Abstract
Background
The serpin (serine protease inhibitor) superfamily constitutes a class of functionally highly diverse proteins usually encompassing several dozens of paralogs in mammals. Though phylogenetic classification of vertebrate serpins into six groups based on gene organisation is well established, the evolutionary roots beyond the fish/tetrapod split are unresolved. The aim of this study was to elucidate the phylogenetic relationships of serpins involved in surveying the secretory pathway routes against uncontrolled proteolytic activity.
Results
Here, rare genomic characters are used to show that orthologs of neuroserpin, a prominent representative of vertebrate group 3 serpin genes, exist in early diverging deuterostomes and probably also in cnidarians, indicating that the origin of a mammalian serpin can be traced back far in the history of eumetazoans. A C-terminal address code assigning association with secretory pathway organelles is present in all neuroserpin orthologs, suggesting that supervision of cellular export/import routes by antiproteolytic serpins is an ancient trait, though subtle functional and compartmental specialisations have developed during their evolution. The results also suggest that massive changes in the exon-intron organisation of serpin genes have occurred along the lineage leading to vertebrate neuroserpin, in contrast with the immediately adjacent PDCD10 gene that is linked to its neighbour at least since divergence of echinoderms. The intron distribution pattern of closely adjacent and co-regulated genes thus may experience quite different fates during evolution of metazoans.
Conclusion
This study demonstrates that the analysis of microsynteny and other rare characters can provide insight into the intricate family history of metazoan serpins. Serpins with the capacity to defend the main cellular export/import routes against uncontrolled endogenous and/or foreign proteolytic activity represent an ancient trait in eukaryotes that has been maintained continuously in metazoans though subtle changes affecting function and subcellular location have evolved. It is shown that the intron distribution pattern of neuroserpin gene orthologs has undergone substantial rearrangements during metazoan evolution.
Background
The serpins represent a superfamily of proteins with a common fold that cover an extraordinary broad spectrum of different biological functions. Most serpins inhibit proteases from one or several different clans of peptidases; some superfamily members, however, exert disparate roles, such as assisting in protein folding or transportation of hormones [1]. This functional diversity is enabled, at least in part, by the unusual structural plasticity of the serpin molecule that, in the native form, often takes a metastable structure. Serpins can perform their activity in the extracellular space or in various subcellular compartments, including the secretory pathway routes [2,3], and they are found in all high-order branches of the tree of life [4]. Deficiency of some serpins, such as antithrombin or neuroserpin, is lethal or may be associated with serious pathology [5,6]. Mutations of the neuroserpin gene for instance may result in formation of intracellular aggregates in the brain causing dementia [6], while wild type neuroserpin provides protection of neuronal cells in cerebral ischemia and other pathologies [7]. Neuroserpin inhibits tissue plasminogen activator (tPA), urokinase-type plasminogen activator, nerve growth factor-γ, and plasmin. These enzymes are also believed to represent physiological targets of the inhibitor [8,9]. Native neuroserpin is found in the medium of some cell lines [9] but also in dense core secretory vesicles of neuronal cells [10,11], suggesting that it could exert a function within the regulated secretory pathway, though there is no experimental evidence for this. Association of neuroserpin with secretory pathway organelles is mediated via a 13 amino acid C-terminal sorting sequence [11]. Recently, serpins equipped with a C-terminal endoplasmic reticulum (ER) retention/retrieval signal that efficiently inhibit furin and/or other members of the proprotein convertase (PC) family have been identified in Drosophila melanogaster [12-15], demonstrating for the first time that serpins with antiproteolytic activity may reside in early secretory pathway organelles.
The elucidation of phylogenetic relationships among animal serpins poses a notorious problem [16]. Serpin genes represent a substantial fraction of metazoan genomes, often amounting to several dozens of members in mammals. In various vertebrate lineages multiple expansions of serpin genes have occurred [17,18] resulting in numerous paralogs. In other lineages, such as fungi, serpins seem to be rare. In some species phylogenetic relationships of serpin genes may be obscured further by a propensity for reciprocal or non-reciprocal exchange of cassette exons coding for the hypervariable reactive site loop region (RSL) [19]. The sequence of this region plays a primary role in determining the specificity of serpin/target enzyme interaction. Inhibition of target proteases involves cleavage of a scissile bond located between positions P1 and P1' of the inhibitor's RSL [1]. Serpins also occur with a patchy distribution in prokaryotes, but the time point of their first emergence is not known [4].
