Abstract
Massively parallel sequencing of millions of <30-nt RNAs expressed in mouse ovary, embryonic pancreas (E14.5), and insulin-secreting beta-cells (βTC-3) reveals that ∼50% of the mature miRNAs representing mostly the mmu-let-7 family display internal insertion/deletions and substitutions when compared to precursor miRNA and the mouse genome reference sequences. Approximately, 12%–20% of species associated with mmu-let-7 populations exhibit sequence discrepancies that are dramatically reduced in nucleotides 3–7 (5′-seed) and 10–15 (cleavage and anchor sites). This observation is inconsistent with sequencing error and leads us to propose that the changes arise predominantly from post-transcriptional RNA-editing activity operating on miRNA:target mRNA complexes. Internal nucleotide modifications are most enriched at the ninth nucleotide position. A common ninth base edit of U-to-G results in a significant increase in stability of down-regulated let-7a targets in inhibin-deficient mice (Inha−/−). An excess of U-insertions (14.8%) over U-deletions (1.5%) and the presence of cleaved intermediates suggest that a mammalian TUTase (terminal uridylyl transferase) mediated dUTP-dependent U-insertion/U-deletion cycle may be a possible mechanism. We speculate that mRNA target site-directed editing of mmu-let-7a duplex-bulges stabilizes “loose” miRNA:mRNA target associations and functions to expand the target repertoire and/or enhance mRNA decay over translational repression. Our results also demonstrate that the systematic study of sequence variation within specific RNA classes in a given cell type from millions of sequences generated by next-generation sequencing (NGS) technologies (“intranomics”) can be used broadly to infer functional constraints on specific parts of completely uncharacterized RNAs.
MicroRNAs, a species of small noncoding RNA, induce post-transcriptional silencing of genes through mRNA decay and/or translational repression in the RNA induced silencing complex (RISC) (Ambros 2001; Bartel and Chen 2004; Wienholds and Plasterk 2005; Carthew 2006). However, more recent reports indicate that miRNAs can also activate or enhance translation (Vasudevan et al. 2007; Place et al. 2008). The active mature miRNA is processed in two steps from a primary miRNA (pri-miR) that is transcribed through Pol II or Pol III promoters. The pri-miR is processed into a precursor miRNA (pre-miR) by the RNaseIII-domain containing RNASEN (also known as DROSHA) enzyme (in the nucleus) and subsequently the pre-miR is processed into a ∼22-nucleotide (nt) dsRNA by another RNaseIII-domain containing enzyme, DICER1, once in the cytoplasm (Ambros 2001; Bartel and Chen 2004; Wienholds and Plasterk 2005; Carthew 2006). The ∼22-nt dsRNAs that are generated by RNASEN (DROSHA)/DICER1 enzymatic action contain 5′-phosphates and 3′ 2-nt overhangs. One of the strands (mature miRNA or guide RNA) is selectively loaded onto the RISC and the other strand (passenger RNA or miR*) is typically degraded. The guide RNA binds mRNA targets within the RISC to establish a miRNA:mRNA duplex. The transcripts that are regulated by a specific miRNA are determined by the binding of its 5′-seed (nucleotides 2–8) and anchor (nucleotides 13–16) with target sequences in the 3′ untranslated region (UTR) of cognate mRNAs (Rajewsky 2006; Grimson et al. 2007). In general, perfect or near-perfect complementarity yields stable duplexes that lead to mRNA decay (Hornstein et al. 2005). In contrast, imperfect miR:mRNA binding represses translation, without destroying the target mRNA.
A number of recent reports reveal that ADAR-mediated A-to-I RNA editing affects the regulation of pri- and pre-miRNA processing (Bass 2006; Yang et al. 2006; Habig et al. 2007; Kawahara et al. 2007; Ohman 2007) and redirects the target repertoire by modifying the 5′-seed (Blow et al. 2006; Liang and Landweber 2007). Recently, high levels of terminal U-insertion and A-insertion were reported in mature miRNAs from a large cohort of 26 cell types (Landgraf et al. 2007). Internal mismatches were reported at a lower frequency. The relatively small number of sequences (∼1300 clones/tissue library) and of mismatches per miRNA per tissue observed in their study made it difficult to distinguish between PCR/sequencing error and other sources of these discrepancies, such as post-transcriptional modification.
Recently developed “next-generation sequencing” (NGS) technologies, including Illumina sequencing (formerly known as Solexa sequencing) and 454 Life Sciences (Roche) pyrosequencing, afford an unprecedented opportunity to uncover novel aspects of miRNA biogenesis, processing, and function. To study the extent of the murine developmental miRNA-transcriptome, we applied Illumina and 454-GS20 amplicon sequencing strategies (Lu et al. 2005; Margulies et al. 2005; Rajagopalan et al. 2006; Ruby et al. 2006; Morin et al. 2008) to characterize small RNAs (<30 nt) expressed in mouse tissues. Ovary (Illumina), mouse embryonic pancreas E14.5 (454), and the insulin-secreting βTC-3 cell line (454) were sequenced (Efrat et al. 1988; Lynn et al. 2007). We then carried out a systematic study of nucleotide changes in mature miRNAs within a cell type through intraspecies comparison. From this work we have discovered RNA editing of internal nucleotides that are constrained in the 5′-seed and cleavage/anchor region nucleotides of predicted miRNA:mRNA duplexes associated with the mmu-let-7 family. The modifications also increase the stability of down-regulated mmu-let-7a:mRNA targets. Our results suggest a dynamic relationship between miRNAs and target genes that may play an important role in establishing the target repertoire and regulating mRNA decay versus translational repression for a given miRNA in a given cell.
