Abstract
Recent genomic sequencing of 10 additional Drosophila genomes provides a rich resource for comparative genomics analyses aimed at understanding the similarities and differences between species and between Drosophila and mammals. Using a phylogenetic approach, we identified 64 genomic elements that have been highly conserved over most of the Drosophila tree, but that have experienced a recent burst of evolution along the Drosophila melanogaster lineage. Compared to similarly defined elements in humans, these regions of rapid lineage-specific evolution in Drosophila differ dramatically in location, mechanism of evolution, and functional properties of associated genes. Notably, the majority reside in protein-coding regions and primarily result from rapid adaptive synonymous site evolution. In fact, adaptive evolution appears to be driving substitutions to unpreferred codons. Our analysis also highlights interesting noncoding genomic regions, such as regulatory regions in the gene gooseberry-neuro and a putative novel miRNA.
Comparative genomics approaches have assumed a central role in the identification of functionally important genomic regions (Kellis et al. 2003; Siepel et al. 2005; Xie et al. 2005; Birney et al. 2007). These approaches are based on the neutral theory prediction that sequences that have been highly conserved over tens of millions of years are either functionally important or are mutational cold spots (although no molecular mechanism for generating cold spots has been proposed). Recent population genetic analyses showed that low-frequency alleles are more common in highly conserved sequences, which supports the idea that such sequences, including those that do not encode proteins, are functionally constrained in multiple lineages (Drake et al. 2006; Asthana et al. 2007; Casillas et al. 2007; Katzman et al. 2007). On the other hand, questions remain about the functional importance of conserved sequences. For example, a recent functional analysis provided no evidence for strong viability selection against four conserved noncoding elements in mice (Ahituv et al. 2007).
The conceptual foundation linking conserved function with conserved sequence ignores the biologically interesting question of how biological functions evolve in different lineages. Indeed, from an evolutionary perspective, understanding the causes of rapid sequence evolution may be at least as interesting as understanding the causes of strong sequence conservation. Of particular relevance for identifying potential major functional changes is the identification of genomic regions that are highly conserved over most of a phylogeny, but that evolve very rapidly in at least one lineage. Such phylogenetically restricted rapid evolution could be due to a dramatic change in functional constraint, an increased mutation rate, or a shift in function, which drives large numbers of substitutions through populations under directional selection (Gillespie 1991).
Although the statistical analysis of heterogeneous rates of coding sequence evolution among lineages has a long history (Zuckerandl and Pauling 1962; Ohta and Kimura 1971; Langley and Fitch 1973, 1974), only recently have genome assemblies and alignments from multiple species (Blanchette et al. 2004; Clark et al. 2007; Stark et al. 2007) permitted such questions to be pursued in a comprehensive manner that is unbiased with respect to genomic feature. For example, Pollard et al. (2006) used alignments of multiple vertebrate species to identify genomic regions that are highly conserved in most vertebrates, but that have evolved rapidly in humans. These human accelerated regions (HARs) are candidates for contributing to human-specific biology. Interestingly, the majority of these regions were noncoding, and many were located near genes functioning in the nervous system. A more recent genomic analysis (Kim and Pritchard 2007) took a similar approach, but broadly investigated heterogeneous rates of evolution for conserved noncoding sequence across vertebrates. They concluded that short bursts of adaptive evolution drive divergence in conserved noncoding sequences.
The recent availability of multiple genome assemblies (Stark et al. 2007) and alignments (Karolchik et al. 2003, 2004; Blanchette et al. 2004) from Drosophila motivates an extension of such approaches to the Drosophila model for three main reasons. First, the experimental power of Drosophila opens up the possibility of detailed, in vivo functional investigation of candidate regions that are generally highly conserved but evolve rapidly in one lineage. Second, the genome organizations of flies and vertebrates are markedly distinct, with flies having much more compact genomes containing less noncoding DNA. This raises interesting questions as to whether the genomic distribution of lineage-specific increases in substitution rates in flies will also be concentrated in noncoding DNA, or whether differences in the biology and/or population genetics of flies and humans lead to different patterns. Finally, the Drosophila melanogaster genome is very well annotated, which facilitates targeted functional studies. Comparison of functional annotations associated with lineage-specific rate increases in different lineages could provide clues as to potential generalities as well as unique biological functions exhibiting these unusual evolutionary patterns.
Results
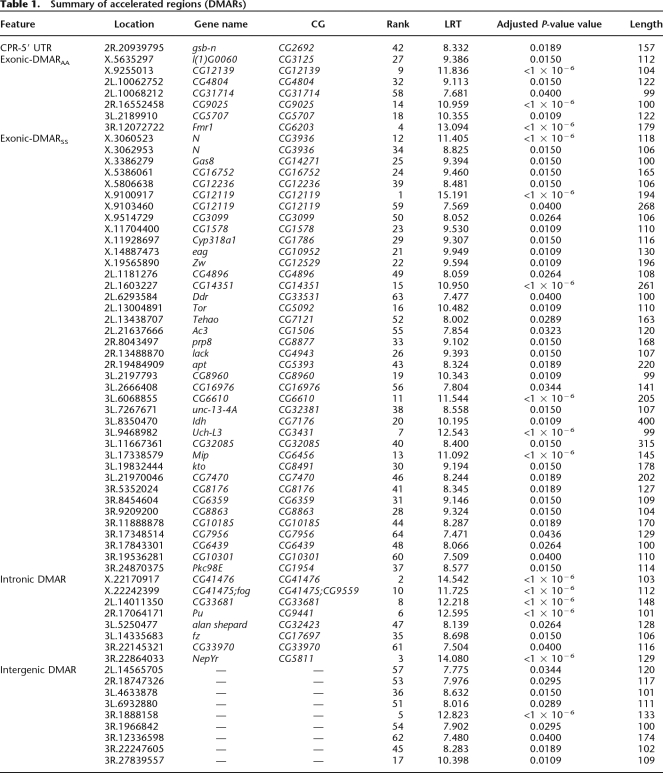
Using whole-genome alignments of 10 Drosophila species to the D. melanogaster reference (Karolchik et al. 2003, 2004; Blanchette et al. 2004), we identified genomic regions that have been highly conserved over tens of millions of years, but show a recent acceleration in the rate of evolution solely along the D. melanogaster branch (Fig. 1A). Genomic regions were defined as conserved if they were 96% similar in sequence between Drosophila simulans, Drosophila yakuba, and Drosophila erecta and were at least 100 bp long. We identified 97,901 conserved regions with a mean (and median) length of 140 bp. Next, we assessed acceleration along the D. melanogaster branch using a likelihood ratio test (LRT) to compare two models of evolution over the Drosophila tree. The three species used to identify conserved regions (D. simulans, D. yakuba, and D. erecta) were excluded from this step in the analysis since, by definition, they were highly conserved. For each candidate region, the LRT compares the likelihood of the multiple alignments under a local null model with no acceleration in D. melanogaster to an alternative model with acceleration. There were 400 accelerated regions with an initial, unadjusted P-value < 0.05. Sixty-four of the conserved regions were determined to have significant acceleration along the D. melanogaster lineage after adjusting for multiple comparisons using the false discovery rate (FDR) (adjusted P-value < 0.05; Table 1). Hereafter, we refer to these as Drosophila melanogaster accelerated regions, or DMARs.
