Abstract
The division of genomes into distinct replication time zones has long been established. However, an in-depth understanding of their organization and their relationship to transcription is incomplete. Taking advantage of a novel synchronization method (“baby machine”) and of genomic DNA microarrays, we have, for the first time, mapped replication times of the entire mouse genome at a high temporal resolution. Our data revealed that although most of the genome has a distinct time of replication either early, middle, or late S phase, a significant portion of the genome is replicated asynchronously. Analysis of the replication map revealed the genomic scale organization of the replication time zones. We found that the genomic regions between early and late replication time zones often consist of extremely large replicons. Analysis of the relationship between replication and transcription revealed that early replication is frequently correlated with the transcription potential of a gene and not necessarily with its actual transcriptional activity. These findings, along with the strong conservation found between replication timing in human and mouse genomes, emphasize the importance of replication timing in transcription regulation.
Replication of the genome occurs during S phase of the cell cycle in a highly regulated process that ensures the reliability of DNA duplication. Each genomic region is replicated at a distinct time during S phase through the activation of an origin of replication (Hand 1978). Detailed analysis of the time of replication (ToR) of individual genes has revealed a striking correlation between expression and early replication (Braunstein et al. 1982; Goldman et al. 1984; Gilbert 1986; Schmidt and Migeon 1990; Hansen et al. 1993). Expressed genes, such as constitutively transcribed housekeeping genes, replicate at early stages of S phase, whereas repressed tissue-specific genes may replicate in most tissues at late stages and become early-replicating only in the expressing tissue (Holmquist 1987; Selig et al. 1992). These developmental changes in ToR occur over large regions (400 kb–1 Mb) and are not restricted to the region in close proximity to the tissue specific gene itself (Selig et al. 1992; Simon et al. 2001; Perry et al. 2004).
Recently, genome-wide ToR in several organisms was determined (for review, see MacAlpine and Bell 2005). Surprisingly, in Saccharomyces cerevisiae, no correlation between ToR and transcriptional activity was observed (Raghuraman et al. 2001; Yabuki et al. 2002). On the other hand, genomic experiments performed in Drosophila and in the human did reveal a significant correlation between early replication and transcriptional activity (Schubeler et al. 2002; MacAlpine et al. 2004; White et al. 2004; Woodfine et al. 2004, 2005; Jeon et al. 2005; Karnani et al. 2007). Nevertheless, the genomic approach also revealed that many genes do not fit into this general scheme. Specifically, many repressed genes replicate early and many active genes replicate late, suggesting a more complex relationship between ToR and transcription. One of the difficulties in the interpretation of results from these whole-genome experiments is the low spatial (insufficient coverage of the genome) and/or temporal (insufficient sampling along S phase) resolution characteristic of most of these studies.
The genomic organization of the replication time zones is not well characterized. How big are the replication zones, and how does transition between adjacent replication time zones happen? Various studies estimate that the typical size of a replication zone is in the range of 0.5–2 Mb (Selig et al. 1992; Simon et al. 2001). This size scale is consistent with the view that each zone is replicated by the activation of several origins of replication, each responsible for the replication of ∼100–200 kb of DNA with a bidirectional fork moving at a rate of 0.6–3.6 kb/min (Berezney et al. 2000). Nevertheless, very large replicons (>400 kb) also exist. These large regions are replicated by activating a single origin, a process that takes several hours to complete (for review, see Berezney et al. 2000). The abundance of the large replicons and their role in the global organization of the replication time zones are not known and cannot be addressed without high resolution genome-wide mapping of the ToR.
In order to overcome the limitations of previous studies, we monitored the ToR of the entire mouse genome using a novel synchronization technique that yields ToR measurements at seven time points along S phase. The combination of a high resolution sampling procedure with the genomic coverage of oligonucleotide microarray technology has allowed the construction of the first genome-wide map of ToR zones in the mouse. Analysis of the results allows the identification and characterization of early middle and late ToR zones, the identification of asynchronous replicating regions, and the characterization of large replicons located at the transition points between early and late replication zones. Transcription analysis revealed that the described correlation between early replication and transcription should be expanded also to genes that are not transcribed in the experimental condition but are known to be transcribed in other conditions; that is, ToR is correlated with transcriptional potential rather than with the transcription status per se.
Results
“Baby machine” based retroactive synchronization
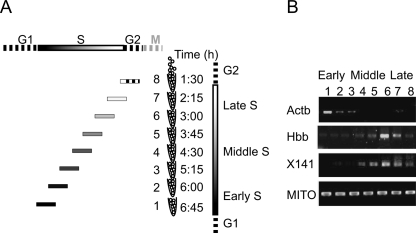
One of the common methods for determining the ToR of a gene is by isolation of newly replicated DNA at different times along S phase of the cell cycle. This procedure involves labeling newly replicated DNA with bromodeoxyuridine (BrdU) at distinct stages of S phase. In order to achieve such synchronization without perturbing the cell cycle we used the “baby machine” device that allows an efficient collection of newborn cells at the end of mitosis (Thornton et al. 2002; Helmstetter et al. 2003). Unsynchronized L1210 cells were pulse labeled by BrdU and immobilized on a membrane, and mitotic cells were collected at different time points. Although all fractions were collected at the end of mitosis, each fraction was exposed to BrdU at a different point in S and thus contains cells with BrdU incorporated in a different part of the genome. In the first sample, only late replicating DNA was labeled, whereas in the last sample, only DNA that replicated during early S phase was labeled (Fig. 1A). BrdU containing DNA was immunoprecipitated from each sample using anti-BrdU antibodies, and the enrichment of certain genomic regions was determined by semi-quantitative PCR. Enrichment of sequences from an early replicating gene, Actb (Holmquist 1987), in the early replicating samples and of late replicating genes, Hbb and X141 regions (Azuara et al. 2003), in the late replicating samples confirms the utility of our approach for studying the replication time zones in the mouse L1210 cell line (Fig. 1B).
Figure 1.
“Baby machine”–based cell cycle synchronization. (A) Schematic representation of the retroactive synchronization method. The cell cycle phases are represented by the bars on the top and right side of the figure. The eight samples collected by the “baby machine” are shown along with the time they were collected (the time after the end of the BrdU labeling is shown). The position and the pattern of the horizontal bars on the left of each sample, represents the portion of S that was BrdU-labeled in each sample. (B) BrdU containing DNA immunoprecipitated from each sample was subjected to semi-quantitative PCR. PCR products were detected for mouse mitochondrial DNA (MITO), demonstrating the uniform loading of newly replicated DNA (Buzina et al. 2005). On the other hand, the early replicating gene (Actb) is enriched in the early samples and the late replicating regions (Hbb and X141) in late samples.
