Abstract
Objective
This study aimed to discover ‘signature pathways’ characterizing biological processes based on genes differentially expressed in the uterine cervix before and after spontaneous labor.
Study Design
The cervical transcriptome was previously characterized from biopsies taken before and after term labor. Pathway analysis was used to study the differentially expressed genes based on two gene-to-pathway annotation databases (KEGG and Metacore™). Over-represented and highly impacted pathways and connectivity nodes were identified.
Results
Fifty-two pathways in the Metacore™ database were significantly enriched in differentially expressed genes. Three of the top 5 pathways were known to be involved in cervical remodeling.Two novel pathways were: plasmin signaling and plasminogen activator urokinase (PLAU) signaling. The same analysis in the KEGG database identified 4 significant pathways, of which impact analysis confirmed. Multiple nodes providing connectivity within the plasmin and PLAU signaling pathways were identified..
Conclusions
Three strategies for pathway analysis were consistent in their identification of novel, unexpected as well as expected networks, suggesting that this approach is both valid and effective for the elucidation of biological mechanisms involved in cervical dilation and remodeling.
Keywords: cervix, cervical dilation, cervical remodeling, labor, microarray, gene signature network, pathway analysis, plasmin, systems biology, parturition
Introduction
Labor, delivery, and the postpartum period are accompanied by dramatic changes in the uterine cervix.1-8 Adverse pregnancy outcome in term (arrest of dilatation) and preterm (cervical insufficiency 9 and preterm labor) gestation may occur as a result of cervical disease. Thus, insights into the processes involved in cervical dilation and remodeling are critical to the understanding of abnormal labor. The current knowledge of the biology of the cervix has been derived from the study of human cervical biopsy specimens using hypothesis-driven research.2;3;10-14 However, the mechanisms which underlie these processes are not completely understood.
Most research on the biology of the uterine cervix during pregnancy has been conducted using a reductionist approach. Reductionism is the study of a phenomenon by identifying the individual components of a system.15 The assumption is that a complex system can be understood by investigating the units of the whole.15 In the case of a biological process, these parts may be represented by the study of individual genes, proteins, carbohydrates, lipids, etc. Although there has been a great deal of progress made to date using a reductionist approach in physics, biology, and medicine, reductionism does not take into account component-component links and the dynamics that result from these interactions. In addition, because a biological function can rarely be attributed to one or a few molecules, there is a need to apply a methodology which can identify and describe the networks of functionally active components such as proteins, transcription factors, signaling pathways, and metabolic networks to provide a comprehensive understanding of a physiologic process or disease.16;17 Network biology, defined as a quantifiable description of the networks that characterize various biological systems, can be used to obtain such an understanding.16
We have used a systems biology approach to the study of the uterine cervix in labor and delivery. As a first step, the gene expression patterns in cervical tissue before and after term parturition using functional genomics were described.18;19 Next, we have undertaken examination of the uterine cervix transcriptome after term labor using Gene Ontology analysis and identified specific biological processes and molecular functions as being involved. Examples include inflammation, response to biotic stimulus, and apoptosis.14 However, merely enumerating the biological processes and molecular function does not provide information about the interactive and dynamic properties of the putative genes and proteins involved.
Recent developments in network biology allow for the construction of maps which display the interaction among genes, proteins, and transcription factors into protein-protein, signaling, metabolic, and transcription-regulatory networks.20 Thus, the third step in our investigation, the use of network and pathway analysis to identify significant ‘signature networks’, allows for a more comprehensive view of the process of cervical change in labor and delivery. The objective of this study was to identify these ‘signature networks’ by applying network and pathway analysis to the observed gene expression changes in the uterine cervix in women not in labor at term (TNL) and in women who underwent spontaneous term labor (TL).
Materials and Methods
Study Design
A cross-sectional study was performed in patients undergoing elective cesarean section with an unripe cervix (term not in labor) and in patients after spontaneous vaginal delivery (term labor). The cervical transcriptome before and after labor was evaluated utilizing cervical biopsies [before labor (n=7) and after labor (n=9)] and hgu133plus2 Affymetrix GeneChip® microarrays. The details of this study have been previously reported.18
Pathway Analysis
In order to identify relevant pathways to cervical biology during parturition, we considered two repositories of pathways and two algorithms for analysis. The two repositories which map genes to pre-defined pathways are: 1) KEGG (Kyoto Encyclopedia of Genes and Genomes), which provides a database of metabolic, regulatory, and disease pathways; and 2) MetaCore™ (St. Joseph, MI, USA), which is a proprietary manually curated database of human protein-protein, protein-DNA and protein compound interactions, metabolic and signaling pathways, and the effects of bioactive molecules. KEGG contains approximately 250 canonical signaling and metabolic pathways while Metacore™ contains approximately 450 such pathways.