Figure 1.
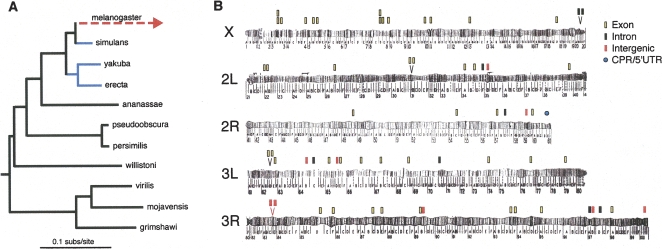
(A) Phylogeny of 11 Drosophila species with genome sequences. Branch lengths are derived from maximum likelihood analysis of all elements conserved throughout the tree. Branches in blue (D. simulans, D. yakuba, and D. erecta) were used to identify the blocks of at least 100 bp with 96% identity between the three species. All other lineages (and the D. melanogaster–D. simulans ancestor) were used to infer whether D. melanogaster had an accelerated rate of evolution relative to the expected rate of evolution based on elements conserved throughout the tree. (B) Locations of D. melanogaster accelerated regions (DMARs). (Stacked bars) Multiple DMARs within a single locus. (Two bars above a “V”) Two DMARs that were within the same chromosomal band. DMARs are found predominately in exons (46/64) and are significantly over-represented on the X chromosome (16/64). Chromosome images adapted from Lefevre (1976).
Table 1.
Summary of accelerated regions (DMARs)
Accelerated rates of evolution could result from multiple single substitution events or they could result from microinversions that would cause a short region of sequence to appear to be rapidly diverged. An analysis of possible microinversions showed that only five substitution pairs could have resulted from this process, which only explains ∼1% of all substitutions in DMARs. Therefore, the substitution process that leads to DMARs predominantly results from multiple single substitution events.
The 64 DMARs were dispersed fairly evenly throughout the major chromosome arms (Fig. 1B). Relative to the proportion of regions identified on the X chromosome as “conserved” in the first step of the analysis (10.5%), DMARs are significantly over-represented on the X chromosome (n = 16, FET two-tailed P-value = 0.0151). If DMARs are driven to fixation by directional selection, more efficient selection on the X chromosome could have led to this finding (for review, see Vicoso and Charlesworth 2006).
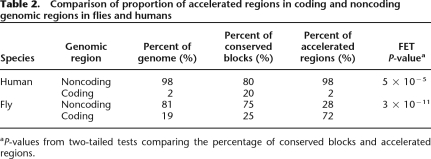
The majority of DMARs (72%) are found in protein-coding regions (Table 1). There were 46 DMARs in exons, nine in intergenic regions, eight in introns, and a single DMAR in a core promoter/5′ untranslated region (UTR). This distribution of DMARs among genomic features contrasts dramatically with regions in the human genome that show evidence of recent acceleration (HARs), which were found primarily in noncoding regions (Table 2; Pollard et al. 2006). The fact that the majority of HARs were found in noncoding regions may not be surprising considering that only 2% of the human genome is protein-coding. Flies have much more compact genomes, with almost 20% of the genome coding for proteins. However, even after considering genomic content in Drosophila, a significant excess of DMARs occur in protein-coding regions (see Table 2).
Table 2.
Comparison of proportion of accelerated regions in coding and noncoding genomic regions in flies and humans
aP-values from two-tailed tests comparing the percentage of conserved blocks and accelerated regions.
Protein-coding DMARs
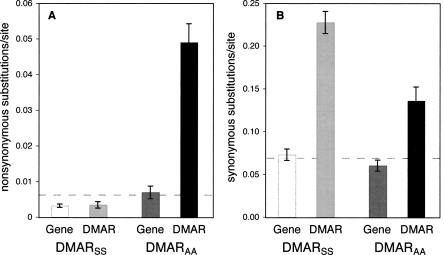
DMARs in coding regions can be divided into two groups based on whether substitutions are found primarily at synonymous sites or nonsynonymous sites (Supplemental Table S1). DMARs with primarily synonymous substitutions (DMARSS) were defined as those with fewer than 25% of substitutions at amino acid changing sites (n = 39); the remaining set (DMARAA) have at least 40% of substitutions at amino acid changing sites (n = 7). This arbitrary definition marks a natural break in the distribution of nonsynonymous substitution rates; DMARs defined as DMARAA have high nonsynonymous substitution rates (0.0334–0.0692 substitutions/site) along the D. melanogaster lineage, whereas nonsynonymous substitution rates in DMARSS are 0.0139–0.0200 substitutions/site (Fig. 2; Supplemental Table S1).
Figure 2.
Nonsynonymous (A) and synonymous (B) substitution rates for DMARs in coding regions and the genes that contain those DMARs. Rates are per nonsynonymous site or synonymous site. Both DMARSS and the genes that contain them (light gray and white bars) have very low levels of amino acid divergence. (Black and dark gray bars) DMARAA have high rates of synonymous and nonsynonymous substitution, but the genes that contain them evolve at similar rates to the genomic average (dashed line).
Acceleration of synonymous site divergence
The DMARSS, by definition, are evolving rapidly at synonymous sites in D. melanogaster, but slowly at amino acid sites—even in comparison to the gene in which they are found (Fig. 2; Supplemental Table S1). The genes that contain these DMARSS are evolving slower at amino acid sites than the genomic average (Fig. 2A), while synonymous site evolution of DMARSS-containing genes is comparable to the genomic average (Fig. 2B). These data suggest that evolutionary rates of DMARSS are not properties of genes, but of small regions within genes.
Rapid synonymous site divergence may indicate a shift in codon usage. Therefore, we examined codon usage in DMARSS, in the genes that contain them, and genome-wide. Our calculation of the number of substitutions to unpreferred codons was based on the mutational opportunity from preferred to unpreferred codons in the inferred ancestor of D. melanogaster and D. simulans (see Methods; Begun et al. 2007). We counted the number of substitutions from preferred to unpreferred codons and divided by the proportion of preferred codons in the inferred ancestor. Genes containing DMARSS have more substitutions to unpreferred codons than do a random selection of genes in the genome (0.0565 vs. 0.0456; permutation test P-value = 0.002). Even more striking is the dramatic skew toward fixation of unpreferred codons in DMARSS compared to the remainder of the gene (0.1689 vs. 0.0565; paired t-test; P-value = 0.0016). Accelerated synonymous site divergence in DMARSS is attributable to fixation of many unpreferred variants.
