Abstract
Cooperatively acting pairs of cis-regulatory elements play important roles in many biological processes. Here, we describe a statistical approach, compositionally orthogonalized co-occurrence analysis (coCOA) that detects pairs of oligonucleotides that preferentially co-occur in pairs of sequence regions, controlling for correlations between the compositions of the analyzed regions. coCOA identified three clusters of oligonucleotide pairs that frequently co-occur at 5′ and 3′ ends of human and mouse introns. The largest cluster involved GC-rich sequences at the 5′ ends of introns that co-occur and are co-conserved with specific AU-rich sequences near intron 3′ ends. These motifs are preferentially conserved when they occur together, as measured by a new co-conservation measure, supporting common in vivo function. These motif pairs are also enriched in introns flanking alternative “cassette” exons, suggesting a role in silencing of intervening exons, and we showed that these motifs can cooperatively silence splicing of an intervening exon in a splicing reporter assay. This approach can be easily generalized to problems beyond RNA splicing.
Expression of most human genes requires extensive splicing of primary RNA transcripts to produce mature protein-coding mRNAs. At the core of pre-mRNA splicing are tandem chemical reactions in which pairs of sequence elements—first the branch point and 5′ splice site (5′ss), then the 5′ss and 3′ splice site (3′ss)—are brought together.
Beyond the motifs associated with the core splice sites, a spectrum of splicing enhancer and silencer elements contribute to the specificity and regulation of the splicing reaction. Such auxiliary splicing elements often interact functionally, mediated through binding to the same factor or to distinct, interacting splicing regulatory factors (for review, see Black 2003; Matlin et al. 2005). However, approaches used for systematic identification of splicing regulatory elements have—largely for technical reasons—sought to identify individual elements in isolation (Fairbrother et al. 2002; Wang et al. 2004; Zhang and Chasin 2004; Smith et al. 2006). By their design, such approaches are incapable of identifying elements that mediate their splicing regulatory activity only in the presence of a second regulatory element. A variety of such “obligatorily cooperative” elements are known (Burge et al. 1998; Frilander and Steitz 1999), and in most of these cases both elements must be present for the corresponding biochemical activity, with little or no activity expected in the presence of either element in isolation.
Motivated by the likelihood that other classes of obligatorily cooperative elements exist but have been refractory to detection by conventional screens, we developed an approach for ab initio identification of pairs of functionally interacting regulatory elements of splicing (or other processes). Several methods have been proposed to address the related problem of identifying pairs of motifs that co-occur in a single sequence region. Some of these methods begin with a defined set of known motifs and ask which pairs preferentially occur together (Pilpel et al. 2001; Hannenhalli and Levy 2002; Kato et al. 2004; Chan et al. 2005; Vardhanabhuti et al. 2007; Sinha et al. 2008). Others identify pairs of co-occurring motifs de novo by building pairs of position-specific scoring matrices using extensions of the Gibbs sampling algorithm (GuhaThakurta and Stormo 2001; Thompson et al. 2004).
We extend this literature with a new statistical approach, called compositionally orthogonalized co-occurrence analysis (coCOA), for identifying pairs of motifs that preferentially co-occur in a set of paired sequence regions while avoiding the types of artifacts that can arise from compositional heterogeneity of the sequences analyzed. Application of coCOA to sequences from the 5′ and 3′ ends of constitutively spliced introns identified pairs of oligonucleotides corresponding to the 5′ss and branch sites of U12-type introns (Burge et al. 1998; Frilander and Steitz 1999). Detection of this known pair of functionally interacting elements demonstrates the high sensitivity of the method, since U12-type introns represent ∼0.2% of all human introns.
coCOA also identified a GC-rich motif near the 5′ss that preferentially co-occurs with an AU-rich motif near the 3′ss of many constitutive introns. Similar pairs of motifs co-occur preferentially near the upstream 5′ss and downstream 3′ss flanking exons that are alternatively included/excluded (skipped). This pattern of co-occurrence suggested that these motifs act cooperatively to direct silencing of intervening exons, an activity which was confirmed using a splicing reporter assay. Finally, to assess the conservation of a pair of motifs when they occur together a generalization of standard single-motif conservation methods called the “co-conservation ratio” (CCR) was developed and applied to the GC-rich/AU-rich motif pair, detecting significant co-conservation. The statistical methods introduced are quite general and should prove equally applicable to cis-regulatory elements involved in other biochemical processes.
Results
Compositional orthogonalization controls for correlated G+C contents
The ends of introns must be brought together by the splicing machinery for splicing to occur, and these regions appear to be enriched for splicing regulatory elements (Yeo et al. 2005; Zhang et al. 2005). We therefore set out to detect pairs of motifs that preferentially co-occur at the beginnings and ends of constitutive human introns. For each pair of k-mers x and y (for k = 4, 5, and 6) we counted the number of introns that contain an occurrence of x within 80 base pairs of the 5′ss and an occurrence of y within 80 base pairs of the 3′ss. We considered comparing this number to the expectation given the marginal rates of occurrence of x and y near the corresponding splice sites, and assumed that these occurrences are independent. In fact, due to the correlation of G+C content at nearby positions in the human genome (Fig. 1A; Federico et al. 2000), these occurrences are not independent, and the overwhelming majority of motif pairs detected by this simple method are false positives (Supplemental material “Simple Co-Occurrence Analysis”; Supplemental Fig. S1). We therefore refined this null hypothesis by calculating the marginal rates of occurrence of x and y conditioned on the G+C contents of the introns near the splice sites, and then adding the expected number of co-occurrences for introns of different G+C contents to get an overall expected number of co-occurrences (Methods). Since this technique restores orthogonality/independence between the intron ends, we refer to it as compositionally orthogonalized co-occurrence analysis (coCOA).