Results
miRNA populations show extensive heterogeneity and nucleotide modifications that exhibit a striking nonrandom spatial distribution
In order to examine the full extent of the mammalian miRNAome, we generated 2,672,643 (133,440; 113,092) sequences from the <30-nt RNA fraction of ovary (E14.5; βTC-3). The pipeline we used to process the data is outlined in the Methods section. Discarding sequences without a perfect 10-nt linker subsequence directly adjoining the insert, of length <10 nt or matching to Escherichia coli, we obtained a yield of 2,652,576 (96,112; 78,061) sequences from ovary (E14.5; βTC-3). Alignment of these sequences with miRNA precursors (pre-miRs) from miRBase 11.0 revealed that 1,331,259 (55,374; 55,589) of ovary (E14.5; βTC-3) aligned with pre-miR sequences (Supplemental Table ST1). Mature miRNAs exhibit a high level of length and sequence heterogeneity in all three tissues. Figure 1 shows an example of this in relation to small RNA sequences isolated from mouse ovary and βTC-3 corresponding to mmu-let-7a. Of the sequences that aligned with pre-miR precursors, 38% (37%; 51%) from ovary (E14.5; βTC-3) aligned exactly with mature miRNA sequences. Another 35% (23%; 25%) exhibited terminal nucleotide additions and/or excisions similar to those previously reported in Caenorhabditis elegans (Ruby et al. 2006) and more recently in human and rodent cell types (Landgraf et al. 2007; Morin et al. 2008). A small number of sequences, 7% (14%; 2%), aligned with the pre-miR outside the mature miRNA sequence and mostly represented the miR* sequence. Approximately, 19% (27%; 22%) of miRNAs exhibit one to three mismatches (“loose matches”) to the mature miRNA sequence. Of the 313 (326; 153) miRNA species that were found to be expressed in ovary (E14.5; βTC-3) we found that 178 (214; 51) were associated with modified sequences as well as the canonical sequence (Supplemental Table ST1). The complete set of canonical and modified sequences aligning with pre-miR precursors from miRBase 11.0 is shown in Supplemental Figures S1–S3. Annotation of these modifications in relation to the pre-miR hairpins is shown in Supplemental Figure S4. The distribution of mismatches with respect to nucleotide position in miRNA populations exhibiting modifications is shown in Figure 2 and Supplemental Tables ST2–ST4. The average frequency distribution of nucleotide modifications in relation to nucleotide position is shown in Figure 3A. Four lines of evidence argue against RT-PCR and sequencing errors as contributing to this discrepancy. First, the linker sequences—themselves subjected to RT-PCR and sequencing in parallel with the inserts—exhibit an error rate on the order of a few tenths of one percent (<0.1%) per base. Second, these changes are seen in sequences generated through two independent NGS technologies (454 and Illumina), as well as standard Sanger sequencing. Third, only 1%–3% of sequences exhibiting internal nucleotide changes matched the mouse genome (mm8) at an alternate location exactly as is expected of novel genomic variants. We also have confirmed that the modifications found in a subset of miRNAs are not present in the constitutional DNA in the heterozygous condition through genomic DNA sequencing (Supplemental Fig. S4). Fourth, and most compelling, is the high degree of positional nonrandomness of nucleotide changes along the length of the miRNA seen in multiple tissues determined through multiple sequencing technologies.
Figure 1.
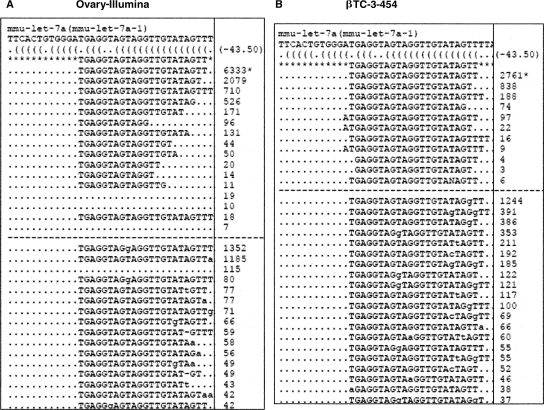
Nucleotide modifications associated with mmu-let-7a in the mouse ovary and βTC-3. This figure shows a heterogeneous collection of sequence signatures that align with the mmu-let-7a-1 precursor from miRBase 11.0 obtained through small RNA (<30 nt) sequencing of adult mouse ovary (A) and βTC-3 mouse insulinoma cell line (B). Copy numbers of each sequence signature in the total population are shown at the right end of each dotted line. Sequences that align with miRNAs mapping to more than one hairpin sequence on the genome (i.e., mmu-let-7a-1 and mmu-let-7a-2) are equally apportioned among the total number of hairpins know to occur. Collectively, the sequences highlight extensive length and sequence heterogeneity associated with mmu-let-7a. Sequence signatures that match the miRBase pre-miR precursor are shown above the dashed line. The signature flagged by an asterisk represents the mature miRNA as it is represented in miRBase 11.0. Loose matches aligning with one to three mismatches to the mmu-let-7a precursor are shown below the dashed line.
Figure 2.
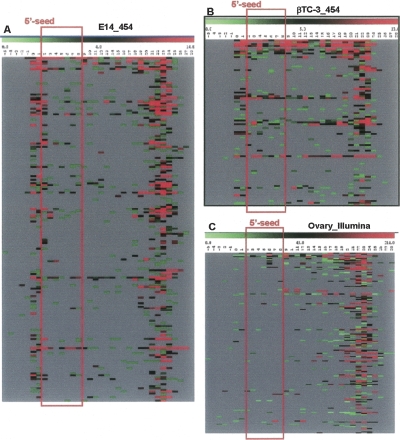
Heat maps summarizing nucleotide modifications across all miRNAs that exhibit “loose matches.” Heat maps were generated by finding all “loose matches” to the pre-miR precursors. Sequence signatures with a copy number ≥ 10 are represented. Each row here represents an individual miRNA. Each column represents a nucleotide position. The values are plotted along a scale of red to green (shown at top), showing the number of changes: (red) high; (black) medium; (green) low; (gray) no changes. (A) Data from E14.5 sequenced through 454; (B) data from βTC-3 sequenced through 454; (C) data from adult mouse ovary sequenced through Illumina.
Figure 3.
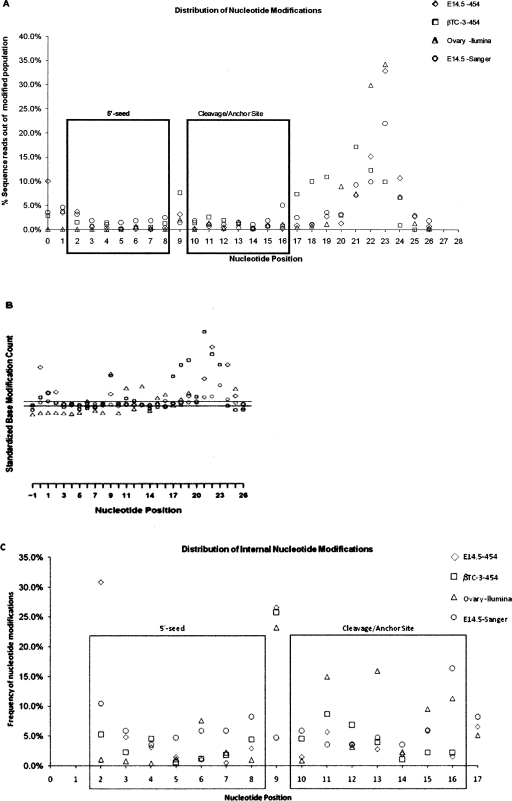
(A) Relative distribution of nucleotide modifications associated with mature miRNAs in relation to nucleotide position. This panel shows the distribution of nucleotide changes along the length of the mature miRNA. Modifications are highly suppressed in nucleotides 2–8 (containing the seed region that is important for miRNA–mRNA binding) and nucleotides 10–15 possibly defining additional determinants in the miRNA–mRNA binding. In sharp contrast, modifications are frequent at nucleotide 9 and the 3′ regions (nucleotides 16 and above). From these analyses we see that nucleotide positions 3–7 in the 5′-seed and nucleotides 10–15 exhibit significant suppression from nucleotide modification. (B) Statistical test for nucleotide modification distribution. This panel shows results from a statistical test used to examine the nature of nucleotide distribution in relation to nucleotide position of the mature miRNA. The method used is described in Methods. Under the null hypothesis where all positions behave the same with respect to base modification, we expect a random distribution of nucleotide modifications in relation to position. In this model we expect this quantity to be approximately normally distributed with a mean of 0 and standard deviation of 1. To ensure a simple plot, the median Z-score across all positions was set to 0 for each sample. The two black horizontal lines delineate the standard deviation around the Z-score. All data points above the upper line reflect modifications that occur at a rate greater than expected by chance. All data points below the lower line reflect modifications that occur at a rate less than expected by chance. From the data presented in this figure we see that the 5′-seed nucleotides 3–7, cleavage site nucleotide 10, and anchor nucleotides 14, 15 are constrained. Nucleotide position 9 is the most enriched internal position for modifications. (C) Relative distribution of internal nucleotide modifications. This panel shows the distribution of nucleotide changes in relation to internal positions (nucleotides 2–17). Modifications are highly suppressed in nucleotides 2–8 (containing the seed region that is important for miRNA–mRNA binding) and nucleotides 10–15 possibly defining additional determinants in the miRNA–mRNA binding. In sharp contrast, modifications are frequent at nucleotide 9 and the 3′-regions (nucleotides 16 and above). From these analyses we see that nucleotide positions 3–7 in the 5′-seed and nucleotides 10–15 exhibit significant suppression from nucleotide modification.