Preferred codons typically end in guanine or cytosine. An overall mutational bias from G|C to A|T could explain increased substitution from preferred to unpreferred codons. Unless the mutational bias was extremely local, it would extend to introns of genes containing DMARSS since they are intercalated among exons. In fact, several studies have found that G+C content was highly correlated between introns and third positions of codons (Kliman and Hey 1994; Heger and Ponting 2007; Vicario et al. 2007). For DMARSS, introns of DMARSS, and introns of all genes in the genome, we calculated the fraction of G|C to A|T substitutions by counting the number of G|C to A|T substitutions and divided that by the sum of all substitutions from ancestrally G|C nucleotides. The average fraction of G|C to A|T substitutions in introns of genes that contain DMARSS was similar to the genome average (0.839 vs. 0.851, respectively). The DMARSS, on the other hand, have a significantly higher fraction of G|C to A|T substitutions than do the introns of the DMARSS-containing genes (0.931 vs. 0.839; paired t-test, t-statistic = 3.00, degrees of freedom [df] = 15, two-tailed P-value = 0.0089), which indicates that a gene-sized local mutational bias does not explain the rapid accumulation of unpreferred codons. This finding contrasts sharply with the substitution bias in HARs. In HARs, there was a preponderance of A|T to G|C substitutions, which indicates that biased gene conversion may be driving HAR substitutions.
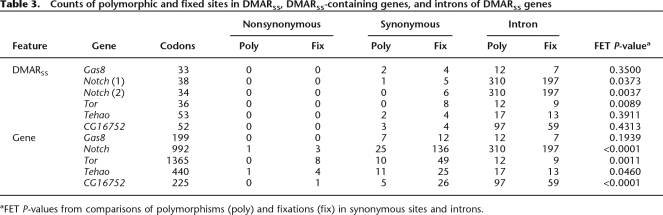
A second hypothesis for the rapid synonymous site divergence in DMARSS is that directional selection has fixed these substitutions. Recent work has shown that short introns (<80 bp) have very low levels of constraint (Halligan et al. 2004), which suggests they are composed primarily of neutral sites. In a modified version of the McDonald-Kreitman test (McDonald and Kreitman 1991), we compared ratios of polymorphism and divergence in short introns to synonymous sites in DMARSS. For six of the DMARSS (two of which are in Notch), we have polymorphism data from the DPGP D. melanogaster resequencing project (http://www.dpgp.org/melanogaster/). We found that three out of six DMARSS show a significant excess of synonymous site fixation, which suggests the action of directional selection (Table 3). We also performed this test on the remainder of the gene (without the DMAR) and found that four out of five show evidence of adaptive synonymous site evolution (Table 3). However, as noted previously, codon usage is significantly different between the DMARSS and the remainder of the gene, with DMARSS fixing significantly more unpreferred codons (paired t-test for six genes with polymorphism data; P-value = 0.0078). This difference in substitution pattern may indicate that different mechanisms of evolution are acting on synonymous sites in DMARSS compared to synonymous sites in regions of the gene that do not have recent accelerations. The identification of DMARSS may have drawn attention to a class of genes with multiple evolutionary pressures driving synonymous substitution.
Table 3.
Counts of polymorphic and fixed sites in DMARSS, DMARSS-containing genes, and introns of DMARSS genes
aFET P-values from comparisons of polymorphisms (poly) and fixations (fix) in synonymous sites and introns.
In earlier work, one DMARSS-containing gene, Notch, was found to harbor a region with rapid synonymous site evolution that overlaps one of the DMARSS (DuMont et al. 2004). In agreement with our findings for many DMARSS, intensive investigation of the Notch region with rapid synonymous site evolution led to the conclusion that directional selection was acting on synonymous sites (DuMont et al. 2004).
Acceleration of amino acid divergence
In genes that contain DMARAA, the rate of amino acid and synonymous site divergence is similar to the genomic average (Fig. 2). In contrast, the DMARAA are evolving rapidly not only at amino acid changing sites (Fig. 2A), but also at synonymous sites (twofold higher than the genomic average) (Fig. 2B). The genes containing DMARAA do not differ significantly from the genomic average with respect to substitutions to unpreferred codons (0.0588 vs. 0.0456; permutation test P-value = 0.067). The small sample size (n = 7) may increase variance in the permutation test and make rejecting the null hypothesis of no difference between DMARAA genes and the genomic average difficult. Regardless, like DMARSS, the proportion of substitutions to unpreferred codons in DMARAA is significantly higher than in the remainder of the gene (0.1384 vs. 0.0588; paired t-test, df = 6, t-statistic = 6.338, P-value = 7.2 × 10−4).
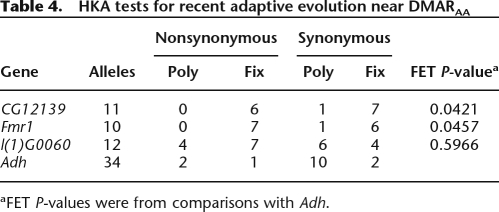
In order to address whether directional selection may have acted to fix amino acid substitutions of DMARAA, we collected sequence data from D. melanogaster inbred lines for three genes [Fmr1, l(1)G0060, and CG12139] (Table 4). The DMARAA and surrounding sequence for Fmr1 and CG12139 have very little polymorphism, which could indicate the action of recent directional selection. In fact, in comparison to the levels of synonymous polymorphism and divergence at the Adh locus (polymorphism data from the DPGP D. melanogaster resequencing project; http://www.dpgp.org/melanogaster/), there are fewer polymorphic synonymous sites than would be expected under a neutral model for both Fmr1 and CG12139 (Table 4; Hudson et al. 1987). For l(1)G0060, polymorphism relative to divergence was not significantly different from the neutral expectation.
Table 4.
HKA tests for recent adaptive evolution near DMARAA
aFET P-values were from comparisons with Adh.
Ontology
Two biological processes (cell–cell signaling and cell communication) and two molecular functions (signal transducer activity and receptor activity) are over-represented among protein-coding genes containing DMARs (permutation test P-value < 0.01). The biological process signal transduction was also slightly over-represented (permutation test P-value = 0.038). There is notable overlap of genes among these terms. In fact, six genes are associated with at least four of these ontology terms (Supplemental Table S3). One other biological process, catabolism, is also significantly over-represented among DMARs in coding regions, but this ontology category does not overlap extensively with the aforementioned. Interestingly, catabolism and several specific types of receptor activity also appear to be enriched in the set of protein-coding genes with significantly accelerated amino acid evolution in D. melanogaster (see Table S21 in Begun et al. 2007). In comparison, in HARs, DNA binding and transcriptional regulation of genes near HARs were over-represented, which, once again, highlights the different biological processes and mechanisms that drive recent accelerations in the human and fly lineages. For DMARs, the biological significance of accelerated evolution in cell signaling genes is an interesting topic for future investigations.
