Abstract
We have determined the complete genome sequences of a host-promiscuous Salmonella enterica serovar Enteritidis PT4 isolate P125109 and a chicken-restricted Salmonella enterica serovar Gallinarum isolate 287/91. Genome comparisons between these and other Salmonella isolates indicate that S. Gallinarum 287/91 is a recently evolved descendent of S. Enteritidis. Significantly, the genome of S. Gallinarum has undergone extensive degradation through deletion and pseudogene formation. Comparison of the pseudogenes in S. Gallinarum with those identified previously in other host-adapted bacteria reveals the loss of many common functional traits and provides insights into possible mechanisms of host and tissue adaptation. We propose that experimental analysis in chickens and mice of S. Enteritidis–harboring mutations in functional homologs of the pseudogenes present in S. Gallinarum could provide an experimentally tractable route toward unraveling the genetic basis of host adaptation in S. enterica.
Zoonotic pathogens, particularly those associated with veterinary animals in the human food chain, are some of the most important causes of infectious diseases in humans. Pathogens associated with zoonotic infections exhibit a promiscuous phenotype in that they maintain the ability to colonize and potentially cause infections in more than one host species. In contrast, some pathogenic agents are significantly host restricted, or adapted, and are normally only able to cause disease in one host. Salmonella enterica is a single bacterial species that includes examples of both promiscuous and host-adapted pathotypes. Isolates from serovars such as S. enterica serovar Typhimurium and S. Enteritidis predominantly retain the ability to infect more than one mammalian host, including humans, whereas serovars such as S. enterica serovars Typhi and S. Gallinarum are restricted to humans and chickens, respectively. The ability to transmit between and within particular host populations is centrally important in dictating the epidemiology of infections and the emergence of new diseases.
Before the mid-1980s, S. Enteritidis was regarded as an S. enterica serovar of minor public health significance, but subsequently this serovar became dominant in terms of human food poisoning in many parts of the world (Rodrigue et al. 1990). National and international legislation regarding the reporting of disease incidence, improved hygiene and biosecurity (Barrow 2000), and vaccination have contributed to controlling S. Enteritidis levels in poultry and consequently in man in Europe, but levels of infection remain significant. Most recent isolates of S. Enteritidis are regarded as promiscuous in the sense that they can cause infections in mice, retain the ability to colonize the tissues of chickens, and cause gastroenteritis in man.
S. Gallinarum, the causative agent of fowl typhoid, is a predominantly avian-restricted serovar (Shivaprasad 2000). Interestingly, in common with the human-restricted serovar S. Typhi, the chicken-adapted S. Gallinarum causes an invasive typhoid-like disease. Thus, here host adaptation appears to have coevolved with loss of the intestinal lifestyle and the acquisition of the ability to cause systemic infection. S. Gallinarum still causes a disease of worldwide economic significance, and although it has been largely controlled in countries with strong health control policies, largely through serology-based test and slaughter schemes, it remains a problem elsewhere. Multi locus enzyme electrophoresis analyses of isolates of S. Enteritidis and S. Gallinarum indicate that, together with isolates of S. Dublin and S. Pullorum, they form a related strain cluster that share the same lipopolysaccharide-based O structure (O-1, 9, 12 characteristic of serogroup D). The nonmotile S. Gallinarum and S. Pullorum were previously suggested to have split independently from a motile ancestor related to S. Enteritidis (Li et al. 1993; McMeechan et al. 2005). Nonmotility in S. Gallinarum has been partially attributed to mutations in the flagellin subunit gene fliC gene (Kilger and Grimont 1993), which would normally express the phase 1 g, m antigens characteristic of S. Enteritidis. Nonmotility may enhance the ability to invade systemically from the gut by avoiding the TLR-5-induced pro-inflammatory responses of the host (Kaiser et al. 2000; Iqbal et al. 2005).
Here we report the full genome sequences of representative isolates of S. Enteritidis and S. Gallinarum and provide a detailed comparative genomic analysis of the two serovars. These data have been used to provide insight into the biology, mechanisms of host/tissue adaptation, and evolutionary relationships of these important pathogens.
Results and Discussion
General features of the S. Enteritidis PT4 strain P125109 and S. Gallinarum strain 287/91 genomes
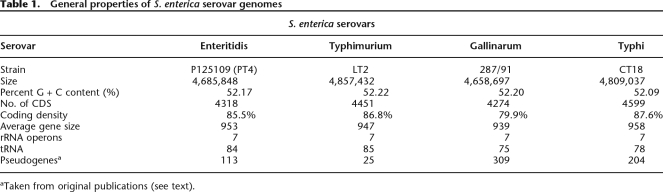
The complete genome sequences of the promiscuous S. Enteritidis PT4 strain P125109 (hereafter S. Enteritidis PT4; EMBL accession no. AM933172) and the highly host-adapted chicken pathogen S. Gallinarum strain 287/91 (hereafter S. Gallinarum 287/91; EMBL accession no. AM933173) were determined and annotated. The main features are summarized in Table 1 and Figure 1, where they are compared with S. Typhimurium strain LT2 (hereafter S. Typhimurium LT2) (McClelland et al. 2001). The most striking feature of the analysis is the predominant similarity and synteny of core regions of the genomes, including many of the Salmonella pathogenicity islands (SPI). Indeed, this comparative analysis highlights an extremely close relationship between the genomes of S. Enteritidis and S. Gallinarum, suggesting the latter is a direct evolutionary descendent of the former. However, in comparison to S. Enteritidis PT4, S. Gallinarum 287/91 harbors a significantly higher number of predicted pseudogenes. Although the number of pseudogenes in S. Enteritidis PT4 is slightly higher than reported for S. Typhimurium LT2, it is in line with levels described in other broad host range enteric pathogens such as Yersinia enterocolitica (Thomson et al. 2006). In contrast, the number of pseudogenes in S. Gallinarum 287/91 is closer to that of the human-restricted S. Typhi CT18 (204 pseudogenes) (Parkhill et al. 2001) and S. enterica serovar Paratyphi A (173 pseudogenes) (McClelland et al. 2004).
Table 1.
General properties of S. enterica serovar genomes
aTaken from original publications (see text).
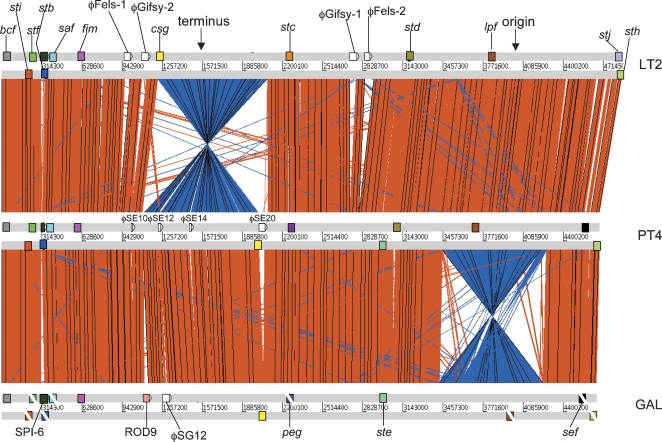
Figure 1.
Global comparison between S. Typhimurium, S. Enteritidis, and S. Gallinarum. ACT comparison (http://www.sanger.ac.uk/Software/ACT) of amino acid matches between the complete six-frame translations (computed using TBLASTX) of the whole-genome sequences of S. Typhimurium LT2 (LT2), S. Enteritidis PT4 (PT4), and S. Gallinarum 287/91 (GAL). Forward and reverse strands of DNA are shown for each genome (light gray horizontal bars). The red bars between the DNA lines represent individual TBLASTX matches, with inverted matches colored blue. The position of all the fimbrial operons in these three genomes are marked as colored boxes positioned on the forward and reverse strands of DNA. Analogous fimbrial operons are colored the same. The boxes of fimbrial operons that include pseudogenes are crossed with a white line. Other genomic features are only shown if they constitute breaks in synteny between genomes. The position of the origin and terminus are marked (solid black arrows).