The distribution of base substitutions/insertions-deletions across the miRNA sequence was examined through statistical testing to evaluate the differences in the extent of sequence variation across the length of the mature miRNA sequence. Under the null hypothesis we assume that nucleotide modifications are random with respect to position and that all positions behave the same with respect to nucleotide modification. Under this null hypothesis we are able to standardize the observed counts of nucleotide modifications by subtracting the expected number of changes under the null model and scaling by the expected standard deviation. We expect the standardized number of changes at each position to be approximately normally distributed with a mean of 0 and standard deviation of 1. Statistical testing on the rate of nucleotide changes along the length of the miRNA sequence reveals a striking nonrandom character of base changes with respect to nucleotide position (Fig. 3B). Modifications in nucleotide positions 3–7 (5′-seed) and 10, 14, and 15 (cleavage site/anchor sites) of the mature miRNA are significantly suppressed.
Ninth nucleotide modifications in mmu-let-7a increase the stability of predicted miRNA:mRNA duplexes
Figure 3A shows the distribution of nucleotide modifications across the length of the mature miRNA for the entire set of miRNAs in ovary, E14.5, and βTC-3. The ninth nucleotide position appears to be especially enriched and statistically significant since the modifications occur above the standard deviation around the Z-score of 0 in Figure 3B. Figure 3C shows the distribution of internal nucleotide modifications encompassing nucleotides 2–17. The ninth nucleotide position occurs in between the 5′-seed (nucleotides 2–8) and anchor (nucleotides 13–16) that specify the minimal requirements for miR:mRNA binding (Rajewsky 2006; Grimson et al. 2007). Therefore, the ninth nucleotide is highly likely to occur in a bulge region of the miR:mRNA duplex. A U-residue at the ninth nucleotide bulge must be paired with a U-residue or a C-residue. The G-residue that replaces the U-residue is able to base-pair with either a U-residue or a C-residue and therefore has a high likelihood of stabilizing the duplex. The majority of the ninth nucleotide edits can be attributed to the let-7 family, which represents one of the most abundant miRNAs in all three cell types. A U-to-G modification at the ninth nucleotide position of mmu-let-7a is a commonly found edit in all three tissues (Supplemental Figs. S1–S3).
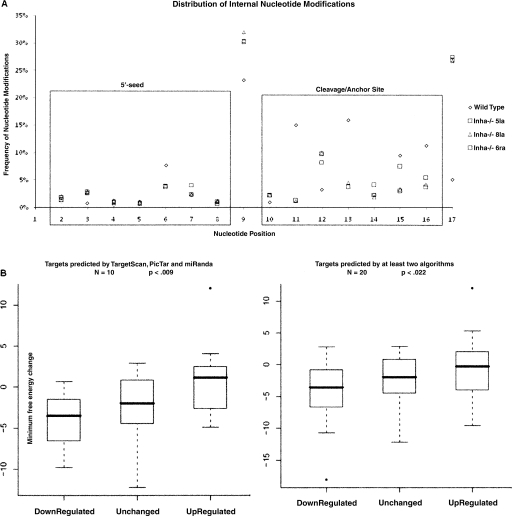
In order to explore the impact of internal edits on the duplex stability of predicted mRNA targets of mmu-let-7a we compared ovaries from wild-type mice and inhibin alpha mutants (Inha−/−) generated by Matzuk et al. (1992). Three independent Inha−/− mutant ovary samples (5la, 6ra, and 8la) were compared with wild type. Figure 4A shows the distribution of internal (nucleotides 2–17) edits across the length of the mature miRNA in relation to Inha−/− 5la, Inha−/− 6ra, Inha−/− 8la, and wild type. The data used to construct Figure 4A are shown in Supplemental Table ST5. The Inha−/− mutants exhibit a significant increase in frequency of ninth nucleotide modifications (average 31%) as compared to wild type (23%). Once again mmu-let-7a showed the most significant changes in the mutants. In order to examine the effect of this increase in ninth nucleotide modifications on target genes we measured minimum free energy changes (Δmfe) in edited mmu-let-7a:target mRNA duplexes as compared to canonical mmu-let-7a:target mRNA duplexes. The fold changes associated with mmu-let-7a target genes predicted by TargetScan, miRanda, and PicTar (Lewis et al. 2003; John et al. 2004; Grün et al. 2005) in the three Inha−/− mutant ovaries are shown in Supplemental Table ST6. Twenty genes each from down-regulated, up-regulated, and unchanged in fold-changes as compared with wild type were analyzed. We selected 10 target genes predicted by all algorithms (TargetScan, PicTar, and miRanda) and 10 additional genes predicted by at least two algorithms. mmu-let-7a is increased by ∼11-fold in the mutant ovaries (Supplemental Table ST6). The average minimum free energy change (Δmfe) for the most common edit (U-to-G modification occurring at the ninth nucleotide position) in mmu-let-7a was measured in relation to mRNA targets. Using the criteria outlined in the probability of interaction by target accessibility (PITA) method (Chitwood and Timmermans 2007; Kertesz et al. 2007), we selected in each case the predicted 3′-UTR target sequence together with 3 nt of upstream and 15 nt of downstream flanking sequence for the analysis. The 3′-UTR sequence of each target gene was first folded with the wild-type unedited mmu-let-7a using the RNAHybrid program (Rehmsmeier et al. 2004). The fold-changes in expression, the Δmfe of mmu-let-7a:target duplexes, and the weighted average Δmfe are shown in Supplemental Table ST7. Analysis of Δmfe shown in Figure 4B shows a monotone trend among the up-regulated, unchanged, and down-regulated transcripts in the Inha−/− mutant. For a set of 20 down-regulated genes we find an average decrease in Δmfe of −3.9 kcal/mol in 10 down-regulated target genes predicted by all three algorithms, TargetScan, PicTar, and miRanda, and Δmfe of −4.2 kcal/mol in 10 down-regulated target genes predicted by at least two algorithms. In contrast, for a set of 20 up-regulated genes we find an average increase in Δmfe of +1.16 kcal/mol in 10 up-regulated target genes predicted by all three algorithms, TargetScan, PicTar, and miRanda, and Δmfe of −0.8 kcal/mol in 10 up-regulated target genes predicted by at least two algorithms. To test the significance of the difference between the up- and down-regulated gene sets, we performed a Wilcoxon signed rank test on the Δmfe values. We found highly significant differences between up- and down-regulated genes sets in terms of Δmfe change. We calculated a Wilcoxon P-value of P < 0.009 for 10 target genes predicted by all three algorithms and P < 0.022 for 20 target genes predicted by at least two algorithms. We were able to find 20 target genes predicted by all three algorithms and unchanged in fold-change in the Inha−/− mutants. For this set we find an average decrease in Δmfe of −2.1 kcal/mol. The differences between the unchanged let-7a targets and the other two groups were not significant. From the data presented in this section we predict that internal RNA editing of mature miRNAs may occur in the miR:mRNA target duplex and functions to decrease mfe and stabilize the duplex.