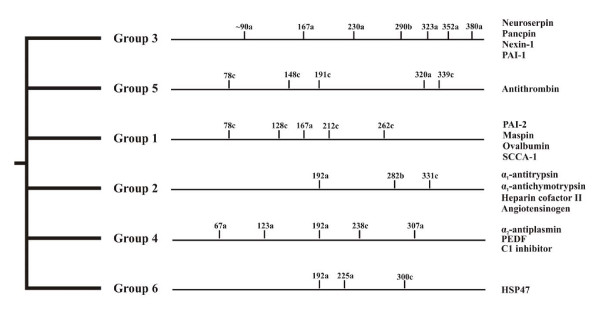
In metazoans, serpin genes display highly variant exon-intron patterns that, however, may be strongly conserved within some taxons. Gene architecture and other rare genetic characters constitute a robust basis to group vertebrate serpins [20-22]. Grounded on number, positions, and phases of introns, serpins have been classified into six groups maintained at least since the fish/tetrapod split (Figure 1). Vertebrate serpin genes with equivalent gene structures often tend to be organised in clusters [22-24]; however, close physical linkage is not always found. Interestingly, none of altogether 24 intron positions mapping to the core domain of vertebrate serpins is shared by all of these six gene groups; however, characteristic amino acid indels provide some further cues for unraveling phylogenetic relationships [20]. None of the group-specific vertebrate gene architectures is found in earlier diverging animal taxons, though a few vertebrate-specific intron positions are present in a scattered fashion in some basal metazoans. Another classification system groups vertebrate serpins into nine clades [1]. However, a deeper rooting, resilient phylogenetic classification of metazoan serpins is not available and their evolutionary roots are unresolved. In addition, there is no data indicating when and how the highly conserved exon-intron patterns of the paralogous vertebrate serpin gene groups arose. Here, data are presented that reveal a deeply rooting, continuous lineage of secretory pathway-associated serpins in metazoans that provide a surveillance and controlling function against proteolytic activity within the major cellular export/import routes.
Figure 1.
Gene structure-based phylogenetic classification of vertebrate serpins. Positions of introns refer to the human α1-antitrypsin sequence. A two amino acid indel present between positions 173 and 174 (α1-antitrypsin numbering) suggests that groups 1, 3, and 5 are more closely related to each other than to the other groups. Gene groups 2, 4, and 6 lack the 173/174 indel and depict an intron at position 192a, implying shared ancestry. Some group 1 members contain an additional intron at position 85c (not shown). For further details see references 20 and 21.
Results
Chromosomal arrangement of genes coding for neuroserpin homologs along the lineages leading to vertebrates
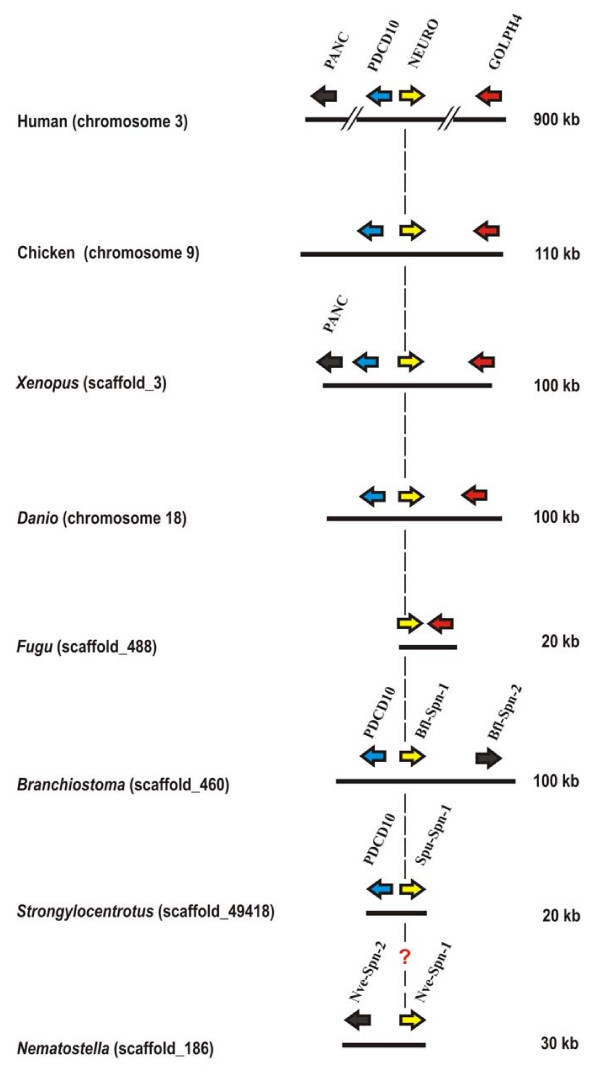