Whole-genome comparisons of S. Enteritidis PT4 and S. Typhimurium LT2
Initially, the genome of S. Enteritidis PT4 was compared with that of S. Typhimurium LT2, a well-characterized and fully sequenced S. enterica isolate. S. Enteritidis PT4 and S. Typhimurium LT2 are both representatives of serovars able to cause enteritis in a broad range of hosts and produce murine typhoid, but they also show significant phenotypic differences, including serovar type. An alignment of the genome of S. Enteritidis PT4 with that of S. Typhimurium LT2 revealed colinearity except for an inversion about the terminus in S. Typhimurium LT2 (Fig. 1) (McClelland et al. 2001), with >90% of coding sequences (CDS) forming an extensive core gene-set (Figs. 2, 3). The average nucleotide identity between the shared orthologs is 98.98% compared with 99.7% between those of LT2 and a second fully sequenced S. Typhimurium strain SL1344 (data not shown). The genes that are only present either in S. Enteritidis PT4 or S. Typhimurium LT2 form 6.4% and 9.6% of their respective genomes (Fig. 3). The majority of S. Enteritidis PT4 unique CDS are in clusters from >3 kb up to >40 kb, but there are very few indels of <3 kb (Fig. 2; Table 2). We refer to these nonshared gene clusters as regions of difference (ROD). CDS present in S. Enteritidis PT4 but absent from S. Typhimurium LT2 are dominated by prophage-related functions, although other functional classes are represented (Fig. 3).
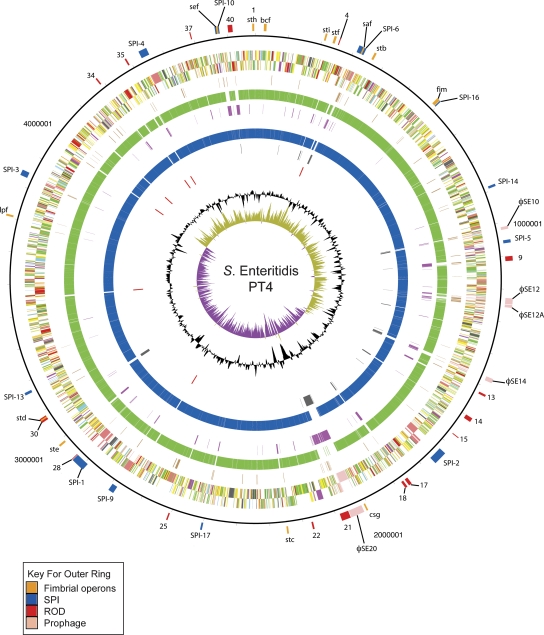
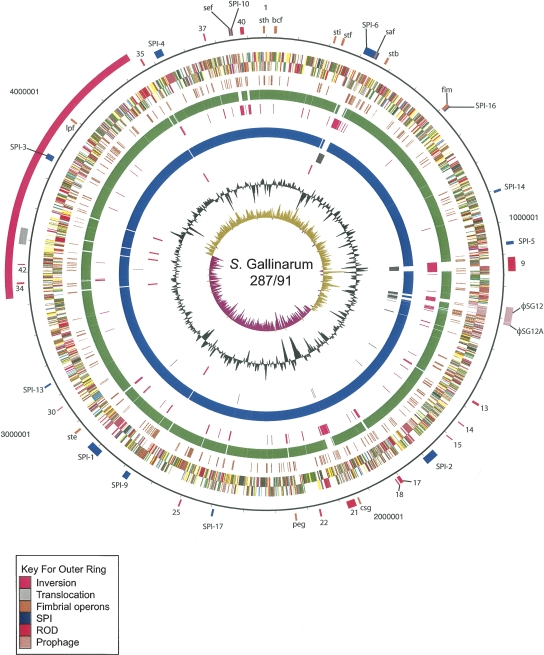
Figure 2.
Circular representation of the S. Enteritidis PT4 chromosome. From the outside in, the outer circle 1 marks the position of regions of difference (mentioned in the text) and is detailed in Table 2. Circle 2 shows the size in base pairs. Circles 3 and 4 show the position of CDS transcribed in a clockwise and anti-clockwise direction, respectively (for color codes see below); circle 5 shows the position of S. Enteritidis PT4 pseudogenes. Circles 6 and 8 show the position of S. Enteritidis PT4 genes that have orthologs (by reciprocal FASTA analysis) in S. Typhimurium strain LT2 (all CDS colored green) and S. Gallinarum strain 287/91 (all CDS colored blue), respectively. Circles 7 and 9 show the position of S. Enteritidis PT4 genes that lack orthologs in (by reciprocal FASTA analysis) in S. Typhimurium strain LT2 (all CDS colored pink) and S. Gallinarum strain 287/91 (all CDS colored gray), respectively. Circle 10 shows the position of S. Enteritidis PT4 rRNA operons (red). Circle 11 shows a plot of G + C content (in a 10-kb window). Circle 12 shows a plot of GC skew ([G − C]/[G + C]; in a 10-kb window). Genes in circles 3 and 4 are color-coded according to the function of their gene products: dark green, membrane or surface structures; yellow, central or intermediary metabolism; cyan, degradation of macromolecules; red, information transfer/cell division; cerise, degradation of small molecules; pale blue, regulators; salmon pink, pathogenicity or adaptation; black, energy metabolism; orange, conserved hypothetical; pale green, unknown; and brown, pseudogenes.
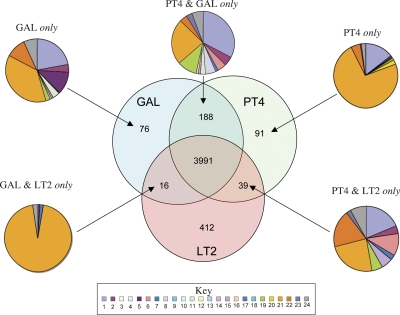
Figure 3.
Distribution of orthologous CDS in S. Enteritidis PT4, S. Typhimurium LT2, and S. Gallinarum 287/91. The Venn diagram shows the number of genes unique or shared between two other S. enterica serovars (see Methods). The associated pie charts show the breakdown of the functional groups assigned for CDS in relevant sections of the Venn diagram. Color code for the pie charts is as follows: (1) hypothetical proteins, (2) conserved hypothetical proteins, (3) chemotaxis and motility, (4) chromosomal replication, (5) chaperones, (6) protective responses, (7) transport and binding proteins, (8) adaptations to atypical conditions, (9) cell division, (10) macromolecule degradation, (11) synthesis and modification of macromolecules, (12) amino acid biosynthesis, (13) biosynthesis of cofactors, prosthetic groups and carriers, (14) central intermediary metabolism, (15) small-molecule degradation, (16) energy metabolism, (17) fatty acid biosynthesis, (18) nucleosides and nucleotide biosynthesis and metabolism, (19) periplasmic/exported/lipoproteins, (20) ribosomal proteins, (21) laterally acquired (including prophage CDS), (22) pathogenicity and virulence, (23) general regulation, and (24) miscellaneous function. PT4 indicates S. Enteritidis PT4; LT2, S. Typhimurium LT2; and GAL, S. Gallinarum 287/91
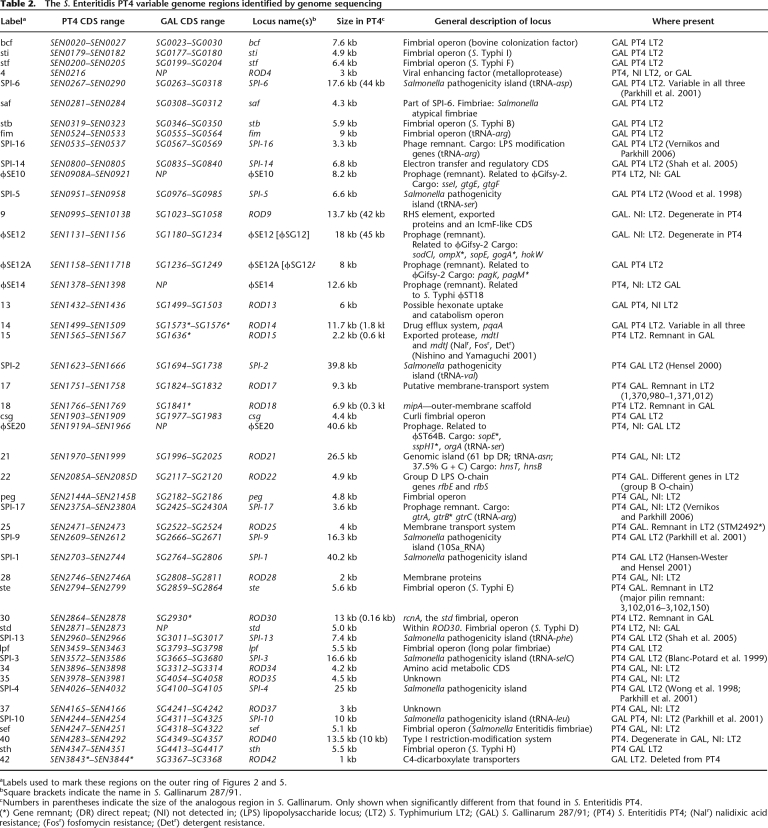
Table 2.
