hundreds of years ago—corresponding to only a few thousand generations, assuming 50 generations/yr for lager brewing conditions; thus, we are most likely observing among the very earliest genome rearrangements that occur after an interspecific hybridization event. It is possible that we are observing the early stages of genome loss and shuffling after a secondary whole-genome duplication event (i.e., between species that have already undergone genome duplication), and that the loss of much of the S. cerevisiae genome in the Group 1 strains could be analogous to the proposed rapid and precipitous gene loss that occurred in the ancestor of the Saccharomyces clade immediately after it experienced its (presumably) first whole-genome duplication (Byrnes et al. 2006; Scannell et al. 2006).
Note, however, that Scannell et al. (2006) and Byrnes et al. (2006) predict that the post-duplication gene loss would be passive and involve single genes scattered individually across the genome, whereas we see mostly losses of large contiguous regions. It is possible that what we observe here is the earliest mechanism of gene loss, which would eventually be followed by the type of scattered gene loss described by these investigators, resulting in something similar to the “interleaved” gene loss pattern of the current S. cerevisiae genome. Similarly, a recent allopolyploid event among two Zygosaccharomyces species has been characterized (Gordon and Wolfe 2008); the investigators show that there are no losses of regions (large or small) of either subgenome and conclude that this hybridization event may have happened so recently that genome rearrangement and shuffling has not had time to occur. Our hybrids may thus represent a middle point in the disintegration of a hybrid genome into what may eventually be construed as a normal diploid genome. Further sequencing, especially whole-genome, of representative lager strains from each group would be of great benefit in shedding light on the status of gene duplications and gene loss in these “early” allopolyploids.
Origin(s) of lager yeast strains
As shown in the Results section, hierarchical clustering of the genome rearrangements detected by our aCGH studies revealed two distinct groups of S. pastorianus strains based on their shared sets of rearrangements. DNA sequencing of the strains further shows that DNA differences segregate almost perfectly with the two aCGH-based groupings. There are two possible interpretations of these sequence data. Either the S. pastorianus strains that we examined had two independent origins, one giving rise to the Group 1 strains and the other to the Group 2 strains, or there was a single origin, followed by an evolutionary bottleneck that resulted in Group 1 and Group 2 strains. We view the first hypothesis, that of multiple origins, more likely, as the second requires that following the initial hybridization event there was a period of sequence divergence, then, after the hypothetical bottleneck, strains in the different groups would have remained largely unchanged as evidenced by the fact that the single nucleotide polymorphisms (SNPs) (with two exceptions mentioned above) are uniformly shared among all members within a group. In addition, the different distribution of transposons within each of the two groups (Liti et al. 2005) makes it more likely that there was more than one independent origin of the Group 1 and Group 2 strains. Finally, we have shown evidence, based on aCGH data and on genomic DNA content studies, that there are differences in the number of S. cerevisiae genomes present in the two groups, with Group 1 containing one (or less, due to aneuploidies) S. cerevisiae genome and Group 2 containing two genomes; while both groups apparently contain one S. bayanus genome. Taking all this evidence together, the most parsimonious explanation is that there were separate origins for Group 1 and Group 2, albeit with closely related strains serving as the S. cerevisiae parent in each case: there is just 0.3% nucleotide variation (14 SNPs in 4710 bp) between S. cerevisiae portions of the two lager groups.
Our sequencing results also suggest that the S. cerevisiae parent, for both Group 1 and Group 2, may have been an ale strain; this would not be surprising as the progenitor hybrid lager yeasts almost certainly arose in a brewery where ale was produced, making it likely that the S. cerevisiae parent would be an ale strain. These data do have to be interpreted cautiously, though, given the relatively low bootstrap values for the phylogenetic tree; note, however, that despite the low bootstrap values within the branch containing the ale and lager strains, this particular group of strains remained together (with different sub-branch structures) in all trees generated during the bootstrap process. The other strains besides ale strains that are retained within this group are YIIc17 E5 (a wine yeast), YJM789 (a clinical isolate of yeast), and NCYC361 (a beer spoilage yeast); the significance of their inclusion in this group is unclear, although the inclusion of the latter yeast, isolated from spoiled beer, is intriguing. Clearly, significant additional sequence data will be required to unequivocally establish the provenance of the S. cerevisiae moiety of the lager genome. Our data, however, are consistent with the data of Legras et al. (2007), who investigated 651 different S. cerevisiae strains from a wide variety of sources using allelic variation at 12 microsatellite loci and showed that the S. cerevisiae portion of 15 different lager yeasts clustered most strongly with the eight ale strains they investigated. Together, these data suggest that the hypothesis that lager strains arose from ale yeast hybrids is likely correct.