Genome-wide mapping of the mouse replication time zones
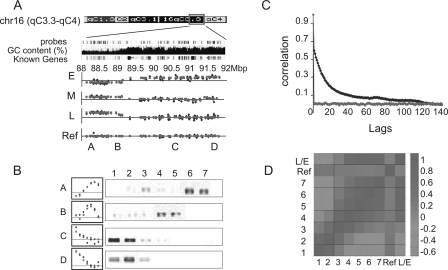
In order to obtain a high resolution map of the replication time zones of the mouse genome, immunoprecipitated DNA from each sample was amplified, labeled, and hybridized to oligonucleotide microarrays. All samples were hybridized against a reference sample of immunoprecipitated DNA from cells in which the entire genome was labeled with BrdU (by a 10-h BrdU pulse). The relative enrichment of a genomic region in each sample reflects the ToR of this region; early replicating regions should be enriched in DNA precipitated from sample 1, whereas late replicating DNA should be enriched in samples 6 or 7. Analysis of the relative enrichment of each of the sequences on the array revealed clear early, middle, and late replicating domains. For example, analysis of a large genomic region on chromosome 16 suggests that it is composed of two large replication time zones (Fig. 2A), a conclusion confirmed by semi-quantitative PCR analysis (Fig. 2B). As expected, the observed ToR is a regional phenomenon with adjacent probes showing similar enrichments (Fig. 2A). This is confirmed using the autocorrelation function, which calculates the correlation between adjacent data points (Fig. 2C). Further confirmation of the data quality is obtained by comparing the enrichment values observed at adjacent time points. As expected from the partial overlap between the BrdU-labeled regions of adjacent samples (Fig. 1A), we observe a high correlation between adjacent samples whereas distant samples are anti correlated (Fig. 2D).
Figure 2.
Genome-wide measurements of the time of replication. (A) A large genomic region (4 Mb) of chromosome 16 is shown along with the probes of the Agilent aCGH microarray, its GC content, and the “Known Genes” track (taken from the UCSC Genome Browser). BrdU enrichment raw (blue) and processed (red; see Methods) data is shown for samples 1,3,6 and the control experiments (E, M, L, and ref, respectively). A, B, C, and D mark the regions that were validated in B. (B) BrdU enrichment, as measured by the arrays (left; red and blue as in A) and by semi-quantitative PCR (right) for regions A–D. Note strong agreement between the data from the microarray and the gene-specific validation. (C) Autocorrelation of all probes in sample 1, with their neighboring probes sorted by their chromosomal position (red) or location on the array (blue) at increasing distances (lag). Similar results were obtained for all seven samples (Supplemental Figure S1). Note that strong correlation is observed for up to at least 100 neighboring sequences (lag = 100). However, no significant autocorrelation is evident for data sorted by array location. (D) Pearson correlation was calculated between each pair of samples. L/E represents a control experiment in which sample 6 was hybridized against sample 1. Note the high correlation around the diagonal, demonstrating the positive correlation between neighboring fractions.
Identification of replication time zones
Data analysis was performed in two steps. First we analyzed the data in each sample by using an optimal segmentation algorithm (Lipson et al. 2006) that defines the optimum segment size for each region and thus overcomes the need to select a set window size for the smoothing algorithm (Fig. 2A; Supplemental Fig. S2, red dots; see Methods). Next we combined the information from the seven samples to define a distinct ToR to each genomic locus. To this end we used two approaches: (1) calculate the time in which half of the replication occurs (TR50) (Jeon et al. 2005), and (2) fit each of the replication vectors (defined by the seven enrichment values) to several predefined distinct replication patterns using a supervised clustering approach (Spellman et al. 1998; Whitfield et al. 2002). Similar results were obtained by both analysis methods (Supplemental Fig. S3). For the rest of this article, we will present the results of the second method since it more easily captures regions that exhibit asynchronous replication.
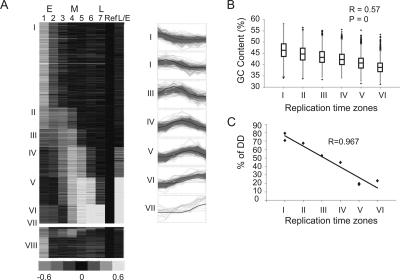
Using supervised clustering, a ToR (I-VII) was assigned to each genomic region (Fig. 3A; Supplemental Fig. S2; see also Methods). We were able to determine the ToR of a total of 1.9 Gb in the mouse genome, which accounts for more than 80% of its currently sequenced part. Hybridizing the same DNA to a denser array (in which a 14-Mb genomic region was probed every 1 kb) improved the coverage to 98% due to a better definition of the borders between adjacent time zones. While the use of a denser array is useful for increasing the coverage, its contribution to the accuracy is limited. For the regions mapped by the two arrays, we obtained almost identical ToR—65% of the sequences were mapped to exactly the same ToR, and another 30% were assigned to a similar ToR (±45 min) (Supplemental Fig. S4).
Figure 3.
Genomic replication timing clustering and validation. (A) The seven predefined patterns (blue) and the probes assigned to each pattern (red, shown on right). A heat-map representation of the genomic data arranged according to the seven clusters (I–VII) is shown on the left. Asynchronous probes that were not assigned to any cluster (VIII) are shown at the bottom. (B) Box plot representation (the box marks the first, second and third quartiles; the whiskers mark values of ±1.5 interquartile range [IQR]; and outliers are marked by black dots) of the GC content of all regions assigned to each cluster. Cluster VII was excluded from this and subsequent analyses since it contains a small number of regions. The correlation to a regression line (R) and the P-value calculated by the F-statistics are shown. (C) FISH-based validation of replication timing. The ToR of eight regions as determined by the percentage of S-phase nuclei in which both alleles are seen as doublets (%DD) is drawn against the ToR of those probes as determined by the array. Additional FISH results are provided in Supplemental Figure S7.
Three lines of evidence support the accuracy of our genome-wide mapping. First, we correctly map the ToR for most (66/81 genes) of the previously measured genes (Supplemental Table S1). Second, early replicating regions highly correlate with high GC content regions, high gene density, and light staining Giemsa bands, as has been previously suggested (Fig. 3B; Supplemental Fig. S5) (Drouin et al. 1994; Woodfine et al. 2004). Third, we validated the replication time of several probes using an independent technique (fluorescence in situ hybridization [FISH]), which resulted in exactly the same results (Fig. 3C). The complete map of the mouse replication time zones is available as an Excel data sheet and as a UCSC genome browser custom track (Supplemental material).