The S. Enteritidis PT4 variable genome regions identified by genome sequencing
aLabels used to mark these regions on the outer ring of Figures 2 and 5.
bSquare brackets indicate the name in S. Gallinarum 287/91.
cNumbers in parentheses indicate the size of the analogous region in S. Gallinarum. Only shown when significantly different from that found in S. Enteritidis PT4.
(*) Gene remnant; (DR) direct repeat; (NI) not detected in; (LPS) lipopolysaccharide locus; (LT2) S. Typhimurium LT2; (GAL) S. Gallinarum 287/91; (PT4) S. Enteritidis PT4; (Nalr) nalidixic acid resistance; (Fosr) fosfomycin resistance; (Detr) detergent resistance.
Gene sets common to both S. Enteritidis PT4 and S. Typhimurium LT2
Within the core genes, there are many of the functions associated with virulence and host interactions and include SPIs and fimbrial operons. With the exception of SPI-6, SPI-9, and SPI-10, the other SPIs in S. Enteritidis PT4 are closely related to their equivalents in S. Typhimurium LT2 (Fig. 2) (McClelland et al. 2001). Of the three SPIs that vary, S. Enteritidis PT4 SPI-10 only encodes the sef fimbrial operon, consistent with this region being mosaic in isolates of different serovars (Edwards et al. 2000; Collighan and Woodward 2001; Bishop et al. 2005). The SPI-9 CDS SE2609, encoding a large repetitive exported protein, appears intact in S. Enteritidis PT4 unlike the ortholog, STM2689, in S. Typhimurium LT2. The SPI-6 region of S. Enteritidis PT4 is 22 kb in size compared with 47 kb in S. Typhimurium LT2. SPI-6 varies markedly in size in all the other sequenced Salmonella, including S. Typhi (McClelland et al. 2001, 2004; Parkhill et al. 2001; Chiu et al. 2005). Of the other known SPIs, SPI-8, SPI-7, and SPI-15 are absent from S. Enteritidis PT4 (Parkhill et al. 2001; Vernikos and Parkhill 2006). Conversely, SPI-17 is present in S. Enteritidis PT4 but absent from S. Typhimurium LT2. The S. Enteritidis PT4 SPI-17 is a degenerate prophage encoding CDS known to be involved in O-antigen conversion in other systems (Vernikos and Parkhill 2006).
S. Enteritidis PT4 harbors 13 fimbrial clusters, 10 of which are highly conserved in S. Typhimurium LT2 with orthologous genes sharing >97% nucleotide identity and inserted at the same sites in both genomes (Fig. 1; Table 2). The only exceptions to this are safA, safB, and stdA, where the S. Enteritidis PT4 and S. Typhimurium LT2 orthologs show 81%, 87%, and 89% nucleotide identity, respectively. The S. Enteritidis PT4 fimbrial clusters not found in S. Typhimurium LT2 include a novel cluster we have termed peg, which is inserted at the same location as the S. Typhimurium LT2 stc operon and is so far restricted to S. Enteritidis, S. Gallinarum 287/91, and S. Paratyphi A. The peg fimbrial proteins show 58%–64% identity with their predicted functional equivalents in the S. Typhimurium LT2 stc cluster (Table 2). Of the remaining fimbrial clusters, ste is absent from S. Typhimurium LT2, but there is a deletion remnant of the ste major pilin subunit remaining at the analogous site (S. Typhimurium LT2; positions 3,102,016–3,102,150 bps). Fimbrial operon stj is present in S. Typhimurium LT2 and replaces a gene of unknown function still present in S. Enteritidis PT4 (SEN4331A). Thus, in common with other promiscuous salmonellae, S. Enteritidis PT4 harbors multiple functional fimbrial operons (Townsend et al. 2001).
In addition to the gene remnants found for the ste fimbrial operon S. Enteritidis PT4, RODs ROD17, ROD25, ROD34, ROD35, and ROD37 (Table 2) that fall outside of the core gene-set also have discernable remnants in S. Typhimurium LT2 or are conserved in other S. enterica and so are likely to have been present in a precursor of S. Enteritidis PT4, shared with S. Typhimurium LT2, and subsequently deleted from S. Typhimurium LT2.
Gene sets only present in S. Enteritidis PT4 but not S. Typhimurium LT2
By analyzing the genetic context of the S. Enteritidis PT4 specific RODs, we distinguish between those likely to have been acquired independently from those that may have been deleted from S. Typhimurium LT2 (discussed above). Examples of likely acquisitions include SPI-17, ROD4, ROD9, ROD13, ROD21, ROD22, ROD28, ROD40, ϕSE14, and ϕSE20 (Table 2). RODs unique to S. Enteritidis PT4 include potentially mobile genomic islands, clusters of genes encoding metabolic functions and prophage-like elements, as well as a variable assortment of fimbrial operons already described (summarized in Table 2).
Genomic islands
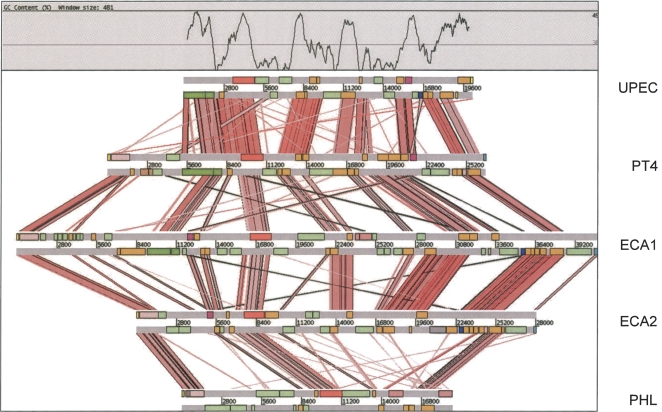
ROD21 is the only S. Enteritidis PT4 genomic island not found in S. Typhimurium LT2 and has features characteristic of mobile genetic elements (Table 2). ROD21 shares significant structural conservation with conserved genomic loci present in a range of bacteria. These islands all display an unusual G + C profile, whereby regions conserved between islands show a higher G + C content compared with the variable island-specific regions (Fig. 4) (Williamson and Free 2005; this study). Surprisingly, most of the ROD21-related islands encode paralogs of H-NS (hnsB) and/or an H-NS antagonist, hnsT (Williamson and Free 2005; Navarre et al. 2006; Doyle et al. 2007). These paralogs may play a role in relieving any potential fitness burden associated with sequestering H-NS to these low G + C DNA elements (Doyle et al. 2007).
Figure 4.
Comparison of the ROD21 locus of S. Enteritidis with related genomic islands. ACT comparison (http://www.sanger.ac.uk/Software/ACT) of amino acid matches between the complete six-frame translations (computed using TBLASTX) of ROD21 compared with related loci uropathogenic E. coli strain CFT073 (UPEC), Erwinia carotovora sbsp. atroseptica strain SCRI1043 (two loci: ECA1 and ECA2), and Photorhabidus luminescens,sbsp. laumondii TT01 (PHL; see Methods). The red bars spanning between the genomes represent individual TBLASTX matches. CDS are marked as colored boxes positioned on the horizontal gray DNA bars: (orange) genes conserved in two or more of the genomic islands; (light green) variable genes of unknown function; (dark pink) hnsB; (dark blue) hnsT; (light pink) integrase; (dark green) type IV pilin-associated genes; (red) plasmid-related mobility functions; (salmon pink) transposase-related genes; (yellow) tRNA genes; (light blue)repeats. The G + C profile for the UPEC loci is shown above. The scale is marked in base pairs.