Thus, it appears that at least two events occurred to lead to the current set of lager strains that we studied. We propose that the ancestor of the Group 1 strains arose by the fusion of a haploid ale S. cerevisiae yeast spore with a haploid S. bayanus spore to produce the Group 1 ancestral allo-diploid S. pastorianus hybrid; this was followed by the rapid loss of large portions of the S. cerevisiae genome (whole chromosomes VI and XII and segmental losses on chromosomes IV, XIII, and XV) (see Fig. 2) to give the immediate shared common ancestor of Group 1 strains. This loss of large amounts of the S. cerevisiae genome may have been precipitated by a strong selective event, such as very cold fermentation temperatures, leaving intact the genome of the cryo-tolerant S. bayanus. Subsequent losses of large portions of the S. cerevisiae genome continued to occur, especially in the ancestor of the Saaz subgroup, leading to the current strains we observe in Group 1. For Group 2 strains, the ancestor appears to have arisen as the fusion of a completely homozygous diploid S. cerevisiae cell with a haploid S. bayanus cell; perhaps, due to loss of one of its MAT loci, it had become a “2N haploid” (e.g., as proposed by Scannell and Wolfe 2008) and was thus able to mate to a haploid S. bayanus spore to create an allo-triploid hybrid. The S. cerevisiae and S. bayanus parents in this case would have been distinct from the parents of the Group 1 ancestor, but extremely closely related, showing only 0.3% and 0.1% sequence divergence, respectively. There was no immediate large-scale loss of either genome, unlike that seen in Group 1; instead, entire versions of each S. cerevisiae chromosome can still be seen in at least one of the members of the extant Group 2. Subsequent rearrangements were much less severe than in Group 1, although compared to the S. bayanus genome, the S. cerevisiae genome has undergone more loss (of either one or both copies of large segments of a chromosome, or more rarely of an entire chromosome). It is not known why the genomes behave somewhat differently in the two groups; perhaps the presence of an extra S. cerevisiae genome buffers against loss, or perhaps the earliest selection regime was very different between the two groups. Again, cold temperatures may have driven the retention of the S. bayanus genome and loss of portions of the S. cerevisiae genome in Group 2; flocculation and other brewing-related selections undoubtedly have also played a role in both groups. The few deletions and amplifications in the S. cerevisiae genome that are shared across the great majority of the strains do appear to mostly involve flocculation and sugar metabolism and transport, as well as nitrogen and metal metabolism and transport; all of these genes presumably play important roles during fermentation of the wort substrate.
In conclusion, we have shown that similarities in gross chromosomal rearrangements in hybrid organisms alone can be used to infer similar phylogeny, with our aCGH grouping correlating very well with the strain grouping we obtained using DNA sequence data, the more traditionally used method for determining relatedness among strains. Furthermore, we provide evidence that interspecific hybrid formation among the Saccharomyces sensu stricto group has occurred in breweries multiple times, and that the S. cerevisiae parents of the hybrid lager yeasts were likely to be ale yeasts. Additionally, the surprising amount of variation in genomic architecture that we see among these allopolyploid hybrid yeasts may be indicative of the early stages of genome shuffling that is thought to occur after a whole-genome duplication event.
Methods
Strains used
Table 1 shows a list of the S. pastorianus yeast strains used in this study. In addition to these strains, two additional strains were used both in the designing of the two-species microarrays and as reference DNA in the microarray hybridizations: the S288C S. cerevisiae strain, one of the most widely used laboratory “wild-type” yeast strains, has been previously described (Mortimer and Johnston 1986) and is also the strain background for the sequenced S. cerevisiae strain, while strain CBS7001 is the sequenced strain of S. bayanus var. uvarum (Cliften et al. 2003; Kellis et al. 2003).
Multispecies microarray design
Microarrays to simultaneously detect DNA copy number for both S. cerevisiae and S. bayanus var. uvarum were designed using the ArrayOligoSelector (Bozdech et al. 2003). Briefly, both the S. cerevisiae genome and the S. bayanus var. uvarum contigs (obtained from Paul Cliften and Mark Johnston, Washington University, St. Louis) were split up into 2-kb fragments, with each fragment overlapping the previous one by 1 kb. In addition, nine control sequences, described in van de Peppel et al. (2003), were also split into similar fragments. ArrayOligoSelector (Bozdech et al. 2003) was then used to find two 60-mer oligonucleotides for each fragment, using a combined file of the S. cerevisiae genome, the S. bayanus var. uvarum contigs, and the control sequences as a mask file, such that chosen oligonucleotides would not have significant cross-hybridization potential. As a subsequent check, all selected oligonucleotides were then compared to this combined file, using BLAST (Altschul et al. 1990), and any oligonucleotides showing significant similarity to regions of either genome other than that for which they were chosen to represent were discarded. From the remaining oligonucleotides, the ones with the most negative ΔG were chosen preferentially, with the aim of having ∼5000 oligonucleotides each for S. cerevisiae and S. bayanus var. uvarum, as evenly spaced as possible. The final array design included 4961 S. bayanus var. uvarum oligonucleotides, 5122 S. cerevisiae oligonucleotides, and two oligonucleotides for each control sequence, each of which were replicated 10 times. After testing of this array, the 491 worst-performing S. bayanus var. uvarum oligonucleotides and the worst-performing 617 S. cerevisiae ones were replaced. Custom microarrays containing the final oligonucleotide set were manufactured by Agilent Technologies.
