Abstract
Researchers, supported by data from polyploid plants, have suggested that whole genome duplication (WGD) may induce genomic instability and rearrangement, an idea which could have important implications for vertebrate evolution. Benefiting from the newly released amphioxus genome sequence (Branchiostoma floridae), an invertebrate that researchers have hoped is representative of the ancestral chordate genome, we have used gene proximity conservation to estimate rates of genome rearrangement throughout vertebrates and some of their invertebrate ancestors. We find that, while amphioxus remains the best single source of invertebrate information about the early chordate genome, its genome structure is not particularly well conserved and it cannot be considered a fossilization of the vertebrate preduplication genome. In agreement with previous reports, we identify two WGD events in early vertebrates and another in teleost fish. However, we find that the early vertebrate WGD events were not followed by increased rates of genome rearrangement. Indeed, we measure massive genome rearrangement prior to these WGD events. We propose that the vertebrate WGD events may have been symptoms of a preexisting predisposition toward genomic structural change.
Researchers have proposed that whole genome duplication (WGD) events may increase the genomic rearrangement rate, possibly directly accelerating evolution itself (Otto 2007). This idea has received some support from estimates of genome rearrangement in plant polyploids (Song et al. 1995; Pontes et al. 2004), and around a WGD event in teleost fish (Semon and Wolfe 2007). In the later case, Semon and Wolfe measured an increased rate of rearrangement at the crown of the tetrapod–teleost lineages, but, lacking an appropriate root species, they could not distinguish whether this increased rearrangement rate occurred around the teleost WGD or in the early branch of tetrapods. In addition, comparisons between human and mouse genomes have revealed that most breakpoints occur close to clusters of tandem gene duplications and large segmental duplications, suggesting that local duplication can promote rearrangement (Armengol et al. 2005). While these data are suggestive, the relation of WGD events to genome rearrangement remains largely untested in animals.
Vertebrate genomes show evidence of widespread gene duplication compared to invertebrate genomes, leading Ohno (1970) to propose the existence of two rounds of WGD during early vertebrate evolution, now known as the 2R hypothesis. This has been a hotly debated topic—in large part because early phylogeny-based approaches could not distinguish WGD from other gene duplication models (Gibson and Spring 2000; Hughes et al. 2001). However, analysis of conserved gene order, or synteny, within complete vertebrate genome sequences has provided an increasing body of evidence supporting the 2R hypothesis (McLysaght et al. 2002; Panopoulou et al. 2003; Vandepoele et al. 2004; Dehal and Boore 2005). These studies led Kasahara (2007) to declare that “there is now incontrovertible evidence supporting the 2R hypothesis.” Fascination with this hypothesis has endured because of the dramatic consequences such events could have had for the evolution of vertebrates. It is clear that WGD events provide a quick and easy way to produce vast numbers of duplicate genes, creating a genetic reservoir from which innovations can arise. However, it is not clear whether these WGD events also sparked widespread genome rearrangement.
The cephalochordate amphioxus (Branchiostoma floridae) genome may be a key source of information about the evolution of the early vertebrate genome. Cephalochordates, together with the tunicates, are the closest living relatives of vertebrates (Delsuc et al. 2006), and both taxa separated from the vertebrate lineage prior to the widespread gene duplications. In contrast to tunicates, which are exceptionally diverged in many regards, genome sequence and fluorescent in situ hybridization (FISH) mapping of selected amphioxus regions indicate that amphioxus shares some genomic features with vertebrates (Abi-Rached et al. 2002; Castro and Holland 2003; Hughes and Friedman 2005). Hence, it is thought to be the best preserved preduplication genome. Its genome sequence has recently been completed, and analysis published by the amphioxus genome project has provided what appears to be the most compelling support in favor of the 2R hypothesis to date (Putnam et al. 2008). With this evidence before us, we are presented with a unique opportunity to reconstruct the evolutionary history of the early vertebrate genome.
However, even with the amphioxus genome present, estimating genome rearrangement rates in the ancient vertebrate lineages is not trivial. Algorithms exist that can reconstruct the most likely series of rearrangement events between two closely related species; however, these methods are only effective for the cases where genomes are fully assembled, and breakpoints are not heavily saturated (Nadeau and Taylor 1984; Nadeau and Sankoff 1998; Pevzner and Tesler 2003). Over longer evolutionary times, or when genomes of interest have only scaffold-level assemblies, exact reconstructions of rearrangement events are impossible. In response, authors have derived more flexible approaches to inferring rates of genome rearrangement. Semon and Wolfe used a parsimony-based model to score gene proximity conservation; however, their model required prior knowledge of the locations of WGD events, and the authors warned that it may produce abnormally high rearrangement estimates for genomes with scaffold-level assemblies (Semon and Wolfe 2007). Smith and Voss (2006) used a chromosome association-based score originally suggested by Housworth and Postlethwait (2002) to measure rates of synteny loss through vertebrates; however, once again this measure is only appropriate for fully assembled genomes and only estimates the amount of interchromosomal rearrangement. No method has been shown to reliably estimate the amount of rearrangement between genomes in scaffold-level assemblies, such as the current amphioxus genome assembly.
Here we have completed a survey of the conservation of gene synteny throughout vertebrates and their invertebrate ancestors, using a proximate gene pair method derived from our previous work (Panopoulou et al. 2003). Clustering of syntenic gene segments clearly illustrates the traces of the vertebrate WGD events. Moreover, we use this synteny data to develop a new, simple metric for syntenic conservation, and thereby estimate rates of synteny loss throughout the vertebrate lineages, and among some of their invertebrate ancestors. Our results reveal that amphioxus genomic structure is not exceptionally conserved. Moreover, we find that the vertebrate WGD events were not followed by increased rearrangement rates; the teleost WGD occurred during a period of relatively low synteny loss, and the early vertebrate WGD events appear to have been preceded by a spike in synteny loss, and then followed by a low rate of synteny loss.
Results
Identifying conserved gene synteny among vertebrates and their ancestors
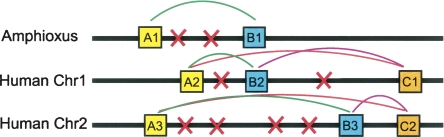
Syntenic gene pairs are the smallest detectable unit of syntenic conservation, and as such, we reasoned that they may provide a suitable foundation for estimating genomic synteny conservation even when studying heavily fragmented genome assemblies. Since the amount of synteny conservation between two species is reduced by genomic rearrangement, genomic rearrangement rates can be inferred from synteny data. To this end, we have adapted our previous gene pair synteny method to detect synteny conservation between pairs of genomes (Fig. 1; Methods). Briefly, for each comparison between two genomes we first group genes into orthologous families, and then identify cases where genes from a pair of gene families are observed in close proximity, which we call a “family combination” (Fig. 1). Family combinations which have proximate gene pairs in more than one location, either across the two genomes of interest or within a single genome, are assumed to have evidence of syntenic conservation. Genes are defined to be in close proximity if they have no more than 10 intervening genes, a threshold which previous simulations have shown is appropriate for identifying true syntenic gene relationships (Panopoulou et al. 2003). These syntenic family combinations can then be assembled into segments of syntenic genes (Fig. 1; Methods).