Figure 1.
coCOA detects three clusters of motif pairs that co-occur at 5′ and 3′ ends of human introns. (A) G+C content in the first 80 nt (x-axis) and last 80 nt (y-axis) of introns is correlated. A density plot of intron co-GC content is shown for a set of 53,326 constitutive human introns, with the darker/lighter squares corresponding to higher/lower intron density, respectively. The diagonal line y = x is shown for reference. (B) co-GC shuffling. (Above) Two hypothetical introns, A and B, with 5′/3′ ends a5/a3 and b5/b3. Intron A has high G+C content at both ends (thick lines). Intron B has high G+C content at the 5′ end, but lower G+C content near the 3′ end (thin solid line). Since the introns have similar G+C content at their 5′ ends, these ends can be swapped. (Below) Co-GC shuffled introns. The beginning of intron B (b5) is now paired with the end of intron A (a3), and the beginning of intron A (a5) is now paired with the end of intron B (b3). Overall co-GC content of the set of introns is preserved. (C–E) Preferentially co-occurring k-mer pairs detected by coCOA are shown for k = 4, 5, and 6 at P ≤ 4−2k, corresponding to a single expected false positive for each value of k. In each panel, k-mers occurring in the first 80 nt of introns are shown at left under “5′SS”; those occurring in the last 80 nt are shown at right under “3′SS”. The co-occurrences could all be grouped into three clusters, denoted I1, I2, and I3, with the 5′ss and 3′ss motifs designated A and B, respectively.
coCOA identifies three clusters of k-mer pairs that preferentially co-occur at opposite ends of introns
coCOA was applied to the data set of 5′ and 3′ ends of constitutive human introns, for k = 4, 5, and 6. At P-value cutoffs of 4−2k in each analysis, 87, 124, and 15 significantly co-occurring k-mer pairs were detected for k = 4, 5, and 6, respectively, well above the null expectation of ∼1 pair at each value of k.
To estimate the rate of false positives for this method, coCOA was also applied to a “co-GC shuffled” set of intron termini. In this procedure, the G+C contents of the beginning and end of each intron are considered, and the intron termini are re-paired in such a way as to preserve the total number of introns with each pair of G+C contents (Fig. 1B). This preserves the degree of correlation in G+C content of the original set but results in pairs in which the 5′ ends of introns are paired with the 3′ ends of unrelated introns. Strikingly, no significant co-occurring pairs were observed in the co-GC shuffled sets for k = 4, 5, or 6, at P = 4−2k, demonstrating that coCOA has a low false-positive rate. To further assess the appropriateness of the P-values generated by coCOA, the fraction of significantly co-occurring k-mer pairs (out of the 42k possible pairs) was plotted as a function of the P-value cutoff. For the co-GC shuffled data, this yielded a plot which was close to a 45° line, indicating that the expected number of false positives is accurately estimated (Supplemental Fig. S2B). For small P-values less than ∼0.01, the controls showed somewhat fewer significant co-occurrences than expected (Supplemental Fig. S2B, lower), suggesting that in this regime the method is actually somewhat conservative. These data show that coCOA effectively controls for the extreme G+C heterogeneity of human introns, producing only the expected number of false positives or fewer in control data sets with the compositional complexity of human introns.
Examining the motif pairs detected by coCOA in constitutive human introns, we noted that the co-occurring pairs for k = 4 and 5 formed two clear clusters, connected by common sequences at the 5′ or 3′ ends of introns (Fig. 1C,D). For k = 6, four isolated pairs were identified, as well as one clear cluster (Fig. 1E). This cluster was named coCOA-I1 (I for intronic) or I1 for short. Pairs of 6-mers in this cluster matched almost perfectly to the 5′ss/branch signal consensus sequences which define the rare class of U12-type introns, which have 5′ss consensus /RUAUCCUU (where / indicates the splice junction and R represents A or G), and branch signal consensus CCUURAC (branch adenosine underlined) (Burge et al. 1998). These distinctive 5′ss/branch motifs can function together in splicing by the U12-dependent spliceosome, but U12-type 5′ss are incompatible with U2-type branch sites, and vice versa, so these motifs are truly obligatorily cooperative. The detection of the signature motifs of U12-type introns in a generic set of human introns demonstrates the high sensitivity of the coCOA method, since U12-type introns represented only ∼0.2% of the introns in the input data set (which is representative of human introns overall). Probably because of the rarity of U12-type introns and the lengths of the core motifs (≥6 nt), motif pairs related to U12-type introns were not detected for k = 4 or 5 at the P-value cutoff used. However, at both of these sizes, medium-sized clusters consisting of C-rich k-mers near the 5′ss that co-occur with C-rich k-mers near the 3′ss were identified; since the sequence pairs identified at k = 4 and 5 were very similar to each other and to one of the 6-mer pairs, these clusters are collectively referred to as coCOA-I3 (Fig. 1C–E).
For both k = 4 and 5, the largest cluster identified involved GC-rich sequences near the 5′ss co-occurring with AU-rich sequences near the 3′ss; three of the co-occurring 6-mer pairs also had similar sequences (Fig. 1C–E). We call this cluster I2 and refer to the 5′-end- and 3′-end-associated motifs as I2A and I2B, respectively. This cluster features a variety of GC-rich oligonucleotides at the 5′ ends of introns that co-occur with a variety of AU-rich sequences at intron 3′ ends, with some apparent preference for stretches of A or U. Typical k-mer pairs representing this cluster for k = 4, 5, and 6 are: CGCG/AAUU, which co-occurred in 336 introns, ∼1.5-fold higher than the expected value of 210; CGCGG/UUUAA, which co-occurred in 90 introns, ∼2.0-fold more than expected (44); and GGGCGC/UUAAAA, which co-occurred in 37 introns, >3-fold more than expected (10.7).
To be certain that these signals were not related to the canonical splicing elements, we repeated the analysis omitting the first and last 20 nt of every intron. As expected, coCOA-I2 and coCOA-I3 motif still significantly co-occurred. coCOA-I1 (U12-type), which only functions when I1A occurs at the beginning of an intron, was no longer detected (Supplemental Fig. S3).
Both the I2A and I2B motifs appeared quite variable in sequence. Similarly degenerate motifs are known to play important roles in splicing, e.g., a wide variety of purine-rich or AC-rich sequences appear to function as exonic splicing enhancers (Coulter et al. 1997; Liu et al. 1998), and a very wide range of pyrimidine-rich sequences can function as the polypyrimidine tract element of the 3′ss. As noted above, no similar motif pairs were detected in the co-GC shuffled introns. It was also notable that no “reversed” versions of the motif pair, i.e., with the AU-rich motif at the 5′ss and the GC-rich motif at the 3′ss, were detected, suggesting that whatever function these motifs have is specific to the “canonical” 5′-I2A/3′-I2B orientation. The counts of all significant co-occurring pairs in constitutive human introns for k = 4, 5, and 6 are listed in Supplemental Tables S1, S2, and S3, respectively.