Of the other S. Enteritidis PT4-specific RODs, ROD13 encodes five CDS displaying sequence similarities and synteny with genes associated with the uptake and catabolism of the hexonate sugar acid L-idonate encoded by the gntII locus of Escherichia coli (Table 2) (Bausch et al. 1998). Although the substrate for this system is unclear, it is known that colonic mucus contains several sugar acids that represent an important source of nutrients and that E. coli mutants unable to utilize them are unable to colonize the mouse large intestine (Sweeney et al. 1996). Moreover, genes involved in the transport of gluconate and related hexonates are up-regulated in S. Typhimurium in macrophage, suggesting that they may also be an important source of carbon for intracellular bacteria (Eriksson et al. 2003).
The S. Enteritidis PT4 RODs also include loci that are highly variable in the salmonellae and, for ROD40, between the wider Enterobacteriaceae. ROD40 locus encodes a Type I restriction/modification system and is analogous to the variable E. coli immigration control region (ICR) (Raleigh 1992; Titheradge et al. 1996).
Prophage
Prophage are known to drive diversity in S. enterica, and thus, it is not surprising that many S. Enteritidis PT4–specific RODs are prophage-like elements, including ϕSE10, ϕSE12, ϕSE12A, ϕSE14, and ϕSE20 (Fig. 2; Table 2) (Thomson et al. 2004; Cooke et al. 2007). All these prophage regions are related and carry the same cargo genes as prophage found previously in other S. enterica (see Table 2), including genes encoding type three secretion system (TTSS) effector proteins—sseK3, sspH2, gogA, sseI, and sopE; the PhoPQ-activated genes pagK and pagM; as well as sodCI encoding a Cu/Zn superoxide dismutase known to be an important colonization factor for S. Typhimurium (Stanley et al. 2000; Figueroa-Bossi et al. 2001; Mmolawa et al. 2003; Thomson et al. 2004). Of the six prophage-related regions, only ϕSE20 appears intact and probably represents a recent insertion event, whereas remnants of ϕSE12A are also present at the same location in S. Typhimurium LT2 and probably represent the most ancient phage insertion that has been maintained in these two Salmonella lineages. However, the number of remnants and intact cargo genes found on the S. Enteritidis PT4 prophage highlights the importance of these elements for gene sampling and increasing the overall diversity and even pathogenic potential of salmonellae.
Whole-genome sequence of S. Gallinarum 287/91 and comparisons with S. Enteritidis PT4 and S. Typhimurium LT2
One of the striking features of the S. Gallinarum 287/91 genome is the high simi larity with S. Enteritidis PT4 compared with S. Typhimurium LT2 (Figs. 3, 5). The average nucleotide identities of orthologs shared between S. Gallinarum 287/91 and S. Enteritidis PT4 were higher (99.7%) than those found in LT2 (98.93%). Another obvious feature is the massive accumulation of pseudogenes in S. Gallinarum 287/91 compared with S. Enteritidis PT4 and S. Typhimurium LT2. The genome of S. Gallinarum 287/91 is slightly smaller than S. Enteritidis PT4, carries significantly fewer tRNA genes (Table 1), and is colinear except for a single inversion (817 kb; about the rRNA operons) and translocation of a region (49 kb) located between two different rRNA operons (Figs. 1, 5).
Figure 5.
Circular representation of the S. Gallinarum 287/91 chromosome. For a full description of this figure, see legend to Figure 2. The exception is that circle 5 shows the position of S. Gallinarum 287/91 pseudogenes. Circles 6 and 8 show the position of S. Gallinarum 287/91 genes which have orthologs (by reciprocal FASTA analysis) in S. Typhimurium LT2 (all CDS colored green) or S. Enteritidis PT4 (all CDS colored blue), respectively. Circles 7 and 9 show the position of S. Gallinarum 287/91 genes that lack orthologs in (by reciprocal FASTA analysis) in S. Typhimurium strain LT2 (all CDS colored pink) and S. Enteritidis PT4 (all CDS colored gray), respectively. Circle 10 shows the position of the S. Gallinarum 287/91 rRNA operons (red).
The number of CDS unique to S. Gallinarum 287/91 (76) or shared exclusively between S. Gallinarum 287/91 and S. Typhimurium LT2 (16) was small and predominantly phage-associated (Figs. 3, 5). Moreover, genes from both of these categories all fell within regions (SPI-6, ROD9, ROD42, and ϕSG12) that are present in S. Enteritidis PT4 but appear to be in the process of being lost. Consequently, these genes are unlikely to be recent acquisitions by S. Gallinarum 287/91.
Of the 130 CDS specific to S. Enteritidis PT4, compared with S. Gallinarum 287/91, those associated with ROD4 and prophages ϕSE10, ϕSE14, and ϕSE20 (82 CDS) appear to be recent acquisitions with no evidence of them ever being present in S. Gallinarum 287/91(Fig. 3; Table 2). Of the remaining 48 CDS in this category, 21 were located on loci (ROD15, ROD18, and ROD30) (Table 2) almost entirely deleted from S. Gallinarum 287/91. The others were located on shared loci such as ROD14 and SPI-6 that are degenerate in both serotypes, compared with S. Typhimurium LT2. The functions that these RODs encode in S. Enteritidis PT4 are summarized in Table 2.
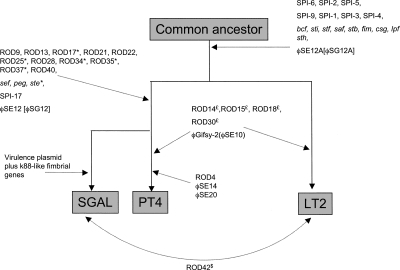
Thus, we provide compelling genetic evidence that S. Enteritidis and S. Gallinarum are recently diverged clones. On this conclusion, we have plotted the most parsimonious explanation for the observed gene flux following the divergence of S. Typhimurium LT2, S. Enteritidis PT4, and S. Gallinarum 287/91 (Fig. 6).
Figure 6.
Line diagram to represent the whole-genome differences of S. Enteritidis PT4, S. Typhimurium strain LT2, and S. Gallinarum strain 287/91. A summary of the observed loss and gain of RODs described in Table 2. The diagram is based on the assumption that following the divergence of PT4 and LT2 from a common ancestor PT4 and GAL have subsequently diverged. Branches are not intended to infer phylogenetic distance. Evidence that a locus was once present in LT2, PT4, or GAL (see legend to Figure 1) but has subsequently been deleted from that genome is marked by the suffix *, $, or £, respectively. Brackets indicate the name for locus in GAL. Parentheses indicate the name for locus in PT4. Dotted arrows mark the position in the pseudo-tree at which that ROD(s) appears
Functional gene loss and pseudogene formation
In addition to the large scale deletion, there is further evidence of reductive evolution in S. Gallinarum 287/91 in the form of 309 putative pseudogenes that carry frameshifts or premature stop codons or that are remnants of genes present in other bacteria. Remarkably, this represents ∼7% of the total coding capacity of the genome and includes genes from many functional categories, including metabolism and virulence (for a full list, see Supplemental Table 1).
S. Gallinarum 287/91 has lost several pathways, which are likely to narrow the spectrum of substrates available for use as sources of carbon and energy. These include malS encoding a periplasmic alpha-amylase required for growth on long chain maltodextrins (Schneider et al. 1992), those affecting the catabolism of (D)-glucarate (gudD) and the mutation in the hyaF gene (within the hydrogenase 1 [hya] gene cluster).
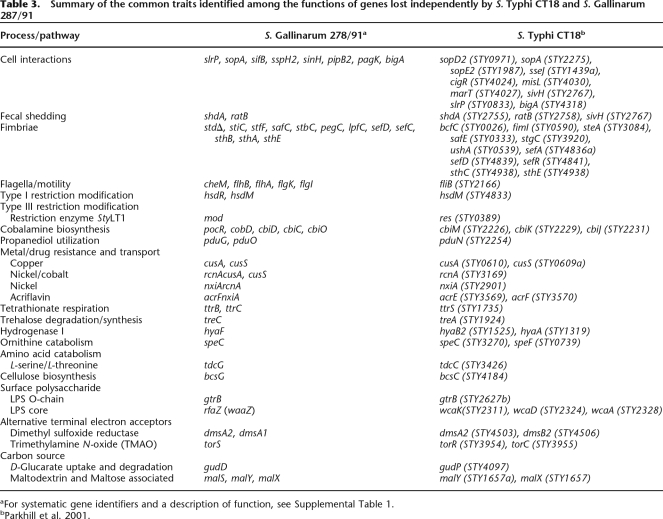
S. Gallinarum 287/91 also possesses multiple mutations in genes within all three operons required for the breakdown 1,2-propanediol: ttr, cbi, and pdu operons directing tetrathionate respiration; coenzyme B12 biosynthesis (B12; cobalamine); and 1,2-propanediol degradation, respectively (Supplemental Table 1) (Roth et al. 1996). 1,2-Propanediol is an important source of energy for S. Typhimurium, and cbi mutants are significantly attenuated in their ability to grow in macrophages (Klumpp and Fuchs 2007). Consequently, for most of the salmonellae, the ability to degrade propanediol is the likely selective pressure maintaining the cbi and ttr genes, and the loss of function of any of these three pathways was probably the precursor to the degeneration of the others in S. Gallinarum 287/91. Mutations within the cbi, pdu, and ttr genes was also a feature of the S. Typhi CT18 genome (Table 3), suggesting that these mutations may be characteristic of more invasive Salmonella serotypes.