Group 3 of mammalian serpin genes contains five members (plasminogen activator inhibitor-1/SERPINE1, nexin-1/SERPINE2, SERPINE3, neuroserpin/SERPINI1, pancpin/MEPI/SERPINI2) that share a highly conserved group-specific exon-intron pattern characterised by the presence of six introns at equivalent positions. Another probably homologous intron mapping to the N-terminal region cannot be positioned unambiguously due to alignment problems [20]. In the human genome, the genes coding for neuroserpin and pancpin are co-localized in opposite directions on chromosome 3 http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=9606. Between these two serpin genes and immediately adjacent to the neuroserpin gene, but in inverse orientation, the PDCD10 (programmed cell death 10) gene is found (Figure 2). PDCD10 is a strongly conserved gene with orthologs in both vertebrates and invertebrates. In humans, the gene product has been shown to be part of a signaling complex involved in vascular development [25]; however, the exact function is unclear and paralogs are not known [26]. Mutations in the PDCD10 gene cause cerebral cavernous malformations (CCM), a syndrome associated with seizures and neurological deficits due to focal haemorrhages [26]. Downstream from the neuroserpin gene and in inverse orientation, the GOLPH4 marker is found that codes for a transmembrane protein (GPP130) involved in endosome-to-Golgi traffic of proteins [27-29].
Figure 2.
Genomic coordinates of the genes coding for neuroserpin homologs and flanking genes in metazoans. A vertical dashed line indicates neuroserpin (NEURO) orthologs. The genes coding for orthologs of neuroserpin and PDCD10 are consistently arranged in a head-to-head orientation at least since divergence of vertebrates and sea urchins. Orthologs are represented in identical colors. Serpin paralogs are represented as black arrows. The genes coding for neuroserpin and pancpin (PANC) share the characteristic intron distribution pattern of group 3 serpins maintained at least since the fish/tetrapod split.
Studying the serpin complement of various metazoans we noted that linkage of the PDCD10-neuroserpin-GOLPH4 triad is maintained in the genomes of chicken, the clawed frog (Xenopus tropicalis), and the zebrafish (Danio rerio). Current genome sequence releases also revealed synteny of the pancpin gene with these three genes in the genome of the clawed frog (Xenopus tropicalis), and conservation of the close head-to-head association of the neuroserpin/GOLPH4 gene pair in the Japanese pufferfish (Fugu rubripes) (Figure 2) and in Tetraodon nigroviridis. Orthology of neuroserpin genes in these species is corroborated by the highly conserved group 3 specific exon-intron gene architecture (Figure 3), and a C-terminal extension (Figure 4) that targets neuroserpin to large dense core vesicles in mammals [11].
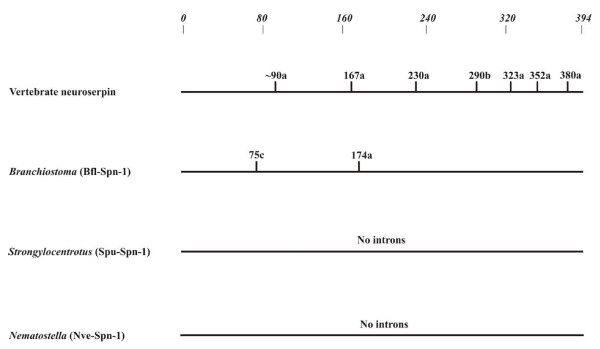
Figure 3.