Further clues to the function of the I2 pair came from analyses of a variety of other sequence sets involving pairs of regions adjacent to authentic or decoy 5′ss and 3′ss (Fig. 2). These sets included pairs of the 5′ss region upstream and the 3′ss region downstream of alternatively spliced (“cassette”) exons, and analogous regions upstream and downstream of constitutive exons. In addition, two “control” sets were constructed, consisting of authentic 5′ss paired with “decoy” 3′ss and of decoy 5′ss paired with authentic 3′ss, respectively. Here, as is customary, decoy 5′ss or 3′ss were defined as intronic sequences with high scores as potential 5′ss or 3′ss, which completely lack transcript evidence of usage as splice sites (Methods). Application of coCOA to the two control sets for k = 4, 5, and 6 (a total of six analyses, using a significance cutoff corresponding to one expected false positive per analysis) yielded a total of only four significantly co-occurring k-mer pairs. These data indicate that the I2A/B motif pair is specifically associated with authentic 5′ss/3′ss pairs and suggest that this motif pair might help to distinguish authentic from inauthentic 5′ss/3′ss pairs.
Figure 2.
Co-occurring motif pairs flanking alternative and constitutive exons and controls. Diagrams of the intron/exon data sets analyzed are shown at left, with exons shown as white boxes, introns as horizontal lines, and locations of the analyzed 80-nt regions shown as gray boxes. Splicing patterns are shown by angled lines; brackets indicate decoy splice sites. Representation of co-occurring k-mer pairs and P-value cutoffs as in Figure 1C–E. Numbers in parenthesis denote the number of significant k-mer pairs in each data set.
Analysis of 5′/3′ intron ends flanking constitutive or alternative exons identified a number of motif pairs resembling the I2A/B motif pair at k = 4 and 5 (Fig. 2). No significant co-occurring pairs matching the GC-rich/AU-rich pattern were observed at k = 6 in either set, possibly as a result of the reduced statistical power for analysis of 6-mers in these smaller data sets. Interestingly, as many or more significantly co-occurring pairs of GC-rich/AU-rich k-mers were detected flanking alternative exons as constitutive exons for both k = 4 (31 for alternative and seven for constitutive, plus one pair that did not fit the GC-rich/AU-rich pattern) and for k = 5 (seven for both sets), despite the reduced statistical power resulting from smaller data set size (N = 12,778 for the alternative exon set compared to 27,788 for the constitutive exon set). Indeed, the 31 4-mer pairs were more than twice as likely to co-occur in splice site pairs flanking alternative exons than those flanking constitutive exons; the counts of all significant co-occurring 4-mers and 5-mers flanking alternative exons are listed in Supplemental Tables S4 and S5, respectively. This observation suggests that I2-related motif pairs may be capable of mediating suppression of intervening exons, e.g., perhaps by defining the upstream 5′ss/downstream 3′ss as an authentic splice site pair. The co-occurrence of the motif pair flanking some constitutive exons might reflect contamination of the constitutive exon set by alternative exons for which transcript evidence has not yet been seen in the EST databases (Yeo et al. 2005), or might indicate other functions.
Similar pairs of motifs preferentially co-occur in mouse introns
To ask whether the I2 and other motif pairs were conserved outside of human, coCOA analysis was applied to a set of 68,998 constitutively spliced mouse introns (Methods). For k = 4 this analysis yielded 109 significantly co-occurring pairs (Supplemental Fig. S4). Of these, 36 formed a cluster very similar to the I2A/B cluster observed in human, and 23 pairs were identical to I2A/B pairs that significantly co-occurred in human. We refer to these 23 pairs as the human/mouse-I2 or HM-I2 set (Fig. 3B). Other clusters observed in mouse had sequences resembling the C-rich pairs forming the I3 cluster and the U12-type intron-related I1 cluster observed in human introns. A fourth cluster, I4, involving co-occurrences between pairs of purine-rich or G-rich 4-mers and 5-mers, was also observed in mouse. This cluster had no apparent human counterpart in the analysis of Figure 1, but 19 of the 28 4-mer pairs in this cluster (68%) significantly co-occurred in the human analysis at P ≤ 0.01, suggesting that purine-rich motifs also act cooperatively in human.
Figure 3.
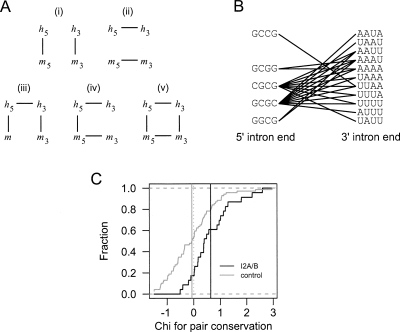
The motif pair I2A/B co-occurs in mouse as well as human and is preferentially co-conserved. (A) Representation of five possible models for co-conservation. Lines represent dependencies that are modeled; absence of a line indicates assumption of independence. Model (v) is the maximum entropy model used to define the co-conservation rate. (B) The 23 HM-I2 tetramer pairs that significantly co-occur between the beginning/end of constitutive mouse as well as human introns are shown. (C) I2A/B motif pairs are more conserved than expected. Empirical cumulative distribution functions of chi statistic (higher values indicate increased conservation) for HM-I2 tetramer pairs and control pairs. Controls have similar numbers of co-occurrences in human and mouse constitutive introns as HM-I2 pairs. Vertical black and gray lines indicate mean of statistic over HM-I2 pairs and control pairs, respectively.