Figure 1.
Identifying and grouping pairs of syntenic genes. Syntenic gene pairs were identified in three steps. First, genes from the two genomes are grouped into orthologous families (A, B, and C). Next, both genomes are searched for syntenic “family combinations”—pairs of genes from two ortholog families that are found in close proximity in more than one genomic location. Genes are in close proximity if they have no more than 10 intervening genes (shown here as red x’s). In this example, three syntenic family combinations are identified (A–B, A–C, and B–C). The A–B combination is present in both amphioxus and humans, while the A–C and B–C combinations are only present in humans. These syntenic gene pairs can then be merged to generate segments of syntenic genes (A1–B1, A2–B2–C1, and A3–B3–C2). In the human genome these syntenic segments form a “synteny group,” which contains three gene families and is present on two chromosomes.
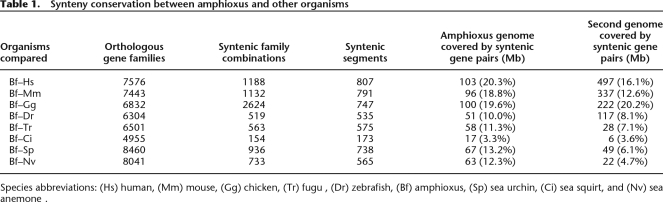
While amphioxus shares the most family combinations with chicken (2644), the family combinations shared with the human genome (1888) assemble into more syntenic segments that cover slightly more of the amphioxus genome (Table 1). The majority of these 807 amphioxus–human syntenic segments include one pair of syntenic genes: 52.6% include two genes, 17% include three genes, and 14.4% include four genes. In total, 490.96 Mb of the human genome is covered by gene pairs with synteny conservation in amphioxus (Supplemental Fig. S1). The equivalent regions in amphioxus extend over 103.02 Mb (Table 1). Supplemental Figure S2 shows the 20 amphioxus scaffolds with the most syntenic gene pairs and their association with the human chromosomes.
Table 1.
Synteny conservation between amphioxus and other organisms
Species abbreviations: (Hs) human, (Mm) mouse, (Gg) chicken, (Tr) fugu , (Dr) zebrafish, (Bf) amphioxus, (Sp) sea urchin, (Ci) sea squirt, and (Nv) sea anemone.
A small set of family combinations are conserved across multiple genomes, possibly identifying cases where strong purifying selection has protected a gene cluster from rearrangement throughout chordates. We identified 167 family combinations that are conserved between amphioxus, human, zebrafish, fugu, and chicken (Supplemental Table S2). These widely conserved genes include the large Hox and Histone syntenic clusters, as well as several ancient tandemly duplicated genes that have preserved their linkage during evolution, including: (1) SMAD2/3–SMAD6/7, (2) gamma-aminobutyric-acid receptors, GABRB3–GABRA5, (3) neuronal voltage-gated calcium channels, CACNG5/7–CACNG4/8, and (4) SIX1/2 and SIX4/5, the last two being proximate in amphioxus, human, and fish, but not in chicken (Kawakami et al. 2000).
Groups of related syntenic segments support the 2R hypothesis
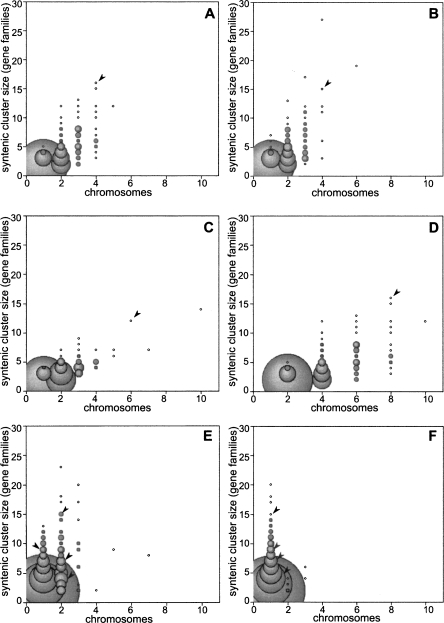
By merging our syntenic gene pairs into groups of related syntenic segments we receive a clear illustration of the genomic duplications events that have occurred during vertebrate evolution. For this analysis, we grouped together syntenic segments that have undergone duplication since an organism’s divergence from the second species used in the synteny comparison (Fig. 1; Methods). By plotting the chromosome coverage of these “synteny groups,” versus their size, we receive a simple illustration of genomic duplication histories (Fig. 2). Vertebrate synteny groups, since divergence from amphioxus, are spread over several chromosomes, reflecting the well-known widespread gene duplication in the vertebrate lineage (Fig. 2A–C). Within the human or chicken genomes, the largest of these synteny groups are present on four chromosomes (Fig. 2A,B), consistent with two WGD events. In zebrafish, the synteny groups spread over a greater number of chromosomes, compatible with an additional WGD in the fish lineage (Fig. 2C). As a control, artificial duplication of the human synteny groups produces a similar spread (Fig. 2D). Furthermore, zebrafish synteny groups built by comparison to the human genome peak at two chromosomes, supporting a single WGD event in the teleost lineage (Fig. 2E). However, many synteny groups are present on three chromosomes, possibly indicating additional duplication mechanisms at work. Conversely, human synteny groups, since divergence from zebrafish, peak sharply at one chromosome, the expected distribution when no WGD has occurred (Fig. 2F).
Figure 2.
Synteny group distributions reveal vertebrate duplication events. The size of each syntenic group, measured by the number of gene families in the group, is plotted against the number of chromosomes over which the group is spread. The bubble sizes are proportional to the number syntenic groups at each point. (A–C) Vertebrate synteny groups built by comparison to amphioxus: (A) human, (B) chicken, and (C) zebrafish. In A and B, the largest groups, with the most conserved synteny, are present on four chromosomes, and a steady reduction in chromosome coverage is seen as the group size decreases. (C) The zebrafish groups are spread out past four chromosomes. (D) As a control, the chromosome coverage of the human groups shown in A was doubled, creating a simulation of a new WGD event on top of the early chordate duplications. This plot shows a similar chromosome spread to C, and post-WGD gene loss in zebrafish could account for the sparser plot. (E) Zebrafish synteny groups, built by comparison to the human genome, show that the largest synteny groups cover two to three chromosomes. (F) Human synteny groups, built by comparison to the zebrafish genome, show a strong peak at one chromosome, as expected in the absence of WGD events. Arrowheads within the plots indicate the bubbles that contain the Hox clusters. In comparisons between amphioxus and vertebrates (A–D), the Hox genes form a single synteny group, while, in comparisons between fish and tetrapods (E–F), they subdivide into four separate groups, indicating that the cluster duplicated twice within the early vertebrate lineage.