Array-CGH protocol
Array Comparative Genomic Hybridization (“aCGH,” also called “microarray karyotyping”) was performed with the S. cerevisiae and S. bayanus var. uvarum two-species microarrays described above. For the two reference strains, S. cerevisiae S288C and S. bayanus var. uvarum CBS7001, genomic DNA was isolated as described (Treco 2003) and then sonicated to an average size of 500–1000 bp. For the S. pastorianus strains, genomic DNA was prepared with YeaStar columns (Zymo Research) and then cut with HaeIII (New England Biolabs). Note that we saw no significant changes in aCGH results when we performed comparative experiments using our reference DNAs prepared and cut in either of the ways described above. The reference DNA in all experiments was prepared by mixing equivalent genomes-worth amounts of genomic DNA prepared from the two strains. We used these strains for the reference DNAs because they were the sequenced strains from which the oligos used on the microarrays were generated; Cy3-labeled reference DNA should thus give a green signal in every channel. After isolation and cutting, 350–400 ng of DNA per sample was directly labeled with fluorescently tagged nucleotides (Perkin-Elmer), usually Cy3-dCTP for the reference strain mix and Cy5-dCTP for the S. pastorianus strains, using the BioPrime random-prime labeling system (Invitrogen); dye swap experiments were performed for several of the S. pastorianus strains, and no significant differences in results were seen. After labeling, the reactions were heat-inactivated, the Cy5- and Cy3-labeled DNAs were mixed and purified away from unincorporated label using Zymo Clean&Concentrate columns (Zymo Research), and then hybridized to the microarrays according to the aCGH protocol given by Agilent Technologies. Arrays were scanned with the Agilent microarray scanner, and the data were analyzed with Agilent’s “Feature Extraction” software. All raw and normalized microarray data for this paper are available from the Stanford Microarray Database (Demeter et al. 2007), where they can be retrieved for further analysis. In addition, they have also been deposited in GEO (Edgar and Barrett 2006), under accession number GSE12177. For all data analysis described, the data were filtered by only excluding automatically flagged spots or spots for which the green channel’s net intensity was not “positive and significant” according to the Agilent software; this filtering was fairly permissive in order to allow truly deleted genes (i.e., no red signal at all) to be detected.
Mapping S. bayanus var. uvarum contigs onto a whole-chromosome scaffold
We obtained a set of sequencing contigs of S. bayanus var. uvarum (strain CBS7001) from Paul Cliften and Mark Johnston and mapped them to the S. cerevisiae genome in order to create Caryoscope images for visualization of our two-species microarray data (see below). Using the chromosomal feature coordinates downloaded from SGD (Nash et al. 2007), we first aligned the contigs with their counterpart regions of the S. cerevisiae chromosomes, after which we were able to infer the chromosomal distances separating each adjacent contig. The contigs were then assembled into whole chromosomes, and each of our S. bayanus var. uvarum microarray probes was assigned a chromosomal position. Note that we assembled the S. bayanus contigs to reflect the actual configuration of the three chromosomes known in S. bayanus to have undergone reciprocal translocations relative to their counterpart S. cerevisiae chromosomes (chromosomes II and IV, chromosomes VI and X, and chromosomes VIII and XV) (Cliften et al. 2003; Kellis et al. 2003).
Creating “Caryoscope” images from microarray data
Microarray CGH data were visualized by generating “Caryoscope diagrams” using the Caryoscope program (Awad et al. 2004). To make a Caryoscope diagram, the log10 of the red/green ratio for each array spot is mapped onto its corresponding chromosomal position for the relevant reference strain (i.e., either S. cerevisiae or S. bayanus var. uvarum). The ratio is depicted as a red or green vertical bar the height of which is proportional to the log10 ratio; if positive (higher red than green signal), the bar is red and protrudes up above the baseline; if negative, the bar is green and protrudes below the baseline. Thus, for the two-species microarrays, the Caryoscope is composed of two separate images, one showing all 16 chromosomes of S. cerevisiae with red or green bars showing regions either amplified or depleted, respectively, with respect to the reference S288C of S. cerevisiae strain. The other image is similar, but represents the amplified or depleted regions relative to the S. bayanus var. uvarum strain. Regions with little or no change in copy number have a log ratio close to 0, and their bars are thus very short and do not protrude much in either direction from the baseline.
Microarray data clustering
To determine the relatedness among strains, microarray data were clustered in SMD (Demeter et al. 2007), which uses XCluster, using the Pearson correlation as the metric of similarity.