DMARs in noncoding DNA
Intergenic and intron accelerated regions
Annotation of the D. melanogaster genome was used to determine the location of DMARs. Therefore, it is possible that the intergenic DMARs are actually protein-coding regions in other species and that D. melanogaster has lost one or more genes (or exons). The accelerated rate of evolution in a putatively intergenic region would then be due to relaxation of purifying selection in D. melanogaster. We investigated whether DMARs in intergenic regions were predicted to be protein-coding genes in D. simulans, D. yakuba, or D. erecta (Stark et al. 2007). In fact, none of the intergenic sequences were parts of predicted proteins in any of those three species. Additionally, we found that none of the DMARs fall within noncoding RNAs included in release 5.2 of the D. melanogaster annotation. However, two intergenic DMARs are near genes and may serve some cis-regulatory function. DMAR 2R.18747326 is 1009 bp from the 5′-UTR of inaD, and DMAR 3R.4633878 is 559 bp from the 3′-end of CG13716. There is no annotated 3′-UTR for CG13716. It is possible that DMAR 3R.4633878 is part of the CG13716 3′-UTR given that the average length of 3′-UTRs in Drosophila is 318 bp and 3′-UTRs > 500 bp are not uncommon.
Intronic DMARs are found primarily in first introns (five of eight), and the remaining DMARs are in the largest introns of the gene. Introns often harbor regulatory elements, and it is possible that these DMARs serve some regulatory function. However, there are no known regulatory elements in intronic DMARs (FlyReg 2.0 [Bergman et al. 2005]; REDFly [Gallo et al. 2006]).
Intergenic and intronic DMARs could be unannotated noncoding RNAs. We took two approaches to address this question. First, we investigated whether whole-genome tiling-array experiments on total RNA (Stolc et al. 2004) revealed expression in the regions of any intronic or intergenic DMARs. In fact, two are expressed (DMAR X.22170917 and DMAR 3R.22145321). These expression profiles are based on total RNA; therefore, it is possible that unprocessed RNA was detected (Stolc et al. 2004). Second, we examined the predicted secondary structure of intergenic and intronic DMARs using EvoFold (Pedersen et al. 2006). All species for which there was available sequence for each DMAR were used in analyses. We also used RNAfold from the Vienna RNA package v1.6.4 (Hofacker et al. 1994) to compare the optimal secondary structures of D. melanogaster and D. simulans. Supplemental Figures S1–S17 show the optimal secondary structures as well as plots of base-pairing for the minimum free energy structure (lower left) and the probability of base-pairing (upper right) for each DMAR. Supplemental Table S2 shows numerical results from both EvoFold and RNAfold. There were four DMARs—X.22170917 (which also shows evidence of transcription), 3L.6932880, 3R.1888158, and 3R.1966842 (Supplemental Table S2; Supplemental Figs. S1, S9, S11, S12, respectively)—with high folding potential scores from the EvoFold analysis, which could indicate secondary structure. Predictions from RNAfold do not show any convincing secondary structure for three of these DMARs. However, in D. melanogaster, intergenic DMAR 3R.1966842 folds into a single hairpin (Supplemental Fig. S18), much like an miRNA, whereas D. simulans has a Y-shaped optimal structure (Supplemental Fig. S12). Drosophila simulans has a much weaker hairpin structure when forced onto the D. melanogaster optimal structure. Three substitutions along the D. melanogaster lineage increase complementary base-pairing in the hairpin (Supplemental Fig. S18).
Using miRScan (Lim et al. 2003a, b), we found that 3R.1966842 has significant potential for being an miRNA, with a total score (11.48) similar to the scores from known miRNAs in vertebrates and Caenorhabditis elegans and substantially higher than those of most non-miRNAs (Lim et al. 2003a, b). The Heidelberg RNA study (Hild et al. 2003) shows expression in the region of DMAR 3R.1966842, with the expressed probe residing within the DMAR. Population data from lines that are isogenic for chromosome 3 (n = 75) show that the DMAR sequence is fixed in D. melanogaster, except for three singleton polymorphisms that do not influence secondary structure (see Supplemental Fig. S18). Expression data are needed to validate whether the mature RNA is of the appropriate size to be considered a miRNA.
EvoFold (Pedersen et al. 2006) has been used to detect secondary structure throughout the genomes of flies (Stark et al. 2007), and these predictions are available as tracks on the UCSC Genome Browser (Karolchik et al. 2003, 2004). We found that there are nine EvoFold predictions that overlap with DMARs (Supplemental Table S4). However, only five of these have substitutions within the DMAR, and of these only one DMAR, X.22170886, which is contained within the intron of CG41476, has convincing secondary structure. Given this analysis and the analysis of entire DMAR sequences, it seems unlikely that substitutions within DMARs for changes in secondary structure would be a general driving force in the evolution of DMARs.
Acceleration in a regulatory region
The gooseberry-neuro (gsb-n) gene contains a DMAR in the core promoter (102 bp) that extends into the 5′ UTR (55 bp). This gene is a tandem duplicate and is transcribed in the opposite direction from its partner, gooseberry; both are transcription factors that are expressed during early development (Baumgartner et al. 1987; Gutjahr et al. 1993). The two genes have nonoverlapping regulatory modules (Li et al. 1993; Li and Noll 1994a, b), but do have partially redundant function; gooseberry regulates gsb-n and is able to perform the functions of gsb-n (Gutjahr et al. 1993). Both of these genes have well-characterized regulatory regions. Unfortunately, comparative expression data from D. melanogaster and D. simulans are not available for the appropriate developmental stage. Functional investigation of changes in the timing, levels, and spatial patterns of expression are warranted and will be a target of future studies.
Discussion
We identified 64 genomic regions that have been highly conserved over many millions of years, but that have recently experienced a burst of evolution along the D. melanogaster lineage. Protein-coding regions harbor the majority of DMARs, and rapid synonymous site evolution was the most common source of divergence. Synonymous site substitutions were overwhelmingly skewed toward unpreferred codons. We ruled out the possibility of a local mutation bias by comparing the substitution bias in DMARSS and their associated introns. Comparisons of polymorphism and divergence in DMARSS and nearby introns suggest that directional selection may be the driving force behind these rapid bursts of evolution at synonymous sites. An alternative hypothesis is that rapidly evolving mutation rates can explain these highly unusual genomic regions. In this scenario, DMARSS and the population genetic evidence for their adaptive divergence could be explained by a recent increase in mutation rate and bias along the branch leading to D. melanogaster, followed by a second, more recent change back to ancestor-like mutation rates and patterns. The finely tuned requirements for the timing of these changes make this hypothesis less parsimonious, but given that these are some of the most unusual genomic regions in D. melanogaster, the possibility cannot be ruled out.
Rapidly evolving D. melanogaster genes often have lower levels of codon bias (Akashi 1994, 1995; Akashi et al. 2007), but, in general, this is not associated with adaptive evolution (Akashi 1995, 1996; Singh et al. 2007). In fact, fixation of unpreferred codons is attributed to the reduced efficacy of selection in D. melanogaster due to smaller population sizes (Akashi 1995, 1996; Vicario et al. 2007). However, a genome-wide computational analysis of unpreferred codon usage of mRNAs in flies, yeast, and bacteria showed that some unpreferred codons are fixed by directional selection in both bacteria and flies (Neafsey and Galagan 2007). Interestingly, in that study, none of the DMARs genes were identified as having evidence of directional selection acting on unpreferred codon usage. In a second genomic study (Singh et al. 2007), only the Notch gene showed evidence of selection for unpreferred codon usage. Most likely, these analyses identify a different set of loci from our study because analysis of the entire gene would miss DMARs-like short stretches of unpreferred codon usage.