Exon-intron organisation of the neuroserpin gene lineage. The Nematostella vectensis serpin gene Nve-Spn-1 is included, though orthology with the deuterostome counterparts is currently only supported by protein-based signature sequences. Specifications for intron positions and their phasing refer to mature human α1-antitrypsin. Only introns mapping to the serpin core domain (residues 33 to 394 of the reference) are considered.
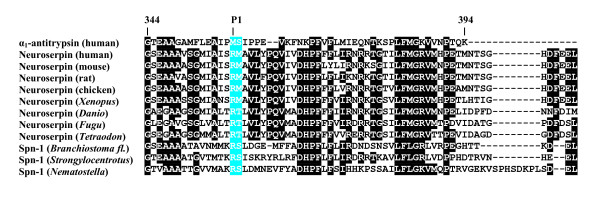
Figure 4.
C-terminal sequences of neuroserpin orthologs from deuterostomes and serpin Spn-1 from Nematostella vectensis. The numbering of amino acids refers to human α1-antitrypsin (top). Amino acids flanking the (putative) scissile bond are marked in turquoise, and the P1 position is indicated. Residues conserved in at least 70% of sequences are reproduced in white-on-black.
Extension of microsynteny analysis to lancelets (Branchiostoma floridae) and sea urchins (Strongylocentrotus purpuratus) showed that a serpin gene is present in either of these species in close vicinity to the PDCD10 gene (Figure 2). As in vertebrates, these genes are arranged in a head-to-head orientation. Sequence comparisons corroborated that the Branchiostoma floridae serpin adjacent to PDCD10, denominated Bfl-Spn-1, is the ortholog of the previously characterised serpin gene Spn-1 from the closely related Branchiostoma lanceolatum (92.2% sequence identity for the C-terminal 385 amino acids) that was recently shown to inhibit proprotein convertases [30]. Each of these serpins contains a highly conserved RSL region (positions P5 – P1': NMMKR ↓ S), and a C-terminal ER retention/retrieval signal (KDEL) (Figure 4). The presence of an N-terminal signal peptide in lancelet Spn-1 mediating access to the secretory pathway is supported by cDNA sequence analysis and expression studies [30]. The gene cluster harbouring the PDCD10/Spn-1 gene pair includes a closely related paralog (Bfl-Spn-2) of B. floridae Spn-1 (Figure 2) that also has a counterpart in B. lanceolatum (not shown).
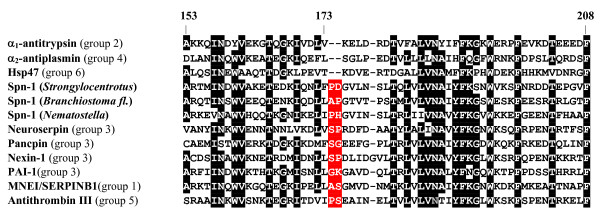
Similar to lancelets, the genome of the sea urchin Strongylocentrotus purpuratus revealed linkage of the PDCD10 gene to an inversely oriented serpin gene, named Spu-Spn-1 (accession number: XP_001186705) within a 20 kb DNA segment. Apart from synteny of its gene with PDCD10, Spu-Spn-1 shares a signal peptide, a conserved RSL region including the dibasic KR motif preceding the inhibitor's putative scissile bond (P5-P1': TMTKR ↓ S), and a variant (HEEL) of the canonical KDEL signal with the corresponding lancelet serpin (Figure 4). The HEEL motif was recently shown to mediate ER retention in transfected HeLa cells [31]. Another feature corroborates evolutionary continuity extending from mammalian neuroserpin via Branchiostoma Spn-1 to Spu-Spn-1 from the sea urchin. Groups 1, 3 and 5 of vertebrate serpins are discernible from groups 2, 4, and 6 by a two amino acid indel following residue 173 (α1-antitrypsin numbering, ref. [20]). The discriminating dipeptide sequence (previously assigned adjacent to position 171, due to use of a different set of aligned serpin sequences) is also found in Branchiostoma Spn-1, and in Spu-Spn-1 from Strongylocentrotus purpuratus (Figure 5). Collectively, these data suggest that a secretory pathway-associated serpin that already existed at least since the emergence of deuterostomes gave rise to mammalian neuroserpin.
Figure 5.