Asynchronous replication
A significant part (Supplemental Fig. S6) of the mapped genome (91%) showed high correlation (≥0.7) to at least one of the replication time patterns (Fig. 3A), strongly suggesting that most of the mouse genome is replicated at a distinct time in S phase. On the other hand, 9% of the genome did not fit into any of the seven patterns (cluster VIII in Fig. 3A). These regions frequently show two peaks of replication, which can be explained either by heterogeneity in the cell population, with some cells replicating a region at early S phase while others replicating the same region later in S phase, or by asynchronous replication of two different alleles in all the cells. A common way to distinguish between these two possibilities is FISH. In FISH experiments, a replicated allele appears as a double dot, whereas an allele that has not yet replicated appears as a single dot. Thus, asynchronous replication will appear as a large proportion of nuclei that exhibit a singlet-doublet (SD) pattern (Kitsberg et al. 1993; Simon et al. 1999, 2001). Performing FISH on the L1210 cells revealed that the asynchronous replication observed in probes from cluster VIII is indeed allelic. All the FISH probes mapped to cluster VIII showed a high percentage (>30%) of SD, whereas probes from the other clusters replicate synchronously (%SD < 10) (Supplemental Fig. S7).
Genomic organization of replication zones
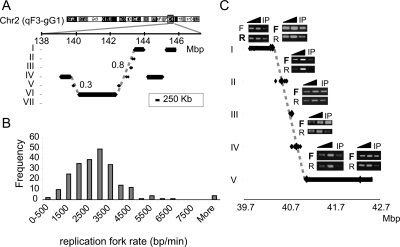
Inspection of the ToR maps revealed many cases in which a gradual change in the time of replication is observed (Fig. 4A; Supplemental Fig. S8). In order to test the extent of this phenomenon, we tested the length of monotonically increasing (or decreasing) sequences of replication zones in the data and compared it to the length observed in shuffled data. The highly significant results (P-value = 5 × 10−43, t-test) (Supplemental Fig. S9) suggest that a significant portion of the genome is arranged in this way. This observation can be explained either by a higher order organization of the replication time zones, a notion that has not been suggested before, or by the existence of very large replicons (>400 kb) in which the time of replication is dependent not only on the time of origin activation but also on the distance from the origin (Ermakova et al. 1999). Four lines of evidence support the large replicon explanation. First, the replication zones in such regions are usually very small (<100 kb). The time each of these small regions replicate fits the assumption (root mean square deviation [RMSD] < 1; see Methods) that a single replication fork proceeds along this region. (Fig. 4A; Supplemental Fig. S8). Second, the replication fork rate (Fig. 4B) deduced from the gradually changing regions (under the assumption of large replicons, see Methods) agrees with previous estimates (Edenberg and Huberman 1975; Housman and Huberman 1975; Yurov 1980). Moreover, in agreement with earlier reports (Housman and Huberman 1975), we found a correlation between the replication fork rate and the ToR. The average fork rate calculated from large replicons starting at time zones I, II, III, and IV was 2.2, 2.6, 3.2, and 3.3 kb/min, respectively. Finally, we analyzed the replication fork direction (see Methods) in four such gradually changing regions. If a region is replicated by a single replication fork, we expect a uniform fork direction along the entire region, whereas if a region contains origins, the fork direction in that region should be bidirectional. We found that in all four regions examined, all the probes (overall 15 probes were examined) exhibit uniform fork direction (P = 3 × 10−5, binomial distribution test; Fig. 4C; Supplemental Fig. S10). Taken together, these results strongly suggest that regions with the gradual ToR variation represent large replicons.
Figure 4.
Large replicons. (A) Large replication segments (>250 kb; black bars) and small replication segments (<250 kb; each flanked by two dots) are shown for a small region in chromosome 2. The vertical position of each line represents its assigned ToR, and the horizontal position represents its location on chromosome 2. Dashed gray lines represent the predicted replication timing of the region assuming a movement of a single replication fork from the early to the late region. The numbers next to the dashed lines represent the RMSD values between the dots and the line. Note the good agreement between the predicted replication time (dashed line) and the actual timing (black dots). Additional examples of such regions are shown in Supplemental Figure S7. (B) Histogram of the replication fork rate, as deduced from the analysis of regions containing large replicons. (C) Replication fork direction was analyzed by measuring the relative amounts of the two strands in BrdU-labeled cells and treated with emetine, which inhibits lagging strand synthesis (see Methods). The ToR of a region on chromosome 1 is shown as in A. The regions for which replication fork direction was analyzed are marked by large diamonds. For each such region, linear PCR with either a forward (F) or reverse (R) primer, followed by semi-quantitative PCR was performed (two dilutions of total DNA [triangles] and emetine-resistant BrdU-labeled DNA [IP]). The direction of the replication fork can be deduced from the relative amount of the reverse and forward reactions. Note that all five regions assayed along the diagonal line show the same direction of the fork (marked by bold letters), strongly suggesting that this entire region is replicated by a single fork.
In light of this interpretation, we divided the genome into two types of replication zones (see Methods): (1) large regions with a uniform ToR—these regions contain origins of replication (although the origins cannot be precisely located with the current array resolution) since a single replication fork cannot replicate such a large region during 45 min; and (2) large regions that show a gradual change in the ToR. Our analysis suggests that these regions are large replicons in which the ToR is defined by the distance from a remote origin (Fig. 4A). As postulated before, our data confirm that most of the genome (90%) consists of large replicons with a uniform ToR. However, our analysis also identified many examples (464 regions covering ∼10% of the genome) of large replicons in which replication is initiated at a distant origin.