Prior intensive investigation at the Notch locus has identified regions with patterns of substitution similar to our findings for DMARs with rapid synonymous site evolution (DuMont et al. 2004; Nielsen et al. 2007). In fact, the Notch locus contained two DMARs with rapid synonymous site evolution, and one DMAR (X.3062953) is located in the region noted as the “3′ region” in DuMont et al. (2004). That study found an excess of unpreferred codon fixation and ruled out the possibility that changes in mutation rate and/or low levels of recombination could explain the pattern completely (DuMont et al. 2004). They concluded that directional selection on synonymous sites has driven the fixation of these unpreferred codons. Our results for Notch DMARs and synonymous site DMARs are in agreement with their findings.
Preferred codons are thought to be favored by selection on translational accuracy (e.g., fidelity of translation) (Akashi 1994), efficiency (e.g., tRNA abundance) (Akashi 2001, 2003), and/or robustness (e.g., proper folding despite mistranslation) (Drummond et al. 2005, 2006). In the case of translational efficiency, experimental work has shown that the use of unpreferred codons reduced the rate of translation in yeast (Purvis et al. 1987), Drosophila (Carlini and Stephan 2003), Escherichia coli (Parker 1989; Andersson and Kurland 1990; Komar et al. 1999), and humans (Kimchi-Sarfaty et al. 2007).
While unpreferred codons that reduce translational efficiency would typically be selected against, in some cases selection may act to reduce rates of protein translation (Konigsberg and Godson 1983; Purvis et al. 1987; Andersson and Kurland 1990; Thanaraj and Argos 1996; Komar et al. 1999). For example, protein folding often occurs before the completion of protein synthesis; pausing caused by the use of rare codons can allow for proper protein folding (Purvis et al. 1987; Thanaraj and Argos 1996; Komar et al. 1999). Directional selection may also act to fix unpreferred (or rare) codons to reduce translational efficiency and therefore protein levels (Konigsberg and Godson 1983; Andersson and Kurland 1990). The two hypotheses for how selection favors unpreferred codons make different predictions for the distribution of unpreferred codon usage. In cases of selection for reduced translational efficiency acting on overall protein abundance, we might expect unpreferred substitutions to be distributed throughout the mRNA. On the other hand, short segments of a coding region that use a high proportion of unpreferred codons may be more effective in causing sufficient ribosomal pausing at a particular position to induce proper folding (Purvis et al. 1987; Thanaraj and Argos 1996; Komar et al. 1999). That is, the physical proximity of unpreferred codons may have a multiplicative effect on translation rates (Purvis et al. 1987). One example of this phenomenon is in the pyruvate kinase gene in yeast. Five rare codons are used just before a predicted fold and are hypothesized to cause a pause in protein synthesis specifically at this location (Purvis et al. 1987). We hypothesize that ribosomal pausing for proper protein folding is a more tenable mechanism for explaining the abundance of DMARs with fixations of unpreferred codons than the alternative of reducing translation efficiency. Why the demands of protein folding would change between two closely related Drosophila species remains an open question.
Adaptive protein evolution is pervasive in Drosophila (Smith and Eyre-Walker 2002; Eyre-Walker 2006; Begun et al. 2007); thus, it is not surprising that two of three DMARs with multiple nonsynonymous substitutions showed evidence of directional selection in our population data. These DMARAA are also evolving more rapidly at synonymous sites than the genome-wide average, which could be due to Hill-Robertson interference (Hill and Robertson 1966). Interference would reduce the efficacy of selection against unpreferred codons. However, this phenomenon is more commonly observed in regions of reduced recombination, and DMARs are not restricted to regions of reduced recombination.
Although the majority of DMARs are located in coding regions, this genomic study is unbiased with respect to genomic location and was not restricted to highlighting unusual patterns of evolution solely in known protein-coding regions. In fact, one interesting finding from this study has been the identification of a putative novel miRNA in D. melanogaster. Our study also identified several other genomic regions that will be the focus of future investigations, such as the core promoter and 5′ UTR of the gooseberry-neuro gene.
Conclusions
This comprehensive investigation of genomic elements that have been conserved over long periods of evolutionary time but that have had a recent burst of evolution in the D. melanogaster lineage suggests that DMARs may result from adaptive evolution. Intriguingly, many DMARs are attributable to recent accelerated synonymous site divergence and the accumulation of unpreferred codons. Population genetic evidence suggests that directional selection on synonymous sites plays a role in this phenomenon; though unusual, nonequilibrium mutational variation is not ruled out. Our findings reveal that DMARs contrast sharply in location, mechanism, and functional properties compared to HARs, which indicates that the biological and ecological differences between humans and flies are important factors in driving the evolutionary properties of genomes. Functional characterization of DMARs is now necessary to determine how radical changes in genotype are reflected in phenotype.
Methods
Genome alignments
MULTIZ alignments that were made in December 2006 using the D. melanogaster Release 5 assembly as the reference sequence (http://www.fruitfly.org/sequence/README.RELEASE5) were downloaded from the UCSC Genome Browser (Karolchik et al. 2003, 2004; Blanchette et al. 2004; http://genome.ucsc.edu/admin/cvs.html). The multiple alignments were generated from high-quality pairwise alignments produced by UCSC’s chaining and netting pipeline (Kent et al. 2003), which uses conserved synteny to ensure orthology of aligned regions. Repeat regions and regions of low complexity were masked prior to alignment. The resulting 15-way alignments included D. melanogaster, D. simulans, Drosophila sechellia, D. yakuba, D. erecta, Drosophila ananassae, Drosophila pseudoobscura, Drosophila persimilis, Drosophila willistoni, Drosophila mojavensis, Drosophila virilis, and Drosophila grimshawi, as well as sequences of Anopheles gambiae, Apis mellifera, and Tribolium castaneum. We removed the D. sechellia sequence before analysis because of the low coverage of the genome. The mosquito, bee, and beetle sequences were also removed before analysis. We also deleted gaps that were inserted due to the non-fly and D. sechellia genome sequences and removed any blocks that overlapped a known transposable element annotated in Release 5.1 of the D. melanogaster genome.
Identification of conserved regions
Conserved blocks were defined as those that were at least 100 bp long and had at least 96% sequence similarity between D. simulans, D. yakuba, and D. erecta. We used mafBlocker (Pollard et al. 2006) to identify conserved blocks. Conserved blocks that included sequence data from at least two additional species outside of the melanogaster subgroup were retained.