Overall, these findings support the existence of two rounds of WGD in early vertebrates (commonly known as the 2R hypothesis), and one additional WGD in teleost fish, in agreement with other recent findings (for review, see Kasahara 2007). In the lineages that are hypothesized to have experienced WGD, as the synteny group size decreases, the number of chromosomes also decreases, indicating that the majority of the observed duplications in these genomes were generated by a mechanism that retains synteny, such as WGD (Fig. 2A–D). If the duplications were generated solely by many local tandem duplications, as suggested by Friedman and Hughes (2001), we would expect to see the opposite trend: Local duplications would be synteny disrupting, so regions duplicated by this mechanism would retain the least synteny. Indeed, this pattern is observed in the human lineage since its divergence from fish, where local segmental duplication has occurred at an increased rate (Fig. 2F) (Bailey and Eichler 2006). These results support data from the amphioxus genome project, which found striking fourfold synteny conservation between the amphioxus and vertebrate genomes (Putnam et al. 2008). Together, these results seem to confirm the emerging consensus in support of the 2R WGD hypothesis.
Syntenic genes maintained in duplicate after WGD are enriched for specific functional classes
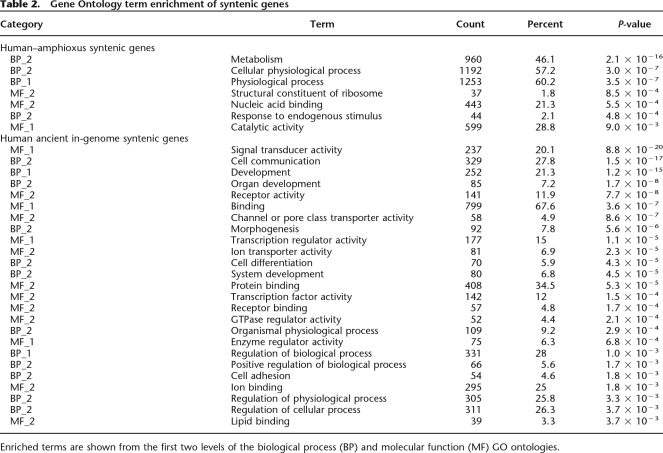
Other reports have indicated that genes retained in duplicate after WGD events often show enrichment for particular functional classes. To test whether the syntenic gene segments that are maintained in duplicate after WGD events show similar functional biases, we used Gene Ontology (GO) term enrichment to compare two sets of human genes—those that show syntenic conservation with amphioxus, and those genes that show duplicate in-genome synteny that was likely to have been created by the early vertebrate WGD events (Methods). The genes in the amphioxus–human syntenic pairs tend to function in metabolism and cellular physiological processes, while the anciently duplicated in-genome human syntenic genes are enriched for a variety of terms related to development, morphogenesis, transcription factor activity, signaling, and regulation (Table 2), indicating that different evolutionary forces may be selecting for synteny conservation and duplicate retention. Previous studies in Arabidopsis and within several vertebrate genomes have similarly observed that genes retained after WGD are enriched for signaling, transcription regulation, and development, indicating that this may be a general consequence of WGD events in plants and animals (Blanc and Wolfe 2004; Maere et al. 2005; Blomme et al. 2006; Brunet et al. 2006).
Table 2.
Gene Ontology term enrichment of syntenic genes
Enriched terms are shown from the first two levels of the biological process (BP) and molecular function (MF) GO ontologies.
Rates of synteny loss did not increase after the vertebrate WGD events
With the existence of the 2R WGD events seemingly well-established, we assessed how these WGD events may have affected rates of synteny loss in vertebrates. We quantified the amount of synteny conservation between two species by calculating the number of shared syntenic gene pairs between the species, and then dividing this value by the number of gene pairs which could be shared if the genomes were ideally arranged (Fig. 3). Because both of these values are reduced by genome fragmentation and gene family loss, in general this syntenic pair measure is largely independent of genome assembly quality. Additional data filters are employed to help correct for structural differences in the genomes (Methods).
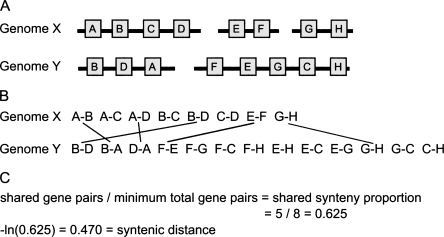
Figure 3.
Estimating the amount of synteny conservation between two genomes. This figure illustrates the method we use to calculate the “syntenic distance” between pairs of genomes. (A) Two genomes, X and Y, share eight orthologous genes, present on three genome fragments in the genome X, and two in genome Y. (B) These orthologous genes can be decomposed into proximate gene pairs (see Methods and Fig. 1). (C) From these gene pairs, we can calculate the shared synteny proportion and convert this proportion into a time-linear distance measure by taking the negative natural logarithm. This is a highly simplified case—for analyses of real genomes, genome fragments are required to have at least 10 genes.
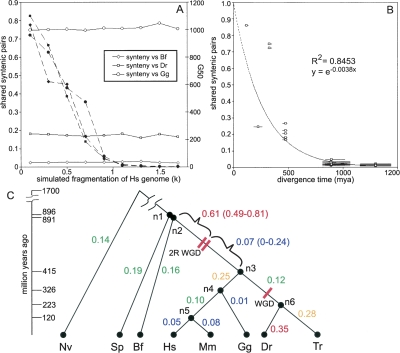
To show that our metric of synteny conservation is indeed robust to genome fragmentation we simulated increasing fragmentation of the human genome, and measured the amount of synteny conservation with the amphioxus, fugu, or chicken genomes (Fig. 4A). In each simulation the increasing fragmentation of the human genome is quantified by measuring the artificial assembly’s “G50” value—the gene number such that 50% of the assembled genome lies in scaffolds containing at least G50 genes. The measured conserved synteny values are highly consistent down to G50 values ∼10–20, after which the variability increases slightly. In general, this indicates that our measures should be robust for our genomes of interest—the amphioxus genome assembly has a G50 value of 90, and the fugu and anemone assemblies both have G50 values of 52. Estimates within the sea urchin lineage should be regarded with the most suspicion since the current sea urchin genome assembly has a G50 value of only three. However, even at this level of fragmentation, our simulated estimates are reasonably reliable.
Figure 4.
Rates of synteny loss throughout vertebrates and their ancestors. (A) Estimates of conserved synteny are robust to genome fragmentation. The human genome was artificially fragmented into scaffolds of random lengths according to a Pareto distribution where k satisfies the equation, scaffold size = 1/U1/k, and U is a random number between 0 and 1. These fragmented human genomes were then compared to amphioxus (Bf), chicken (Gg), or zebrafish (Dr). As k increases, the G50 size of the fragmented human genome decreases (dashed lines), but the syntenic shared pair metric remains relatively consistent (solid lines). G50 is the gene number such that 50% of the assembled genome lies in scaffolds containing at least G50 genes. (B) Conserved synteny was measured between all pairwise combinations of human, fugu, zebrafish, chicken, mouse, amphioxus, sea urchin, and sea anemone and then plotted relative to the divergence age of the comparison. The values are well fit by an exponential curve. (C) Syntenic distances were apportioned to the known species tree and then divided by the estimated evolutionary time in each branch to obtain rates of synteny loss. Internal nodes are labeled n1–n6. The highest rates of loss are observed in the period after the vertebrate divergence from amphioxus but before the early vertebrate WGD events (n2–2R WGD), and in the terminal zebrafish lineage (n6–Dr). Species abbreviations are human (Hs), mouse (Mm), chicken (Gg), fugu (Tr), zebrafish (Dr), amphioxus (Bf), sea urchin (Sp), sea anemone (Nv).