Relative proportion of S. cerevisiae:S. bayanus genomes
After being filtered and processed by Agilent “Feature Extraction” software as described above, the microarray data for the 17 lager strains were used to determine the relative intrastrain genome proportions of S. cerevisiae to S. bayanus. The red channel readings (the background-subtracted intensities only, not the red:green ratios) were divided into two groups, either S. cerevisiae probes or S. bayanus probes, and an arithmetic mean was taken for each group. The ratio of the S. cerevisiae red intensity mean to the S. bayanus red intensity mean (“Sc:Sb” ratio) was calculated and is shown in Table 2 for each lager strain. Additionally, for each lager Group (i.e., the Saaz and non-Saaz subgroups of Group 1 and Group 2), an average ratio (calculated as the geometric mean of the ratios for each member of the group) was determined. Since the red intensities for each lager strain must be proportional to the relative amounts of each genome within that strain, this “Sc:Sb” ratio represents a completely internally controlled measurement of the relative proportions of each component genome for a given strain. Note that there may be a slight bias for higher average red intensities for the S. cerevisiae probes (owing to the known lesser similarity of the S. bayanus portion of the lager genome to the S. bayanus var. uvarum probes on the microarray), but the same bias will exist for all the strains.
Reconstruction of the last common ancestor of the S. cerevisiae portion of the S. pastorianus genome
By iteratively examining the karyotypes of the strains within Group 1, from the leaves of the tree toward the root, we generated ancestral karyotypes for each node in the tree, combining karyotypes for strains or nodes until we reached the node representing Group 1. We assumed that S. cerevisiae DNA sequences can only be lost, not gained de novo. Where there was an absence of segment(s) in one strain/group and presence in the other, we took the log ratio values for the region that was present; when there was presence of segments in both groups but with differing values (e.g., apparently one copy of the segment in one group and multiple copies in the other group), we took the values for the region that appeared to be equivalent to one copy, as this would presumably represent the ancestral state. This process was iterated using the reconstructed ancestor and the next most closely related strain, and so on.
Analysis of rearrangement breakpoint positioning
Rearrangement breakpoints were determined by manual examination of the Caryoscope images, selecting the two probes that span the breakpoint and taking the midpoint location between the two probes as the breakpoint location. Because our probes are located 2 kb apart on average, our detection resolution is ∼1 kb. Based on the aCGH, ploidy, and DNA sequence evidence presented in this study, we deemed that the S. cerevisiae genomes of Groups 1 and 2 were independently derived and thus any rearrangement breakpoints in these two strains, even if occurring at the same point (within our detection resolution), are independently derived. However, we do not have DNA sequence evidence that the same is true for the S. bayanus portions of the Group 1 and 2 strains, so we have conservatively kept them together in our breakpoint analysis. Breakpoints with different chromosomal coordinates (i.e., differing flanking probes) are always considered to be independently derived.
To determine whether the rearrangement breakpoints we observed were nonrandomly positioned across the genome, we first concatenated the entire genome and mapped all the breakpoints onto it. We then used the R statistics package’s (http://www.r-project.org/) randpois function to generate random Poisson-distributed “breakpoints” (repeating 1000 times) for varying numbers of subsegments (n) of the whole genome, and compared the output to the random distribution of the actual breakpoints, calculating variance and P-values. If the actual breakpoints were distributed significantly differently from the randomly generated ones (i.e., if the P-value was <0.05), they were considered to be statistically nonrandom in their distribution. In fact, at values of “n” subsegments above 30 (up to at least 100), the actual breakpoint distribution was highly significantly different from a random Poisson distribution, and the actual breakpoints were thus deemed to be occurring in some number of “clusters.”
To determine where the actual clusters occurred, we performed K-means clustering for each chromosome, using the K-means function in the R statistics package (the script used is available in the Supplemental material). For each chromosome, we used values of k from 2 to (Kmax − 1), where Kmax is the total number of breakpoints we observed on the chromosome, and for each value of k, we ran K-means 10 times. We then calculated the aggregate sum of the “within-sum-of-squares” (“withinss”) values for each of the 10 different runs for each K-value (number of clusters). Because the K-means test does not allow duplicate values for the breakpoint locations, in those cases where we observed independent breakpoints at the same location, we assigned breakpoint values differing by 1 kb. We then took the minimum aggregate sum found for each value of k as the best grouping for that k. To determine whether these clusters were significant, we performed the same procedure on randomly permuted breakpoint data to generate a null distribution, similar to the manner used in the Gap statistic (Tibshirani et al. 2001). We determined how many times the random data gave a minimum aggregate sum of the “within-sum-of-squares” less than that generated from the real data, which was used to calculate a P-value. For a given chromosome, thus, we obtained (Kmax − 2) P-values, one for each number of clusters from K = 2 to K = Kmax − 1. Those chromosomes that had no significant P-values (i.e., <0.05) for any K were deemed to have no significant clustering of breakpoints. For chromosomes with one significant P-value, the K-value associated with that P-value was taken as the number of statistically significant clusters of breakpoints on that chromosome. For chromosomes with more than one significant P-value, the K-value associated with the lowest non-zero P-value was taken as the number of statistically significant clusters of breakpoints on that chromosome. (Note that P-values of exactly zero were found to arise artificially when that particular K-value forced the splitting of identically located breakpoints into different clusters.)