The two algorithms for pathway analysis were: 1) statistical analysis for overrepresentation 21 and 2) impact analysis (http://vortex.cs.wayne.edu/ontoexpress). The first method assesses the level of agreement between the observed proportion of genes that can be mapped on a given pathway in the differentially expressed set and the reference set (hgu133plus2 Affymetrix® array). A Fisher exact test was performed in order to test for the equality of proportions. P-values for significance of the over-representation for each pathway tested were derived using R (www.r-project.org). In contrast, impact analysis performed with Pathway Express (http://vortex.cs.wayne.edu/ontoexpress) takes into consideration: a) the number of differentially expressed genes on each pathway, b) the position of the genes within the pathway, and c) the signaling interactions between various genes as described by the pathway. Signaling interaction refers to the situation in which the change in the activity of a given gene affects the expression of another gene in a consistent way. Signaling pathways from KEGG define a number of signaling interactions including: activation, repression, inhibition, phosphorylation, etc. The exact definitions for all these types of signaling interactions can be found at: http://www.genome.jp/kegg/. An impact value and p-value are assigned to each pathway. The p-values obtained from both analyses were adjusted using the false discovery rate method, 22 p<0.05 was considered statistically significant. The position of a gene (node) in the pathway refers to the number and the magnitude of signals that feed in and out of a given node. Genes located in entry points in the pathway are more likely to impact the pathway than leaf nodes.
Network Connectivity Analysis
An additional analysis was carried out to evaluate the potential importance of individual nodes in protein interaction networks for providing connectivity among differentially expressed genes.
To identify such “topologically significant” proteins the set of differentially expressed genes was used to construct the shortest path network connecting corresponding nodes in the global database of protein interactions (MetaCore™). Next, the number of all paths traversing each node in this shortest path network specific for genes differentially expressed in labor was computed. For each node this number was compared to the total number of all paths going via the same node in the global network. Using these numbers, as well as the relative size of the differential gene set, p-values were calculated for each node in the shortest path network. A conservative Bonferroni correction was used to correct for multiple testing and a p<0.01 was deemed significant. A conservative Bonferroni correction was used to correct for multiple testing, and a significance threshold of 0.01 was chosen. The p-value for a node demonstrates the statistical significance for providing connectivity among the original set of differentially expressed genes. The nodes that are deemed significant by this method are displayed on pathways maps where their functional roles can be further evaluated. A detailed description of the connectivity analysis is available as supplementary material on the web site of the Journal.
Real time Quantitative RT-PCR assays
Quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) assays of selected genes were performed on a different set of cervical biopsy samples from those used in the microarray analysis. Patients included those undergoing elective cesarean section with an unripe cervix (TNL) as well as patients after spontaneous vaginal delivery (TL). A detailed description of the methodology and analysis is available as supplementary material on the web site of the Journal.
Results
Pathway Analysis
Over-representation method on signaling and metabolic pathways
Fifty-two of the 450 pathways in the Metacore™ database were significant based on the over-representation algorithm (p<0.05). The top 20 of these are listed in Table I, the entire list can be found as supplementary material on the web site of the Journal. The five most significant were: 1) chemokines and adhesion, 2) extracellular matrix remodeling, 3) plasmin signaling, 4) plasminogen activator, urokinase (PLAU) signaling (Figure 1), and 5) vascular endothelial growth factor (VEGF)-family signaling. The figures of the remaining pathways can be found as supplementary material on the web site of the Journal.