Table 3.
Summary of the common traits identified among the functions of genes lost independently by S. Typhi CT18 and S. Gallinarum 287/91
aFor systematic gene identifiers and a description of function, see Supplemental Table 1.
Uniquely among the Salmonella, S. Gallinarum 287/91 and S. Pullorum are unable to make glycogen (Supplemental Table 2) (McMeechan et al. 2005). The genome sequence data revealed that mutations in the glycogen biosynthetic pathway are extensive in S. Gallinarum 287/91, including glgA, glgB, and glgC, which are responsible for all steps in biosynthesis. Although the significance of these mutations are not clear, they may explain in part the poor survival of this bacterium outside of the host (McMeechan et al. 2005).
Several of the identified S. Gallinarum 287/91 pseudogenes lie in amino acid catabolic or biosynthetic pathways. Salmonella encodes three pathways for arginine degradation (Reitzer 2003), and S. Gallinarum 287/91 carries mutations in two of these: There are deletions or frameshifts in the genes of the arginine N-succinyltransferase (AST) (astA) and arginine deiminase (ADI) (arcA) pathways, respectively. S. Gallinarum 287/91 also carries a mutation in speC encoding the ornithine decarboxylase, making the one remaining intact arginine catabolic pathway, arginine decarboxylase pathway (ADC), an essential biosynthetic route for putrescine. The mutation in speC is also likely to explain the inability of S. Gallinarum to decarboxylate ornithine, a defining feature of this Salmonella serovar (Supplemental Table 2) (Crichton and Old 1990).
Pseudogenes potentially involved in virulence and host adaptation
S. Gallinarum 287/91 is defined as being nonmotile. S. Gallinarum 287/91 carries 50 genes associated with motility and chemotaxis, distributed over three loci. Of these, five genes, present in two loci, carry mutations that explain the nonmotile phenotype, including cheM, flhA, flhB, flgK, and flgI (Supplemental Table 1).
Of the 13 fimbrial operons detected in S. Enteritidis PT4, the std fimbrial operon is not present in S. Gallinarum 287/91 (see ROD30 above). The remaining 12 S. Gallinarum 287/91 fimbrial operons are identical to those in S. Enteritidis PT4 except for the mutations in genes within operons sti, stf, saf, stb, peg, lpf, sef, and sth (see Supplemental Table 1). The level of pseudogene formation within these 12 fimbrial gene clusters (16%) is over twice that of the genome average (7%). Only operons fim, bcf, csg, and ste remain undisrupted on the S. Gallinarum 287/91 chromosome (summarized in Fig. 1). However, both S. Enteritidis and S. Gallinarum carry fimbrial operons on their virulence plasmids. The S. Enteritidis virulence plasmid carries five genes—pefA, pefB, pefC, pefD, and pefR—highly conserved with those of S. Typhimurium LT2 (Woodward and Kirwan 1996). The pef operon is not present on the S. Gallinarum 287/91 plasmid; in its place are three intact fimbrial genes displaying sequence similarity with those of the E. coli K88 fimbrial gene cluster (Rychlik et al. 1998). This fimbrial operon represents the only evidence of S. Gallinarum 287/91 having acquired new functions since the split from S. Enteritidis. It would be interesting to know if any isolates of S. Enteritidis carry such a fimbrial operon and whether this is a common characteristic of all S. Gallinarum strains. Significantly, the host-adapted S. Typhi harbors novel fimbrial genes, including Type IV pili associated with the acquisition of mobile elements (Pickard et al. 2003), and like S. Gallinarum 287/91, there is an elevated level of mutation in fimbrial genes (14% compared with the genome average of 4.4%), again suggesting parallel paths toward host adaptation (Table 3).
Salmonella can express several paralogous TTSS effector proteins, which show a degree of functional redundancy, for example, sopE and sopE2 (Friebel et al. 2001). Other effectors related by sequence include pipB and pipB2, and sifA and sifB. S. Gallinarum 287/91 has lost one of each of these parlogous pairs. Other S. Gallinarum 287/91 TTSS effector genes that carry mutations include sopA, which has been implicated in S. Typhimurium–induced intestinal inflammation (Zhang et al. 2006). Using an antibody against the C-terminal portion of SopA, we detected a secreted protein of the expected size in S. Enteritidis PT4 but not S. Gallinarum 287/91 (data not shown), consistent with the location of a stop codon prior to the mAb-binding region in S. Gallinarum SopA predicted by the genome sequence (Supplemental Table 2). SopA influences Salmonella-induced enteritis, and taken together with the attrition of other Type III secreted effectors, this may partially dictate the differential virulence of the serovars in mammalian hosts.
As well as additional pseudogenes associated with cell interactions, again like S. Typhi, S. Gallinarum 287/91 carries mutations in genes also associated with shedding (shdA and ratB), drug resistance, DNA restriction/modification, and protective responses (Supplemental Table 1). The majority of S. Enteritidis isolates can produce a biofilm, of which cellulose is a key component. While mutations in biofilm production may not measurably affect virulence, they are significantly less resistant to chemical and mechanical stress. Consequently, this is likely to be an adaptation by Salmonella to survival in the environment but has also been suggested to prolong retention in the gut (Solano et al. 2002). We have shown experimentally that S. Gallinarum 287/91 is unable to make cellulose, and this is likely to be explained by a mutation in bcsG (Supplemental Table 2). This is consistent with the reduced ability of S. Gallinarum to colonize the gut compared with S. Enteritidis.
Conclusions
The data presented in this report provide several clear messages, some of which may be experimentally tractable. Comparative analysis of the genomes of S. Enteritidis PT4 and S. Gallinarum 287/91 shows that representative strains of these two S. enterica serovars are highly related and that S. Gallinarum may be a direct descendant of S. Enteritidis. Importantly, S. Enteritidis is promiscuous, being able to colonize and infect multiple hosts, including chickens, cattle, mice, and humans, in addition to producing murine typhoid. Whereas S. Gallinarum is highly restricted to causing a typhoid-like disease in avian species, it is relatively noninfectious in other hosts, including mice, and does not colonize the gut of animals. Thus, we suggest that there is an experimental opportunity to use genetic approaches to define the genetic basis of host restriction by directly comparing the pathogenicity of strains of S. Enteritidis and S. Gallinarum in murine and chicken models.
Previous genome analyses on host-restricted salmonellae has involved human-restricted serovars, including S. Typhi and S. Paratyphi, limiting experimental tractability. Nevertheless genome comparisons of host-restricted/adapted S. enterica serovars, and indeed of other pathogens, indicate that loss of gene function may be a common evolutionary mechanism through which host adaptation occurs. Gene loss not only may limit the interhost promiscuity of the pathogen but also is likely to restrict the potential pathogenicity in the host to a more limited set of interactions. We hypothesize that gene loss may be a mechanism of targeting the invading pathogen preferentially to particular tissues or host cells and avoiding the potential stimulation of nonspecific inflammation. An example here would be the loss of flagella or fimbriae, which can mediate attachment and invasion of cell surfaces and may activate pattern recognition molecules. In addition, gene loss can influence the ability of the pathogen to survive in the external environment or even in stressful situations within the host. Table 3 provides a list of some of the common traits identified among the functions of genes lost independently by S. Typhi and S. Gallinarum. Some of the overlaps are striking, including the loss of common TTSS effectors and genes involved in common metabolic processes such as cobalamine and propanediol utilization, tetrathionate respiration, sugar uptake and utilization, hydrogenase activity, cellulose production, ornithine decarboxylase activity, and electron transport acceptor function. Some of these common traits have also been noted to have changed in representatives of gut adapted (Y. enterocolitica) versus systemic (Y. pestis) yersiniae, and again in this system, gene loss may be involved in the adaptation from a gut to a systemic lifestyle. We believe that further studies analyzing the contribution of pseudogenes and their functional alleles to host adaptation and tissue specificity and, in particular, the parallel but overlapping degradative evolutionary pathways followed by different organisms adapting to different hosts will lead to significant understanding of the mechanisms of host adaptation and host restriction and could be applicable to the less tractable human-adapted organisms, such as S. Typhi.