A discriminatory indel supports relationships of neuroserpin and homologs from sea urchins, lancelets, and Nematostella. Human representatives of vertebrate serpin groups 1, 3, and 5 containing the indel (marked in red), and from groups 2, 4 and 6 that lack the indel, are shown. The numbering of positions shown above the alignment refers to the sequence of mature human α1-antitrypsin. Positions conserved in at least 70% of sequences are represented in white-on-black printing.
Inspection of genomes from deeper rooting metazoans revealed PDCD10 orthologs in Drosophila melanogaster and in C. elegans (Figure 6), depicting 49% and 39% sequence identity at the protein level with their human counterpart [26], but close linkage of this marker to a serpin gene is neither evident in the fruit fly nor in the worm. In Drosophila melanogaster, genes of unknown functions flank PDCD10, and in the nematode genome, the PDCD10 microenvironment differs from that of both Drosophila and that of vertebrates (see Additional file 1), making identification of neuroserpin orthologs in these species more difficult. Some data, however, suggest the existence of a neuroserpin ortholog at least since the divergence of sponges and eumetazoans, believed to have occurred at least 650 to 700 million years ago [32-34]. To date, three serpin genes have been identified in the genome of the sea anemone Nematostella vectensis [34], one of which (accession number: XP_001627732; http://genome.jgi-psf.org/Nemve1/Nemve1.home.html: estExt_fgenesh1_pg.C_1860016) displays features suggesting shared ancestry with neuroserpin orthologs. These features include a dibasic amino acid sequence motif preceding the putative scissile bond, and a C-terminal extension ending with the tetrapeptide sequence SDEL, a functional variant of the canonical ER retention/retrieval signal [31]. This serpin (as its sea anemone paralogs) also possesses the dipeptide indel adjacent to position 173; however, further sequence-independent data are needed to firmly establish the presumed type of kinship.
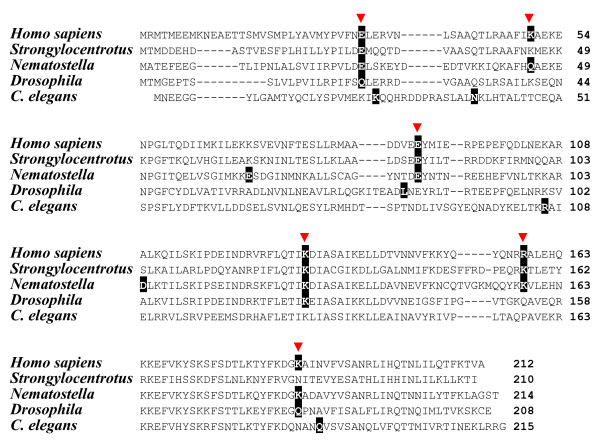
Figure 6.
Intron positions of PDCD10 genes in metazoans. Intron positions (white-on-black printing, phasing not indicated) were identified with GENEWISE and mapped onto the protein sequences. Intron positions conserved in at least two species are marked with an arrow head. Accession numbers for PDCD10 sequences: AAH16353 (human); XP_001186662 (Strongylocentrotus purpuratus); EDO34838 (Nematostella vectensis); AAF55190 (Drosophila melanogaster); CAA90115 (C. elegans).
The exon-intron structures of the adjacent neuroserpin and PDCD10 gene orthologs underwent different fates during deuterostome evolution
Though microsynteny and signature sequences strongly argue in favour of a common ancestor giving rise to mammalian neuroserpin, Spu-Spn-1 from Strongylocentrotus purpuratus, and Spn-1 from Branchiostoma, their genes depict quite different patterns of intron distribution (Figure 3). The sea urchin Spu-Spn-1 gene does not contain any intron mapping to the serpin core domain, and the single (correctly predicted?) intron resides in the sequence coding for the signal peptide (accession number: NW_001288761). The Spn-1 gene from lancelets harbours introns at positions 75c and 174a (α1-antitrypsin numbering). This intron-poor gene architecture contrasts with the mammalian neuroserpin gene that depicts the characteristic group 3 exon-intron structure with introns at positions 167a, 230a, 290b, 323a, 352a, and 380a (the first intron of the neuroserpin gene mapping to the serpin core domain, tentatively assigned to position ~90a, cannot be assigned reliably, due to alignment ambiguities). Strikingly, none of these intron positions is conserved among neuroserpin orthologs from lancelets, sea urchins or vertebrates. There is also no congruence of the introns at positions 75c and 174a in the lancelet Spn-1 gene with any of the other vertebrate serpin genes (Figure 1). Obviously, massive changes have occurred along the neuroserpin gene lineage concerning exon-intron organisation since divergence of echinoderms, cephalochordates and vertebrates. The Spn-1 gene from Nematostella vectensis also does not contain an intron mapping to the serpin body (Figure 3).