Assessment of significant acceleration
For all conserved blocks, we used likelihood ratio tests (LRTs) to determine whether the D. melanogaster branch had a significantly faster rate of evolution than expected. We excluded D. simulans, D. yakuba, and D. erecta from the LRT so that results were independent of the initial identification of the conserved regions. For the LRT, we used D. melanogaster, the inferred D. melanogaster–D. simulans ancestor, and at least two species from the following: D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi. The D. melanogaster–D. simulans ancestor was used as a node on the tree (with 0 branch length) so that we could ascribe evolutionary changes specifically to the D. melanogaster branch. The ancestral state was derived by a majority rule parsimony analysis of the D. melanogaster, D. simulans, and D. yakuba trio; instances of no majority were called “N.”
For all conserved blocks, we used phyloFit to estimate two models of evolution (Siepel and Haussler 2004). The null model was derived by rescaling branch lengths from 15-species whole-genome MULTIZ alignments so that relative substitution rates remain constant across branches, but each conserved region has its own rate (branch lengths represented in Fig. 1A). Estimates of base frequencies and the substitution matrix were also taken from the combined 15-species whole-genome MULTIZ alignments. The alternative model included the same rescaling plus allowed the D. melanogaster branch to have an accelerated rate of evolution.
We assessed the statistical significance of regions identified as accelerated along the D. melanogaster branch by simulation using parametric bootstrapping. First, we generated 1 million alignments based on parameters from the 15-species whole-genome MULTIZ alignments. The simulated null alignments were 140 bp, which was the mean (and median) of the conserved regions we identified. The LRT statistic was then computed for each alignment. For each of the conserved elements we identified in step 1, the empirical P-value is equal to the proportion of simulated data sets with a larger LRT statistic. Based on the number of simulated data sets, the smallest P-value that can be estimated is P = 1 × 10−6. Empirical P-values were adjusted for multiple comparisons by the method of Benjamini and Hochberg (Benjamini and Hochberg 1995), which controls the false discovery rate (FDR). Any region with FDR adjusted P-value ≤ 0.05 was taken as having a significant acceleration along the D. melanogaster branch. We, therefore, expect that the proportion of false positives in this set is <5%. For each DMAR with an FDR adjusted P-value ≤ 0.05, we ensured that each DMAR was the reciprocal best BLAST hit with D. simulans.
Release 5.1 of the D. melanogaster annotation was used to determine whether DMARs were located in intergenic, coding, intron, or UTR sequence. Initial identification of conserved regions required that they were at least 100 bp long in D. simulans, D. yakuba, and D. erecta. Some DMARs may be shorter than 100 nt for two reasons. First, there may have been deletions along the D. melanogaster lineage. Second, DMARs were placed in categories (e.g., coding, intron, UTR) based on the location of the majority of nucleotides, which, in a small number of cases, resulted in a few conserved nucleotides being trimmed from one end. This only occurred when the DMAR was located primarily in a coding region, and estimates of polymorphism and divergence of synonymous and nonsynonymous sites would have been compromised by including noncoding nucleotides.
Molecular methods
D. melanogaster population data for Fmr1, l(1)G0060, and CG12139 (n = 9–11 alleles) were from isofemale lines from Malawi, Africa. For population sampling of DMAR 3R.1966842 (n = 75), we used D. melanogaster lines that were collected by A.G. Clark (Maryland); these lines are isogenic for chromosome 3. DNA was PCR-amplified using Promega GoTaq Flexi DNA polymerase (Promega) for Fmr1 and CG12139 and AmpliTaq (Applied Biosystems) for l(1)G0060 and DMAR 3R.1966842. PCR products were ligated into a PCR4 TOPO vector (Invitrogen). Ligations were transformed and plated, with the resulting colonies subjected to PCR using vector primers with AmpliTaq (Applied Biosystems). One clone was randomly selected from each line for sequencing. Colony PCR products were purified and sequenced at the University of California, Davis, College of Biological Sciences DNA Sequencing Facility. Sequences were submitted to GenBank under accession nos. EU588685–EU588714. Information for substitutions in the population sample of DMAR 3R.1966842 is in Supplemental Figure S18.
D. melanogaster population data for Gas8, Notch, Tor, Tehao, and CG16752 were obtained from the Drosophila Population Genomics Project (http://www.dpgp.org/). DPGP data serve as a community resource and consist of 7 Mb of population data for 40 U.S. strains and 10 African strains that were resequenced using array-based sequencing technology (Affymetrix GeneChip CustomSeq Resequencing Arrays). Singleton single-nucleotide polymorphisms were eliminated before analysis. Data are available at http://www.dpgp.org/melanogaster.
Sequence analysis
D. melanogaster divergence from the inferred D. melanogaster/D. simulans ancestor was estimated using gestimator from the libsequence C++ library (Thornton 2003). The expected nucleotide heterozygosity (π) was estimated as the average pairwise difference between D. melanogaster alleles (Nei 1987; Weir 1990). For coding regions, the numbers of synonymous and nonsynonymous sites were counted using the method of Nei and Gojobori (1986). The pathway between two codons was calculated as the average number of synonymous and nonsynonymous changes from all possible paths between the pair. Substitutions to/from G|C from/to A|T were counted using the inferred D. melanogaster/D. simulans sequence described above. Substitutions to/from preferred and unpreferred codons in D. melanogaster were also estimated from the inferred D. melanogaster/D. simulans ancestor (Begun et al. 2007).
Polarized McDonald-Kreitman tests (McDonald and Kreitman 1991) used D. melanogaster polymorphism data and D. simulans and D. yakuba reference sequences to infer the D. simulans/D. melanogaster ancestral state. We took the conservative approach of using the pathway between codons that minimized the number of nonsynonymous substitutions along the D. melanogaster lineage. A Perl script for McDonald-Kreitman tests is available from the Corresponding Author. Hudson-Kreitman-Aguade tests (Hudson et al. 1987) were carried out using DnaSP version 4 (Rozas et al. 2003).
Gene Ontology
We used Gene Ontology terms from the Flybase Gene Ontology terms (http://flybase.org/genes/lk/function) in combination with the generic Gene Ontology Slim set of ontology terms (http://geneontology.org/GO.slims.shtml#avail). The proportion of genes containing a DMAR was calculated for each ontology term. We determined whether each ontology term had a higher proportion of genes with DMARs than would be expected from the empirical distribution. We derived the empirical distribution for each ontology term by drawing the same number of genes that were annotated with each term from all genes that were present in conserved blocks. We used only genes contained in blocks previously identified as conserved in case there was some bias present in the set of genes contained within conserved regions. We then calculated the proportion in the resampled data set that contained DMARs. We used 10,000 resampled data sets to derive the empirical distribution for each term.
Secondary structure analysis
We estimated the secondary structure of DMARs using EvoFold (Pedersen et al. 2006) and RNAFold (Hofacker et al. 1994). Additionally, we uploaded the coordinates of DMARs as a custom track on the UCSC Genome Browser to determine whether there were any predicted smaller regions of secondary structure that would not have been identified in examination of the secondary structure of the entire DMAR sequences.