Figure 4.
(A) Relative distribution of internal nucleotide modifications associated with miRNAs from wild-type and Inha−/− mutant ovaries. This panel shows the distribution of nucleotide changes in relation to internal positions (nucleotides 2–17) associated with miRNAs from wild-type and mutant ovaries. Modifications are highly suppressed in nucleotides 2–8 (containing the seed region that is important for miRNA–mRNA binding) and nucleotides 10–15 in all four samples. In sharp contrast, modifications are frequent at the ninth position. In addition, there is a significant increase in ninth position modifications in all three mutants (average frequency of 31%) as compared to wild type (23%). (B) Impact of ninth nucleotide U-to-G modification on mmu-let-7a targets in the Inha−/− mutants. This panel shows a comparison of Δmfe among three groups of mmu-let-7a targets that exhibit down-regulation, no change, and up-regulation, respectively, in the Inha−/− mutant ovaries as compared with wild type. The Inha−/− mutants show on the average a ∼11-fold increase in expression as compared to wild type and an increase in the frequency of ninth nucleotide modification from 23% (wild type) to 31% (mutants) (Supplemental Table ST5). In this experiment we selected 20 genes each from the list of target genes shown in Supplemental Table ST6 to represent down-regulated, unchanged, and up-regulated categories. All mmu-let-7a target site sequences were analyzed against the canonical mmu-let-7a miRNA sequence and compared with mmu-let-7a miRNA containing the ninth nucleotide edit. The minimum free energy changes (Δmfe) associated with canonical versus edited duplexes are shown in Supplemental Table ST7. The left panel shows results from 10 target genes that are predicted by all three algorithms, and the right panel shows 10 genes predicted by all three algorithms and 10 genes predicted by at least two algorithms. A Wilcoxon test specifically comparing the up-regulated targets versus down-regulated targets (including all data points) shows that down-regulated targets exhibit a Δmfe (mmu-let-7a wild type vs. mmu-let-7a-edited) that is significantly lower (P < 0.009 and P < 0.022) than the up-regulated targets. Although the unchanged targets show an average Δmfe of −2.1, this difference is not significant from up- or down-regulated targets.
TUTase-mediated dUTP-dependent U-insertion/U-deletion cycle: One possible mechanism for internal editing
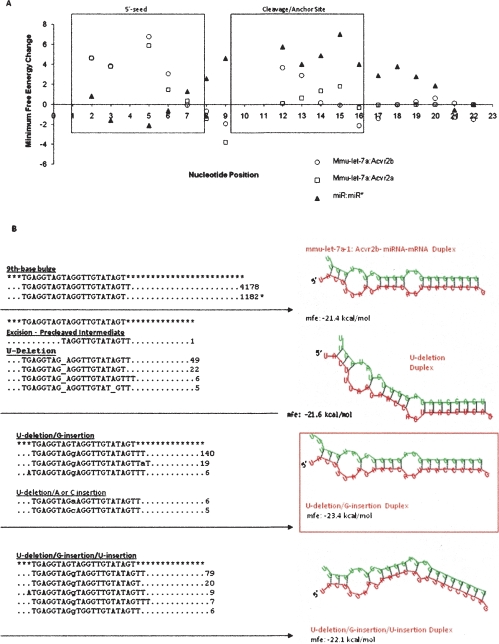
Nucleotide changes also exhibit nonrandom character in relation to the types of modifications. Across the three tissues examined, we observe an excess of U-insertions (14.8%) over U-deletions (1.5%) (Supplemental Fig. S5; Supplemental Tables ST8–ST10). In order to explore possible mechanisms that may underlie RNA editing of internal nucleotides we examined in detail the complete set of modified and partial products associated with mmu-let-7a from ovary, E14.5, and βTC-3. For this analysis we selected two predicted target genes, Acvr2a and Acvr2b, that play important roles in the ovary and pancreas. Autocrine/paracrine signaling of activin, a member of the TGF-beta superfamily, through activin receptor Acvr2a is fundamentally important for ovarian follicle development (Matzuk et al. 1995; Lovell et al. 2007; Pangas et al. 2007). In addition, the activin receptors Acvr2b and Smad2 have been shown to converge in cell signaling pathways essential for pancreas islet formation (Goto et al. 2007). We examined the mfe of mmu-let-7a-1:Acvr2a/b duplexes in the presence and absence of observed nucleotide modifications. These results are summarized in Figure 5A and show that the majority of nucleotide modifications are restricted to bulge regions of the duplex and yield a decrease in the average Δmfe (Fig. 5B). The unmodified let-7a-1 has a minimum free energy (mfe) of −18.5 kcal/mol and −21.4 kcal/mol with respect to Acvr2a and Acvr2b 3′-UTR sequences, respectively. Supplemental Table ST11A,B shows the results of mfe changes (Δmfe) in the let-7a-1:Acvr2a and let-7a-1:Acvr2b duplexes in relation to the complete set of mmu-let-7a-1 species identified with a single internal edit. The results are summarized in Figure 5A. Once again the ninth nucleotide edits exhibited the most striking decrease in Δmfe. In contrast, the miR:miR* duplex exhibits a significant increase in Δmfe at this location. The minority of modifications located in duplexed regions of the predicted miR:mRNA complexes yield an increase in Δmfe and destabilizes the duplex.
Figure 5.