CGH-Miner
The CGH-Miner program (Wang et al. 2005) was installed and run as described in the CGH-Miner User guide and Manual. The plot shown in Supplemental Figure 3 was generated by the program using the parameters for BAC analysis, but changing the moving window size to one in order to see significant copy number changes at single probes. Three separate “self–self” hybridizations—that is, where equimolar amounts of S. cerevisiae and S. bayanus genomic DNA were mixed together and labeled with either Cy3-dCTP (green) or Cy5-dCTP (red) as described above, then the two differentially labeled reactions were mixed and hybridized to the arrays—were used as “NN” controls in the CGH-Miner program, giving a robust baseline to determine the shared copy number changes among the lager strains. When the CGH-Miner program generated the consensus plot shown in Supplemental Figure 3, it simultaneously created a spreadsheet file showing the genes that are significantly altered in copy number relative to the “normal” (self–self) arrays; this list, along with the normalized array data used for the CGH-Miner program, is given in Supplemental Table 3.
DNA sequencing
PCR primers (Supplemental Table 2) were designed to be specific for amplifying intron and intron-adjacent regions from only the S. cerevisiae portion of the genome by selecting 100 bp of sequence adjacent to the desired intron (obtained from SGD) (Nash et al. 2007), then splitting the sequence into 20-bp segments that overlapped by 10 bp (for a total of nine segments per 100 bp). Each 20-bp segment was then analyzed by BLAST analysis against both the S. cerevisiae genomic DNA and the S. bayanus genomic DNA sequences. Forward and reverse primers were selected from these candidates by choosing the segments that showed a single identical hit to the S. cerevisiae genome (with only low similarity alternatives) and no high-similarity hits to the S. bayanus genome; primer lengths were adjusted to give matching melting temperatures for a primer pair. In most cases, primers were designed to amplify the intron plus adjacent sequence, but for longer introns, primer pairs were chosen that would amplify only intronic sequences. Uncut genomic DNA, isolated as described above for microarrays, was used as template DNA for all amplifications; note that S. bayanus genomic DNA was included as a template for each primer pair to detect any nonspecific amplification; if any was seen, the melting temperature was adjusted upward and/or primers were redesigned until only one expected-size band was seen with S. cerevisiae and S. pastorianus template DNAs and zero bands were seen with S. bayanus template DNA. The reactions were then purified over Zymo Research Clean&Concentrate-5 columns, 200 μg of the product was mixed with either the forward primer or the reverse PCR primer at 0.5 μΜ final concentration, and then BigDye 3.0 sequencing reactions (ABI) were performed according to the manufacturer. The sequencing reactions were purified over 96-well format Sephadex columns, and then run on an ABI 3100 sequencer by the Stanford PAN facility.
Multiple sequence alignment and sequence tree construction
Assemblies for S. cerevisiae strains were obtained from the Saccharomyces Genome Resequencing Project (http://www.sanger.ac.uk/Teams/Team71/durbin/sgrp/index.shtml) from their FTP site (ftp://ftp.sanger.ac.uk/pub/dmc/yeast/latest/cere_assemblies.tgz), which was last updated on February 21, 2008. Regions corresponding to the genes of interest were identified using BLAST (Altschul et al. 1990) and extracted from these assemblies using custom Perl scripts. Multiple alignments for each locus were produced using version 3.7 of MUSCLE (Edgar 2004) with default parameters. These alignments were then trimmed and concatenated. The phylogenetic tree was generated using the dnapars program from the PHYLIP package, version 3.67 (http://evolution.genetics.washington.edu/phylip.html). To generate bootstrap values, we used seqboot to generate 100 bootstrap data sets, and consense to derive a consensus tree with bootstrap values, also from the PHYLIP package. The resulting tree was then rerooted at its midpoint for display purposes, using the retree program, also from the PHYLIP package.
Acknowledgments
We thank Fiona Wainwright for assistance with genomic DNA preparations, Gianni Liti and Vincent Robert for strains and useful discussions, Tom Pugh for strains and comments on the manuscript, and Rob Tibshirani and Daniela Witten for help with statistics. This work was supported by NSF ADVANCE grant DBI-0340856 to B.D. and NIH grant R01 HG00332 to G.S.