Contrasting with the neuroserpin gene lineage, comparably few changes are evident in the architecture of the immediately adjacent PDCD10 gene since the split of sea urchins and mammals (Figure 6). The PDCD10 genes from humans and Strongylocentrotus have four out of six intron positions in common. Two introns (positions 50c and 186b, numbering based on the human sequence) seem to have been lost in the sea urchin, since they are present in the earlier diverging cnidarian, Nematostella vectensis. The sea anemone PDCD10 gene contains eight introns, six of which are found at equivalent positions in the human homolog. Nematostella vectensis genes were recently demonstrated to share the majority of intron positions with their mammalian counterparts [34]. None of the PDCD10 introns of C. elegans superimposes on an intron found in the orthologs from humans, the sea urchin, or the sea anemone.
Discussion
The findings here reveal a clear history of neuroserpin, a prominent group 3 vertebrate serpin. Features derived from the genomic, gene and protein level provide ample discriminatory data to enable drawing of a reliable kinship history of its previously unknown origin. Microsynteny analysis proved to be especially illuminating, demonstrating that rare genomic characters can provide very useful information for decoding of bonds in protein families with intricate evolutionary history. Recent investigations provide a plausible explanation for the strongly conserved syntenic association of PDCD10 and neuroserpin orthologs during diversification of deuterostomes. Apparently, expression of the head-to-head arranged genes is controlled by a bi-directional, asymmetrically acting promoter region inserted within the ~0.9 kb intergenic region separating the transcription units coding for PDCD10 and neuroserpin [35]. Dependence on the common regulatory region thus may have forced the maintenance of linkage of these genes. The rapidly increasing flood of data from genome sequencing projects will certainly continue to provide further discriminatory information from multiple, independent levels of biological organisation, such as codon usage dichotomy [36], to enable robust classification of other metazoan serpins.
Neuroserpin orthologs from early diverging deuterostomes, like Strongylocentrotus or Branchiostoma, contain classical ER retention signals (KDEL or HEEL) at their C-terminal ends, and the Nematostella Spn-1 sequence terminates with SDEL, which functions as an autonomous ER retention/retrieval signal in HeLa cells, when hooked to a reporter protein [31]. The C-terminal end of neuroserpin from mammals, chicken, and Xenopus is HDFEEL (Figure 4). In HeLa cells, which express three different KDEL receptors with overlapping, but not identical passenger specificities, the FEEL sequence targets attached passenger proteins primarily to the Golgi, though some 25% of cells depict ER localisation [31]. In transfected COS cells, intracellular neuroserpin localises to either the ER or Golgi [11]; in cells with a regulated secretory pathway, however, neuroserpin resides in large dense core vesicles, mediated by a C-terminal extension encompassing the last 13 amino acids, including the FEEL sequence [11]. Collectively, these data are compatible with the view that, in an ancient ortholog of neuroserpin, a two amino acid insertion (FE) gave rise (in combination with additional residues?) to a modified sorting signal enabling a more specialised subcellular localisation. Irrespective of the still fragmentary data concerning the phylogenetic classification of Spn-1 from the sea anemone, it is clear that surveillance of the secretory pathway routes by serpins is an ancient and conserved trait in eukaryotes. Whether the C-terminal extensions of neuroserpin orthologs from fishes (Figure 4) are functional secretory pathway address signals remains to be determined.
The regional changes of placement within the secretory route may have come along with diversifications associated with the inhibitors' functions due to changes within the RSL region. Neuroserpin from vertebrates is believed to interact with its preferred target enzyme, tPA, via the single Arg residue (P1 position) in the RSL region [9]. In lancelets, the scissile bond is preceded by the dipeptide motif Lys-Arg (KR), which is characteristic for substrates and inhibitors of proprotein convertases, which indeed, have been identified as target enzymes of lancelet Spn-1 [30]. Similar biochemical properties are expected for Spn-1 from the sea urchin, and Spn-1 from the sea anemone (Figure 4). The physiological interaction partners of these inhibitors have not yet been identified.