EvoFold identifies functional RNA structures in multiple sequence alignments using a probabilistic model that takes into account evolutionary relationships between species in the alignment (Pedersen et al. 2006). RNAfold uses a dynamic programming algorithm to predict structures with minimum free energies and computes the equilibrium partition functions and base-pairing probabilities (Zuker and Stiegler 1981; McCaskill 1990; Hofacker et al. 1994).
Acknowledgments
We thank Angie Hinrichs at UCSC for providing the 15-way whole-genome MULTIZ alignments, Ryan Bickel and Mia Levine at UC Davis for valuable comments and suggestions, Melissa Eckert at UC Davis for laboratory advice and assistance, Elizabeth Milano and Umbreen Arshad at UC Davis for laboratory assistance, and Hiram Clawson at UCSC for help with installation of the Kent library. We also thank four anonymous reviewers for comments that improved this work. A.K.H. and D.J.B. were funded by NIH Grant R01-GM071926 to D.J.B.; A.S. was funded by NSF Faculty Early Career Development grant DBI-0644111.
Footnotes
[Supplemental material is available online at www.genome.org. Sequence data have been submitted to GenBank under accession nos. EU588685–EU588714.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.077131.108.
References
- Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M., Holt A., Afzal V., Pennacchio L.A., Rubin E.M., Afzal V., Pennacchio L.A., Rubin E.M., Pennacchio L.A., Rubin E.M., Rubin E.M. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Molecular evolution between Drosophila melanogaster and D. simulans: Reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics. 1996;144:1297–1307. doi: 10.1093/genetics/144.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164:1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H., Goel P., John A., Goel P., John A., John A. Ancestral inference and the study of codon bias evolution: Implications for molecular evolutionary analyses of the Drosophila melanogaster subgroup. PLoS One. 2007;2:e1065. doi: 10.1371/journal.pone.0001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S.G., Kurland C.G., Kurland C.G. Codon preferences in free-living microorganisms. Microbiol. Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S., Noble W.S., Kryukov G., Grant C.E., Sunyaev S., Stamatoyannopoulos J.A., Noble W.S., Kryukov G., Grant C.E., Sunyaev S., Stamatoyannopoulos J.A., Kryukov G., Grant C.E., Sunyaev S., Stamatoyannopoulos J.A., Grant C.E., Sunyaev S., Stamatoyannopoulos J.A., Sunyaev S., Stamatoyannopoulos J.A., Stamatoyannopoulos J.A. Widely distributed noncoding purifying selection in the human genome. Proc. Natl. Acad. Sci. 2007;104:12410–12415. doi: 10.1073/pnas.0705140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S., Bopp D., Burri M., Noll M., Bopp D., Burri M., Noll M., Burri M., Noll M., Noll M. Structure of two genes at the gooseberry locus related to the paired gene and their spatial expression during Drosophila embryogenesis. Genes & Dev. 1987;1:1247–1267. doi: 10.1101/gad.1.10.1247. [DOI] [PubMed] [Google Scholar]
- Begun D.J., Holloway A.K., Stevens K.S., Hillier L.W., Poh Y., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C., Holloway A.K., Stevens K.S., Hillier L.W., Poh Y., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C., Stevens K.S., Hillier L.W., Poh Y., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C., Hillier L.W., Poh Y., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C., Poh Y., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C., Nista P.M., Jones C.D., Kern A.D., Dewey C., Jones C.D., Kern A.D., Dewey C., Kern A.D., Dewey C., Dewey C., et al. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bergman C.M., Carlson J.W., Celniker S.E., Carlson J.W., Celniker S.E., Celniker S.E. Drosophila DNase I footprint database: A systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Snyder M., Dermitzakis E.T., Thurman R.E., Dermitzakis E.T., Thurman R.E., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlini D.B., Stephan W., Stephan W. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics. 2003;163:239–243. doi: 10.1093/genetics/163.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas S., Barbadilla A., Bergman C.M., Barbadilla A., Bergman C.M., Bergman C.M. Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol. Biol. Evol. 2007;24:2222–2234. doi: 10.1093/molbev/msm150. [DOI] [PubMed] [Google Scholar]
- Clark A.G., Eisen M.B., Smith D.R., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Eisen M.B., Smith D.R., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Smith D.R., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Markow T.A., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Kaufman T.C., Kellis M., Gelbart W., Iyer V.N., Kellis M., Gelbart W., Iyer V.N., Gelbart W., Iyer V.N., Iyer V.N., et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Drake J.A., Bird C., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Bird C., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., Attar H., Antonarakis S.E., Dermitzakis E.T., Antonarakis S.E., Dermitzakis E.T., Dermitzakis E.T., et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat. Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Drummond D.A., Bloom J.D., Adami C., Wilke C.O., Arnold F.H., Bloom J.D., Adami C., Wilke C.O., Arnold F.H., Adami C., Wilke C.O., Arnold F.H., Wilke C.O., Arnold F.H., Arnold F.H. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.A., Raval A., Wilke C.O., Raval A., Wilke C.O., Wilke C.O. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- DuMont V.B., Fay J.C., Calabrese P.P., Aquadro C.F., Fay J.C., Calabrese P.P., Aquadro C.F., Calabrese P.P., Aquadro C.F., Aquadro C.F. DNA variability and divergence at the notch locus in Drosophila melanogaster and D. simulans: A case of accelerated synonymous site divergence. Genetics. 2004;167:171–185. doi: 10.1534/genetics.167.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. The genomic rate of adaptive evolution. Trends Ecol. Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Gallo S.M., Li L., Hu Z., Halfon M.S., Li L., Hu Z., Halfon M.S., Hu Z., Halfon M.S., Halfon M.S. REDfly: A regulatory element database for Drosophila. Bioinformatics. 2006;22:381–383. doi: 10.1093/bioinformatics/bti794. [DOI] [PubMed] [Google Scholar]
- Gillespie J.H. The causes of molecular evolution. Oxford University Press; New York: 1991. [Google Scholar]
- Gutjahr T., Patel N.H., Li X., Goodman C.S., Noll M., Patel N.H., Li X., Goodman C.S., Noll M., Li X., Goodman C.S., Noll M., Goodman C.S., Noll M., Noll M. Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and activates gooseberry neuro. Development. 1993;118:21–31. doi: 10.1242/dev.118.1.21. [DOI] [PubMed] [Google Scholar]