Footnotes
[Supplemental material is available online at www.genome.org. The microarray data from this study have been submitted to Gene Expression Omnibus under accession no. GSE12177.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.076075.108.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Awad I.A., Rees C.A., Hernandez-Boussard T., Ball C.A., Sherlock G., Rees C.A., Hernandez-Boussard T., Ball C.A., Sherlock G., Hernandez-Boussard T., Ball C.A., Sherlock G., Ball C.A., Sherlock G., Sherlock G. Caryoscope: An Open Source Java application for viewing microarray data in a genomic context. BMC Bioinformatics. 2004;5:151. doi: 10.1186/1471-2105-5-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U., Neal C., Donnelly D., James T.C., Neal C., Donnelly D., James T.C., Donnelly D., James T.C., James T.C. Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Curr. Genet. 2004;45:360–370. doi: 10.1007/s00294-004-0504-x. [DOI] [PubMed] [Google Scholar]
- Bozdech Z., Zhu J., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L., Zhu J., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L., Joachimiak M.P., Cohen F.E., Pulliam B., DeRisi J.L., Cohen F.E., Pulliam B., DeRisi J.L., Pulliam B., DeRisi J.L., DeRisi J.L. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes J.K., Morris G.P., Li W.H., Morris G.P., Li W.H., Li W.H. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol. Biol. Evol. 2006;23:1136–1143. doi: 10.1093/molbev/msj121. [DOI] [PubMed] [Google Scholar]
- Casaregola S., Nguyen H.V., Lapathitis G., Kotyk A., Gaillardin C., Nguyen H.V., Lapathitis G., Kotyk A., Gaillardin C., Lapathitis G., Kotyk A., Gaillardin C., Kotyk A., Gaillardin C., Gaillardin C. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int. J. Syst. Evol. Microbiol. 2001;51:1607–1618. doi: 10.1099/00207713-51-4-1607. [DOI] [PubMed] [Google Scholar]
- Casey G.P. Molecular and genetic analysis of chromosomes X in Saccharomyces carlsbergensis. Carlsberg Res. Commun. 1986;51:343–362. [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Corran H.S. A history of brewing. David and Charles; Newton Abbot, North Pomfret, VT: 1975. [Google Scholar]
- de Barros Lopes M., Bellon J.R., Shirley N.J., Ganter P.F., Bellon J.R., Shirley N.J., Ganter P.F., Shirley N.J., Ganter P.F., Ganter P.F. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 2002;1:323–331. doi: 10.1111/j.1567-1364.2002.tb00051.x. [DOI] [PubMed] [Google Scholar]
- Demeter J., Beauheim C., Gollub J., Hernandez-Boussard T., Jin H., Maier D., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Beauheim C., Gollub J., Hernandez-Boussard T., Jin H., Maier D., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Gollub J., Hernandez-Boussard T., Jin H., Maier D., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Hernandez-Boussard T., Jin H., Maier D., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Jin H., Maier D., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Maier D., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Matese J.C., Nitzberg M., Wymore F., Zachariah Z.K., Nitzberg M., Wymore F., Zachariah Z.K., Wymore F., Zachariah Z.K., Zachariah Z.K., et al. The Stanford Microarray Database: Implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M.J., Badrane H., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D., Badrane H., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D., Ferea T., Adams J., Brown P.O., Rosenzweig F., Botstein D., Adams J., Brown P.O., Rosenzweig F., Botstein D., Brown P.O., Rosenzweig F., Botstein D., Rosenzweig F., Botstein D., Botstein D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Barrett T., Barrett T. NCBI GEO standards and services for microarray data. Nat. Biotechnol. 2006;24:1471–1472. doi: 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt B. Cluster analysis 122. Heinemann; London: 1974. [Google Scholar]
- Giudici P., Caggia C., Pulvirenti A., Rainieri S., Caggia C., Pulvirenti A., Rainieri S., Pulvirenti A., Rainieri S., Rainieri S. Karyotyping of Saccharomyces strains with different temperature profiles. J. Appl. Microbiol. 1998;84:811–819. doi: 10.1046/j.1365-2672.1998.00416.x. [DOI] [PubMed] [Google Scholar]
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Hoheisel J.D., Jacq C., Johnston M., Jacq C., Johnston M., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gonzalez S.S., Gallo L., Climent M.A., Barrio E., Querol A., Gallo L., Climent M.A., Barrio E., Querol A., Climent M.A., Barrio E., Querol A., Barrio E., Querol A., Querol A. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int. J. Food Microbiol. 2007;116:11–18. doi: 10.1016/j.ijfoodmicro.2006.10.047. [DOI] [PubMed] [Google Scholar]