A method to detect preferential co-conservation of a pair of motifs
Functional elements in genomic sequences are very often subject to negative (“purifying”) selection, resulting in higher rates of sequence conservation than surrounding sequences. A variety of methods in common use assess whether occurrences of an individual sequence element are conserved more often than expected, including the “conservation rate” (Xie et al. 2005) and the “conserved occurrence rate” (Wang et al. 2006) measures. For paired motifs, we are interested in the number of conserved co-occurrences ncc. For example, in the case of motif pairs occurring near intron ends, ncc is defined as the number of orthologous intron pairs in which motif x appears near the beginning and y appears near the end of both the mouse and human introns. We would like to calculate the expected number of conserved co-occurrences, E[ncc], under an appropriate null model. For such a pair of obligatorily cooperative elements, one would not necessarily expect the elements to be conserved when they occur in isolation—only when they co-occur. However, it is not enough to simply compare the frequency of conserved co-occurrence (“co-conservation”) of a pair of motifs to the product of the conservation frequencies of the individual motifs (Fig. 3Ai) because this estimate ignores the bias introduced if the pair preferentially co-occurs in one or both genomes. On the other hand, one might compare the frequency of co-conservation to the product of the frequencies of co-occurrence in the two genomes (Fig. 3Aii). This would instead ignore the bias introduced by the individual conservation rates of the two motifs. Indeed there are four pairwise marginal frequencies that should be controlled for: within-genome co-occurrence frequencies in human and mouse and between-genome conservation rates of both motifs (Fig. 3Av). However, it is possible to write down a simple expression for E[ncc] only for those models that control for at most two (as above) or three of the four marginal frequencies (Fig. 3Aiii, i; see Supplemental Methods). In order to control for all four pairwise frequencies, we applied the maximum entropy principle (MEP), which states that the least biased estimate of a distribution based on partial information (such as marginal frequencies) is that which maximizes the Shannon entropy given the constraints imposed by that information (Jaynes 1957; Yeo and Burge 2004). This approach has been applied widely in information processing and geophysics applications, and more recently to sequence motif modeling (Yeo and Burge 2004). Thinking of our model as a probability distribution over the 16 binary 4-tuples indicating presence or absence of the corresponding motifs at the ends of human and mouse introns, we applied the MEP to the set of constraints consisting of all four pairwise marginal distributions, and used this distribution to determine E[ncc]. We refer to the ratio of the observed to expected conserved co-occurrences ncc/E[ncc] as the “co-conservation ratio” (CCR).
Evidence for co-conservation of I2A/B motif pairs between human and mouse
To assess the co-conservation of I2A/B pairs, we mapped the set of mouse introns analyzed above to orthologous human introns, yielding a set of 24,503 constitutive human/mouse ortholog pairs. For each of the 23 HM-I2 4-mer pairs, the number of conserved co-occurrences and the corresponding expected distribution were determined (Supplemental Figs. S6, S7). More than 80% (19 of 23) of such pairs had CCR > 1, indicating a tendency toward higher co-conservation than expected. As anticipated, for control sets of 4-mer pairs the numbers of pairs with CCR above and below 1 were essentially equal. To assess significance, for each pair a signed χ-value was calculated, measuring the degree of difference between the observed and expected co-conservation counts, with positive or negative sign depending on whether the observed was greater or less than the expected value, respectively (Methods). The mean χ-value for I2A/B pairs (0.64) was significantly greater than 0 at P = 4.8 × 10−4 by a one-sided t-test, while the mean for control pairs (−0.07) was not significantly different from 0 (P = 0.51 by a two-sided t-test) (Fig. 3C). Since the most similar pairs do not have similar χ-values (Supplemental Fig. S8) the statistical significance of the mean χ-value is not due to a possible lack of independence between similar pairs. Thus, CCR analysis indicated that HM-I2 pairs are more conserved when they co-occur in the same intron, supporting a conserved cooperative function of these motif pairs in mammalian introns.
I2A/B motif pairs can suppress splicing of an intervening exon
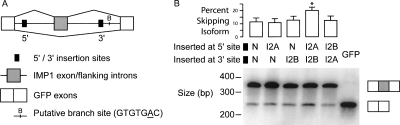
The common co-occurrence of I2 motif pairs in constitutive introns of both human and mouse, their preferential co-conservation, and their increased co-occurrence flanking alternative/skipped exons suggested that these pairs might cooperate to define splice site pairs and/or to silence splicing of intervening exons. To test this hypothesis, a three-exon minigene was used, adapted from that described by Wang et al. (2004) (Fig. 4A). In this reporter, the middle (test) exon, derived from exon 12 of the human IGF2BP1 gene (alias IMP-1) (Yeo et al. 2004; Kol et al. 2005), is skipped at a basal level of ∼10%. The 12-mers GGGCGCGGGCGC and TTAAAATTAAAA—tandem duplicates of the most significantly co-occurring 6-mer pairs in the human constitutive intron set (Fig. 1E)—were chosen to represent the I2A and I2B motifs, and these 12-mers were inserted near the 5′ss and 3′ss, respectively. A “neutral” 12-mer, CGGTTACGAGTA, was used as a control. This sequence has balanced base composition (50% C and G bases) and was designed to avoid matches to known splicing regulatory elements (Methods). The representative I2A and I2B sequences were inserted into the reporter singly or in combination, with the neutral motif used as a control so that all tested constructs had identical size and spacing of elements (Fig. 4A). Following transient transfection of each construct into HeLa cells, splicing was assayed by semi-quantitative RT-PCR (Fig. 4B). Insertion of the I2A and I2B motifs together in canonical 5′-I2A/3′-I2B order resulted in a ∼1.8-fold increase in exon skipping relative to controls (P = 1.1 × 10−5 level by one-sided t-test), strongly supporting the hypothesis that these motifs can function together to silence intervening exons. Neither the I2A motif nor the I2B motif by itself had an appreciable effect on the level of exon skipping relative to the control motif, nor did the I2A/B pair in reversed order. These controls demonstrate that the I2A/B motifs are not conventional intronic splicing regulatory elements (Ladd and Cooper 2002; Matlin et al. 2005). Instead, as predicted from the coCOA and CCR analyses, this pair appears to function in exon silencing/splice site pairing in a manner that is obligatorily cooperative and sensitive to motif order.
Figure 4.