Synteny conservation was measured between all pairwise combinations of sea anemone, sea urchin, amphioxus, human, mouse, chicken, fugu, and zebrafish. This species set was chosen to provide representative species on both sides of the 2R WGD events, while maintaining a set of species where all shared some clear aspects of synteny conservation. Each measure is bootstrapped giving both a 95% confidence estimate on the measure and showing that the measures are robust to significant incompleteness in the genomes (Supplemental Table S1; Methods). Not surprisingly, these measures show an exponential trend when plotted against evolutionary divergence time (Fig. 4B). Over increasing evolutionary distances, the number of remaining syntenic pairs decreases, and therefore the probability that a new rearrangement event disrupts an existing syntenic pair also decreases, creating an exponential decay process. As such, we converted our shared pairs proportions to a linear measure of “syntenic distance” by taking the negative natural logarithm. The largest syntenic distance among these organisms was observed between sea anemone and zebrafish: 4.976 (4.735–5.330). Using randomly shuffled sea anemone and zebrafish genomes, syntenic distances always exceeded 6.368 (from 50 iterations), indicating that even in our most extreme comparison the amount of conserved synteny is well above that expected by chance (P = 3.2 × 10−31).
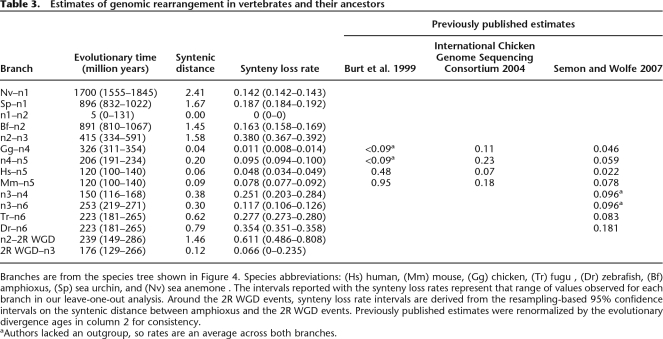
These syntenic distance measures can then be fit onto the known species tree using an additive tree model, and converted to synteny loss rates by dividing by the evolutionary time in each branch (Table 3; Fig. 4C). Because the tree model used to estimate the branch-based estimates of synteny loss could produce skewed results if individual organisms have erroneous data, such as bad gene orthology mapping or inconsistent estimates of divergence age, we tested the robustness of our estimates by eliminating each organism from our data, one at a time, and recalculating synteny loss for every branch possible (i.e., a leave-one-out analysis). Estimated branch lengths do appear to be robust; the range of values received is reported in Table 3. To estimate synteny loss rates around the early vertebrate WGD events, we reconstructed a set of syntenic gene pairs that were likely to have existed in the pre-2R ancestral genome, and used this information to measure the syntenic distance between amphioxus and the 2R WGD events (Methods). We then reallocated the synteny loss in branch n2–n3 before and after the 2R events (Table 3; Fig. 4C).
Table 3.
Estimates of genomic rearrangement in vertebrates and their ancestors
Branches are from the species tree shown in Figure 4. Species abbreviations: (Hs) human, (Mm) mouse, (Gg) chicken, (Tr) fugu , (Dr) zebrafish, (Bf) amphioxus, (Sp) sea urchin, and (Nv) sea anemone. The intervals reported with the synteny loss rates represent that range of values observed for each branch in our leave-one-out analysis. Around the 2R WGD events, synteny loss rate intervals are derived from the resampling-based 95% confidence intervals on the syntenic distance between amphioxus and the 2R WGD events. Previously published estimates were renormalized by the evolutionary divergence ages in column 2 for consistency.
aAuthors lacked an outgroup, so rates are an average across both branches.
Despite the simplicity of our method, we find that our synteny loss rates are generally consistent with published rearrangement rate estimates generated from more complicated models, indicating that gene pair-based synteny loss is a reasonable estimator of genomic rearrangement rates (Table 3). We agree with previous reports that mice have experienced more synteny loss than humans or chicks in their terminal lineages (Burt et al. 1999; International Chicken Genome Sequencing Consortium 2004), and that the rate of rearrangement between the tetrapod radiation and the mammalian radiation (nodes n4–n5) exceeded that in the human and chick lineages. Moreover, our estimates agree with Semon and Wolfe (2007) that fish have experienced more synteny loss in their terminal branches than tetrapods.
In the tested invertebrate lineages—amphioxus, sea urchin, and sea anemone—we find that all three have experienced a similar moderate rate of synteny loss (Fig. 4C). Interestingly, our estimates indicate that urchin has nearly as much conserved synteny with vertebrates as amphioxus, something which has not been previously appreciated in the literature, perhaps because of the extreme fragmentation of the current urchin genome assembly. While it appears that amphioxus currently is the best single source of invertebrate information about the early chordate genome, our data indicate that its genome structure is not particularly strongly conserved, and therefore it cannot be assumed to be uniquely representative of the ancestral chordate genome.
Intriguingly, the vertebrate 2R WGD were followed by a period of relatively low synteny loss (0.07, Fig. 4C). In fact, we estimate that the rate of synteny loss preceding the 2R WGD events was more than eight times higher than the rate afterward. Indeed, the pre-2R WGD period had the highest synteny loss rate observed in our analysis (0.61, Fig. 4C). Some of this synteny loss will have occurred between the two WGD events; however, this period is believed to have been relatively brief, and as such is unlikely to fully account for the observed synteny loss spike (Gibson and Spring 2000; Furlong and Holland 2002; Vandepoele et al. 2004). Overall, these results indicate that the 2R WGD events did not increase the rate synteny loss, and may have occurred during a preexisting period of intense genome rearrangement.
We measure relatively low rates of synteny loss around the teleost WGD (0.12, Fig. 4C). While this result seems to disagree with Semon and Wolfe (2007), these authors lacked a suitable invertebrate outgroup, and as such could not separate the rates of rearrangement in the early tetrapod lineage (n3–n4) from those in the early fish lineage (n3–n6). Our estimates show that the rate of synteny loss in the early tetrapod lineage was more than twice as high as the rate in the early fish lineage (0.25 vs. 0.12). With this new information, it appears that the rearrangement rate around the teleost WGD event was lower than in most neighboring branches. In an attempt to determine the synteny loss rate before and after the teleost WGD event, we employed the same method of ancestral reconstruction used for the 2R WGD events (Methods). Unfortunately, the error in our estimates exceeded the total amount of syntenic loss within this branch. While we note that the synteny loss appears higher after the teleost WGD, the large overlapping confidence intervals on these estimates make it impossible to draw any reliable conclusions (n3–WGD synteny loss = 0.06, 95% confidence = 0–0.15; WGD–n6 synteny loss = 0.38, 95% confidence = 0–0.71).