Table I.
| Map Name | Number of Genes in Reference Array | Number of Genes in Differentially Expressed List | P Adjusted |
|---|---|---|---|
| Chemokines and adhesion | 206 | 20 | 3.85E-06 |
| ECMremodeling | 60 | 12 | 3.85E-06 |
| Plasmin signaling | 47 | 10 | 1.42E-05 |
| PLAU signaling | 46 | 9 | 6.64E-05 |
| VEGF-family signaling | 45 | 9 | 6.64E-05 |
| HGF signaling pathway | 43 | 8 | 0.0003 |
| NAD metabolism | 54 | 8 | 0.0011 |
| Role of AP-1 inregulation of cellular metabolism | 43 | 7 | 0.0016 |
| VEGF signaling via VEGFR2-generic cascades | 43 | 7 | 0.0016 |
| IL6 signaling pathway | 32 | 6 | 0.0026 |
| FGF-family signaling | 34 | 6 | 0.0031 |
| MIF in innate immunity response | 57 | 7 | 0.0061 |
| Estrone metabolism | 14 | 4 | 0.0064 |
| HETE and HPETE diosynthesis and metabolism | 42 | 6 | 0.0064 |
| Role of PBX in fibroblasts signaling pathways | 26 | 5 | 0.0064 |
| Transcription regulation of amino acid metabolism | 42 | 6 | 0.0064 |
| IFN gamma signaling pathway | 63 | 7 | 0.0071 |
| PH proteins participation in RTKs adaptor complexes formation and Intermediation with focal adhesion complex. Part2 | 63 | 7 | 0.0071 |
| Antiapoptotic Function of TRADD/TRAF2 complex | 31 | 5 | 0.0096 |
| Prostagland in 1 biosynthesis and metabolism | 17 | 4 | 0.0096 |
* Number of genes in the array = approximately 20,000
Figure 1.
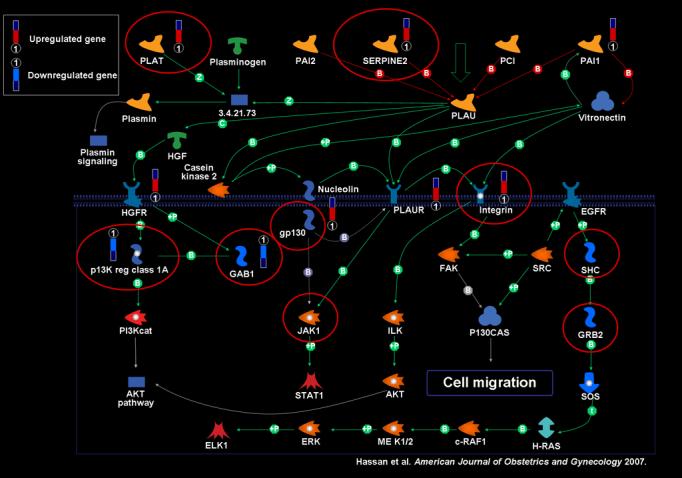
Display of differentially expressed genes in the uterine cervix after term spontaneous labor mapped on the PLAU signaling pathway. ‘Thermometer’ represents differentially expressed gene.
Connectivity Analysis results: Several nodes in the PLAU signaling pathway were found to be significant in providing connectivity among the differentially expressed genes. Significant nodes are encircled in red.
When the KEGG gene-to-pathway annotation was used, four significant pathways were identified: cytokine-cytokine receptor interaction, complement and coagulation cascades, calcium signaling pathway, and arginine and proline metabolism (Table II).
Table II.
| Map Name | Number of Genes in Reference Array | Number of Genes in Differentially Expressed List | P Adjusted |
|---|---|---|---|
| METACORE | |||
| Chemokines and adhesion | 206 | 20 | 3.85E-06 |
| Extracellular matrix remodeling | 60 | 12 | 3.85E-06 |
| Plasmin signaling | 47 | 10 | 1.42E-05 |
| PLAU signaling | 46 | 9 | 6.64E-05 |
| VEGF-family signaling | 45 | 9 | 6.64E-05 |
| KEGG | |||
| Cytokine-cytokine interaction | 519 | 44 | 2.33E-08 |
| Complement and coagulation | 139 | 17 | 2.76E-05 |
| Calcium signaling pathway | 600 | 6 | 0.0392 |
| Arginine and proline | 110 | 9 | 0.0493 |
* Number of genes in the array = approximately 20,000
Impact analysis on KEGG signaling pathways
Impact analysis restricted to KEGG signaling pathways performed using Pathway Express identified 7 significant pathways among which 2 pathways that were not noted using the overrepresentation method: leukocyte transendothelial migration and epithelial cell signaling (Table III).
Table III.