I2A/B motif pairs can suppress splicing of an intervening exon. (A) Mini-gene construct for interrogating I2A/B motif pair, constructed by inserting exon 12 of the human IGF2BP1 gene and its flanking introns into the middle of the ORF in an eGFP expression construct. (B) The I2A/B motif pair promotes exon skipping in HeLa cells. The five indicated constructs containing I2A, I2B, or neutral (N) motifs inserted near the 5′ss or 3′ss were transfected into HeLa cells. Twenty-four hours later RNA was extracted and semi-quantitative RT-PCR using primers targeted to reporter exons 1 and 3 was performed to assay for relative isoform levels. (Top) Quantization of skipping levels. Data shown are mean + SEM for eight replicates—two PCRs for each of four transfection experiments. *I2A/B motif pair shows significantly more skipping than N/N control (P = 1.1 × 10−5 by one-sided t-test; 1.75-fold increase in skipping). At the 5% level, none of the other skipping levels is significantly greater than that of N/N. (Bottom) Representative gel showing levels of inclusion isoform (upper band) and skipping isoform (lower band). Last lane, intronless GFP control.
Discussion
The universe of cis-regulatory elements can be divided into those which can function in relative isolation and those that require additional element(s) for activity. The former class of elements has received the lion’s share of attention. The typical paradigm for detection of such motifs involves application of one or more motif-finding algorithms such as the Gibbs Sampler (Lawrence et al. 1993) or MEME (Bailey and Elkan 1994) to detect short motifs that are statistically enriched in an input sequence set of interest. Sequence conservation and/or experimental manipulation of motifs detected in this manner are then used to assess potential in vivo function. Because standard single-motif search methods may miss motif pairs that function in an obligatorily cooperative fashion, we have developed an alternative paradigm focused specifically on identifying pairs of cis-elements that function when both elements occur together. This paradigm involves application of the pair-motif finding algorithm coCOA to sets of sequence pairs of interest, followed by analysis of the co-conservation of identified motif pairs using the CCR statistic and/or experimental tests of the activity of the identified motifs singly and in combination.
The fundamental principle underlying our approach is that, when two motifs function in concert, they should experience different (stronger) selective pressure when they occur together than when they occur separately. Thus, the primary signal for detection of such cooperatively active motifs is not enrichment or conservation of the individual motifs per se, but an excess of counts and conservation of pairs of elements relative to that expected based on the counts and conservation of the elements separately. We have shown that, in order to avoid artifacts arising from correlations in the base composition of nearby regions of the human genome, it is necessary to apply the “compositional orthogonalization” technique we call coCOA. This approach identified the 5′ss and branch motifs of U12-type introns, a known pair of obligatorily cooperative elements, while also detecting two other clusters of k-mer pairs at the 5′/3′ ends of constitutive human introns. For all three of these clusters, similar clusters of co-occurring k-mers were observed in mouse introns, suggesting evolutionarily conserved cooperative functions.
The largest of the identified clusters, I2, involved GC-rich I2A motifs near the 5′ end and AU-rich I2B motifs near the 3′ end of introns. For this cluster, preferential co-occurrence at the upstream 5′ss and downstream 3′ss flanking alternatively spliced exons suggested a role in silencing of intervening exons. For a representative pair of GC-rich and AU-rich motifs such a role was confirmed in a splicing reporter assay. Because by themselves neither of these motifs affected splicing of the test exon, this pair appears to function in an obligatorily cooperative manner, perhaps explaining why these motifs had not been highlighted previously in screens for splicing regulatory elements. Presence of such motif pairs flanking many introns might facilitate the evolution of new alternatively spliced exons, by allowing constitutive introns to accept insertion of sequences containing pseudoexons or even authentic exons without losing expression of the original message.
The sheer size of many human introns must present a significant challenge to the splicing machinery, which must accurately bring together intron ends that can be tens of kilobases apart, while avoiding recognition of the many pairs of decoy splice sites (“pseudoexons”) that occur in increasing numbers as intron size increases. Interestingly, pairs of the I2 cluster were found to significantly co-occur at the ends of the longest introns (>1775 nt, not shown), where a role in juxtaposition of intron ends and/or suppression of intervening pseudoexons would be particularly beneficial. One possible mechanism by which this might occur would be by co-transcriptional splicing repression. The factor that binds I2A (even perhaps at the DNA level) might then be recruited by the elongating transcriptional complex, maintaining it in a splicing-incompetent state until an I2B motif and, possibly, 3′ss are recognized. Very little is known about the splicing of long mammalian introns. However, in Drosophila, splicing of very long introns (≥10 kb) appears to require special elements not required for splicing of average-sized introns (Burnette et al. 2005). Thus, I2 motif pairs might represent a mammalian adaptation to facilitate proper intron end pairing, especially in long introns.
As a rough estimate of the number of human introns whose splicing is likely to be regulated by I2A/B motif pairs, we determined that at least six pairs of I2A/B 5-mers occur flanking 1364 constitutive human introns, an excess of >450 introns over the corresponding number, 905, for co-GC shuffled introns. Furthermore, 718 alternative exons were flanked by one or more pairs of I2A/B 5-mers, an excess of >200 introns over the 500 observed in the co-GC shuffled intron set. Thus, at least several hundred human introns are likely to be regulated by I2A/B motif pairs. The identity(ies) of the involved trans-acting factors and their mechanism of action is not clear. However, a number of factors capable of binding specifically to AU-rich RNA sequences are known, including TIA1, HNRNPD (formerly known as AUF1), and hnRNPs A1 and C (Hamilton et al. 1993; Zhang et al. 1993; Del Gatto-Konczak et al. 2000). For hnRNP A1, a function in looping out of introns (leading to suppression of intervening splice sites or exons) has been proposed (Blanchette and Chabot 1999; Nasim et al. 2002). Perhaps I2A/B pairs can mediate suppression of intervening exons by a similar mechanism. Interestingly, genes containing alternative exons flanked by multiple I2A/B pairs are enriched for the Gene Ontology process “RNA splicing” (Table 1), hinting that the trans-factor(s) that mediate I2A/B activity may in fact regulate the processing of their own messages, as has been seen for many other splicing factors (Stoilov et al. 2004; Wollerton et al. 2004; Dredge et al. 2005; Lareau et al. 2007).
Table 1.