Replication time and transcription
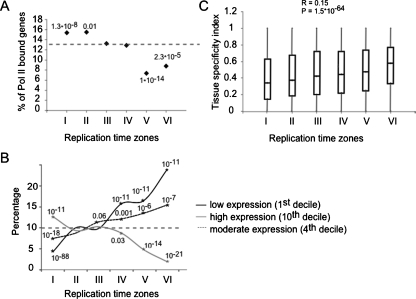
Previous studies suggested that active genes reside in early replicating regions (Braunstein et al. 1982; Goldman et al. 1984; Gilbert 1986; Schmidt and Migeon 1990; Hansen et al. 1993). However, genomics studies in Drosophila and human cells identified many examples of expression of genes residing in late replicating genomic regions (MacAlpine et al. 2004; White et al. 2004; Woodfine et al. 2004, 2005; Jeon et al. 2005). We confirmed these findings in our system by monitoring RNA polymerase II (Pol II) promoter occupancy, using ChIP-chip (Ren et al. 2000), and measuring transcript abundance using expression arrays. Indeed, early replicated regions were enriched with transcribed genes, whereas late replicating regions contained significantly less transcribed genes (Fig. 5A; Supplemental Fig. S11). However, the existence of active late replicating genes and inactive early replicating genes suggests that the relationship between transcription and replication is complex. In order to further characterize the type of genes that reside in each replication time zone, we performed three types of bioinformatics analyses.
Figure 5.
Coordination between replication and transcription. (A) The percentage of genes in each replication time (I–VI) associated with RNA polymerase II (log2 ratio > 0.5) in a ChIP-chip experiment is shown along with the hypergeometric test P-values for cases enriched (clusters I and II) or depleted (clusters V and VI) over the expected average Pol II association (dashed line). (B) The percentage of genes in each replication time zone that fall into the first, fourth, and 10th expression deciles in blood cells. Significant enrichments or depletions over the expected value of 10% (dashed horizontal line) are shown (asterisks and hypergeometric P-values). Early clusters are enriched with highly expressed genes (10th decile) and depleted with low expressed genes (first decile) while late clusters show the opposite pattern. Middle clusters are enriched with genes expressed at moderate levels (fourth decile). (C) Box plot representation of the tissue specificity index distribution for each cluster. Early clusters tend to contain more house-keeping genes (index close to 0) while late clusters tend to contain more tissue-specific genes (index close to 1).
First, we examined the association between transcript levels and ToR, using the L1210 expression data. All transcripts were divided into deciles according to their transcription levels, and each replication time zone was analyzed for the abundance of genes within each decile. We observed a strong correlation between ToR and transcript level; highly transcribed genes (10th decile) are enriched in the early replicating group and depleted from late groups, whereas genes that are barely transcribed (first decile) show the opposite pattern (Fig. 5B). We also observed that genes that replicate in middle S phase are enriched for moderate expression levels (fourth decile). Second, we analyzed the association between the pattern of expression of the genes across various tissues and its ToR. To this end we used a tissue specificity index, a measure of how ubiquitous the expression of a gene is (Yanai et al. 2005). The highly significant correlation (R = 0.15, P < 10−63) between the ToR and the degree of tissue specificity (Fig. 5C), goes beyond the early and late stages and extends also to the middle replicating genes. It should be noted that the expression level in lymphocytic cells was not included in the determination of the tissue specificity index, and, thus, the strong correlation between ToR in lymphocytes (L1210 cells) and the tissue specificity index suggests that ToR is correlated with the overall transcription regulation of a gene and not with its actual transcription in a given cell. Further support for this conclusion comes from the strong correlation between the average gene expression level (across many types of tissues) (Su et al. 2004) and replication time (Supplemental Fig. S12).
Finally, we analyzed the genes in each replication time zone, according to their biological process using GO annotation (Supplemental Table S2). We found the late and middle ToR regions to be enriched for various categories of tissue-specific genes that are not expressed in lymphocytes such as midbrain development (P = 10−4), sensory perception of smell (P = 10−14) and taste (P = 10−6), and keratinization (P = 10−10). On the other hand, the early replicating regions were enriched mainly for housekeeping gene categories such as metabolism (P = 10−60), transport (P = 10−14), transcription (P = 10−14), and cell cycle (P = 10−7), but also for the lymphocytic specific categories, such as immune response (P = 10−7) and lymphocyte activation (P = 10−5). We also found among the early replicating genes, enrichment for the stress response (P = 10−7), and the apoptosis (P = 10−7) categories. The genes in these two latter categories are not expressed under normal growth conditions in L1210 cells but need to be ready to be transcribed quite rapidly in response to certain stimuli. Enrichment of these categories among the early genes further supports the notion that early replication is correlated with transcriptional potential rather than with transcription per se.
Human and mouse conservation
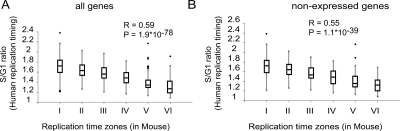
Intuitively, if ToR is an important feature of the genome, then it should be conserved during evolution. In order to address this question, we took advantage of the only other work that profiled the ToR of an entire vertebrate genome. This work profiled the human genome ToR in lymphocytes using BAC arrays by measuring the differences in S/G1 DNA content (Woodfine et al. 2004). The ToR of homologous genes in mouse and human lymphocytes was compared, and the results are shown in Figure 6A. The high correlation between the human and the mouse ToR (R = 0.59, P < 10−77) goes beyond the conservation of GC content and the transcription level of homologous genes, since it is observed even for genes that reside in regions with different GC content or that are not transcribed (Fig. 6B; Supplemental Fig. S13).
Figure 6.
Conservation in replication timing between human and mouse. Box plot representation of the correspondence between human and mouse replication time for all genes (A) and for not expressed genes (B). The human replication time is shown as S:G1 ratio values (Woodfine et al. 2004), in which higher values correspond to early replication. The regression value (R) and the P-values (F-statistics) are shown.
Discussion
We have mapped for the first time the replication time zones of the entire mouse genome using a novel synchronization method that allows immunoprecipitation of newly replicated DNA followed by genome-wide microarray hybridization. Using this method, we were able to obtain high temporal resolution without introducing any perturbation of the cell cycle. Our results are consistent with previous knowledge both in terms of the general features of the replication time zones (Figs. 2C, 3B) and the ToR of individual regions (Supplemental Table S1). Moreover, the ToR of several regions was confirmed by two independent techniques (FISH and semi-quantitative PCR) with high agreement between the results (Figs. 2B, 3C). The accurate high resolution ToR map allows us to tackle issues that were not fully addressed by previous genomic studies.