Pathway Express Ranked by Impact Factor
| Pathway Name | Corrected p-value |
|---|---|
| Cell adhesion molecules (CAMs) | 2.74E-14 |
| Cytokine-cytokine receptor interaction | 8.15E-09 |
| signaling system | 1.87E-06 |
| Complement and coagulation cascades | 0.0145 |
| Leukocyte transendothelial migration | 0.0176 |
| Focal adhesion | 0.0235 |
| Epithelial cell signaling related to specific infections | 0.0449 |
Plasmin/PLAU signaling pathways
The plasmin and PLAU signaling pathways were among the most highly significant. Further investigation with the Metacore™ database identified multiple transcription factors that activate the expression of PLAT: JunB, JunD, FOSL2, FosB, CREM, and c-FOS (Figure 2). The PLAU signaling pathway (Figure 1) displays plasminogen activator urokinase binding to its receptor on the cell surface, subsequent binding to JAK1, and phosphorylation of STAT1 leading to activation. The pathway is truncated at this point in the Metacore™ pathway database. Thus, the relationship between activation of STAT1 and the differentially regulated genes in the uterine cervix before and after labor was explored using network analysis. Network analysis indicated that activation of STAT1 is linked to regulation in expression of several genes listed on the right side of figure 3. JAK1 provides an essential network conduit between plasminogen activator urokinase receptor and several differentially expressed targets of STAT1.
Figure 2.
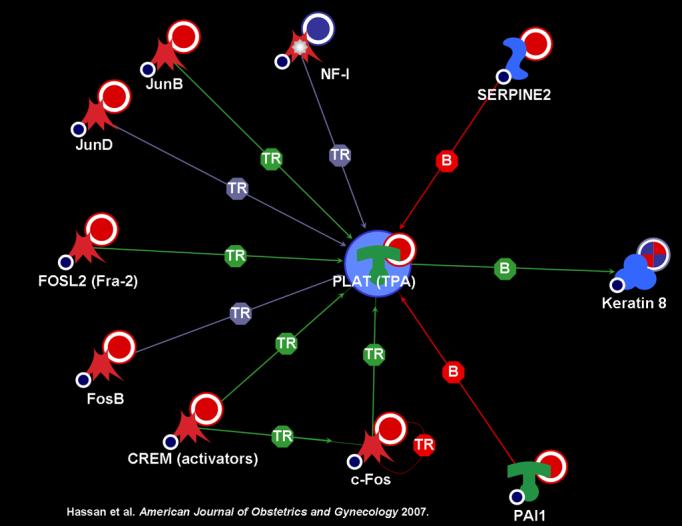
Network of transcription factors that activate the expression of PLAT
Figure 3.
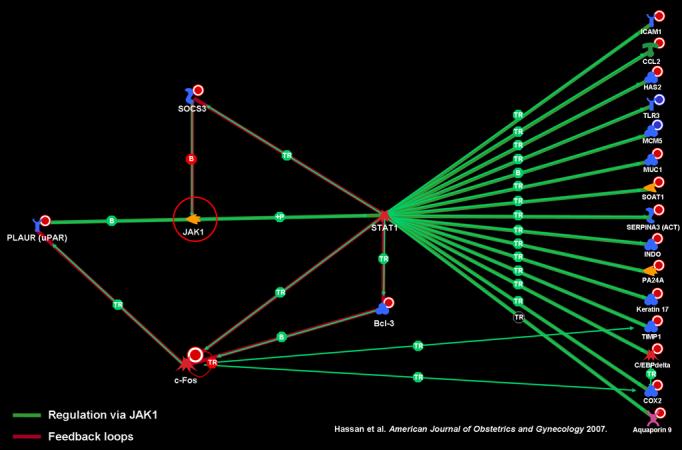
JAK1 provides essential network conduit between PLAUR and many differentially expressed targets of STAT1
Metacore™based on connectivity
Multiple nodes within the plasmin and PLAU signaling pathways were found to be topologically significant based on their function of providing connectivity (see Figure 1).
qRT-PCR
qRT-PCR assays were performed to measure mRNA levels of selected genes involved in the plasmin and PLAU signaling pathways. PLAUR (plasminogen activator, urokinase receptor), SERPINE2 (plasminogen activator inhibitor type 1), LEKT1 (serine protease inhibitor), FGF2 (fibroblast growth factor 2) were selected for further study. When examining the top 5 pathways FGF2 and LEKT1 were found to be specific to the plasmin and/or PLAU signaling pathways.
Consistent with the microarray analysis, qRT-PCR revealed FGF2, PLAUR, and SERPINE2 to be significantly up-regulated in term labor patients when compared to term no labor. LEKT1 was significantly downregulated after term labor (Figure 4).
Figure 4.