(A) Impact of internal modifications on the let-7a-1:Acvr2a/b duplexes. This panel summarizes the average minimum free energy change (Δmfe) that results from the full set of nucleotide edits at that position. The complete data that were used to compute the averages can be found in Supplemental Table ST11A,B. Here we see that modifications in the duplexed regions (nucleotides 2–8, 10–15) are rare and result in a net increase in mfe and decrease in stability. In contrast, nucleotide edits in bulge regions, including nucleotide positions 9 and 16 and above, result in a net decrease in mfe and increase in stability. In contrast, we see an increase in mfe at the ninth position in the miR:miR* duplex. We do not observe any edits for positions 4, 10, or 11 in mmu-let-7a, and hence these positions do not have associated values in the graph. (B) Intermediates and final products of ninth nucleotide edits associated with the mmu-let-7a-1:Acvr2. This panel shows the minimum free energy and secondary structure of let-7a-1 species that possibly represent intermediates and end products of a U-deletion/insertion reaction. Edited sequences with copy numbers denoted at the end of the dotted line and the wild-type unedited miRNA denoted by an * are shown on the left panel. We find sequences that are consistent with excision intermediates, U-deletion products following religation, U-deletion/G-insertion products, and U-deletion/G-insertion/U-insertion products. All of these products exhibit a lower duplex mfe than the mature miRNA without edits (mfe = −21.4 kcal/mol). The U-deletion/G-insertion product that occurs at the highest copy number is also the most stable, with an mfe of −23.4 kcal/mol.
Figure 5B shows mmu-let-7a-1 miRNA species that could potentially fit a U-deletion/insertion cycle. These include precleaved intermediates: U-deletions, U-deletion/G-insertions, and U-deletion/G-insertions/U-insertions. Most of the modified miRNAs exhibit increased stability reflected by a decrease in free energy. U-deletion products exhibit a slight increase in stability (mfe = −21.6 kcal/mol). U-deletion followed by G-insertion is the most common modification and leads to considerable stabilization of the miRNA:mRNA duplex (mfe = −23.4 kcal/mol). A U-deletion/G-insertion/U-insertion also results in an increase in stability (mfe = −22.1 kcal/mol). The complete analysis of all mmu-let-7a species with a single modification is shown in Supplemental Fig. S6.
Discussion
Massively parallel sequencing of small RNA populations from multiple mouse tissues reveals length and sequence heterogeneity that is consistent with extensive post-transcriptional RNA editing of ∼10%–20% of specific populations of mature miRNAs associated with the mmu-let-7 family. The majority of modifications reported in this manuscript are distinct from the classical A-to-I editing reported previously. A-to-I editing acts on pri- and pre-miRNA substrates and does not discriminate between 5′-seed versus non-seed nucleotides (∼50% of modifications are in the 5′-seed) (Blow et al. 2006). In contrast, nucleotide modifications uncovered through “deep sequencing” reported here exhibit significant suppression in the 5′-seed and anchor nucleotides that are typically base-paired in the miR:mRNA duplex. This is “mirrored” by the high degree of sequence conservation observed in the 5′-seed sequences among variant miRNA families and target sites in 3′-UTR regions of cognate mRNAs (Chen and Rajewsky 2007). Recently, nucleotides 13–16 were reported to contain additional determinants (anchor) that may be essential to miRNA–mRNA association (Grimson et al. 2007). Our data extend that region to include nucleotide 10, which also includes the mRNA cleavage site (10–12 nt) (Ebert et al. 2007). Recently published small RNA sequencing data from Landgraf et al. (2007) and Cummins et al. (2006) do not report internal nucleotide changes or U-insertion/deletions. Landgraf et al. (2007) examined ∼1300 clones/sample, and Cummins et al. (2006) examined ∼11,000 sequences/sample. Our data shown in Figure 3A–C show that 1100 sequences from E14.5 obtained through Sanger sequencing reported here exhibit significant levels of terminal nucleotide changes similar to Landgraf et al. (2007). However, our Sanger data do not reveal a significantly high level of internal modifications. We believe that both the Landgraf et al. (2007) and Cummins et al. (2006) studies did not have sufficient sequencing depth of coverage to identify internal editing events that we see only at a scale of 55,000–1,300,000 sequences/sample. Although the edited populations occur in lower abundance than the canonical form (10%–20%), they may have a high impact on specific sets of target genes with which they form more stable duplexes.
Since the constrained regions (5′-seed and anchor) define the minimal binding requirements, it is also likely that they occur in duplexed regions and therefore are protected from modification. In support of this model we also find the ninth nucleotide position that is located between the 5′-seed (2–8 nt) and cleavage/anchor (10–16) sites is especially vulnerable to RNA editing in the mmu-let-7 family. Given that the 5′-seed cleavage and anchor sites are meaningful only in the context of the miR:mRNA duplex when the miRNA is bound to 3′-UTR elements of target genes, we suggest that RNA editing of internal nucleotides occurs on the miR:mRNA duplex. We find a significant increase in levels of mmu-let-7a containing the ninth position U-to-G edit in Inha−/− mutants. In these mutants we also find a significant decrease in the Δmfe (−4.0 kcal/mol on average) and increase in stability of down-regulated targets of mmu-let-7a. Target genes that are up-regulated and therefore most likely represent false positives in relation to the conventional 3′-UTR targeting exhibit a negligible change in the Δmfe (+0.18 kcal/mol on average) and a decrease in stability. Target genes that are unchanged that could contain a mix of “true” target genes that are simply translationally repressed and false “positives” also exhibit an average decrease in the Δmfe (−2.1 kcal/mol on average) and increase in stability. Intentionally designed bulges at positions 9–12 have been used to protect RNA interference-type cleavage and degradation of concatenated 3′-UTR target sites used for competitive inhibition studies (Ebert et al. 2007). Based on these results we propose a model in which the ninth nucleotide position of the mmu-let-7 family in the absence of any functional constraints may largely occur in “bulge regions” of a subset of let-7:mRNA duplexes that are selected for mRNA decay over translational repression. mRNA template-directed RNA editing may drive the “ninth nucleotide bulge” to undergo base-pairing and increase stability of the duplex. Increased complementarity near the cleavage site (nucleotides 10–12) may favor mRNA cleavage over translational repression, resulting in the down-regulation of those targets. These results make a compelling case for mRNA-directed RNA editing of bulge nucleotide in the miR:mRNA duplex, possibly due to increased access and susceptibility to RNA-editing enzymes. We suggest that loose mmu-let-7:mRNA associations that are initially precipitated through 5′-seed and anchor binding may be reinforced and stabilized through mRNA template-directed RNA editing of mismatched miRNA nucleotides in bulge regions.