Discussion
Our data indicate that the early vertebrate whole genome duplication events (2R WGD) did not spark an increase in genomic rearrangement, but were in fact preceded by intense genome rearrangement. These findings contrast with previous studies in plant polyploids (Song et al. 1995; Pontes et al. 2004), indicating that there is not a simple cause-and-effect relationship between WGD events and genome rearrangement. In fact, in vertebrates the opposite may be true: WGD events may be symptoms of existing genome instability.
Naturally, these conclusions are based on our assumptions regarding the existence and timing of the 2R WGD events. A body of evidence from multiple genomes now seems to provide compelling evidence in support of the existence of the 2R WGD events (McLysaght et al. 2002; Panopoulou et al. 2003; Vandepoele et al. 2004; Dehal and Boore 2005; Putnam et al. 2008). We have assumed that these events were relatively closely spaced and centered on the divergence of the lamprey/hagfish lineage from jaw-vertebrates, as indicated by the most recent published reports (Nakatani et al. 2007; Putnam et al. 2008). Assuming an older age, closer to the cephalochordate divergence, merely exaggerates our conclusions, creating a more intense rearrangement spike prior to the WGD events. In the other direction, if we move the 2R WGD events as recent as the divergence of cartilaginous fish (525 million years ago)—a conservative lower bound for their age—there is still more rearrangement prior to the 2R WGD than after (0.40 vs. 0.24). While information from additional chordate genomes will continue to refine these conclusions, it appears quite clear that there was not an increase in synteny loss after the 2R WGD events.
The early vertebrate diversification appears to have been a hot spot of genome structural change. In addition to the two WGD events and the intense prior genome rearrangement, current evidence suggests that the lamprey and hagfish genomes, which diverged from jawed vertebrates around the time of the 2R WGD, may have undergone additional WGD events (Fried et al. 2003; Stadler et al. 2004). This indicates that these lineages may have also possessed a preposition toward structural genome change. From this data it is impossible to determine the cause of this evolutionary hot spot. However, because this time period coincides with an amazing phylogenetic diversification, it is tempting to speculate that there may have been selective pressures favoring structural genome change, possibly as a way of creating functional diversity or sparking speciation. Nonetheless, it is also possible that evolutionarily neutral processes, such as environmental changes or genetic mutations, led to a general increase in genomic instability. Future research will be needed to resolve these issues, and genome sequence from organisms closer to the WGD events, such as lamprey and hagfish, could provide new insights.
Our estimates of the genomic rearrangement rates around the teleost WGD event are somewhat less clear. A previous report indicated an increased rate of synteny loss in the early tetrapod and fish lineages (n3–n4 + n3–n6) (Semon and Wolfe 2007). By including invertebrate genomes in our analysis, we are able to divide the synteny loss in these branches, revealing that this increased rate is due to intense rearrangement in the early tetrapod lineage. Hence, the teleost WGD event appears to have occurred during a period of relatively low genome rearrangement (Fig. 4C). Nonetheless, we were unable to reliably determine the synteny loss rate before and after the teleost WGD event, so it remains possible that synteny loss did increase after the teleost WGD event. However, Nakatani et al. (2007) observed that the teleost WGD event was preceded by a period of intense chromosome fusions, possibly suggesting that the teleost WGD event also followed on the heels of prior genome instability.
In addition to the polyploidy events observed in plants, WGD has been associated with genome rearrangement in another setting—cancer oncogenesis (for review, see Ganem et al. 2007). Genome instability, characterized by frequent aneuploidy and genome rearrangement, is a common feature of cancerous cells and is believed to play a key role in creating oncogenic genetic changes. Studies have associated tetraploidy with early-stage cancer; however, these abnormal tetraploidy events are generally associated with dysfunction in genes like TP53 (also known as p53) and RB1 (also known as Rb), key regulators of genomic integrity (Galipeau et al. 1996; Olaharski et al. 2006). Indeed, within p53-null cells, artificially induced genome duplication can trigger genome instability and oncogenesis (Fujiwara et al. 2005). Hence, while tetraploidy can play a role in oncogenesis, it generally appears to first require changes in the genes that regulate genome stability. In support of this notion, many normal differentiating human cells undergo endoreplication—DNA replication without cell division—showing that polyploidy does not induce genome instability in normal cellular contexts (Ravid et al. 2002). While cancer oncogenesis and phylogenetic diversification occur over very different time scales, we may see a general theme emerging—WGD events alone are not sufficient to trigger genome instability or rearrangement, but instead often appear to associate with prior events that decrease overall genome stability.
Methods
Common names are used for the following organisms: human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), zebrafish (Danio rerio), fugu (Takifugu rubripes), amphioxus (Branchiostoma floridae), sea urchin (Strongylocentrotus purpuratus), sea anemone (Nematostella vectensis), sea squirt (Ciona intestinalis).
Orthology detection
Gene lists and genomic locations were extracted from Ensembl release 42 for all the vertebrate organisms, JGI v1.0 for amphioxus (B. floridae) and anemone (N. vectensis), and Spur v2.1 with Gnomon gene predictions for sea urchin (S. purpuratus). Orthology was assigned using the BLAST-based method Inparanoid, which creates groups of genes between two species that are likely to be related to a single gene in their common ancestor (Remm et al. 2001).
Identifying syntenic gene pairs
When searching for syntenic gene pairs between two genomes, we first grouped genes into orthologous families and then identified cases where genes from a pair of gene families (a “family combination”) were observed in close proximity in more than one location (Fig. 1). Genes were defined to be in close proximity if they had no more than 10 intervening genes. When defining the intervening genes between potential syntenic gene pairs, only protein-coding genes were counted, and genes were treated as one-dimensional objects located at the genes’ predicted start sites. Family combinations which have proximate gene pairs in more than one location, either across the two genomes of interest or within a single genome, are assumed to have evidence of syntenic conservation. Family combinations that only have in-genome synteny are required to have gene pairs on at least two chromosomes, helping to remove groups created by recent local duplication.
Synteny groups were created by exhaustively merging all syntenic family combinations that had gene pairs which shared a gene. Family combinations with in-genome synteny within the target-genome and/or cross-genome synteny with the reference-genome were used. For these analyses we eliminated all vertebrate gene families with 95 or more genes, to prevent these gene families from forming spurious linkages. This threshold eliminated only two classes of genes: olfactory receptor genes, which have seen dramatic expansion in tetrapods, and zebrafish LINE transposable elements.
When compared to our 2003 results, we found that cross-genome amphioxus–human synteny reveals a set of syntenic regions, which are largely different from the syntenic regions we previously identified by human in-genome synteny (Panopoulou et al. 2003). Hence, by combining both sources of information we greatly improve our ability to detect conserved synteny. In the present analysis, in-genome human synteny defines syntenic segments that cover 652.8 Mb of the human genome, while amphioxus–human synteny defines segments that cover 496.7 Mb (Supplemental Fig. S1). These two sets overlap by only 62.1 Mb, defined by 84 syntenic segments containing 44 different family combinations. Supplemental Figure S2 shows a Cohen-Friendly association plot summarizing the synteny association between human chromosomes and amphioxus scaffolds (Cohen 1980; Friendly 1992).