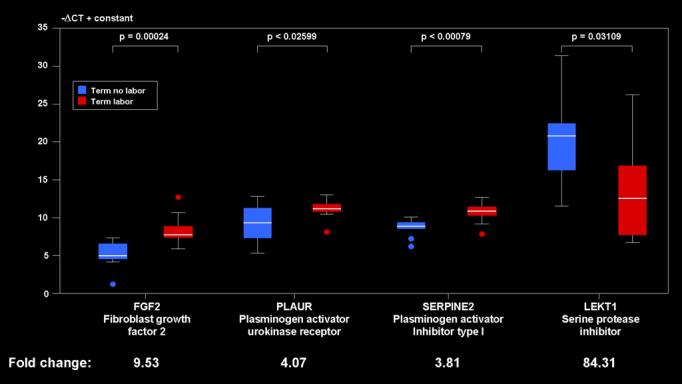
qRT-PCR results for FGF2, PLAUR, SERPINE2, and LEKT1 The boxes encompass 50% of the data from the 1st quartile up to the 3rd quartile. The middle line represents the median value (50% quantile). The whiskers extend to the most extreme data point which is no more than 1.5 times the inter-quartile range from the box.
Comment
Principal findings of the study
1) Cervical dilatation and remodeling after term labor is associated with specific gene signature networks; 2) Fifty-two Metacore™ metabolic and signaling pathways involve the activity of the genes of the cervical transcriptome of spontaneous term labor; 3) the five most significant of these pathways were a) chemokines and adhesion, b) extracellular matrix remodeling, c) plasmin signaling, d) PLAU signaling, and e) vascular endothelial growth factor (VEGF)-family signaling; 4) A network analysis identified multiple transcription factors that activate the expression of PLAT; 5) The same network analysis also indicated that JAK1 provides an essential network conduit between plasminogen activator urokinase receptor and several differentially expressed targets of STAT1; and 6) qRT-PCR confirmed the involvement of the plasminogen/plasmin system in the process of cervical dilation and remodeling in labor.
Meaning of the study
The use of pathway analysis to derive gene ‘signature networks’ allows for the following: 1) the ability to transition from biology at the molecular level to a more global systems approach to disease/biological processes; 2) the identification of key regulators or transcription factors that may not have been identified by microarray analysis; and 3) further interpretation of gene expression data by providing information on the protein-protein interaction, metabolic, signaling, and transcription-regulatory networks.17;23-25
It has been proposed that cervical changes during pregnancy occur in four phases: softening, ripening, dilation, and changes which occur after parturition.26 We present a unique report of the signaling and metabolic pathways involved in phase four of this process: cervical dilation and remodeling after term labor. In the current study, 52 pathways documented by the Metacore™ database were noted to be significant. Some of them represent canonical pathways that can be found also in KEGG, while others are proprietary. The use of novel pathway analysis methods [Impact Factor (Pathway Express) and Connectivity (Metacore™)] confirmed the involvement of several of these signaling pathways and genes leading to a consistent result of what represents the processes involved in cervical dilation and remodeling in term spontaneous labor.
Several pathways that were found to be significant (chemokine and adhesion, extracellular matrix remodeling, cytokine-cytokine interaction, VEGF-family signaling) contain genes that have previously been suspected as involved in cervical dilation and remodeling.27-35 Different database and pathway analysis methods confirmed these findings.
Of interest was the finding that the complement and coagulation cascades (determined by Pathway Express), which include portions of the PLAU signaling and plasmin signaling pathways (determined by Metacore™), are involved in cervical dilation and remodeling after labor. The plasminogen activation cascade is a proteolytic enzyme system which converts plasminogen to the active serine protease plasmin. Plasmin degrades most extracellular matrix proteins.31 Plasminogen activation plays a role in the remodeling of the extracellular matrix in human amnion, choriodecidua, and placenta during and after labor.34 In addition, plasminogen activator inhibitor activity is increased in the serum of pregnant women near term when compared to non-pregnant women and decreased after delivery.29 Degradation of the cervical extracellular matrix may occur as a result of the activation of the plasminogen system. The finding of up-regulation of multiple transcription factors involved in the activation of PLAT provides further support for the involvement of this pathway in cervical dilation and remodeling after term labor.