Though the data clearly indicate that the roots of mammalian neuroserpin may be traced back far in the history of animals, unequivocal support for a neuroserpin ortholog in arthropods is still lacking. Several labs have provided evidence for a serpin (Spn4) with furin inhibiting activity and containing a canonical ER targeting signal in Drosophila [13-15], and a similar protein has been detected in Anopheles [37]. However, caution should be advised, because homoplasy due to convergent evolution currently cannot be excluded. The Spn4 gene is prone to recombination events, especially in the regions coding for the RSL region [19]. Unraveling the relationships of the Spn4 gene from fruit flies and neuroserpin orthologs from deuterostomes requires further investigation.
The history of the neuroserpin/PDCD10 gene pair reveals some remarkable insights into the evolution of the exon-intron structure of metazoan genes. Even closely adjacent genes that are physically linked at least since divergence of echinoderms and chordates may be subject to quite different trends affecting the intron distribution patterns. Comparably few changes in the exon-intron architecture have happened in PDCD10 orthologs since divergence of lineages leading to sea anemones and vertebrates (Figure 6). In PDCD10 genes, six out of eight intron positions occurring in humans or in the cnidarian are conserved. This is in accordance with findings demonstrating that the majority of genes from early diverging present-day eumetazoans are intron-rich with most introns apparently maintained since ancient times [34,38]; for serpin genes, however, the situation appears to be different. Regardless of the still rudimentary evidence for the putative sea anemone neuroserpin ortholog, the available data show that serpin genes in Nematostella vectensis are intron-poor. The sea anemone Spn-1 gene does not contain any introns mapping to the serpin body, and the single serpin core intron identified in one (accession number: XP_001627750) of the currently known three Nematostella vectensis serpin genes maps to residue 42c (α1-antitrypsin numbering; not shown). Looking up at deuterostomes, the sea urchin neuroserpin ortholog Spu-Spn-1 is also devoid of introns within the region coding for the serpin core. In contrast, the Spn-1 genes from Branchiostoma floridae (Figure 3) and its close relative, Branchiostoma lanceolatum [30] each depict two introns mapping to identical sites within the serpin body. Their positions, however, are not congruent with any of the introns of mammalian neuroserpin, the prototype group 3 vertebrate serpin gene or with any other intron location known from vertebrate serpin genes [20]. Therefore it must be considered that, in the serpin lineage leading to mammalian neuroserpin, an appreciable fraction of introns is not ancient, but may have been acquired during metazoan evolution; however, it cannot be excluded that intron paucity in present-day serpin genes of cnidarians (and in neuroserpin orthologs from sea urchins and lancelets) is due to massive intron loss, in contrast to most other introns that have survived hundreds of millions of years in these creatures. Intron gain is possibly not as rare as sometimes believed [39], however, it could be confined to certain gene families and/or to discrete evolutionary phases [40], for as yet unexplored reasons. Several types of processes have been proposed that may explain how introns may be acquired, but definite answers are still awaited.
Conclusion
In this study, we analysed and resolved the evolutionary roots of neuroserpin, a secretory-pathway associated mammalian serpin. Insight into the intricate history of the multi-membered serpin superfamily beyond the fish/tetrapod split was obtained by showing that orthologs of neuroserpin exist at least since the emergence of deuterostomes and probably already since divergence of eumetazoans and Bilateria. The continuous presence of neuroserpin orthologs equipped with C-terminal signal sequences assigning residence within the secretory pathway documents that serpins functioning as guards of the cellular export/import routes represent an ancient trait. This surveillance role has been subject to subtle functional and local variances during evolution as evidenced by changes within the RSL and the subcellular address signal. In contrast to many other, even closely linked genes, in which the majority of intron positions has been conserved for hundreds of millions of years, the intron distribution pattern of neuroserpin gene orthologs has experienced massive changes, perhaps dominated by intron gain.