Gene Ontology analysis
The DAVID Bioinformatics Resource 2007 was used to look for enriched GO terms in different classes of human genes with ancient synteny (Table 2) (Dennis et al. 2003). The human–amphioxus gene list includes all human genes contained within proximate gene pairs present in both the human and amphioxus genomes. The human ancient in-genome syntenic gene list includes all human genes contained within in-genome syntenic pairs where phylogeny-based evidence indicates that duplication happened prior to the fish–tetrapod divergence (described in more detail in the last Methods section). The background population was the union of both gene lists—representing the set of all human genes for which we have evidence of ancient syntenic conservation. P-values reported are multiple-test corrected according to the Benjamini-Hochberg method implemented in DAVID. We report all enriched terms from levels 1 and 2 of the molecular function and biological process ontologies with corrected P-values < 0.01.
Measuring rates of synteny loss
Cross-genome syntenic family combinations between each pair of genomes were identified as previously described, with two additional filters that help correct for structural biases in the genomes. First, gene duplications can mask rearrangement events, creating biased estimates of synteny, especially when measuring across WGD events. Therefore, orthology groups are required to have only a single gene within vertebrate genomes. We do not similarly filter orthology groups in amphioxus, and other invertebrates, since these organisms’ current genome builds contain mixtures of haplotypes, which create the appearance of far more gene duplicates than truly exist. Genuine gene duplication in these invertebrate lineages may mask some rearrangement, so we consider our calculated rates of synteny loss in these lineages to be minimum estimates. Second, highly fragmented genome assemblies may contain a disproportionately high number of gene pairs that are immediately adjacent, and these adjacent pairs are more likely to be conserved through evolution than pairs with several intervening genes. Hence, we exclude all chromosomes or scaffolds with less than 10 genes prior to synteny calculations.
To calculate synteny conservation, we count the number of family combinations with proximate genes in at least one location in each genome (total proximate family combinations), and then count the number of family combinations with proximate genes in both genomes (shared family combinations). The shared synteny proportion is calculated as the number of “shared family combinations” divided by the smaller of the “total proximate family combinations” from the two genomes. Each measure is bootstrapped giving both a 95% confidence estimate on the measure and showing that the measures are robust to significant incompleteness in the genomes (Supplemental Table S1). In each iteration, 50% of the family combinations in the smaller genome are removed randomly, and a new proportion is calculated. Confidence intervals shown are each based on 1000 iterations.
The shared synteny proportion is the product of an exponential decay process which can be described by the following formula, where Nt is the observed number of shared family combinations, N0 is the maximum number that could be shared, λ is the rate of synteny loss, and t is the evolutionary divergence time.
Hence, we can calculate a time linear “syntenic distance” (λt), by taking the negative natural logarithm of our syntenic shared pair values.
These syntenic distance measures are then used to calculate the amount of synteny loss on each branch of the species tree using the least-squares-based Fitch-Margoliash method implemented by PHYLIP (Fitch and Margoliash 1967; Retief 2000). This method has the advantage of not relying on ancestral reconstructions at each node and has a long history of use in distance-based phylogenetics. Branch estimates of synteny loss can then be converted to synteny loss rates (λ) by dividing by the amount of evolutionary time in each branch. All synteny loss rates presented in the paper have been multiplied by 100, to improve readability. Divergence times were based on the best nuclear gene estimates in the current literature (Hedges et al. 2004; Blair and Hedges 2005; Blair et al. 2005; Hurley et al. 2007). The 2R WGD events were assumed to be centered on the divergence of the lamprey lineage from jaw-vertebrates (652 million years ago), as indicated in other reports (Nakatani et al. 2007; Putnam et al. 2008). C. intestinalis was not included in our tree-based rate estimates because of concerns about the exact placement of tunicates on the species tree (addressed in Putnam et al. 2008); however, its genome is clearly exceptionally rearranged, confirming previous observations (Ikuta et al. 2004). When compared to amphioxus it has a syntenic distance of 4.83, exceeding the syntenic distance measured between amphioxus and anemone, despite the fact that their divergence is many hundreds of millions of years older.
Synteny conservation was measured between all pairwise combinations of sea anemone, sea urchin, amphioxus, human, mouse, chicken, fugu, and zebrafish. This species set was chosen to provide representatives on both sides of the 2R WGD events, which also satisfied two simple criteria: (1) The species must have publicly available genome sequence, and (2) the species must show aspects of clear synteny conservation with vertebrates. In phyla, where the amount of conserved synteny approaches the amount expected by chance, our syntenic distance metric begins to saturate, leading to increasingly large 95% confidence intervals. This concern led us to specifically exclude from our analysis nematodes, insects, and tunicates.
All of the syntenic distance estimates presented here defined gene proximity using the 10-gene interval shown to be appropriate in our previous work (Panopoulou et al. 2003); however, synteny loss calculations using intervals of five and 15 genes produced highly similar results (data not shown). In both cases the estimated branch lengths had a Pearson correlation exceeding 0.99 when compared to the 10-gene interval values reported here.
We verified the robustness of our branch estimates with a leave-one-out analysis (see Results and Table 3); however, some concern was raised that this analysis might be skewed by the greater number of vertebrates relative to invertebrates. In response to this concern, we made a series of phylogenetically balanced trees that included the three available invertebrates and a selection of three vertebrate genomes. For these vertebrate genomes, we tested all combinations that (1) included at least one tetrapod and one fish and (2) did not include both humans and mice, since these genomes are quite closely related (seven total possibilities). Among these trees, synteny loss rates varied from 0.366 to 0.392 for branch n2–n3, 0.159 to 0.166 for Bf–n2, 0.182 to 0.191 for Sp–n1, and 0.142 to 0.143 for Nv–n1. These intervals are highly similar to those already reported in Table 3 for the leave-one-out analysis, and exactly the same for the sea anemone lineage (0.367–0.392, 0.158–0.169, 0.184–0.192, 0.142–0.143, respectively). These values indicate that the branch estimates are not skewed by organism distribution, and validate the leave-one-out analysis.
Measuring syntenic distance between amphioxus and the 2R WGD events
To identify a set of syntenic gene pairs that were present in the vertebrate genome during the 2R WGD events, we began by identifying family combinations in human, fugu, or zebrafish that have conserved in-genome synteny, i.e., proximate gene pairs have been preserved in multiple places in the same genome. We then built maximum likelihood phylogenetic trees for all the gene families in these family combinations, using the amphioxus orthologs to root the trees (Schmidt et al. 2002; Frickey and Lupas 2004), and subsequently selected family combinations where the genes within the in-genome gene pairs were generated by duplication prior to the tetrapod–teleost split, and declared these ancient vertebrate pairs. If we assume that the majority of gene duplication occurring prior to the tetrapod–teleost split was generated by the vertebrate 2R WGD events—an assumption supported by our synteny group analysis (Fig. 2)—then we can infer that the majority of these ancient vertebrate gene pairs will have been present in the vertebrate genome during 2R WGD events. This process identifies 419 family combinations in the human genome, 113 in fugu, and 70 in zebrafish. Together, this makes 513 qualifying ancient vertebrate pairs, of which only 28 are conserved in amphioxus. The syntenic distance between the amphioxus genome and these pre-2R ancient pairs was estimated to be 2.91 (2.17–3.38).