It is noteworthy that the MIF (macrophage migration inhibiting factor) in innate immune response pathway was significant. MIF encodes a lymphokine involved in cell-mediated immunity, immunoregulation, and inflammation. It plays a role in the regulation of the macrophage function in host defense through the suppression of anti-inflammatory effects of glucocorticoids.27 Elevated amniotic fluid concentrations of MIF are associated with intraamniotic inflammation, histologic chorioamnionitis, and shorter amniocentesis-to-delivery interval in patients in preterm labor.28
Interestingly, this study has identified Shc and GRB2 as significant nodes. Both are important mediators in MAPK-related signal transduction. Both mediate response to various growth factors and inflammatory response. The involvement of MAPK cascade during parturition has recently been demonstrated.36 The role of these pathways in the mechanisms involved in cervical dilation and remodeling in term labor require further investigation.
The understanding of the gene signature pathways before and after labor in the uterine cervix is central to the molecular elucidation of the mechanisms responsible for cervical insufficiency, preterm labor, and arrest of dilatation. Thus far these common complications of pregnancy have eluded pathophysiologic definition.
Strengths and weaknesses of the study
A major strength of this study is that this report represents the first pathway analysis of the uterine cervix before and after human term labor and delivery. In addition, several significant pathways were noted to be consistently important when evaluated by several methodologies. The results of these analyses are consistent with previous results. However, we highlight pathways that had not been previously implicated. The report of the plasminogen activation cascade and the pathways of innate immunity playing substantial roles in cervical dilation and remodeling is novel. Some of these findings reported herein were confirmed by qRT-PCR.
A limitation of this investigation is that we were unable to temporally follow the changes seen in an individual's cervix as labor progresses due to the obvious constraints of human research. Of note, the two patient groups differ in their obstetrical history. While it is not certain that this could account for any changes seen in gene expression, we wish to point out this difference.
Comparison with previous reports
Pathway analysis has been utilized to examine the changes in myometrium in animal models. Salomonis et al examined the mouse myometrium in the non-pregnant, mid-gestation, late gestation, postpartum states and defined gene expression changes under these conditions. HOPACH (hierarchical ordered partitioning and collapsing hybrid) and GenMAPP 2.0 pathway analysis identified term quiescence, term activation, and postpartum involution expression patterns.32 There are no previous reports of the mechanisms involved in cervical dilation and remodeling after spontaneous labor and delivery using pathway analysis.
Unanswered questions and proposed future research
The processes of cervical dilation and remodeling are the result of the activation of several pathways that have been implicated in the common terminal pathway of parturition. The pathways involved in this complex process include networks involved in chemokine and adhesion activation, cytokine-cytokine activity, extracellular matrix remodeling, the plasminogen system, recognition by the innate immune system, and the complement and coagulation cascade.
This investigation provides a unique global view of the changes seen in the uterine cervix in term labor and delivery. Unanswered questions remain which include the timing of the activation of each cascade, the role of each pathway in patients with cervical disease resulting in abnormal term (arrest of dilatation) and preterm birth (preterm labor, cervical insufficiency), and effective treatment strategies for cervical disease in pregnancy. The understanding of the pathways that lead to the changes in the cervix during labor and delivery may be critical to unraveling the solutions for the treatment of cervical disease in pregnancy. Future investigation of effective treatments for cervical disease in pregnancy should be targeted to the processes that have been identified as playing a critical role in the metabolic and signaling pathways identified. In addition, future studies should focus on the remaining three components of system-level understanding: system dynamics, control, and design.
Acknowledgment
This research was supported in part by the Intramural Research Program of the National Institute of Child Health and Human Development, NIH, DHHS.
Footnotes
Condensation: Cervical remodeling after term labor is associated with the gene pathways of chemokines and adhesion, extracellular matrix remodeling, VEGF-family signaling, and unsuspected pathways such as plasmin signaling.
Presented at the 27th annual meeting of the Society for Maternal-Fetal Medicine, February 5th − 10th, 2007, San Francisco, California.