Middle S replication
Measuring the ToR at multiple time points allowed us to characterize middle S replication. Earlier works based on [3H]thymidine incorporation into DNA of populations of synchronized cells observed a decline in replication during middle S phase. This decline was interpreted as a cessation in DNA replication at the middle of S phase (Holmquist et al. 1982). In contrast to this interpretation, we did observe replication in middle S phase (Fig. 3A). Analyses of the correlation between ToR and GC content, gene density, transcript level, and tissue specificity index indicate that the strong correlation observed for all these features goes beyond the early and late replicating genes and extends to genes from all stages of S phase. These results suggest that the common view of dividing the genome into early and late replicating regions does not capture the full picture, and actually the ToR is a continuous feature with characteristics specific to regions replicating at each time point along S phase. These findings are in line with observations regarding the distinct chromatin structure and nuclear localization of regions replicating at middle S phase (Wu et al. 2005).
Asynchronous replication
Measuring ToR at high temporal resolution allows us to address asynchronous replication on a genome-wide level. We found that ∼9% of the mapped mouse genome replicates asynchronously with two distinct peaks of BrdU incorporation (cluster VIII in Fig. 3A; Supplemental Fig. S7). We also found many genomic regions with a broad replication peak (see Methods) corresponding to cases in which the two alleles replicate asynchronously with a smaller gap between the ToR of each allele (Simon et al. 2001). Indeed, analysis of such regions revealed that they are highly enriched for olfactory receptor genes (20%, P < 10−10) and X-linked genes (8%, P < 10−8) that are known to replicate asynchronously (Chess et al. 1994; Hansen et al. 1996). These two categories of asynchronously replicating regions together cover ∼20% of the mouse genome. Our genome-wide analysis confirms previous estimates (Gimelbrant and Chess 2006) and provides detailed information about the asynchronously replicated regions across the entire mouse genome.
Genomic organization of replication zones
Our analysis suggests that a large portion of the genome (∼10%) is replicated through the activation of a distal origin. This observation allowed us to examine the transition between early and late replication zones. Early replication forks may pause and wait for convergence with late forks. Alternatively, an early fork can continue its replication over a long distance (a process that takes a lot of time) until it will converge with a late fork. Inspection of the ToR maps reveals that almost always the transition between early and late replication zones occurs gradually, and abrupt transitions are quite rare. This observation is not surprising since replication pause sites are more susceptible to DNA breaks and rearrangements (Bierne and Michel 1994; Verbovaia and Razin 1997; Rothstein et al. 2000), and, thus, a gradual transition between replication zones is preferable. A previous report of such transition regions suggested that they are enriched with disease related genes (Watanabe et al. 2002). In our data we did not find any difference in the distribution of single nucleotide polymorphisms (SNPs) or DNA rearrangements between the transition regions and other types of replication zones (data not shown). However, we cannot rule out the possibility that large replicons are more susceptible to DNA damage and thus will have a higher frequency of chromosomal aberrations in early stages of cancer transformation, as has been suggested for fragile sites regions (Tsantoulis et al. 2007).
Replication and transcription
Our data confirmed previous results (Schubeler et al. 2002; MacAlpine et al. 2004; White et al. 2004; Woodfine et al. 2004, 2005; Jeon et al. 2005; Karnani et al. 2007) of an incomplete correlation between early replication and gene expression, suggesting that the relationship between transcription and replication is complex.
In considering the control of gene expression one has to distinguish between two concepts—actual gene transcription and the transcriptional potential of a gene. Thus, inducible genes have high transcription potential even under conditions in which they are not transcribed. Similarly, a silent lineage-specific gene in ES cells has higher transcriptional potential than the same gene in terminally differentiated tissue. A recent study found that many silent lineage-specific genes replicate early in ES cells and later in differentiated cells (Azuara et al. 2006), suggesting that the transcriptional potential of a gene is relevant to its ToR. Our finding of strong enrichment for the apoptosis and stress response GO categories among the early replicating genes further supports this idea and expands it to include terminally differentiated cells. Moreover, the strong correlation found between the ToR and the tissue specificity index of a gene (Fig. 5C), supports the conclusion that the relationship between replication and transcription is not limited to transcription of the gene itself but rather relates to the overall regulation of expression of the gene.
Does replication timing affect transcription?
The causal relationship between ToR and transcription is not clear. Two working models have been proposed to describe this relationship. One model proposes that silent regions are less accessible to replication factors, and therefore their ToR is delayed. An alternative model, supported by microinjection experiments (Zhang et al. 2002), suggests that DNA synthesized at different stages of S phase is exposed to different chromatin components. According to this model, the ToR determines the chromatin structure and therefore the transcription potential of a region (Gilbert 2002; Goren and Cedar 2003). Although it is impossible to determine the causal relationship between replication and transcription just by analyzing correlation data, several aspects of our analysis do support the latter model, indicating that the ToR is not a mere consequence of transcription. First, the observed correlation between the ToR and the transcription potential suggests that transcription per se is not necessary for the determination of the ToR of a region. Second, we identified multiple regions that replicate much later than the time their origin is activated (large replicons). These regions can help distinguish between the two models since according to the first model, in such regions no correlation between ToR and transcription is expected, since as they harbor no origin, the chromatin structure in these regions should not affect their ToR. We found a strong correlation between ToR and transcription, even in these regions (Supplemental Fig. S14), supporting the second model. Finally, we observed a strong conservation of ToR between the human and mouse genome. This conservation was found even in nontranscribed regions, strongly suggesting that ToR is directly regulated and not a mere consequence of transcription.
Methods
“Baby machine”–based synchronization
Newborn cells were isolated from the “baby machine” device, as previously described (Thornton et al. 2002). Cells were pulse labeled with 1 μM BrdU for 45 min and collected for 45 min, starting 90 min after the BrdU chase. Synchronization of the cells isolated from the baby machine was assessed using a ZB Coulter electronic particle counter and flow cytometry (Supplemental Fig. S15). BrdU incorporation was assessed using a quantitative dot blotting method (Supplemental Fig. S16) (Ueda et al. 2005). Based on this analysis, we excluded sample 8 from further experiments since it contained minimal amounts of BrdU.
Isolation of BrdU-labeled DNA by immunoprecipitation
lmmunoprecipitation of BrdU-labeled DNA was carried out essentially as described (Azuara et al. 2003) using 2 μg of anti-BrdU monoclonal antibody (cat. no. 347580, Becton-Dickinson). The specificity of the immunoprecipitation was assessed by mixing BrdU-labeled mouse DNA with sixfold excess of unlabeled human DNA. The relative amounts of mouse and human DNA were measured by semi-quantitative PCR using species specific primers after immunoprecipitation. Almost no human DNA was detected after the immunoprecipitation (Supplemental Fig. S17).