Gene Ontology categories enriched for I2A/B-flanked alternative exons
The remaining cluster, I3, consisted of pairs of C-rich motifs that co-occur at the ends of constitutive introns. Short runs of C were previously identified as candidate intronic splicing enhancers in mammalian introns based in part on their enrichment in introns adjacent to weak splice sites (Yeo et al. 2004). Preferential co-occurrence of similar C-rich motifs suggests that these elements may function cooperatively. The similarity between the motifs identified at intron 5′ and 3′ ends for this cluster suggests that these motifs may be bound by (separate molecules or domains of) the same factor. Candidate trans-factors include members of the poly(C) binding protein family, which contains five members in human and mouse, including hnRNPs K/J and the alphaCPs, PCBP1–4 (also known as alphaCP1–4), which may bind cooperatively to multiple C-rich regions (Makeyev and Liebhaber 2002; Paziewska et al. 2004).
Detection of preferential co-conservation of a pair of motifs is substantially more complex than the corresponding problem for individual motifs. We have developed a statistic called CCR based on the MEP that effectively measures co-conservation while controlling for the conservation levels of the individual motifs and potentially biased co-occurrence in the respective genomes. This method supports preferential co-conservation of the I2A/B motifs in mammalian introns. Just as the “conservation rate” of individual motifs can be used by itself to detect functional elements without use of a conventional motif finder (Lewis et al. 2005; Xie et al. 2005), one could imagine using the CCR method by itself (i.e., without coCOA) to identify cooperatively active pairs of elements based on co-conservation alone. Another angle that we have not explored in depth would be to search for preferentially avoided pairs of motifs using coCOA.
Since the assumptions that the coCOA and CCR methods are based on are quite general and not specifically related to pre-mRNA splicing, this approach should be equally applicable to analysis of transcription or other biochemical processes involving cooperatively active motif pairs. For example, 5C is a new technology that allows high-throughput mapping of pairs of physically interacting regions of chromatin (Dostie et al. 2006). coCOA would be an ideal method for the identification of DNA sequences that mediate these and other long-range interactions.
Methods
Sequence data sets
The sequence sets used in this study were derived using SpliceGraph, a software toolbox that generates for each gene a graph-based representation of its transcript variants (R. Sandberg and M.B. Stadler, unpubl.). SpliceGraph databases for human and mouse genes were constructed using spliced alignments of cDNAs and ESTs to the human and mouse genomes from the University of California, Santa Cruz genome website (http://genome.ucsc.edu; Kent et al. 2002). In brief, the transcripts that shared splice sites were first clustered into gene models, which were processed to define sets with specific splicing patterns such as constitutive and alternative exons, introns, etc.
The human and mouse constitutive intron data sets used in this study were derived from transcript-supported introns identified by SpliceGraph, using a rather stringent definition of constitutive, requiring that all transcripts that aligned to at least one upstream exon and at least one exon downstream of the intron have an alignment precisely spanning the intron, i.e., using the same 5′ss and 3′ss. All introns used were required to be at least 160 nt long (so that the 80-nt regions at the 5′ and 3′ ends would not overlap). The resulting sequence set contained 68,363 intron start/end pairs. After filtering of sequences that overlap known repetitive elements, or that have close paralogs (see below), the set contained 53,331 start/end pairs.
The set of human/mouse orthologous intron pairs was created by using the liftOver tool from the UCSC Web site to map the mouse constitutive introns onto the human genome, and taking only those that mapped exactly to human constitutive introns.
The human alternative exon data set used in the analyses of Figure 2 was defined as the set of SpliceGraph human exons for which at least one transcript supported inclusion of the exon and at least one transcript supported precise skipping of the exon. (The exon was required to be at least 80 nt as a filter for alignment quality.) The regions used in the analysis of Figure 2 were the first 80 nt of the upstream intron, and the last 80 nt of the downstream intron. For consistency with the constitutive intron data set, both introns were required to be at least 160 nt long. The initial set contained 16,794 sequence pairs, and the filtered set (see below) contained 12,778 sequence pairs. The human constitutive exon set used in Figure 2 was derived similarly from SpliceGraph constitutive internal exons at least 80 nt in length for which both flanking introns were at least 160 nt long. The initial sequence set contained 34,914 pairs before and 27,788 pairs after repeat/paralog filtering (see below).
The authentic 5′ss/decoy 3′ss data set was constructed from the human constitutive intron data set by searching the intron for decoy 3′ss located at least 160 nt downstream from the 5′ss (so that the two 80-nt regions would not overlap). A decoy 3′ss was defined as a stretch of 23 nt which scored at least as high as the natural 3′ss of the intron by the Maximum Entropy model (Yeo and Burge 2004), which models compositional biases and statistical dependencies between positions in the last 20 nt of introns (including the polypyrimidine tract and AG and the first 3 nt of the exon). The resulting set contained 42,070 pairs and the filtered set (see below) contained 20,489 pairs. The decoy 5′ss/authentic 3′ss set was defined analogously and contained 42,070 and 26,591 sequence pairs before and after filtering, respectively.
The sequence sets are available in Supplemental material.
Sequence set filtering
After creating the unfiltered sequence sets described above, any sequences that overlapped with annotated repeats (also from http://genome.ucsc.edu; Kent et al. 2002) were removed. These included both interspersed repetitive elements such as Alus and long interspersed nuclear elements (LINEs) and shorter simple sequence repeats.
Next, these data sets were purged of highly similar (paralogous) subsets of sequences. To do this, three similarity graphs were made for each set. These graphs had the same node set, one node for each sequence pair in the set. The first graph had an edge for each significant BLASTN hit of nucleotide +10 to +85 from the 5′ss, and the second graph the same for nucleotide −100 to −25 from the 3′ss (i.e., overlapping extensively with the regions analyzed in this study, but excluding the core splice site motifs). The third graph had as its edge set the intersection of the first two graphs, so that two sequence pairs were connected if and only if both regions showed sequence similarity. Greedy node removal was applied to this intersection graph, iteratively removing the node of highest degree (and any attached edges) until no edges remained, so that there were no two sequence pairs with both ends homologous. The sequence pairs corresponding to the remaining nodes formed the input to coCOA and other analysis algorithms.