The significant excess of U-insertions (14.8%) over U-deletions (1.5%) coupled with the presence of partial cleavage products that occur at U-residues suggests that TUTase-mediated RNA editing found in trypanosomatid mitochondria may be one possible mechanism (Smith et al. 1997). The trypanosome RET1 TUTase typically acts on mismatched bases in a duplex that is established between a small guide RNA (g-RNA) that imperfectly matches the mRNA to catalyze a dUTP-dependent U-insertion/U-deletion cycle that generates significantly higher U-insertions than U-deletions (Sacharidou et al. 2006; Zhelonkina et al. 2006). A mismatched bulge region in a miRNA:mRNA duplex could mimic a g-RNA:mRNA duplex. Argonaute-2 (Ago-2) has been shown to be the major catalytic enzyme that mediates cleavage of the mRNA target at the cleavage site (nucleotides 10–12) of the miRNA in the miRNA:mRNA duplex (Liu et al. 2004). It is possible that Ago-2 also is responsible for cleaving mismatched bulge regions in the miRNA strand of the duplex immediately abutting the cleavage site. A cycle that involves excision of the mismatched base followed by U-insertion/deletion and/or substitution could explain the high level of U-to-G substitutions associated with mmu-let-7a. Human U6-TUTase, the only TUTase identified in mammals, has been shown to act exclusively on U6 snRNA and appears to be significantly divergent from the trypanosome TUTase (Aphasizhev and Aphasizheva 2008). However, HeLa cell extracts have been found to have high levels of TUTase activity that can modify a range of RNAs in vitro (Aphasizhev and Aphasizheva 2008). Although some of the putative intermediates such as the partial cleavage products may represent artifacts that are difficult to map with confidence and occur in low abundance, it is possible that a novel mammalian TUTase-like enzyme is responsible for some of the internal nucleotide modifications associated with miRNAs.
RNA-editing and RNA-silencing pathways have been previously shown to converge during gene silencing. The RNA-editing enzyme APOBEC3G was shown to antagonize miRNA-mediated repression of genes (Huang et al. 2007). The RNA-editing pathway has also been found to antagonize the RNAi pathway (Scadden and Smith 2001). Silencing RNAs (siRNAs) are restricted in both target repertoire (based on perfect complementarity with target) and silencing strategy (mRNA decay). In contrast, most miRNAs have several hundred predicted mRNA targets that are silenced both through mRNA decay and/or translational repression. It has been shown that an increase in complementarity between miRNA and target sequence can convert silencing through translational repression to mRNA cleavage (Hutvágner and Zamore 2002). We propose that mRNA-directed RNA editing of miRNA:mRNA duplex bulges in the mmu-let-7 family functions both to expand the target repertoire and regulate the relative ratio of mRNA cleavage versus translational inhibition. The former may be achieved through the stabilization of “loosely” associated miR:mRNA duplexes and the latter by increasing or decreasing duplex complementarity to favor cleavage or translational repression, respectively. The evolution from a more restricted siRNA-based RNA interference system to a miRNA-based RNA interference system that is fluid enough to capture a large repertoire of targets and balance mRNA decay versus translational repression through the cooption of RNA-editing pathways may be fundamental to the exquisite miRNA/mRNA balance during embryonic development and organismal evolution.
Finally the work presented here highlights the tremendous potential of deep sequencing efforts to uncover novel, unique, and previously unimaginable routes to learning vital new information about noncoding RNAs. This “comparative RNA intranomics,” or intranomics, approach allows us to infer regions of functional importance in previously uncharacterized RNAs as well as potential regulatory mechanisms by intraspecies comparison without the need to compare different individuals or species and without relying on underlying mechanisms.
Methods
Small RNA cloning
Embryos from CD-1 mice were removed from pregnant mothers at E14.5, and embryonic pancreata were dissected and isolated from the spleen and other surrounding tissue; βTC-3 was grown in culture. RNA was then isolated using TRIzol and the manufacturer’s protocols (Invitrogen), with the exception that the RNA pellet was not washed with 70% ethanol following isopropanol precipitation. We obtained 189 μg of RNA from 118 pancreata. RNA was isolated from βTC-3 cells (passage 34) using TRIzol as previously described, and 600 μg of total RNA was used in the cloning procedure. miRNA cloning was carried out as previously described by Lau et al. (2001); briefly, linkers were then ligated onto the 3′- and 5′-ends of size-fractionated RNA, RNA was reverse-transcribed and amplified using PCR and Pfu polymerase (20 cycles; Stratagene). 454 sequencing primer binding sites were added using another round of PCR with the following primers: 5′-GCCTCCCTCGCGCCATCAGGAATTCCTCACTAAA-3′ and 5′-GCCTTGCCAGCCCGCTCAGGAATTCGCGGTTAAA-3′. Mouse ovaries were isolated and processed to isolate total RNA using Ambion’s mirVana kit. Sequencing was carried out using 454 or Illumina technology.
454/Illumina read processing
Out of a total of several million reads, we discarded any reads without a perfect 10-nt linker subsequence directly adjoining the insert, yielding 2,672,643 (133,440; 113,092) sequences recovered from ovary (E14.5; βTC-3) of length 10 nt or longer that were subject to further processing. We discarded all full-length, exact sequence matches to E. coli (k12, o157:h7, o157:h7 edl933, cft073) to eliminate possible sequence artifacts arising from the amplification process. Following the application of these filters, a population of 2,635,770 (96,309; 78,187) from ovary (E14.5; βTC-3) was recovered.
Statistical test to determine nature of distribution of nucleotide modification in relation to nucleotide position
Statistical hypothesis tests were established to determine the distribution of base substitutions/indels across the miRNA sequence for each of the four samples (ovary, Illumina; E14.5, 454; βTC-3, 454; and E14.5, Sanger). Standardized base change count scores were determined for each position for a total number of 28 positions (nucleotides −1, 0, 1–26) in each sample. The rate of base modification for each nucleotide position was calculated as total number of edits at a given position/(28 × total number of reads). Under the null hypothesis where all positions behave the same with respect to base modification, we would expect the number of changes at each position to be the total number of reads × uniform rate of base modification, which, in this instance, is total edits/28 × total reads. The expected number of edits at any single position is therefore total edits/28. To compute the standardized score we take the observed number of edits at each position, subtract the expected number of edits, and divide by the square root (expected). The denominator is determined assuming a Poisson model for the number of edits and taking the observed rate as the Poisson rate parameter. Under the assumptions that edit events are independent as are reads, we would expect the standardized counts to be approximately normally distributed with a mean of 0 and a standard deviation of 1 around the mean. To ensure a simple plot, the median Z-score across all positions was set to 0 for each sample. Results from this analysis are presented in Figure 3B.