- Gonzalez S.S., Barrio E., Querol A., Barrio E., Querol A., Querol A. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 2008;74:2314–2320. doi: 10.1128/AEM.01867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J.L., Wolfe K.H., Wolfe K.H. Recent allopolyploid origin of Zygosaccharomyces rouxii strain ATCC 42981. Yeast. 2008;25:449–456. doi: 10.1002/yea.1598. [DOI] [PubMed] [Google Scholar]
- Groth C., Hansen J., Piskur J., Hansen J., Piskur J., Piskur J. A natural chimeric yeast containing genetic material from three species. Int. J. Syst. Bacteriol. 1999;49:1933–1938. doi: 10.1099/00207713-49-4-1933. [DOI] [PubMed] [Google Scholar]
- Hansen E.C. Recherches sur la physiologie et la morphologie des ferments alcooliques V. Methodes pour obtenir des cultures pures de Saccharomyces et de mikroorganismes analogues. C.R. Trav. Lab. Carlsberg. 1883;2:92–105. [Google Scholar]
- Hegarty M.J., Hiscock S.J., Hiscock S.J. Hybrid speciation in plants: New insights from molecular studies. New Phytol. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Holmberg S. Genetic differences between Saccharomyces carlsbergensis and S. cerevisiae. II. Restriction endonuclease analysis of genes in chromosome III. Carlsberg Res. Commun. 1982;47:233–244. [Google Scholar]
- Hornsey I. A history of beer and brewing. RSC Paperbacks; Cambridge, UK: 2003. [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kielland-Brandt C.M., Nillson-Tillgren T., Gjermansen C., Hollenberg S., Pedersen M.B., Nillson-Tillgren T., Gjermansen C., Hollenberg S., Pedersen M.B., Gjermansen C., Hollenberg S., Pedersen M.B., Hollenberg S., Pedersen M.B., Pedersen M.B. Genetics of brewing yeasts. In: Rose A.H., et al., editors. The yeasts. Academic Press; London: 1995. pp. 223–254. [Google Scholar]
- Kodama Y., Kielland-Brandt M.C., Hansen J., Kielland-Brandt M.C., Hansen J., Hansen J. Lager brewing yeast. In: Sunnerhagen P., Piškur J., Piškur J., editors. Comparative genomics. Springer-Verlag; Berlin: 2005. pp. 145–164. [Google Scholar]
- Kuehne H.A., Murphy H.A., Francis C.A., Sniegowski P.D., Murphy H.A., Francis C.A., Sniegowski P.D., Francis C.A., Sniegowski P.D., Sniegowski P.D. Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr. Biol. 2007;17:407–411. doi: 10.1016/j.cub.2006.12.047. [DOI] [PubMed] [Google Scholar]
- Legras J.L., Merdinoglu D., Cornuet J.M., Karst F., Merdinoglu D., Cornuet J.M., Karst F., Cornuet J.M., Karst F., Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Liti G., Peruffo A., James S.A., Roberts I.N., Louis E.J., Peruffo A., James S.A., Roberts I.N., Louis E.J., James S.A., Roberts I.N., Louis E.J., Roberts I.N., Louis E.J., Louis E.J. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast. 2005;22:177–192. doi: 10.1002/yea.1200. [DOI] [PubMed] [Google Scholar]
- Lopandic K., Gangl H., Wallner E., Tscheik G., Leitner G., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Gangl H., Wallner E., Tscheik G., Leitner G., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Wallner E., Tscheik G., Leitner G., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Tscheik G., Leitner G., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Leitner G., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W., Breitenbach M., Prillinger H., Tiefenbrunner W., Prillinger H., Tiefenbrunner W., Tiefenbrunner W. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007;7:953–965. doi: 10.1111/j.1567-1364.2007.00240.x. [DOI] [PubMed] [Google Scholar]
- Martini A.V., Kurztman C.P., Kurztman C.P. Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int. J. Syst. Bacteriol. 1985;35:508–511. [Google Scholar]
- Martini A.V., Martini A., Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Van Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- Masneuf I., Hansen J., Groth C., Piskur J., Dubourdieu D., Hansen J., Groth C., Piskur J., Dubourdieu D., Groth C., Piskur J., Dubourdieu D., Piskur J., Dubourdieu D., Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R.K., Johnston J.R., Johnston J.R. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash R., Weng S., Hitz B., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Weng S., Hitz B., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Hitz B., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Engel S.R., Fisk D.G., Hirschman J.E., Fisk D.G., Hirschman J.E., Hirschman J.E., et al. Expanded protein information at SGD: New pages and proteome browser. Nucleic Acids Res. 2007;35:D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova E.S., Naumov G.I., Masneuf-Pomarede I., Aigle M., Dubourdieu D., Naumov G.I., Masneuf-Pomarede I., Aigle M., Dubourdieu D., Masneuf-Pomarede I., Aigle M., Dubourdieu D., Aigle M., Dubourdieu D., Dubourdieu D. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast. 2005;22:1099–1115. doi: 10.1002/yea.1298. [DOI] [PubMed] [Google Scholar]