Microarray experiments
BrdU-labeled DNA isolated from each sample was amplified using partially degenerate primers as described (Lieb et al. 2001). We used limited amplification cycles (17–21 cycles) that preserve the relative abundance of the different fragments (data not shown). Samples from each time point were labeled and hybridized to Agilent mouse 44k catalog aCGH arrays and to Agilent custom arrays covering a 14-Mb region on chromosome 1 (154 M–168 M) with an average probe spacing of 1 kb using Agilent standard CGH protocols (http://www.chem.agilent.com/scripts/literaturePDF.asp?iWHID=52010&filename=g4410-90010_cgh_protocol_v5.pdf). The array design avoids repetitive regions of the genome to minimize cross-hybridization. All samples were hybridized against a reference sample containing DNA immunoprecipitated from cells labeled with BrdU for a complete cell cycle (10 h) that were processed in the same manner. In addition two controls were included: (1) hybridization of a reference sample against another reference sample, and (2) hybridization of a late sample (sample 6) against an early sample (sample 1). The results of these two control experiments are included in Figures 2D and 3A. The raw data from each sample were processed using a DNA copy number analysis software application (Stepgram) (Lipson et al. 2006), resulting in optimal segmentation of the genome into segments of homogenous enrichment of BrdU containing DNA (see red dots in Fig. 2A and Supplemental Fig. S2). The processed data were then filtered using a three-median filter to remove single probe outliers. Finally, we recentered each sample according to the 0.1 quantile, allowing comparison of the different samples. Raw data were deposited at ArrayExpress (accession no. E-MEXP-1022) and can also be found along with the processed data in the Supplemental material.
Transcription analysis
ChIP-chip was carried out as described (Odom et al. 2004) using Pol II–specific antibody (cat. no. 8WG16 MMS-126R, COVANCE) and microarray containing 13,000 mouse intergenic regions (Mouse promoterChip BCBC-5A, Beta Cell Consortium). Expression profiling was performed on Affymetrix mouse microarrays 430A 2.0. Raw data were deposited at ArrayExpress (accession no. E-MEXP-1681).
Fluorescence in situ hybridization
ToR of selected regions was determined by performing FISH as described previously (Selig et al. 1992; Kitsberg et al. 1993).
Time of replication determination
Two methods were used: (1) TR50 calculation was done as described by Jeon et al. (2005). Briefly, we defined the total replication of the locus as the sum of the enrichment values in all time points (after adjusting each vector so the minimal value is 0), and the time of 50% replication was calculated by linear interpolation. (2) For supervised clustering, we defined seven Gaussian-like vectors of the form [0, 0, 0.2, 0.6, 0.8, 0.6, 0.2] with peaks at the seven different time points and clustered the ToR measurements for each probe to one of them using Pearson correlation. A significant percentage of the probes (84%) showed high correlation (≥0.7) to at least one of the seven vectors (Fig. 3A), an enrichment of 3.2 over 26% of the probes that showed this level of correlation in randomized data (Supplemental Fig. S3). We partitioned the probes into seven replication clusters according to the vector to which they have maximum correlation. Sixteen percent of the probes did not show significant correlation (≥0.7) with any of the clusters and were therefore assigned to a separate cluster (cluster VIII in Fig. 3A). To determine the replication time zones, we combined adjacent probes that were assigned to the same cluster into a single segment. The 42,694 probes covered ∼1.9 Gb of the mouse genome (after excluding 493 segments containing only one probe). For raw and processed data, see Supplemental material.
Genes were considered asynchronous if they fell into one of two categories: either probes assigned to cluster VIII, or probes with three or more of their correlation values higher than 0.7, which implies broad peak of BrdU enrichment. The first group contains 16% of all probes, which correspond to 9% of all regions measured in our array (167 Mb). In the second group, there are 1983 probes, which correspond to 12% of the mapped genome.
Large replicons definition
Replication initiates at origins and propagates at an approximate rate of 3 kb/min. Thus, during 45 min a single replication fork will label ∼135 kb of DNA. Therefore finding much larger fragments (>250 kb) with uniform BrdU labeling is indicative of the presence of at least one origin in this region. Early replicating time zones (cluster I) must contain an origin as well; thus, both early replicating regions and large replication zones were defined by us as origin containing regions (gray bars in Fig. 4A). On the other hand, small fragments may indicate replication by a distant origin (dots in Fig. 4A). We defined as large replicons those small regions that are in close proximity to a linear line drawn between two origin containing regions, which represent the predicted ToR of the region assuming a movement of a single replication fork from the early to the late region (Fig. 4A, dashed lines). The distance between the measured and the predicted replication time was calculated using the RMSD function. Comparison of the RMSD distribution in the real data and in shuffled data revealed that indeed most of the short replication segments reside in close proximity to the hypothetical lines (Supplemental Fig. S18). We defined as large replicons those regions with a RMSD < 1. The replication fork rate was calculated by dividing the total length of a zone by the time differences between the replication of its beginning to its end. Regions with poor coverage were excluded from this analysis.
Replication fork direction determination
Replication fork direction was determined by using the protein synthesis inhibitor, emetine, which selectively inhibits lagging strand replication as described (Goren et al. 2008).
Acknowledgments
We thank Howard Cedar for advice and comments on the manuscript, Zahava Siegfried for critical reading of the manuscript, and Sharon Elizur for assistance in the FISH experiments. This research was supported by the Israel Science Foundation (grant no. 634/06).
Footnotes
[Supplemental material is available online at www.genome.org. Microarray data have been deposited in ArrayExpress under accession nos. E-MEXP-1022 and E-MEXP-1681.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.079566.108.