Reference List
- 1.Junqueira LC, Zugaib M, Montes GS, Toledo OM, Krisztan RM, Shigihara KM. Morphologic and histochemical evidence for the occurrence of collagenolysis and for the role of neutrophilic polymorphonuclear leukocytes during cervical dilation. Am.J.Obstet.Gynecol. 1980;138:273–81. doi: 10.1016/0002-9378(80)90248-3. [DOI] [PubMed] [Google Scholar]
- 2.Kleissl HP, van der RM, Naftolin F, Glorieux FH, de LA. Collagen changes in the human uterine cervix at parturition. Am J Obstet Gynecol. 1978;130:748–53. doi: 10.1016/0002-9378(78)90003-0. [DOI] [PubMed] [Google Scholar]
- 3.Maillot KV, Zimmermann BK. The solubility of collagen of the uterine cervix during pregnancy and labour. Arch Gynakol. 1976;220:275–80. doi: 10.1007/BF00673411. [DOI] [PubMed] [Google Scholar]
- 4.Osman I, Young A, Ledingham MA, Thomson AJ, Jordan F, Greer IA, et al. Leukocyte density and pro-inflammatory cytokine expression in human fetal membranes, decidua, cervix and myometrium before and during labour at term. Mol.Hum.Reprod. 2003;9:41–45. doi: 10.1093/molehr/gag001. [DOI] [PubMed] [Google Scholar]
- 5.Osmers RG, Blaser J, Kuhn W, Tschesche H. Interleukin-8 synthesis and the onset of labor. Obstet.Gynecol. 1995;86:223–29. doi: 10.1016/0029-7844(95)93704-4. [DOI] [PubMed] [Google Scholar]
- 6.Rajabi MR, Dodge GR, Solomon S, Poole AR. Immunochemical and immunohistochemical evidence of estrogen-mediated collagenolysis as a mechanism of cervical dilatation in the guinea pig at parturition. Endocrinology. 1991;128:371–78. doi: 10.1210/endo-128-1-371. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto Y, Moran P, Searle RF, Bulmer JN, Robson SC. Interleukin-8 is involved in cervical dilatation but not in prelabour cervical ripening. Clin.Exp.Immunol. 2004;138:151–57. doi: 10.1111/j.1365-2249.2004.02584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Word RA, Landrum CP, Timmons BC, Young SG, Mahendroo MS. Transgene insertion on mouse chromosome 6 impairs function of the uterine cervix and causes failure of parturition. Biol.Reprod. 2005;73:1046–56. doi: 10.1095/biolreprod.105.042663. [DOI] [PubMed] [Google Scholar]
- 9.Berghella V, Iams JD, Newman RB, Macpherson C, Goldenberg RL, Mueller-Heubach E, et al. Frequency of uterine contractions in asymptomatic pregnant women with or without a short cervix on transvaginal ultrasound scan. Am J Obstet Gynecol. 2004;191:1253–56. doi: 10.1016/j.ajog.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Danforth DNBJCRJW., Jr Connective tissue changes incident to cervical effacement. Am J Obstet Gynecol. 1960;80:939–45. doi: 10.1016/0002-9378(60)90472-5. [DOI] [PubMed] [Google Scholar]
- 11.Petersen LK, Uldbjerg N. Cervical collagen in non-pregnant women with previous cervical incompetence. Eur.J.Obstet.Gynecol.Reprod.Biol. 1996;67:41–45. doi: 10.1016/0301-2115(96)02440-2. [DOI] [PubMed] [Google Scholar]
- 12.Sennstrom MB, Ekman G, Westergren-Thorsson G, Malmstrom A, Bystrom B, Endresen U, et al. Human cervical ripening, an inflammatory process mediated by cytokines. Mol.Hum.Reprod. 2000;6:375–81. doi: 10.1093/molehr/6.4.375. [DOI] [PubMed] [Google Scholar]
- 13.Sennstrom MK, Brauner A, Lu Y, Granstrom LM, Malmstrom AL, Ekman GE. Interleukin-8 is a mediator of the final cervical ripening in humans. Eur J Obstet Gynecol Reprod Biol. 1997;74:89–92. doi: 10.1016/s0301-2115(97)02757-7. [DOI] [PubMed] [Google Scholar]
- 14.Stjernholm-Vladic Y, Stygar D, Mansson C, Masironi B, Akerberg S, Wang H, et al. Factors involved in the inflammatory events of cervical ripening in humans. Reprod.Biol.Endocrinol. 2004;2:74. doi: 10.1186/1477-7827-2-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strange K. The end of “naive reductionism”: rise of systems biology or renaissance of physiology? Am J Physiol Cell Physiol. 2005;288:C968–C974. doi: 10.1152/ajpcell.00598.2004. [DOI] [PubMed] [Google Scholar]
- 16.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat.Rev.Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 17.Bugrim A, Nikolskaya T, Nikolsky Y. Early prediction of drug metabolism and toxicity: systems biology approach and modeling. Drug Discov.Today. 2004;9:127–35. doi: 10.1016/S1359-6446(03)02971-4. [DOI] [PubMed] [Google Scholar]