Methods
Bacterial strains
S. Gallinarum strain 287/91 was isolated from an outbreak of fowl typhoid in brown egg–laying hens by A. Berchieri, University of Sao Paulo, Jaboticabal, Brazil. It is highly virulent (>90% mortality) in susceptible breeds of chickens (P. Barrow and A. Berchier, unpubl.). It was chosen in preference to the well-characterized strain 9 (Smith 1955) because of the length of laboratory passage of the latter strain. S. Enteritidis phage type 4 (PT4) strain P125109 was isolated from an outbreak of human food-poisoning in the United Kingdom that was traced back to a poultry farm. The strain is highly virulent in newly hatched chickens and is also invasive in laying hens, resulting in egg contamination (Barrow 1991; Barrow and Lovell 1991). Biochemical tests for carbohydrate catabolism were performed using api 50 CH according to manufacturer’s instructions (BioMerieux).
Growth and sequencing of S. Enteritidis PT4 and S. Gallinarum 287/91
Methods for sequencing S. Enteritidis PT4 and S. Gallinarum 287/91 were identical unless stated. A single bacterial colony was picked from Congo Red agar and grown overnight in BAB broth with shaking at 37°C. Cells were collected, and total DNA (10 mg) was isolated using proteinase K treatment followed by phenol extraction. The DNA was fragmented by sonication, and several libraries were generated in pUC18 using size fractions ranging from 1.0–2.5 kb.
The whole genome sequenced to a depth of 9× coverage from M13mp18 (insert size 1.4–2 kb) and pUC18 (insert size 2.2–4.2 kb) small insert libraries using dye terminator chemistry on ABI3700 automated sequencers. End sequences from larger insert plasmid (pBACe3.6, 12–30 kb insert size) libraries were used as a scaffold.
The sequence was assembled, finished, and annotated as described previously (Parkhill et al. 2000), using the program Artemis (Berriman and Rutherford 2003) to collate data and facilitate annotation.
The genomes have been submitted to EMBL under the following accession numbers: S. Enteritidis PT4 genome, AM933172; S. Gallinarum 287/91 genome, AM933173.
In silico genome analysis
The genome sequences of S. Typhimurium strain LT2 (McClelland et al. 2001), S. Enteritidis PT4, and S. Gallinarum 287/91 were compared pairwise using the Artemis Comparison Tool (ACT) (Carver et al. 2005). Subsequences taken from the genomes of uropathogenic E. coli strain CFT073 (Welch et al. 2002), Erwinia carotovora sbsp. atroseptica strain SCRI1043 (Bell et al. 2004), and Photorhabidus luminescens,sbsp. laumondii TT01 (Duchaud et al. 2003) were compared with ACT as above and used to construct Figure 4.
Pseudogenes had one or more mutations that would ablate expression; each of the inactivating mutations was confirmed by subsequently rechecking the original sequencing data and where necessary were resequenced.
Orthologous gene sets were identified by reciprocal FASTA searches. Only those pairs of homologous CDS were retained for further analysis where the predicted amino acid identity was ≥40% over 80% of the protein length. These genes were then subject to manual curation using gene synteny to increase the accuracy of this analysis. This strategy was applied to pairwise comparisons of the genomes of S. Typhimurium strain LT2, S. Enteritidis PT4, and S. Gallinarum 287/91.
Cellulose production assay
For preparation and use of Calcofluor plates, Calcofluor white stain was obtained from Sigma as a 0.1% w/v solution. This was added to L-agar at a final concentration of 200 μg/mL as recommended by Solano et al. (2002). Bacterial cultures were inoculated and then left at room temperature for 48 h.
Colony fluorescence was examined by holding the plate over a 366-nm UV transilluminator. Controls used included E. coli C600 (negative control) and S. typhimurium SL1344 (positive control). Colony fluorescence was scored quantitatively using the controls as standards.
Acknowledgments
We thank the core sequencing and informatics teams at the Sanger Institute for their assistance and The Wellcome Trust for its support of the Sanger Institute Pathogen Sequencing Unit. This project was funded through The Wellcome Trust Beowulf Genomics Initiative.
Footnotes
[Supplemental material is available online at www.genome.org. The genome sequence data from this study have been submitted to EMBL under accession nos. AM933172 and AM933173.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.077404.108.
References
- Barrow P. Experimental infection of chickens with Salmonella enteritidis. Avian Pathol. 1991;20:145–153. doi: 10.1080/03079459108418749. [DOI] [PubMed] [Google Scholar]
- Barrow P.A. Rev. Sci. Tech. Vol. 19. 2000. The paratyphoid salmonellae; pp. 351–375. [DOI] [PubMed] [Google Scholar]
- Barrow P.A., Lovell M.A., Lovell M.A. Experimental infection of egg-laying hens with Salmonella enteritidis. Avian Pathol. 1991;20:339–352. doi: 10.1080/03079459108418769. [DOI] [PubMed] [Google Scholar]
- Bausch C., Peekhaus N., Utz C., Blais T., Murray E., Lowary T., Conway T., Peekhaus N., Utz C., Blais T., Murray E., Lowary T., Conway T., Utz C., Blais T., Murray E., Lowary T., Conway T., Blais T., Murray E., Lowary T., Conway T., Murray E., Lowary T., Conway T., Lowary T., Conway T., Conway T. Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli. J. Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K.S., Sebaihia M., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Sebaihia M., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Pritchard L., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Holden M.T., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Hyman L.J., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Holeva M.C., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Thomson N.R., Bentley S.D., Churcher L.J., Mungall K., Bentley S.D., Churcher L.J., Mungall K., Churcher L.J., Mungall K., Mungall K., et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. 2004;101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M., Rutherford K., Rutherford K. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 2003;4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- Bishop A.L., Baker S., Jenks S., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Baker S., Jenks S., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Jenks S., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Fookes M., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Gaora P.O., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Pickard D., Anjum M., Farrar J., Hien T.T., Ivens A., Anjum M., Farrar J., Hien T.T., Ivens A., Farrar J., Hien T.T., Ivens A., Hien T.T., Ivens A., Ivens A., et al. Analysis of the hypervariable region of the Salmonella enterica genome associated with tRNA(leuX) J. Bacteriol. 2005;187:2469–2482. doi: 10.1128/JB.187.7.2469-2482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Potard A.B., Solomon F., Kayser J., Groisman E.A., Solomon F., Kayser J., Groisman E.A., Kayser J., Groisman E.A., Groisman E.A. The SPI-3 pathogenicity island of Salmonella enterica. J. Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Chiu C.H., Tang P., Chu C., Hu S., Bao Q., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Tang P., Chu C., Hu S., Bao Q., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Chu C., Hu S., Bao Q., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Hu S., Bao Q., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Bao Q., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Yu J., Chou Y.Y., Wang H.S., Lee Y.S., Chou Y.Y., Wang H.S., Lee Y.S., Wang H.S., Lee Y.S., Lee Y.S. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 2005;33:1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collighan R.J., Woodward M.J., Woodward M.J. The SEF14 fimbrial antigen of Salmonella enterica serovar Enteritidis is encoded within a pathogenicity islet. Vet. Microbiol. 2001;80:235–245. doi: 10.1016/s0378-1135(01)00309-1. [DOI] [PubMed] [Google Scholar]