References
- Azuara V., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Buckle V., Merkenschlager M., Fisher A.G., Merkenschlager M., Fisher A.G., Fisher A.G. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 2003;5:668–674. doi: 10.1038/ncb1006. [DOI] [PubMed] [Google Scholar]
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Casanova M., Warnes G., Merkenschlager M., Warnes G., Merkenschlager M., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Berezney R., Dubey D.D., Huberman J.A., Dubey D.D., Huberman J.A., Huberman J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- Bierne H., Michel B., Michel B. When replication forks stop. Mol. Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- Braunstein J.D., Schulze D., DelGiudice T., Furst A., Schildkraut C.L., Schulze D., DelGiudice T., Furst A., Schildkraut C.L., DelGiudice T., Furst A., Schildkraut C.L., Furst A., Schildkraut C.L., Schildkraut C.L. The temporal order of replication of murine immunoglobulin heavy chain constant region sequences corresponds to their linear order in the genome. Nucleic Acids Res. 1982;10:6887–6902. doi: 10.1093/nar/10.21.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzina A., Aladjem M.I., Kolman J.L., Wahl G.M., Ellis J., Aladjem M.I., Kolman J.L., Wahl G.M., Ellis J., Kolman J.L., Wahl G.M., Ellis J., Wahl G.M., Ellis J., Ellis J. Initiation of DNA replication at the human beta-globin 3′ enhancer. Nucleic Acids Res. 2005;33:4412–4424. doi: 10.1093/nar/gki747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A., Simon I., Cedar H., Axel R., Simon I., Cedar H., Axel R., Cedar H., Axel R., Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Drouin R., Holmquist G.P., Richer C.L., Holmquist G.P., Richer C.L., Richer C.L. High-resolution replication bands compared with morphologic G- and R-bands. Adv. Hum. Genet. 1994;22:47–115. doi: 10.1007/978-1-4757-9062-7_2. [DOI] [PubMed] [Google Scholar]
- Edenberg H.J., Huberman J.A., Huberman J.A. Eukaryotic chromosome replication. Annu. Rev. Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- Ermakova O.V., Nguyen L.H., Little R.D., Chevillard C., Riblet R., Ashouian N., Birshtein B.K., Schildkraut C.L., Nguyen L.H., Little R.D., Chevillard C., Riblet R., Ashouian N., Birshtein B.K., Schildkraut C.L., Little R.D., Chevillard C., Riblet R., Ashouian N., Birshtein B.K., Schildkraut C.L., Chevillard C., Riblet R., Ashouian N., Birshtein B.K., Schildkraut C.L., Riblet R., Ashouian N., Birshtein B.K., Schildkraut C.L., Ashouian N., Birshtein B.K., Schildkraut C.L., Birshtein B.K., Schildkraut C.L., Schildkraut C.L. Evidence that a single replication fork proceeds from early to late replicating domains in the IgH locus in a non-B cell line. Mol. Cell. 1999;3:321–330. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M. Temporal order of replication of Xenopus laevis 5S ribosomal RNA genes in somatic cells. Proc. Natl. Acad. Sci. 1986;83:2924–2928. doi: 10.1073/pnas.83.9.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D.M. Replication timing and transcriptional control: Beyond cause and effect. Curr. Opin. Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A.A., Chess A., Chess A. An epigenetic state associated with areas of gene duplication. Genome Res. 2006;16:723–729. doi: 10.1101/gr.5023706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist G.P., Gray M.C., Caston L.A., Nag A., Holmquist G.P., Gray M.C., Caston L.A., Nag A., Gray M.C., Caston L.A., Nag A., Caston L.A., Nag A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984;224:686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Goren A., Cedar H., Cedar H. Replicating by the clock. Nat. Rev. Mol. Cell Biol. 2003;4:25–32. doi: 10.1038/nrm1008. [DOI] [PubMed] [Google Scholar]
- Goren A., Tabib A., Hecht M., Cedar H., Tabib A., Hecht M., Cedar H., Hecht M., Cedar H., Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes & Dev. 2008;22:1319–1324. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R. Eucaryotic DNA: Organization of the genome for replication. Cell. 1978;15:317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Canfield T.K., Lamb M.M., Gartler S.M., Laird C.D., Canfield T.K., Lamb M.M., Gartler S.M., Laird C.D., Lamb M.M., Gartler S.M., Laird C.D., Gartler S.M., Laird C.D., Laird C.D. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Canfield T.K., Fjeld A.D., Gartler S.M., Canfield T.K., Fjeld A.D., Gartler S.M., Fjeld A.D., Gartler S.M., Gartler S.M. Role of late replication timing in the silencing of X-linked genes. Hum. Mol. Genet. 1996;5:1345–1353. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- Helmstetter C.E., Thornton M., Romero A., Eward K.L., Thornton M., Romero A., Eward K.L., Romero A., Eward K.L., Eward K.L. Synchrony in human, mouse and bacterial cell cultures—a comparison. Cell Cycle. 2003;2:42–45. doi: 10.4161/cc.2.1.185. [DOI] [PubMed] [Google Scholar]
- Holmquist G.P. Role of replication time in the control of tissue-specific gene expression. Am. J. Hum. Genet. 1987;40:151–173. [PMC free article] [PubMed] [Google Scholar]
- Holmquist G., Gray M., Porter T., Jordan J., Gray M., Porter T., Jordan J., Porter T., Jordan J., Jordan J. Characterization of Giemsa dark- and light-band DNA. Cell. 1982;31:121–129. doi: 10.1016/0092-8674(82)90411-1. [DOI] [PubMed] [Google Scholar]
- Housman D., Huberman J.A., Huberman J.A. Changes in the rate of DNA replication fork movement during S phase in mammalian cells. J. Mol. Biol. 1975;94:173–181. doi: 10.1016/0022-2836(75)90076-5. [DOI] [PubMed] [Google Scholar]
- Jeon Y., Bekiranov S., Karnani N., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Bekiranov S., Karnani N., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Karnani N., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Kapranov P., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Ghosh S., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., MacAlpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Lee C., Hwang D.S., Gingeras T.R., Dutta A., Hwang D.S., Gingeras T.R., Dutta A., Gingeras T.R., Dutta A., Dutta A. Temporal profile of replication of human chromosomes. Proc. Natl. Acad. Sci. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N., Taylor C., Malhotra A., Dutta A., Taylor C., Malhotra A., Dutta A., Malhotra A., Dutta A., Dutta A. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res. 2007;17:865–876. doi: 10.1101/gr.5427007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H., Driscoll D.J., Nicholls R.D., Cedar H., Nicholls R.D., Cedar H., Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Lieb J.D., Liu X., Botstein D., Brown P.O., Liu X., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Lipson D., Aumann Y., Ben-Dor A., Linial N., Yakhini Z., Aumann Y., Ben-Dor A., Linial N., Yakhini Z., Ben-Dor A., Linial N., Yakhini Z., Linial N., Yakhini Z., Yakhini Z. Efficient calculation of interval scores for DNA copy number data analysis. J. Comput. Biol. 2006;13:215–228. doi: 10.1089/cmb.2006.13.215. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Bell S.P., Bell S.P. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Rodriguez H.K., Bell S.P., Rodriguez H.K., Bell S.P., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., Schreiber J., Rolfe P.A., Gifford D.K., Rolfe P.A., Gifford D.K., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P., Sauer S., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Sauer S., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Billon N., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Richardson W.D., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Spivakov M., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Warnes G., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Livesey F.J., Merkenschlager M., Fisher A.G., Azuara V., Merkenschlager M., Fisher A.G., Azuara V., Fisher A.G., Azuara V., Azuara V. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle. 2004;3:1645–1650. [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L., Davis R.W., Brewer B.J., Fangman W.L., Brewer B.J., Fangman W.L., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Michel B., Gangloff S., Michel B., Gangloff S., Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes & Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- Schmidt M., Migeon B.R., Migeon B.R. Asynchronous replication of homologous loci on human active and inactive X chromosomes. Proc. Natl. Acad. Sci. 1990;87:3685–3689. doi: 10.1073/pnas.87.10.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M., Kooperberg C., van Steensel B., Delrow J., Groudine M., van Steensel B., Delrow J., Groudine M., Delrow J., Groudine M., Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat. Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Selig S., Okumura K., Ward D.C., Cedar H., Okumura K., Ward D.C., Cedar H., Ward D.C., Cedar H., Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Tenzen T., Reubinoff B.E., Hillman D., McCarrey J.R., Cedar H., Tenzen T., Reubinoff B.E., Hillman D., McCarrey J.R., Cedar H., Reubinoff B.E., Hillman D., McCarrey J.R., Cedar H., Hillman D., McCarrey J.R., Cedar H., McCarrey J.R., Cedar H., Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Simon I., Tenzen T., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Tenzen T., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Mostoslavsky R., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Fibach E., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Lande L., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Milot E., Gribnau J., Grosveld F., Fraser P., Cedar H., Gribnau J., Grosveld F., Fraser P., Cedar H., Grosveld F., Fraser P., Cedar H., Fraser P., Cedar H., Cedar H. Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B., Sherlock G., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B., Zhang M.Q., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B., Iyer V.R., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B., Anders K., Eisen M.B., Brown P.O., Botstein D., Futcher B., Eisen M.B., Brown P.O., Botstein D., Futcher B., Brown P.O., Botstein D., Futcher B., Botstein D., Futcher B., Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton M., Eward K.L., Helmstetter C.E., Eward K.L., Helmstetter C.E., Helmstetter C.E. Production of minimally disturbed synchronous cultures of hematopoietic cells. Biotechniques. 2002;32:1098–1105. doi: 10.2144/02325rr05. [DOI] [PubMed] [Google Scholar]
- Tsantoulis P.K., Kotsinas A., Sfikakis P.P., Evangelou K., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Kotsinas A., Sfikakis P.P., Evangelou K., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Sfikakis P.P., Evangelou K., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Evangelou K., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Sideridou M., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Levy B., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Mo L., Kittas C., Wu X.R., Papavassiliou A.G., Kittas C., Wu X.R., Papavassiliou A.G., Wu X.R., Papavassiliou A.G., Papavassiliou A.G., et al. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2007;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- Ueda J., Saito H., Watanabe H., Evers B.M., Saito H., Watanabe H., Evers B.M., Watanabe H., Evers B.M., Evers B.M. Novel and quantitative DNA dot-blotting method for assessment of in vivo proliferation. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G842–G847. doi: 10.1152/ajpgi.00463.2004. [DOI] [PubMed] [Google Scholar]
- Verbovaia L.V., Razin S.V., Razin S.V. Mapping of replication origins and termination sites in the Duchenne muscular dystrophy gene. Genomics. 1997;45:24–30. doi: 10.1006/geno.1997.4875. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Fujiyama A., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Fujiyama A., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Ichiba Y., Hattori M., Yada T., Sakaki Y., Ikemura T., Hattori M., Yada T., Sakaki Y., Ikemura T., Yada T., Sakaki Y., Ikemura T., Sakaki Y., Ikemura T., Ikemura T. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: Disease-related genes in timing-switch regions. Hum. Mol. Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- White E.J., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Emanuelsson O., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Scalzo D., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Royce T., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Kosak S., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Oakeley E.J., Weissman S., Gerstein M., Groudine M., Snyder M., Weissman S., Gerstein M., Groudine M., Snyder M., Gerstein M., Groudine M., Snyder M., Groudine M., Snyder M., Snyder M., et al. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc. Natl. Acad. Sci. 2004;101:17771–17776. doi: 10.1073/pnas.0408170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., Perou C.M., Hurt M.M., Brown P.O., Hurt M.M., Brown P.O., Brown P.O., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Debernardi S., Mott R., Dunham I., Carter N.P., Mott R., Dunham I., Carter N.P., Dunham I., Carter N.P., Carter N.P. Replication timing of the human genome. Hum. Mol. Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Woodfine K., Beare D.M., Ichimura K., Debernardi S., Mungall A.J., Fiegler H., Collins V.P., Carter N.P., Dunham I., Beare D.M., Ichimura K., Debernardi S., Mungall A.J., Fiegler H., Collins V.P., Carter N.P., Dunham I., Ichimura K., Debernardi S., Mungall A.J., Fiegler H., Collins V.P., Carter N.P., Dunham I., Debernardi S., Mungall A.J., Fiegler H., Collins V.P., Carter N.P., Dunham I., Mungall A.J., Fiegler H., Collins V.P., Carter N.P., Dunham I., Fiegler H., Collins V.P., Carter N.P., Dunham I., Collins V.P., Carter N.P., Dunham I., Carter N.P., Dunham I., Dunham I. Replication timing of human chromosome 6. Cell Cycle. 2005;4:172–176. doi: 10.4161/cc.4.1.1350. [DOI] [PubMed] [Google Scholar]
- Wu R., Terry A.V., Singh P.B., Gilbert D.M., Terry A.V., Singh P.B., Gilbert D.M., Singh P.B., Gilbert D.M., Gilbert D.M. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N., Terashima H., Kitada K., Terashima H., Kitada K., Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E., Bar-Even A., Horn-Saban S., Safran M., Domany E., Horn-Saban S., Safran M., Domany E., Safran M., Domany E., Domany E., et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yurov Y.B. Rate of DNA replication fork movement within a single mammalian cell. J. Mol. Biol. 1980;136:339–342. doi: 10.1016/0022-2836(80)90378-2. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu F., Hashimshony T., Keshet I., Cedar H., Xu F., Hashimshony T., Keshet I., Cedar H., Hashimshony T., Keshet I., Cedar H., Keshet I., Cedar H., Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]