This syntenic distance is based on a relatively small set of inferred syntenic family combinations, and naturally we were wary that such sets may not produce accurate estimates of syntenic distance. To assure that this type of comparison is valid, we also tested ancestral sets that were inferred to be present at the tetrapod–teleost divergence (node n3), and at the zebrafish–fugu divergence (node n6), and then compared these estimates to the values generated from our previous whole genome comparisons. For the tetrapod–teleost divergence we selected the family combinations common to the human genome and at least one fish genome, a set of 9161 pairs, and compared this set to the amphioxus genome, measuring a syntenic distance of 2.96 (2.88–3.06), close to the distance of 3.02 estimated by our additive tree model (Table 3, Bf–n2 + n2–n3). To obtain a set of ancestral family combinations of similar size to our 2R WGD set, we randomly selected sets of 500 family combinations from these 9161 tetrapod–teleost pairs. In 100 trails, the mean syntenic distance was 2.98, and the estimated 95% confidence intervals contained 3.02 exactly 95% of the time. Similar results were obtained from the ancestral set inferred to exist at the divergence of zebrafish and fugu. For 100 trails of 500 random family combinations, the mean distance to amphioxus was 3.48, and the confidence intervals contained the previous estimate of 3.32 (Table 3, Bf–n2 + n2–n3 + n3–n6) in 91% of the cases. Hence, syntenic distances calculated from ancestral sets are reliable, and the confidence intervals appear to generally account for the increase in uncertainty.
We used similar ancestral reconstruction to try to subdivide the synteny loss before and after the teleost-specific WGD event. The human, fugu, zebrafish, and amphioxus phylogenetic trees were used to identify syntenic gene pairs that duplicated prior to the teleost divergence (n6), but after the tetrapod–fish divergence (n3). This analysis identified 135 family combinations that were likely to be present immediately prior to the teleost WGD event. The syntenic distance between these pairs and the human genome is 0.75 (0.59–0.93). The 95% confidence interval for this measure is narrower than the interval calculated for the 2R WGD reconstruction (0.34 vs. 1.21), but nonetheless, because the total amount of synteny loss around the teleost WGD event is so low (Table 3, n3–n6 = 0.30), the confidence intervals for the resulting synteny loss rates before and after the teleost WGD event are large and overlapping (n3–WGD, synteny loss rate = 0.06, 95% confidence = 0–0.15; WGD–n6, synteny loss rate = 0.38, 95% confidence = 0–0.71). Hence, the relatively low amount of synteny loss in this branch prevents us from determining the synteny loss before and after the teleost WGD with any reliability.
Acknowledgments
We thank N. Putnam from the JGI amphioxus genome project for sharing their unpublished data with us, and S. Haas, P. Polak, and A. Borchers for helpful comments. This work was supported by the Max-Planck Society (Max-Planck Gesellschaft zur Forderung der Wissenschaften e.v.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.080119.108.
References
- Abi-Rached L., Gilles A., Shiina T., Pontarotti P., Inoko H., Gilles A., Shiina T., Pontarotti P., Inoko H., Shiina T., Pontarotti P., Inoko H., Pontarotti P., Inoko H., Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat. Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- Armengol L., Marques-Bonet T., Cheung J., Khaja R., Gonzalez J.R., Scherer S.W., Navarro A., Estivill X., Marques-Bonet T., Cheung J., Khaja R., Gonzalez J.R., Scherer S.W., Navarro A., Estivill X., Cheung J., Khaja R., Gonzalez J.R., Scherer S.W., Navarro A., Estivill X., Khaja R., Gonzalez J.R., Scherer S.W., Navarro A., Estivill X., Gonzalez J.R., Scherer S.W., Navarro A., Estivill X., Scherer S.W., Navarro A., Estivill X., Navarro A., Estivill X., Estivill X. Murine segmental duplications are hot spots for chromosome and gene evolution. Genomics. 2005;86:692–700. doi: 10.1016/j.ygeno.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Eichler E.E., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Blair J.E., Hedges S.B., Hedges S.B. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Blair J.E., Shah P., Hedges S.B., Shah P., Hedges S.B., Hedges S.B. Evolutionary sequence analysis of complete eukaryote genomes. BMC Bioinformatics. 2005;6:53. doi: 10.1186/1471-2105-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H., Wolfe K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T., Vandepoele K., De Bodt S., Simillion C., Maere S., de Van Peer Y., Vandepoele K., De Bodt S., Simillion C., Maere S., de Van Peer Y., De Bodt S., Simillion C., Maere S., de Van Peer Y., Simillion C., Maere S., de Van Peer Y., Maere S., de Van Peer Y., de Van Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet F.G., Crollius H.R., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Crollius H.R., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Paris M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Aury J.M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M., Jaillon O., Laudet V., Robinson-Rechavi M., Laudet V., Robinson-Rechavi M., Robinson-Rechavi M. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Burt D.W., Bruley C., Dunn I.C., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Bruley C., Dunn I.C., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Dunn I.C., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Morrice D.R., Paton I.R., Smith J., Windsor D., Paton I.R., Smith J., Windsor D., Smith J., Windsor D., Windsor D., et al. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- Castro L.F., Holland P.W., Holland P.W. Chromosomal mapping of ANTP class homeobox genes in amphioxus: Piecing together ancestral genomes. Evol. Dev. 2003;5:459–465. doi: 10.1046/j.1525-142x.2003.03052.x. [DOI] [PubMed] [Google Scholar]
- Cohen A. On the graphical display of the significant components in a two-way contingency table. Comm. Statist. Theory Methods. 1980;A9:1025–1041. [Google Scholar]
- Dehal P., Boore J.L., Boore J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H., Brinkmann H., Chourrout D., Philippe H., Chourrout D., Philippe H., Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Fitch W.M., Margoliash E., Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Frickey T., Lupas A.N., Lupas A.N. PhyloGenie: Automated phylome generation and analysis. Nucleic Acids Res. 2004;32:5231–5238. doi: 10.1093/nar/gkh867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried C., Prohaska S.J., Stadler P.F., Prohaska S.J., Stadler P.F., Stadler P.F. Independent Hox-cluster duplications in lampreys. J. Exp. Zoolog. B Mol. Dev. Evol. 2003;299:18–25. doi: 10.1002/jez.b.37. [DOI] [PubMed] [Google Scholar]
- Friedman R., Hughes A.L., Hughes A.L. Pattern and timing of gene duplication in animal genomes. Genome Res. 2001;11:1842–1847. doi: 10.1101/gr.200601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friendly M. Graphical methods for categorical data. SAS User Group International Conference Proceedings. 1992;17:190–200. [Google Scholar]