Co-GC shuffling
For efficiency, the co-GC shuffled sets were generated in “one fell swoop” by iterating through the real sequence pairs, and for each sequence pair of co-GC content (s1, s2) choosing at random, and without replacement, a first sequence (i.e., first in its pair) of GC content s1 and, independently, a second sequence of GC content s2. These together formed one co-GC shuffled sequence pair. This achieves the same outcome but is more computationally efficient than the simpler swapping procedure described in the legend to Figure 1.
coCOA
As described in the Results, sequence pairs are binned in two dimensions according to b1 and b2, the number of G+C in the first and the second sequences of the pair, respectively. Let N(b1,b2) denote the number of sequence pairs in cell (b1, b2), and let C(b1,b2)(x,y) denote the number of sequence pairs in that cell containing k-mers x and y in the first and second sequences, respectively. Define n1b1(x) as the number of first sequences in G+C bin b1 containing x, and define n2b2(y) as the number of second sequences in bin b2 containing y. Letting N1b1 denote the total number of first sequences in G+C content bin b1, and N2b2 the number of second sequences in G+C bin b2, bin-specific (marginal) k-mer frequencies are defined by f1b1(x):= n1b1(x)/N1b1 and f2b2(y):= n2b2(y)/N2b2. Under the null hypothesis that k-mers occur independently in the respective sequences within each cell with frequencies given by the marginal values, we calculate the expected number of co-occurrences of k-mers x and y in a cell as:
Then, using the additive property of the Poisson distribution, the total number of co-occurrences of x,y in the whole data set, C(x,y), should have a Poisson distribution with parameter λx,y, given by
Thus, in this approach, the significance of the observed number of co-occurrences of the k-mers x and y can be estimated as the tail probability of a Poisson (λx,y) random variable. The P-value for coCOA (or simpleCOA) can also be calculated exactly using the hypergeometric distribution, but the implementation is slower and the results are nearly identical (data not shown).
The coCOA Software is available in Supplemental material.
Evaluating the significance of a set of CCR values
The significance of the higher CCR values observed for I2A/B tetramer pairs was evaluated as follows. For each pair, a Z statistic was calculated, defined as Z = (n1111 − E[n1111])/E[n1111] + ((n − n1111) – (n − E[n1111]))2/(n − E[n1111]), defining χ = sign(n1111 − E[n1111]) √X2 so that χ has positive sign for n1111 > E[n1111] and negative sign for n1111 < E[n1111]. Three control sets of 23 matched 4-mer pairs were chosen to match the distribution of co-occurrence counts of the I2-HM sets. The mean CCR for control pairs was 0.97 (not significantly different from 1).
Selection of a neutral motif
The neutral motif used in the splicing reporter experiments was chosen so that when inserted into either cloning site it would not create any known splicing regulatory elements. Our set of “known” splicing regulatory elements was the union of the RESCUE exonic splicing enhancer hexamers (Fairbrother et al. 2002), the FAS-ESS cut2 exonic splicing silencer hexamers (Wang et al. 2004), and a set of putative intronic splicing enhancer and silencer hexamers derived from a RESCUE-based computational screen (X. Xiao and C.B. Burge, unpubl.). The neutral motif chosen has 50% G+C content and does not contain any of these hexamers or overlap with any of them in either of the two contexts into which it was inserted. It was obtained using a Perl script that randomly generated sequences until finding one that met all of these criteria.
Cell culture and transfection
HeLa cell lines were cultured in Dulbecco's modified Eagle's medium, supplemented with 4.5 g/mL glucose and 10% fetal bovine serum. Cells were cultured in six-well plates in a humidified atmosphere at 37°C with 5% CO2. Cells were grown to 70% confluence and transfection was performed using Lipofectamine 2000 (Invitrogen) and 1.0 μg plasmid DNA according to manufacturer's protocol.
RNA extraction and RT-PCR analysis
Twenty four hours after transfection, total RNA was isolated using TRIzol reagent (Invitrogen), then first strand cDNA synthesis was carried out by incubating 1 μg of total RNA with 5 μM oligo(dT) primer for 5 min at 65°C followed by 60 min at 50°C after addition of 5 U SuperScript III Reverse Transcriptase (Invitrogen), 1× Reverse Transcriptase Buffer (Invitrogen), 5 mM DTT, and 0.5 mM dNTPs. Following inactivation at 70°C for 15 min 2 μL of the cDNA was used for 20 cycles of PCR amplification with 1 U Taq polymerase (Invitrogen), 1× supplied buffer (Invitrogen), 1.5 mM MgCl, 0.5 mM dNTPs, 0.5 μM of each primer (forward 5′-TACGTACGTACGTACGT; reverse 5′-TCATGCATGCTGACTG CAT), and 2.5 μCi 32P-alpha-dCTP/reaction. PCR products were separated in 5% TBE gels (Bio-Rad) and quantitated after exposing to a phosphorimager screen using ImageQuant software (Amersham/GE Healthcare) on a 445 SI PhosphorImager (Molecular Dynamics). The level of skipping of exon two was calculated as the background-corrected integrated intensity of the exon two-skipping band divided by the sum of the intensities of the exon two-including and exon two-skipping bands. The identities of these bands were confirmed by sequencing.
Acknowledgments
The problem of identifying significantly co-occurring motif pairs was first brought to our attention by Rodger Voelker (University of Oregon). This work was supported in part by grants from the NIH and NSF (C.B.B.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.080085.108.