- Halligan D.L., Eyre-Walker A., Andolfatto P., Keightley P.D., Eyre-Walker A., Andolfatto P., Keightley P.D., Andolfatto P., Keightley P.D., Keightley P.D. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 2004;14:273–279. doi: 10.1101/gr.1329204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A., Ponting C.P., Ponting C.P. Variable strength of translational selection among 12 Drosophila species. Genetics. 2007;177:1337–1348. doi: 10.1534/genetics.107.070466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild M., Beckmann B., Haas S.A., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Beckmann B., Haas S.A., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Haas S.A., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Koch B., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Solovyev V., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Busold C., Fellenberg K., Boutros M., Vingron M., Sauer F., Fellenberg K., Boutros M., Vingron M., Sauer F., Boutros M., Vingron M., Sauer F., Vingron M., Sauer F., Sauer F., et al. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G., Robertson A., Robertson A. The effect of linkage on limits to artificial selection. Genet. Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P., Bonhoeffer S., Tacker M., Schuster P., Tacker M., Schuster P., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Hudson R.R., Kreitman M., Aguade M., Kreitman M., Aguade M., Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Sugnet C.W., Haussler D., Kent W.J., Haussler D., Kent W.J., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S., Kern A.D., Bejerano G., Fewell G., Fulton L., Wilson R.K., Salama S.R., Haussler D., Kern A.D., Bejerano G., Fewell G., Fulton L., Wilson R.K., Salama S.R., Haussler D., Bejerano G., Fewell G., Fulton L., Wilson R.K., Salama S.R., Haussler D., Fewell G., Fulton L., Wilson R.K., Salama S.R., Haussler D., Fulton L., Wilson R.K., Salama S.R., Haussler D., Wilson R.K., Salama S.R., Haussler D., Salama S.R., Haussler D., Haussler D. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D., Baertsch R., Hinrichs A., Miller W., Haussler D., Hinrichs A., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Pritchard J.K., Pritchard J.K. Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet. 2007;3:1572–1586. doi: 10.1371/journal.pgen.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Calcagno A.M., Ambudkar S.V., Gottesman M.M., Ambudkar S.V., Gottesman M.M., Gottesman M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kliman R.M., Hey J., Hey J. The effects of mutation and natural selection on codon bias in the genes of Drosophila. Genetics. 1994;137:1049–1056. doi: 10.1093/genetics/137.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A.A., Lesnik T., Reiss C., Lesnik T., Reiss C., Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G.N., Godson G.N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc. Natl. Acad. Sci. 1983;80:687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C.H., Fitch W.M., Fitch W.M. The constancy of evolution: A statistical analysis of the alpha and beta haemoglobins, cytochrome c and fibrinopeptide A. In: Morton N.E., editor. Genetic structure of populations. University of Hawaii Press; Honolulu: 1973. pp. 246–262. [Google Scholar]
- Langley C.H., Fitch W.M., Fitch W.M. An estimation of the constancy of the rate of molecular evolution. J. Mol. Evol. 1974;3:161–177. doi: 10.1007/BF01797451. [DOI] [PubMed] [Google Scholar]
- Lefevre G. A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands. In: Ashburner M., Novitski E., Novitski E., editors. The genetics and biology of Drosophila. Academic Press; London: 1976. pp. 31–66. [Google Scholar]
- Li X., Noll M., Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994a;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Noll M., Noll M. Evolution of distinct developmental functions of three Drosophila genes by acquisition of different cis-regulatory regions. Nature. 1994b;367:83–87. doi: 10.1038/367083a0. [DOI] [PubMed] [Google Scholar]
- Li X., Gutjahr T., Noll M., Gutjahr T., Noll M., Noll M. Separable regulatory elements mediate the establishment and maintenance of cell states by the Drosophila segment-polarity gene gooseberry. EMBO J. 1993;12:1427–1436. doi: 10.1002/j.1460-2075.1993.tb05786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Yekta S., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Vertebrate microRNA genes. Science. 2003a;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P., Rhoades M.W., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003b;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- McDonald J.H., Kreitman M., Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Neafsey D.E., Galagan J.E., Galagan J.E. Positive selection for unpreferred codon usage in eukaryotic genomes. BMC Evol. Biol. 2007;7:119. doi: 10.1186/1471-2148-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Nei M., Gojobori T., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bauer DuMont V.L., Hubisz M.J., Aquadro C.F., Bauer DuMont V.L., Hubisz M.J., Aquadro C.F., Hubisz M.J., Aquadro C.F., Aquadro C.F. Maximum likelihood estimation of ancestral codon usage bias parameters in Drosophila. Mol. Biol. Evol. 2007;24:228–235. doi: 10.1093/molbev/msl146. [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M., Kimura M. On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1971;1:18–25. doi: 10.1007/BF01659391. [DOI] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D., Lander E.S., Kent J., Miller W., Haussler D., Kent J., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A., Katzman S., King B., Onodera C., Siepel A., King B., Onodera C., Siepel A., Onodera C., Siepel A., Siepel A., et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Purvis I.J., Bettany A.J., Santiago T.C., Coggins J.R., Duncan K., Eason R., Brown A.J., Bettany A.J., Santiago T.C., Coggins J.R., Duncan K., Eason R., Brown A.J., Santiago T.C., Coggins J.R., Duncan K., Eason R., Brown A.J., Coggins J.R., Duncan K., Eason R., Brown A.J., Duncan K., Eason R., Brown A.J., Eason R., Brown A.J., Brown A.J. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol. 1987;193:413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sanchez-DelBarrio J.C., Messeguer X., Rozas R., Sanchez-DelBarrio J.C., Messeguer X., Rozas R., Messeguer X., Rozas R., Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Siepel A., Haussler D., Haussler D. Phylogenetic estimation of contextdependent substitution rates by maximum likelihood. Mol. Biol. Evol. 2004;21:468–488. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.D., Bauer Dumont V.L., Hubisz M.J., Nielsen R., Aquadro C.F., Bauer Dumont V.L., Hubisz M.J., Nielsen R., Aquadro C.F., Hubisz M.J., Nielsen R., Aquadro C.F., Nielsen R., Aquadro C.F., Aquadro C.F. Patterns of mutation and selection at synonymous sites in Drosophila. Mol. Biol. Evol. 2007;24:2687–2697. doi: 10.1093/molbev/msm196. [DOI] [PubMed] [Google Scholar]
- Smith N.G., Eyre-Walker A., Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Crosby M.A., Rasmussen M.D., Roy S., Rasmussen M.D., Roy S., Roy S. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Hua S., Herreman T., Tongprasit W., Barbano P.E., Herreman T., Tongprasit W., Barbano P.E., Tongprasit W., Barbano P.E., Barbano P.E., et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Thanaraj T.A., Argos P., Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. Libsequence: A C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Vicario S., Moriyama E.N., Powell J.R., Moriyama E.N., Powell J.R., Powell J.R. Codon usage in twelve species of Drosophila. BMC Evol. Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., Charlesworth B. Evolution on the X chromosome: Unusual patterns and processes. Nat. Rev. Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Weir B.S. Genetic data analysis. Sinauer; Sunderland, MA: 1990. [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerandl E., Pauling L., Pauling L. Molecular disease, evolution, and genetic heterogeneity. In: Kasha M., Pullman B., Pullman B., editors. Horizons in biochemistry. Academic Press; New York: 1962. pp. 189–225. [Google Scholar]