- Fujiwara T., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D., Nitta M., Ivanova E.V., Bronson R.T., Pellman D., Ivanova E.V., Bronson R.T., Pellman D., Bronson R.T., Pellman D., Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Furlong R.F., Holland P.W., Holland P.W. Were vertebrates octoploid? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipeau P.C., Cowan D.S., Sanchez C.A., Barrett M.T., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J., Cowan D.S., Sanchez C.A., Barrett M.T., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J., Sanchez C.A., Barrett M.T., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J., Barrett M.T., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J., Levine D.S., Rabinovitch P.S., Reid B.J., Rabinovitch P.S., Reid B.J., Reid B.J. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc. Natl. Acad. Sci. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Storchova Z., Pellman D., Storchova Z., Pellman D., Pellman D. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Gibson T.J., Spring J., Spring J. Evidence in favour of ancient octaploidy in the vertebrate genome. Biochem. Soc. Trans. 2000;28:259–264. doi: 10.1042/bst0280259. [DOI] [PubMed] [Google Scholar]
- Hedges S.B., Blair J.E., Venturi M.L., Shoe J.L., Blair J.E., Venturi M.L., Shoe J.L., Venturi M.L., Shoe J.L., Shoe J.L. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housworth E.A., Postlethwait J., Postlethwait J. Measures of synteny conservation between species pairs. Genetics. 2002;162:441–448. doi: 10.1093/genetics/162.1.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L., Friedman R., Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol. Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- Hughes A.L., da Silva J., Friedman R., da Silva J., Friedman R., Friedman R. Ancient genome duplications did not structure the human Hox-bearing chromosomes. Genome Res. 2001;11:771–780. doi: 10.1101/gr.160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley I.A., Mueller R.L., Dunn K.A., Schmidt E.J., Friedman M., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Mueller R.L., Dunn K.A., Schmidt E.J., Friedman M., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Dunn K.A., Schmidt E.J., Friedman M., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Schmidt E.J., Friedman M., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Friedman M., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Ho R.K., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Prince V.E., Yang Z., Thomas M.G., Coates M.I., Yang Z., Thomas M.G., Coates M.I., Thomas M.G., Coates M.I., Coates M.I. A new time-scale for ray-finned fish evolution. Proc. Biol. Sci. 2007;274:489–498. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T., Yoshida N., Satoh N., Saiga H., Yoshida N., Satoh N., Saiga H., Satoh N., Saiga H., Saiga H. Ciona intestinalis Hox gene cluster: Its dispersed structure and residual colinear expression in development. Proc. Natl. Acad. Sci. 2004;101:15118–15123. doi: 10.1073/pnas.0401389101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Kasahara M. The 2R hypothesis: An update. Curr. Opin. Immunol. 2007;19:547–552. doi: 10.1016/j.coi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Sato S., Ozaki H., Ikeda K., Sato S., Ozaki H., Ikeda K., Ozaki H., Ikeda K., Ikeda K. Six family genes—Structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y., Raes J., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y., Casneuf T., Van Montagu M., Kuiper M., Van de Peer Y., Van Montagu M., Kuiper M., Van de Peer Y., Kuiper M., Van de Peer Y., Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A., Hokamp K., Wolfe K.H., Hokamp K., Wolfe K.H., Wolfe K.H. Extensive genomic duplication during early chordate evolution. Nat. Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Nadeau J.H., Sankoff D., Sankoff D. Counting on comparative maps. Trends Genet. 1998;14:495–501. doi: 10.1016/s0168-9525(98)01607-2. [DOI] [PubMed] [Google Scholar]
- Nadeau J.H., Taylor B.A., Taylor B.A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y., Takeda H., Kohara Y., Morishita S., Takeda H., Kohara Y., Morishita S., Kohara Y., Morishita S., Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer-Verlag; Berlin, New York: 1970. [Google Scholar]
- Olaharski A.J., Sotelo R., Solorza-Luna G., Gonsebatt M.E., Guzman P., Mohar A., Eastmond D.A., Sotelo R., Solorza-Luna G., Gonsebatt M.E., Guzman P., Mohar A., Eastmond D.A., Solorza-Luna G., Gonsebatt M.E., Guzman P., Mohar A., Eastmond D.A., Gonsebatt M.E., Guzman P., Mohar A., Eastmond D.A., Guzman P., Mohar A., Eastmond D.A., Mohar A., Eastmond D.A., Eastmond D.A. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006;27:337–343. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- Otto S.P. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Panopoulou G., Hennig S., Groth D., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Hennig S., Groth D., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Groth D., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Krause A., Poustka A.J., Herwig R., Vingron M., Lehrach H., Poustka A.J., Herwig R., Vingron M., Lehrach H., Herwig R., Vingron M., Lehrach H., Vingron M., Lehrach H., Lehrach H. New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. Genome Res. 2003;13:1056–1066. doi: 10.1101/gr.874803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P., Tesler G., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Comai L., Viegas W., Pikaard C.S., Viegas W., Pikaard C.S., Pikaard C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N.H., Butts T., Ferrier D.E., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Butts T., Ferrier D.E., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Ferrier D.E., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Furlong R.F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J.K., Shoguchi E., Terry A., Yu J.K., Terry A., Yu J.K., Yu J.K., et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Ravid K., Lu J., Zimmet J.M., Jones M.R., Lu J., Zimmet J.M., Jones M.R., Zimmet J.M., Jones M.R., Jones M.R. Roads to polyploidy: The megakaryocyte example. J. Cell. Physiol. 2002;190:7–20. doi: 10.1002/jcp.10035. [DOI] [PubMed] [Google Scholar]
- Remm M., Storm C.E., Sonnhammer E.L., Storm C.E., Sonnhammer E.L., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Retief J.D. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Semon M., Wolfe K.H., Wolfe K.H. Rearrangement rate following the whole-genome duplication in teleosts. Mol. Biol. Evol. 2007;24:860–867. doi: 10.1093/molbev/msm003. [DOI] [PubMed] [Google Scholar]
- Smith J.J., Voss S.R., Voss S.R. Gene order data from a model amphibian (Ambystoma): New perspectives on vertebrate genome structure and evolution. BMC Genomics. 2006;7:219. doi: 10.1186/1471-2164-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T.C., Lu P., Tang K., Osborn T.C., Tang K., Osborn T.C., Osborn T.C. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler P.F., Fried C., Prohaska S.J., Bailey W.J., Misof B.Y., Ruddle F.H., Wagner G.P., Fried C., Prohaska S.J., Bailey W.J., Misof B.Y., Ruddle F.H., Wagner G.P., Prohaska S.J., Bailey W.J., Misof B.Y., Ruddle F.H., Wagner G.P., Bailey W.J., Misof B.Y., Ruddle F.H., Wagner G.P., Misof B.Y., Ruddle F.H., Wagner G.P., Ruddle F.H., Wagner G.P., Wagner G.P. Evidence for independent Hox gene duplications in the hagfish lineage: A PCR-based gene inventory of Eptatretus stoutii. Mol. Phylogenet. Evol. 2004;32:686–694. doi: 10.1016/j.ympev.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., De Vos W., Taylor J.S., Meyer A., de Van Peer Y., De Vos W., Taylor J.S., Meyer A., de Van Peer Y., Taylor J.S., Meyer A., de Van Peer Y., Meyer A., de Van Peer Y., de Van Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]