References
- Bailey T.L., Elkan C., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Chabot B., Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C.B., Padgett R.A., Sharp P.A., Padgett R.A., Sharp P.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- Burnette J.M., Miyamoto-Sato E., Schaub M.A., Conklin J., Lopez A.J., Miyamoto-Sato E., Schaub M.A., Conklin J., Lopez A.J., Schaub M.A., Conklin J., Lopez A.J., Conklin J., Lopez A.J., Lopez A.J. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Elemento O., Tavazoie S., Elemento O., Tavazoie S., Tavazoie S. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 2005;1:e69. doi: 10.1371/journal.pcbi.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter L.R., Landree M.A., Cooper T.A., Landree M.A., Cooper T.A., Cooper T.A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Bourgeois C.F., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Le Guiner C., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Kister L., Gesnel M.C., Stevenin J., Breathnach R., Gesnel M.C., Stevenin J., Breathnach R., Stevenin J., Breathnach R., Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Rubio E.D., Krumm A., Lamb J., Nusbaum C., Krumm A., Lamb J., Nusbaum C., Lamb J., Nusbaum C., Nusbaum C., et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge B.K., Stefani G., Engelhard C.C., Darnell R.B., Stefani G., Engelhard C.C., Darnell R.B., Engelhard C.C., Darnell R.B., Darnell R.B. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B., Yeh R.F., Sharp P.A., Burge C.B., Sharp P.A., Burge C.B., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Federico C., Andreozzi L., Saccone S., Bernardi G., Andreozzi L., Saccone S., Bernardi G., Saccone S., Bernardi G., Bernardi G. Gene density in the Giemsa bands of human chromosomes. Chromosome Res. 2000;8:737–746. doi: 10.1023/a:1026797522102. [DOI] [PubMed] [Google Scholar]
- Frilander M.J., Steitz J.A., Steitz J.A. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes & Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuhaThakurta D., Stormo G.D., Stormo G.D. Identifying target sites for cooperatively binding factors. Bioinformatics. 2001;17:608–621. doi: 10.1093/bioinformatics/17.7.608. [DOI] [PubMed] [Google Scholar]
- Hamilton B.J., Nagy E., Malter J.S., Arrick B.A., Rigby W.F., Nagy E., Malter J.S., Arrick B.A., Rigby W.F., Malter J.S., Arrick B.A., Rigby W.F., Arrick B.A., Rigby W.F., Rigby W.F. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- Hannenhalli S., Levy S., Levy S. Predicting transcription factor synergism. Nucleic Acids Res. 2002;30:4278–4284. doi: 10.1093/nar/gkf535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes E.T. Information theory and statistical mechanics. Phys. Rev. 1957;106:620–630. [Google Scholar]
- Kato M., Hata N., Banerjee N., Futcher B., Zhang M.Q., Hata N., Banerjee N., Futcher B., Zhang M.Q., Banerjee N., Futcher B., Zhang M.Q., Futcher B., Zhang M.Q., Zhang M.Q. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 2004;5:R56. doi: 10.1186/gb-2004-5-8-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol G., Lev-Maor G., Ast G., Lev-Maor G., Ast G., Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- Ladd A.N., Cooper T.A., Cooper T.A. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3:reviews0008.1–reviews0008.16. doi: 10.1186/gb-2002-3-11-reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E., Inada M., Green R.E., Wengrod J.C., Brenner S.E., Green R.E., Wengrod J.C., Brenner S.E., Wengrod J.C., Brenner S.E., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lawrence C.E., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C., Liu J.S., Neuwald A.F., Wootton J.C., Neuwald A.F., Wootton J.C., Wootton J.C. Detecting subtle sequence signals: A Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu H.X., Zhang M., Krainer A.R., Zhang M., Krainer A.R., Krainer A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev A.V., Liebhaber S.A., Liebhaber S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A.J., Clark F., Smith C.W., Clark F., Smith C.W., Smith C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Nasim F.U., Hutchison S., Cordeau M., Chabot B., Hutchison S., Cordeau M., Chabot B., Cordeau M., Chabot B., Chabot B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8:1078–1089. doi: 10.1017/s1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paziewska A., Wyrwicz L.S., Bujnicki J.M., Bomsztyk K., Ostrowski J., Wyrwicz L.S., Bujnicki J.M., Bomsztyk K., Ostrowski J., Bujnicki J.M., Bomsztyk K., Ostrowski J., Bomsztyk K., Ostrowski J., Ostrowski J. Cooperative binding of the hnRNP K three KH domains to mRNA targets. FEBS Lett. 2004;577:134–140. doi: 10.1016/j.febslet.2004.08.086. [DOI] [PubMed] [Google Scholar]
- Pilpel Y., Sudarsanam P., Church G.M., Sudarsanam P., Church G.M., Church G.M. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat. Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- Sinha S., Adler A.S., Field Y., Chang H.Y., Segal E., Adler A.S., Field Y., Chang H.Y., Segal E., Field Y., Chang H.Y., Segal E., Chang H.Y., Segal E., Segal E. Systematic functional characterization of cis-regulatory motifs in human core promoters. Genome Res. 2008;18:477–488. doi: 10.1101/gr.6828808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R., Wang J., Chew S.L., Zhang M.Q., Krainer A.R., Chew S.L., Zhang M.Q., Krainer A.R., Zhang M.Q., Krainer A.R., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Stoilov P., Daoud R., Nayler O., Stamm S., Daoud R., Nayler O., Stamm S., Nayler O., Stamm S., Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- Thompson W., Palumbo M.J., Wasserman W.W., Liu J.S., Lawrence C.E., Palumbo M.J., Wasserman W.W., Liu J.S., Lawrence C.E., Wasserman W.W., Liu J.S., Lawrence C.E., Liu J.S., Lawrence C.E., Lawrence C.E. Decoding human regulatory circuits. Genome Res. 2004;14:1967–1974. doi: 10.1101/gr.2589004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhanabhuti S., Wang J., Hannenhalli S., Wang J., Hannenhalli S., Hannenhalli S. Position and distance specificity are important determinants of cis-regulatory motifs in addition to evolutionary conservation. Nucleic Acids Res. 2007;35:3203–3213. doi: 10.1093/nar/gkm201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B., Yeo G., Tung V., Mawson M., Burge C.B., Tung V., Mawson M., Burge C.B., Mawson M., Burge C.B., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Wang Z., Xiao X., Van Nostrand E., Burge C.B., Xiao X., Van Nostrand E., Burge C.B., Van Nostrand E., Burge C.B., Burge C.B. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Wagner E.J., Garcia-Blanco M.A., Smith C.W., Garcia-Blanco M.A., Smith C.W., Smith C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G., Burge C.B., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo G., Hoon S., Venkatesh B., Burge C.B., Hoon S., Venkatesh B., Burge C.B., Venkatesh B., Burge C.B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl. Acad. Sci. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B., Van Nostrand E., Holste D., Poggio T., Burge C.B., Holste D., Poggio T., Burge C.B., Poggio T., Burge C.B., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Chasin L.A., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wagner B.J., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G., Wagner B.J., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G., Crater D., DeHaven K., Long L., Brewer G., DeHaven K., Long L., Brewer G., Long L., Brewer G., Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Leslie C.S., Chasin L.A., Leslie C.S., Chasin L.A., Chasin L.A. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]