- Nilson-Tillgren T., Gjermansen C., Kielland-Brandt M.C., Petersen J.G.L., Holmberg S., Gjermansen C., Kielland-Brandt M.C., Petersen J.G.L., Holmberg S., Kielland-Brandt M.C., Petersen J.G.L., Holmberg S., Petersen J.G.L., Holmberg S., Holmberg S. Genetic differences between Saccharomyces carlsbergensis and S. cerevisiae. Analysis of chromosome III by single chromosome transfer. Carlsberg Res. Commun. 1981;46:65–76. [Google Scholar]
- Noonan G.J. New brewing lager beer: The most comprehensive book for home- and microbrewers. Brewers Publications; Boulder, CO: 1996. [Google Scholar]
- Ogata T., Izumikawa M., Kohno K., Shibata K., Izumikawa M., Kohno K., Shibata K., Kohno K., Shibata K., Shibata K. Chromosomal location of Lg-FLO1 in bottom-fermenting yeast and the FLO5 locus of industrial yeast. J. Appl. Microbiol. 2008 doi: 10.1111/j.1365-2672.2008.03852.x. [DOI] [PubMed] [Google Scholar]
- Pederson M.B. DNA sequence polymorphisms in the genus Saccharomyces. II Analysis of the genes RDN1, HIS4, LEU2 and Ty transposable elements in the Carlsberg, Tuborg and 22 Bavarian brewing strains. Carlsberg Res. Commun. 1985;50:263–272. [Google Scholar]
- Petersen J.G.L., Nilsson-Tillgren T., Kielland-Brandt M.C., Gjermansen C., Holmberg S., Nilsson-Tillgren T., Kielland-Brandt M.C., Gjermansen C., Holmberg S., Kielland-Brandt M.C., Gjermansen C., Holmberg S., Gjermansen C., Holmberg S., Holmberg S. Structural heterozygosis at genes ILV2 and ILV5 in Saccharomyces carlsbergensis. Curr. Genet. 1987;12:167–174. [Google Scholar]
- Rainieri S., Kodama Y., Kaneko Y., Mikata K., Nakao Y., Ashikari T., Kodama Y., Kaneko Y., Mikata K., Nakao Y., Ashikari T., Kaneko Y., Mikata K., Nakao Y., Ashikari T., Mikata K., Nakao Y., Ashikari T., Nakao Y., Ashikari T., Ashikari T. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 2006;72:3968–3974. doi: 10.1128/AEM.02769-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainieri S., Kodama Y., Nakao Y., Pulvirenti A., Giudici P., Kodama Y., Nakao Y., Pulvirenti A., Giudici P., Nakao Y., Pulvirenti A., Giudici P., Pulvirenti A., Giudici P., Giudici P. The inheritance of mtDNA in lager brewing strains. FEMS Yeast Res. 2008;8:586–596. doi: 10.1111/j.1567-1364.2008.00363.x. [DOI] [PubMed] [Google Scholar]
- Sato M., Kishimoto M., Watari J., Takashio M., Kishimoto M., Watari J., Takashio M., Watari J., Takashio M., Takashio M. Breeding of brewer’s yeast by hybridization between a top-fermenting yeast Saccharomyces cerevisiae and a cryophilic yeast Saccharomyces bayanus. J. Biosci. Bioeng. 2002;93:509–511. doi: 10.1016/s1389-1723(02)80101-3. [DOI] [PubMed] [Google Scholar]
- Scannell D.R., Wolfe K.H., Wolfe K.H. A burst of protein sequence evolution and a period of asymmetric evolution follow gene duplication in yeast. Genome Res. 2008;18:137–147. doi: 10.1101/gr.6341207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Byrne K.P., Gordon J.L., Wong S., Wolfe K.H., Byrne K.P., Gordon J.L., Wong S., Wolfe K.H., Gordon J.L., Wong S., Wolfe K.H., Wong S., Wolfe K.H., Wolfe K.H. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Smart K.A. Brewing yeast genomes and genome-wide expression and proteome profiling during fermentation. Yeast. 2007;24:993–1013. doi: 10.1002/yea.1553. [DOI] [PubMed] [Google Scholar]
- Tamai Y., Momma T., Yoshimoto H., Kaneko Y., Momma T., Yoshimoto H., Kaneko Y., Yoshimoto H., Kaneko Y., Kaneko Y. Co-existence of two types of chromosome in the bottom fermenting yeast, Saccharomyces pastorianus. Yeast. 1998;14:923–933. doi: 10.1002/(SICI)1097-0061(199807)14:10<923::AID-YEA298>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Tibshirani R., Walther G., Hastie T., Walther G., Hastie T., Hastie T. Estimating the number of clusters in a dataset via the Gap statistic. J.R. Statist. Soc. B. 2001;63:411–423. [Google Scholar]
- Treco D.A. Preparation of yeast DNA. In: Ausuble F.M., et al., editors. Current protocols in molecular biology. John Wiley & Sons Inc.; New York: 2003. p. 13.11. [Google Scholar]
- van de Peppel J., Kemmeren P., van Bakel H., Radonjic M., van Leenen D., Holstege F.C., Kemmeren P., van Bakel H., Radonjic M., van Leenen D., Holstege F.C., van Bakel H., Radonjic M., van Leenen D., Holstege F.C., Radonjic M., van Leenen D., Holstege F.C., van Leenen D., Holstege F.C., Holstege F.C. Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep. 2003;4:387–393. doi: 10.1038/sj.embor.embor798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Kim Y., Pollack J., Narasimhan B., Tibshirani R., Kim Y., Pollack J., Narasimhan B., Tibshirani R., Pollack J., Narasimhan B., Tibshirani R., Narasimhan B., Tibshirani R., Tibshirani R. A method for calling gains and losses in array CGH data. Biostatistics. 2005;6:45–58. doi: 10.1093/biostatistics/kxh017. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Ogata T., Ogata T. Chromosomal structures of bottom fermenting yeasts. Syst. Appl. Microbiol. 1999;22:341–353. doi: 10.1016/S0723-2020(99)80041-1. [DOI] [PubMed] [Google Scholar]