- Cooke F.J., Wain J., Fookes M., Ivens A., Thomson N., Brown D.J., Threlfall E.J., Gunn G., Foster G., Dougan G., Wain J., Fookes M., Ivens A., Thomson N., Brown D.J., Threlfall E.J., Gunn G., Foster G., Dougan G., Fookes M., Ivens A., Thomson N., Brown D.J., Threlfall E.J., Gunn G., Foster G., Dougan G., Ivens A., Thomson N., Brown D.J., Threlfall E.J., Gunn G., Foster G., Dougan G., Thomson N., Brown D.J., Threlfall E.J., Gunn G., Foster G., Dougan G., Brown D.J., Threlfall E.J., Gunn G., Foster G., Dougan G., Threlfall E.J., Gunn G., Foster G., Dougan G., Gunn G., Foster G., Dougan G., Foster G., Dougan G., Dougan G. Prophage sequences defining hot spots of genome variation in Salmonella enterica serovar Typhimurium can be used to discriminate between field isolates. J. Clin. Microbiol. 2007;45:2590–2598. doi: 10.1128/JCM.00729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton P.B., Old D.C., Old D.C. Salmonellae of serotypes gallinarum and pullorum grouped by biotyping and fimbrial-gene probing. J. Med. Microbiol. 1990;32:145–152. doi: 10.1099/00222615-32-3-145. [DOI] [PubMed] [Google Scholar]
- Doyle M., Fookes M., Ivens A., Mangan M.W., Wain J., Dorman C.J., Fookes M., Ivens A., Mangan M.W., Wain J., Dorman C.J., Ivens A., Mangan M.W., Wain J., Dorman C.J., Mangan M.W., Wain J., Dorman C.J., Wain J., Dorman C.J., Dorman C.J. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- Duchaud E., Rusniok C., Frangeul L., Buchrieser C., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Rusniok C., Frangeul L., Buchrieser C., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Frangeul L., Buchrieser C., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Buchrieser C., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Givaudan A., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Taourit S., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Bocs S., Boursaux-Eude C., Chandler M., Charles J.F., Boursaux-Eude C., Chandler M., Charles J.F., Chandler M., Charles J.F., Charles J.F., et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- Edwards R.A., Schifferli D.M., Maloy S.R., Schifferli D.M., Maloy S.R., Maloy S.R. A role for Salmonella fimbriae in intraperitoneal infections. Proc. Natl. Acad. Sci. 2000;97:1258–1262. doi: 10.1073/pnas.97.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J.C., Lucchini S., Thompson A., Rhen M., Hinton J.C., Thompson A., Rhen M., Hinton J.C., Rhen M., Hinton J.C., Hinton J.C. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N., Uzzau S., Maloriol D., Bossi L., Uzzau S., Maloriol D., Bossi L., Maloriol D., Bossi L., Bossi L. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 2001;39:260–271. doi: 10.1046/j.1365-2958.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- Friebel A., Ilchmann H., Aepfelbacher M., Ehrbar K., Machleidt W., Hardt W.D., Ilchmann H., Aepfelbacher M., Ehrbar K., Machleidt W., Hardt W.D., Aepfelbacher M., Ehrbar K., Machleidt W., Hardt W.D., Ehrbar K., Machleidt W., Hardt W.D., Machleidt W., Hardt W.D., Hardt W.D. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J. Biol. Chem. 2001;276:34035–34040. doi: 10.1074/jbc.M100609200. [DOI] [PubMed] [Google Scholar]
- Hansen-Wester I., Hensel M., Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- Hensel M. Salmonella pathogenicity island 2. Mol. Microbiol. 2000;36:1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- Iqbal M., Philbin V.J., Withanage G.S., Wigley P., Beal R.K., Goodchild M.J., Barrow P., McConnell I., Maskell D.J., Young J., Philbin V.J., Withanage G.S., Wigley P., Beal R.K., Goodchild M.J., Barrow P., McConnell I., Maskell D.J., Young J., Withanage G.S., Wigley P., Beal R.K., Goodchild M.J., Barrow P., McConnell I., Maskell D.J., Young J., Wigley P., Beal R.K., Goodchild M.J., Barrow P., McConnell I., Maskell D.J., Young J., Beal R.K., Goodchild M.J., Barrow P., McConnell I., Maskell D.J., Young J., Goodchild M.J., Barrow P., McConnell I., Maskell D.J., Young J., Barrow P., McConnell I., Maskell D.J., Young J., McConnell I., Maskell D.J., Young J., Maskell D.J., Young J., Young J., et al. Identification and functional characterization of chicken toll-like receptor 5 reveals a fundamental role in the biology of infection with Salmonella enterica serovar typhimurium. Infect. Immun. 2005;73:2344–2350. doi: 10.1128/IAI.73.4.2344-2350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P., Rothwell L., Galyov E.E., Barrow P.A., Burnside J., Wigley P., Rothwell L., Galyov E.E., Barrow P.A., Burnside J., Wigley P., Galyov E.E., Barrow P.A., Burnside J., Wigley P., Barrow P.A., Burnside J., Wigley P., Burnside J., Wigley P., Wigley P. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology. 2000;146:3217–3226. doi: 10.1099/00221287-146-12-3217. [DOI] [PubMed] [Google Scholar]
- Kilger G., Grimont P.A., Grimont P.A. Differentiation of Salmonella phase 1 flagellar antigen types by restriction of the amplified fliC gene. J. Clin. Microbiol. 1993;31:1108–1110. doi: 10.1128/jcm.31.5.1108-1110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp J., Fuchs T.M., Fuchs T.M. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology. 2007;153:1207–1220. doi: 10.1099/mic.0.2006/004747-0. [DOI] [PubMed] [Google Scholar]
- Li J., Smith N.H., Nelson K., Crichton P.B., Old D.C., Whittam T.S., Selander R.K., Smith N.H., Nelson K., Crichton P.B., Old D.C., Whittam T.S., Selander R.K., Nelson K., Crichton P.B., Old D.C., Whittam T.S., Selander R.K., Crichton P.B., Old D.C., Whittam T.S., Selander R.K., Old D.C., Whittam T.S., Selander R.K., Whittam T.S., Selander R.K., Selander R.K. Evolutionary origin and radiation of the avian-adapted non-motile salmonellae. J. Med. Microbiol. 1993;38:129–139. doi: 10.1099/00222615-38-2-129. [DOI] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Courtney L., Porwollik S., Ali J., Dante M., Du F., Porwollik S., Ali J., Dante M., Du F., Ali J., Dante M., Du F., Dante M., Du F., Du F., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Meyer R., Bieri T., Ozersky P., McLellan M., Bieri T., Ozersky P., McLellan M., Ozersky P., McLellan M., McLellan M., et al. Comparison of genome degradation in Paratyphi A and Typhi: Human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- McMeechan A., Lovell M.A., Cogan T.A., Marston K.L., Humphrey T.J., Barrow P.A., Lovell M.A., Cogan T.A., Marston K.L., Humphrey T.J., Barrow P.A., Cogan T.A., Marston K.L., Humphrey T.J., Barrow P.A., Marston K.L., Humphrey T.J., Barrow P.A., Humphrey T.J., Barrow P.A., Barrow P.A. Glycogen production by different Salmonella enterica serotypes: Contribution of functional glgC to virulence, intestinal colonization and environmental survival. Microbiology. 2005;151:3969–3977. doi: 10.1099/mic.0.28292-0. [DOI] [PubMed] [Google Scholar]