RNA folding and minimum free energy calculations
Specific miR:mRNA duplexes were examined using the RNAHybrid folding algorithm. Free energy changes between the wild-type let-7a-1-mRNA target and edited let-7a-1-mRNA target were compared in order to measure the impact of the edits on the stability of the duplex. The TargetScan prediction of the target site for mmu-let-7a in the 3′ UTR of each target gene was used including flanking 3 nt upstream and 15 nt downstream as described in the PITA method (Chitwood and Timmermans 2007; Kertesz et al. 2007). Each sequence containing a single edit was folded in the context of each target site and flanking nucleotides using the RNAHybrid program (Rehmsmeier et al. 2004). Let-7a edits were grouped by the position of their edit. For each position, the edit-target site free energy was averaged and then subtracted by the wild-type let-7a-target mRNA duplex mfe. This value represents the average change of free energy (Δmfe) that an edit at a certain position is predicted to produce in relation to the miR:mRNA duplex. A positive value designates an edit that is nonbeneficial to the binding of the given target site, whereas a negative value designates an edit with a stabilizing effect. Results from these studies are shown in Supplemental Tables ST7, ST11A,B, and Supplemental Figure S6.
Statistical test to compare impact of edits on target genes
In order to determine the impact of ninth nucleotide U-to-G edit in mmu-let-7a, we compared canonical mmu-let-7a sequence and edited mmu-let-7a sequence in relation to miR:mRNA duplexes in the Inha−/− mutant ovaries where mmu-let-7a shows an 11-fold increase over wild type. The Inha−/− mutant ovaries also showed a significant increase in the frequency of ninth nucleotide edits (Supplemental Table ST5). In this experiment we selected 20 genes each from the list of target genes shown in Supplemental Table ST6 to represent down-regulated, unchanged, and up-regulated categories. All mmu-let-7a target site sequences were analyzed against the canonical mmu-let-7a miRNA sequence and compared with mmu-let-7a miRNA containing the ninth nucleotide edit. The minimum free energy changes (Δmfe) associated with canonical versus edited duplexes are shown in Supplemental Table ST7. The left panel shows results from 10 target genes that are predicted by all three algorithms, and the right panel shows 10 genes predicted by all three algorithms + 10 genes predicted by at least two algorithms. A Wilcoxon test specifically comparing the up-regulated targets versus down-regulated targets (including all data points) shows that down-regulated targets exhibit a Δmfe (mmu-let-7a wild-type vs. mmu-let-7a edited) that is significantly lower (P < 0.009 and P < 0.022) than the up-regulated targets. A one-way ANOVA and t-test of this data yielded similar results, but because of the small sample size and the presence of large deviations in the data set, the nonparametric Wilcoxon test was deemed more appropriate. Although the unchanged targets show an average Δmfe of −2.1, this difference is not significant from up- or down-regulated targets.
βTC-3 cell culture
βTC-3 cells were maintained in high glucose containing DMEM (Invitrogen) supplemented with 15% horse serum (Invitrogen), 2.5% fetal bovine serum (Invitrogen), and penicillin-streptomycin (100 U/mL penicillin, 0.1 mg/mL streptomycin; Invitrogen) at 37°C in humidified air containing 5% CO2. Cells were passaged every 4–5 d or when ∼70% confluent. Cells used in library generation were all passage 34 from initial derivation (Efrat et al. 1988).
Acknowledgments
This work was supported by grants from The Ovarian Cancer Research Fund (OCRF) to P.H.G, M.M.M., J.G.R., and A.K.N.; The Keck Center for Interdisciplinary Bioscience Training to J.G.R.; Larry L Hillblom Foundation grant no. 2002/1(E) and NIH R01DK21344 grant to M.S.G.; Juvenile Diabetes Research Foundation Advanced Fellowship award 10-2007-86 to F.C.L.; Clayton Foundation for Research award to D.B.C; and NIH/NHGRI 2 U54 HG003273-04 grant to R.A.G. We thank Toni Opalt, Andrea Trevino, Dave Hoang, Michael Fountain, Khalid Al-Kalla, Tariq Al-Kalla, Olivia Dziadek, Tariq Dayah, Richard Durante, Quratulanne Jan, Sana Sarfaraz, Poyani Desai, Levent Albayrak, Tom Samuel, Eylem Andogdu, and Maha Khalil for assistance with data analysis. We also thank Karon Cassidy for critically reading this manuscript. This work would not have been possible without University of Houston’s Illumina Genome Analyzer GA-1 acquired through generous funding from the Cullen Foundation to the University of Houston’s Institute for Molecular Design (IMD).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.078246.108.
References
- Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim. Biophys. Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bass B.L. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb. Symp. Quant. Biol. 2006;71:285–292. doi: 10.1101/sqb.2006.71.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M.J., Grocock R.J., Van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M., Grocock R.J., Van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M., Van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M., Dicks E., Futreal P.A., Wooster R., Stratton M., Futreal P.A., Wooster R., Stratton M., Wooster R., Stratton M., Stratton M. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R.W. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Chen K., Rajewsky N., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Timmermans M.C., Timmermans M.C. Target mimics modulate miRNAs. Nat. Genet. 2007;39:935–936. doi: 10.1038/ng0807-935. [DOI] [PubMed] [Google Scholar]
- Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., He Y., Leary R.J., Pagliarini R., Diaz L.A., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Leary R.J., Pagliarini R., Diaz L.A., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Pagliarini R., Diaz L.A., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Diaz L.A., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Bentwich Z., Szafranska A.E., Labourier E., Szafranska A.E., Labourier E., Labourier E., et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M.S., Neilson J.R., Sharp P.A., Neilson J.R., Sharp P.A., Sharp P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efrat S., Linde S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Linde S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Delannoy M., Grant S., Hanahan D., Baekkeskov S., Grant S., Hanahan D., Baekkeskov S., Hanahan D., Baekkeskov S., Baekkeskov S. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc. Natl. Acad. Sci. 1988;85:9037–9041. doi: 10.1073/pnas.85.23.9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Nomura M., Tanaka K., Kondo A., Morinaga H., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E., Nomura M., Tanaka K., Kondo A., Morinaga H., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E., Tanaka K., Kondo A., Morinaga H., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E., Kondo A., Morinaga H., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E., Morinaga H., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E., Yanase T., Nawata H., Takayanagi R., Li E., Nawata H., Takayanagi R., Li E., Takayanagi R., Li E., Li E. Genetic interactions between activin type IIB receptor and Smad2 genes in asymmetrical patterning of the thoracic organs and the development of pancreas islets. Dev. Dyn. 2007;236:2865–2874. doi: 10.1002/dvdy.21303. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P., Garrett-Engele P., Lim L.P., Bartel D.P., Lim L.P., Bartel D.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N., Langenberger D., Gunsalus K.C., Rajewsky N., Gunsalus K.C., Rajewsky N., Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig J.W., Dale T., Bass B.L., Dale T., Bass B.L., Bass B.L. miRNA editing—We should have inosine this coming. Mol. Cell. 2007;25:792–793. doi: 10.1016/j.molcel.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E., Mansfield J.H., Yekta S., Hu J.K., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J., Mansfield J.H., Yekta S., Hu J.K., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J., Yekta S., Hu J.K., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J., Hu J.K., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J., Baskerville S., Bartel D.P., Tabin C.J., Bartel D.P., Tabin C.J., Tabin C.J. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Huang J., Liang Z., Yang B., Tian H., Ma J., Zhang H., Liang Z., Yang B., Tian H., Ma J., Zhang H., Yang B., Tian H., Ma J., Zhang H., Tian H., Ma J., Zhang H., Ma J., Zhang H., Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J. Biol. Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., Zamore P.D., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Aravin A., Tuschl T., Sander C., Marks D.S., Tuschl T., Sander C., Marks D.S., Sander C., Marks D.S., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Iizasa H., Hatzigeorgiou A.G., Nishikura K., Hatzigeorgiou A.G., Nishikura K., Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E., Iovino N., Unnerstall U., Gaul U., Segal E., Unnerstall U., Gaul U., Segal E., Gaul U., Segal E., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Lovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rusu M., Sheridan R., Sewer A., Lovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sheridan R., Sewer A., Lovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Sewer A., Lovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Lovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., Rice A., Kamphorst A.O., Landthaler M., Kamphorst A.O., Landthaler M., Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liang H., Landweber L.F., Landweber L.F. Hypothesis: RNA editing of microRNA target sites in humans? RNA. 2007;13:463–467. doi: 10.1261/rna.296407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Hammond S.M., Joshua-Tor L., Hannon G.J., Joshua-Tor L., Hannon G.J., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lovell T.M., Al-Musawi S.L., Gladwell R.T., Knight P.G., Al-Musawi S.L., Gladwell R.T., Knight P.G., Gladwell R.T., Knight P.G., Knight P.G. Gonadotrophins modulate hormone secretion and steady-state mRNA levels for activin receptors (type I, IIA, IIB) and inhibin co-receptor (betaglycan) in granulosa and theca cells from chicken prehierarchical and preovulatory follicles. Reproduction. 2007;133:1159–1168. doi: 10.1530/REP-06-0179. [DOI] [PubMed] [Google Scholar]
- Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Haudenschild C.D., Meyers B.C., Green P.J., Meyers B.C., Green P.J., Green P.J. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Lynn F.C., Skewes-Cox P., Kosaka Y., McManus M.T., Harfe B.D., German M.S., Skewes-Cox P., Kosaka Y., McManus M.T., Harfe B.D., German M.S., Kosaka Y., McManus M.T., Harfe B.D., German M.S., McManus M.T., Harfe B.D., German M.S., Harfe B.D., German M.S., German M.S. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y., Chen Z., Berka J., Braverman M.S., Chen Y., Chen Z., Braverman M.S., Chen Y., Chen Z., Chen Y., Chen Z., Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk M.M., Finegold M.J., Su J.-G.J., Hsueh A.J.W., Bradley A., Finegold M.J., Su J.-G.J., Hsueh A.J.W., Bradley A., Su J.-G.J., Hsueh A.J.W., Bradley A., Hsueh A.J.W., Bradley A., Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- Matzuk M.M., Kumar T.R., Bradley A., Kumar T.R., Bradley A., Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., Zhao Y., McDonald H., Zeng T., Hirst M., McDonald H., Zeng T., Hirst M., Zeng T., Hirst M., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman M. A-to-I editing challenger or ally to the microRNA process. Biochimie. 2007;89:1171–1176. doi: 10.1016/j.biochi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Pangas S.A., Jorgez C.J., Tran M., Agno J., Li X., Brown C.W., Kumar T.R., Matzuk M.M., Jorgez C.J., Tran M., Agno J., Li X., Brown C.W., Kumar T.R., Matzuk M.M., Tran M., Agno J., Li X., Brown C.W., Kumar T.R., Matzuk M.M., Agno J., Li X., Brown C.W., Kumar T.R., Matzuk M.M., Li X., Brown C.W., Kumar T.R., Matzuk M.M., Brown C.W., Kumar T.R., Matzuk M.M., Kumar T.R., Matzuk M.M., Matzuk M.M. Intraovarian activins are required for female fertility. Mol. Endocrinol. 2007;21:2458–2471. doi: 10.1210/me.2007-0146. [DOI] [PubMed] [Google Scholar]
- Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R., Li L.C., Pookot D., Noonan E.J., Dahiya R., Pookot D., Noonan E.J., Dahiya R., Noonan E.J., Dahiya R., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P., Vaucheret H., Trejo J., Bartel D.P., Trejo J., Bartel D.P., Bartel D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R., Steffen P., Hochsmann M., Giegerich R., Hochsmann M., Giegerich R., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell J., Lee W., Nusbaum C., Ge H., Bartel D.P., Jan C., Player C., Axtell J., Lee W., Nusbaum C., Ge H., Bartel D.P., Player C., Axtell J., Lee W., Nusbaum C., Ge H., Bartel D.P., Axtell J., Lee W., Nusbaum C., Ge H., Bartel D.P., Lee W., Nusbaum C., Ge H., Bartel D.P., Nusbaum C., Ge H., Bartel D.P., Ge H., Bartel D.P., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sacharidou A., Cifuentes-Rojas C., Halbig K., Hernandez A., Dangott L.J., De Nova-Ocampo M., Cruz-Reyes J., Cifuentes-Rojas C., Halbig K., Hernandez A., Dangott L.J., De Nova-Ocampo M., Cruz-Reyes J., Halbig K., Hernandez A., Dangott L.J., De Nova-Ocampo M., Cruz-Reyes J., Hernandez A., Dangott L.J., De Nova-Ocampo M., Cruz-Reyes J., Dangott L.J., De Nova-Ocampo M., Cruz-Reyes J., De Nova-Ocampo M., Cruz-Reyes J., Cruz-Reyes J. RNA editing complex interactions with a site for full-round U deletion in Trypanosoma brucei. RNA. 2006;12:1219–1228. doi: 10.1261/rna.2295706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden A.D., Smith C.W., Smith C.W. RNAi is antagonized by A→I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.C., Gott J.M., Hanson M.R., Gott J.M., Hanson M.R., Hanson M.R. A guide to RNA editing. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J.A., Tong Y., Steitz J.A., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Plasterk R.H., Plasterk R.H. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Seeburg P.H., Shiekhattar R., Nishikura K., Shiekhattar R., Nishikura K., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelonkina A.G., O’Hearn S.F., Law J.A., Cruz-Reyes J., Huang C.E., Alatortsev V.S., Sollner-Webb B., O’Hearn S.F., Law J.A., Cruz-Reyes J., Huang C.E., Alatortsev V.S., Sollner-Webb B., Law J.A., Cruz-Reyes J., Huang C.E., Alatortsev V.S., Sollner-Webb B., Cruz-Reyes J., Huang C.E., Alatortsev V.S., Sollner-Webb B., Huang C.E., Alatortsev V.S., Sollner-Webb B., Alatortsev V.S., Sollner-Webb B., Sollner-Webb B. T. brucei RNA editing: Action of the U-insertional TUTase within a U-deletion cycle. RNA. 2006;12:476–487. doi: 10.1261/rna.2243206. [DOI] [PMC free article] [PubMed] [Google Scholar]