- 18.Hassan SS, Romero R, Haddad R, Hendler I, Khalek N, Tromp G, et al. The transcriptome of the uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol. 2006;195:778–86. doi: 10.1016/j.ajog.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 19.Huber A, Hudelist G, Czerwenka K, Husslein P, Kubista E, Singer CF. Gene expression profiling of cervical tissue during physiological cervical effacement. Obstet.Gynecol. 2005;105:91–98. doi: 10.1097/01.AOG.0000146636.61611.e3. [DOI] [PubMed] [Google Scholar]
- 20.Brazhnik P, de la FA, Mendes P. Gene networks: how to put the function in genomics. Trends Biotechnol. 2002;20:467–72. doi: 10.1016/s0167-7799(02)02053-x. [DOI] [PubMed] [Google Scholar]
- 21.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–70. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav.Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 23.Auffray C, Imbeaud S, Roux-Rouquie M, Hood L. From functional genomics to systems biology: concepts and practices. C.R.Biol. 2003;326:879–92. doi: 10.1016/j.crvi.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 24.Fraser AG, Marcotte EM. Development through the eyes of functional genomics. Curr.Opin.Genet.Dev. 2004;14:336–42. doi: 10.1016/j.gde.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Kitano H. Computational systems biology. Nature. 2002;420:206–10. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 26.Word RA, Li XH, Hnat M, Carrick K. Dynamics of cervical remodeling during pregnancy and parturition: mechanisms and current concepts. Semin.Reprod Med. 2007;25:69–79. doi: 10.1055/s-2006-956777. [DOI] [PubMed] [Google Scholar]
- 27.Calandra T, Bucala R. Macrophage migration inhibitory factor (MIF): a glucocorticoid counter-regulator within the immune system. Crit Rev.Immunol. 1997;17:77–88. doi: 10.1615/critrevimmunol.v17.i1.30. [DOI] [PubMed] [Google Scholar]
- 28.Chaiworapongsa T, Romero R, Espinoza J, Kim YM, Edwin S, Bujold E, et al. Macrophage migration inhibitory factor in patients with preterm parturition and microbial invasion of the amniotic cavity. J.Matern.Fetal Neonatal Med. 2005;18:405–16. doi: 10.1080/14767050500361703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koelbl H, Kirchheimer J, Tatra G. Influence of delivery on plasminogen activator inhibitor activity. J Perinat Med. 1989;17:107–11. doi: 10.1515/jpme.1989.17.2.107. [DOI] [PubMed] [Google Scholar]
- 30.Liggins G. Cervical ripening as an inflammatory reaction. In: Ellwood D, Anderson A, editors. The cervix in pregnancy and labour. Churchill Livingstone; Edinburgh: 1981. pp. 1–9. [Google Scholar]
- 31.Plow EF, Herren T, Redlitz A, Miles LA, Hoover-Plow JL. The cell biology of the plasminogen system. FASEB J. 1995;9:939–45. doi: 10.1096/fasebj.9.10.7615163. [DOI] [PubMed] [Google Scholar]
- 32.Salomonis N, Cotte N, Zambon AC, Pollard KS, Vranizan K, Doniger SW, et al. Identifying genetic networks underlying myometrial transition to labor. Genome Biol. 2005;6:R12. doi: 10.1186/gb-2005-6-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tornblom SA, Klimaviciute A, Bystrom B, Chromek M, Brauner A, Ekman-Ordeberg G. Non-infected preterm parturition is related to increased concentrations of IL-6, IL-8 and MCP-1 in human cervix. Reprod.Biol.Endocrinol. 2005;3:39. doi: 10.1186/1477-7827-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsatas D, Baker MS, Rice GE. Differential expression of proteases in human gestational tissues before, during and after spontaneous-onset labour at term. J Reprod Fertil. 1999;116:43–49. doi: 10.1530/jrf.0.1160043. [DOI] [PubMed] [Google Scholar]
- 35.Young A, Thomson AJ, Ledingham M, Jordan F, Greer IA, Norman JE. Immunolocalization of proinflammatory cytokines in myometrium, cervix, and fetal membranes during human parturition at term. Biol.Reprod. 2002;66:445–49. doi: 10.1095/biolreprod66.2.445. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Stjernholm YV. Plasma membrane receptor mediated MAPK signaling pathways are activated in human uterine cervix at parturition. Reprod Biol Endocrinol. 2007;5:3. doi: 10.1186/1477-7827-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