- Mmolawa P.T., Schmieger H., Tucker C.P., Heuzenroeder M.W., Schmieger H., Tucker C.P., Heuzenroeder M.W., Tucker C.P., Heuzenroeder M.W., Heuzenroeder M.W. Genomic structure of the Salmonella enterica serovar Typhimurium DT 64 bacteriophage ST64T: Evidence for modular genetic architecture. J. Bacteriol. 2003;185:3473–3475. doi: 10.1128/JB.185.11.3473-3475.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W.W., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S.J., Fang F.C., Porwollik S., Wang Y., McClelland M., Rosen H., Libby S.J., Fang F.C., Wang Y., McClelland M., Rosen H., Libby S.J., Fang F.C., McClelland M., Rosen H., Libby S.J., Fang F.C., Rosen H., Libby S.J., Fang F.C., Libby S.J., Fang F.C., Fang F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Nishino K., Yamaguchi A., Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Wren B.W., Mungall K., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Wren B.W., Mungall K., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Mungall K., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Davies R.M., Feltwell T., Holroyd S., Feltwell T., Holroyd S., Holroyd S., et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Mungall K.L., Bentley S.D., Holden M.T., Bentley S.D., Holden M.T., Holden M.T., et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Pickard D., Wain J., Baker S., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Wain J., Baker S., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Baker S., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Line A., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Chohan S., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Fookes M., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Barron A., Gaora P.O., Chabalgoity J.A., Thanky N., Gaora P.O., Chabalgoity J.A., Thanky N., Chabalgoity J.A., Thanky N., Thanky N., et al. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 2003;185:5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E.A. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol. Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Rodrigue D.C., Tauxe R.V., Rowe B., Tauxe R.V., Rowe B., Rowe B. International increase in Salmonella enteritidis: A new pandemic? Epidemiol. Infect. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J.R., Lawrence J.G., Bobik T.A., Lawrence J.G., Bobik T.A., Bobik T.A. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- Rychlik I., Lovell M.A., Barrow P.A., Lovell M.A., Barrow P.A., Barrow P.A. The presence of genes homologous to the K88 genes faeH and faeI on the virulence plasmid of Salmonella gallinarum. FEMS Microbiol. Lett. 1998;159:255–260. doi: 10.1111/j.1574-6968.1998.tb12869.x. [DOI] [PubMed] [Google Scholar]
- Schneider E., Freundlieb S., Tapio S., Boos W., Freundlieb S., Tapio S., Boos W., Tapio S., Boos W., Boos W. Molecular characterization of the MalT-dependent periplasmic alpha-amylase of Escherichia coli encoded by malS. J. Biol. Chem. 1992;267:5148–5154. [PubMed] [Google Scholar]
- Shah D.H., Lee M.J., Park J.H., Lee J.H., Eo S.K., Kwon J.T., Chae J.S., Lee M.J., Park J.H., Lee J.H., Eo S.K., Kwon J.T., Chae J.S., Park J.H., Lee J.H., Eo S.K., Kwon J.T., Chae J.S., Lee J.H., Eo S.K., Kwon J.T., Chae J.S., Eo S.K., Kwon J.T., Chae J.S., Kwon J.T., Chae J.S., Chae J.S. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology. 2005;151:3957–3968. doi: 10.1099/mic.0.28126-0. [DOI] [PubMed] [Google Scholar]
- Shivaprasad H.L. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 2000;19:405–424. doi: 10.20506/rst.19.2.1222. [DOI] [PubMed] [Google Scholar]
- Smith H.W. Observations on experimental fowl typhoid. J. Comp. Pathol. 1955;65:37–54. doi: 10.1016/s0368-1742(55)80005-7. [DOI] [PubMed] [Google Scholar]
- Solano C., Garcia B., Valle J., Berasain C., Ghigo J.M., Gamazo C., Lasa I., Garcia B., Valle J., Berasain C., Ghigo J.M., Gamazo C., Lasa I., Valle J., Berasain C., Ghigo J.M., Gamazo C., Lasa I., Berasain C., Ghigo J.M., Gamazo C., Lasa I., Ghigo J.M., Gamazo C., Lasa I., Gamazo C., Lasa I., Lasa I. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Stanley T.L., Ellermeier C.D., Slauch J.M., Ellermeier C.D., Slauch J.M., Slauch J.M. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer’s patches. J. Bacteriol. 2000;182:4406–4413. doi: 10.1128/jb.182.16.4406-4413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney N.J., Laux D.C., Cohen P.S., Laux D.C., Cohen P.S., Cohen P.S. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 1996;64:3504–3511. doi: 10.1128/iai.64.9.3504-3511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson N., Baker S., Pickard D., Fookes M., Anjum M., Hamlin N., Wain J., House D., Bhutta Z., Chan K., Baker S., Pickard D., Fookes M., Anjum M., Hamlin N., Wain J., House D., Bhutta Z., Chan K., Pickard D., Fookes M., Anjum M., Hamlin N., Wain J., House D., Bhutta Z., Chan K., Fookes M., Anjum M., Hamlin N., Wain J., House D., Bhutta Z., Chan K., Anjum M., Hamlin N., Wain J., House D., Bhutta Z., Chan K., Hamlin N., Wain J., House D., Bhutta Z., Chan K., Wain J., House D., Bhutta Z., Chan K., House D., Bhutta Z., Chan K., Bhutta Z., Chan K., Chan K., et al. The role of prophage-like elements in the diversity of Salmonella enterica serovars. J. Mol. Biol. 2004;339:279–300. doi: 10.1016/j.jmb.2004.03.058. [DOI] [PubMed] [Google Scholar]
- Thomson N.R., Howard S., Wren B.W., Holden M.T., Crossman L., Challis G.L., Churcher C., Mungall K., Brooks K., Chillingworth T., Howard S., Wren B.W., Holden M.T., Crossman L., Challis G.L., Churcher C., Mungall K., Brooks K., Chillingworth T., Wren B.W., Holden M.T., Crossman L., Challis G.L., Churcher C., Mungall K., Brooks K., Chillingworth T., Holden M.T., Crossman L., Challis G.L., Churcher C., Mungall K., Brooks K., Chillingworth T., Crossman L., Challis G.L., Churcher C., Mungall K., Brooks K., Chillingworth T., Challis G.L., Churcher C., Mungall K., Brooks K., Chillingworth T., Churcher C., Mungall K., Brooks K., Chillingworth T., Mungall K., Brooks K., Chillingworth T., Brooks K., Chillingworth T., Chillingworth T., et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titheradge A.J., Ternent D., Murray N.E., Ternent D., Murray N.E., Murray N.E. A third family of allelic hsd genes in Salmonella enterica: Sequence comparisons with related proteins identify conserved regions implicated in restriction of DNA. Mol. Microbiol. 1996;22:437–447. [PubMed] [Google Scholar]
- Townsend S.M., Kramer N.E., Edwards R., Baker S., Hamlin N., Simmonds M., Stevens K., Maloy S., Parkhill J., Dougan G., Kramer N.E., Edwards R., Baker S., Hamlin N., Simmonds M., Stevens K., Maloy S., Parkhill J., Dougan G., Edwards R., Baker S., Hamlin N., Simmonds M., Stevens K., Maloy S., Parkhill J., Dougan G., Baker S., Hamlin N., Simmonds M., Stevens K., Maloy S., Parkhill J., Dougan G., Hamlin N., Simmonds M., Stevens K., Maloy S., Parkhill J., Dougan G., Simmonds M., Stevens K., Maloy S., Parkhill J., Dougan G., Stevens K., Maloy S., Parkhill J., Dougan G., Maloy S., Parkhill J., Dougan G., Parkhill J., Dougan G., Dougan G., et al. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G.S., Parkhill J., Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: Revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Welch R.A., Burland V., Plunkett G., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Burland V., Plunkett G., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Plunkett G., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Buckles E.L., Liou S.R., Boutin A., Hackett J., Liou S.R., Boutin A., Hackett J., Boutin A., Hackett J., Hackett J., et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson H.S., Free A., Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol. Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- Wong K.K., McClelland M., Stillwell L.C., Sisk E.C., Thurston S.J., Saffer J.D., McClelland M., Stillwell L.C., Sisk E.C., Thurston S.J., Saffer J.D., Stillwell L.C., Sisk E.C., Thurston S.J., Saffer J.D., Sisk E.C., Thurston S.J., Saffer J.D., Thurston S.J., Saffer J.D., Saffer J.D. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar typhimurium LT2. Infect. Immun. 1998;66:3365–3371. doi: 10.1128/iai.66.7.3365-3371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.W., Jones M.A., Watson P.R., Hedges S., Wallis T.S., Galyov E.E., Jones M.A., Watson P.R., Hedges S., Wallis T.S., Galyov E.E., Watson P.R., Hedges S., Wallis T.S., Galyov E.E., Hedges S., Wallis T.S., Galyov E.E., Wallis T.S., Galyov E.E., Galyov E.E. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol. Microbiol. 1998;29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]
- Woodward M.J., Kirwan S.E., Kirwan S.E. Detection of Salmonella enteritidis in eggs by the polymerase chain reaction. Vet. Rec. 1996;138:411–413. doi: 10.1136/vr.138.17.411. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Higashide W.M., McCormick B.A., Chen J., Zhou D., Higashide W.M., McCormick B.A., Chen J., Zhou D., McCormick B.A., Chen J., Zhou D., Chen J., Zhou D., Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]