Methods
Identification of serpin DNA and protein sequences and microsynteny analysis
Serpin protein and DNA sequences of various genomes were extracted from publicly accessible databases (see Additional file 2) via the BLAST software package (including PSI-BLAST) using key words or the human α1-antitrypsin sequence for searching. Chromosomal microsynteny analysis was performed using the NCBI Map Viewer [41], the ENSEMBL genome browser [42], the JGI genome browser [43], the Tetraodon genome browser [44], the UCSC genome browser [45], and inspecting the Strongylocentrotus purpuratus genome database [46].
Sequence alignments, gene structure analyses and mapping of intron positions
Alignments of protein sequences were performed with CLUSTAL X [47] and refined manually in GeneDoc [48]. Intron positions were identified and assigned with GENEWISE [49]. Mature human α1-antitrypsin was used as reference for mapping of positions and phasing of introns in serpin genes [20].
Authors' contributions
AK carried out analyses. HR conceived and supervised the project and wrote the paper. Both authors have read and approved the final manuscript.
Supplementary Material
Genes flanking PDCD10 orthologs in D. melanogaster and C. elegans. Genes flanking PDCD10 orthologs in D. melanogaster and C. elegans. Neighbouring genes of PDCD10
Sources of data for genomes investigated in this study. Sources of data for genomes investigated in this study. Web adresses of genomes
Contributor Information
Abhishek Kumar, Email: abhishek.abhishekkumar@gmail.com.
Hermann Ragg, Email: hr@zellkult.techfak.uni-bielefeld.de.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft, Graduate Program 'Bioinformatics' at the University of Bielefeld.
References
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, Irving JA, Bird PI. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci. 2004;61:301–325. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragg H. The role of serpins in the surveillance of the secretory pathway. Cell Mol Life Sci. 2007;64(21):2763–70. doi: 10.1007/s00018-007-7157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TH, Hejgaard J, Saunders NF, Cavicchioli R, Curmi PM. Serpins in unicellular Eukarya, Archaea, and Bacteria: sequence analysis and evolution. J Mol Evol. 2004;59:437–447. doi: 10.1007/s00239-004-2635-6. [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Kojima T, Kadomatsu K, Nakayama Y, Takagi A, Suzuki M, Takeda N, Ito M, Yamamoto K, Matsushita T, Kusugami K, Muramatsu T, Saito H. Complete antithrombin deficiency in mice results in embryonic lethality. J Clin Invest. 2000;106:873–878. doi: 10.1172/JCI10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas DA, Carrell RW. Serpinopathies and the conformational dementias. Nat Rev Genet. 2002;3:759–768. doi: 10.1038/nrg907. [DOI] [PubMed] [Google Scholar]
- Miranda E, Lomas DA. Neuroserpin: a serpin to think about. Cell Mol Life Sci. 2006;63:709–722. doi: 10.1007/s00018-005-5077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings GA, Coleman TA, Haudenschild CC, Stefansson S, Smith EP, Barthlow R, Cherry S, Sandkvist M, Lawrence DA. Neuroserpin, a brain-associated inhibitor of tissue plasminogen activator is localized primarily in neurons. J Biol Chem. 1997;272:33062–33067. doi: 10.1074/jbc.272.52.33062. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Cinelli P, Baici A, Pennella A, Krueger SR, Schrimpf SP, Meins M, Sonderegger P. The axonally secreted serine proteinase inhibitor, neuroserpin, inhibits plasminogen activators and plasmin but not thrombin. J Biol Chem. 1998;273:2312–2321. doi: 10.1074/jbc.273.4.2312. [DOI] [PubMed] [Google Scholar]
- Parmar PK, Coates LC, Pearson JF, Hill RM, Birch NP. Neuroserpin regulates neurite outgrowth in nerve growth factor-treated PC12 cells. J Neurochem. 2002;82:1406–1415. doi: 10.1046/j.1471-4159.2002.01100.x. [DOI] [PubMed] [Google Scholar]
- Ishigami S, Sandkvist M, Tsui F, Moore E, Coleman TA, Lawrence DA. Identification of a novel targeting sequence for regulated secretion in the serine protease inhibitor neuroserpin. Biochem J. 2007;402:25–34. doi: 10.1042/BJ20061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger O, Ladewig J, Köster K, Ragg H. Widespread occurrence of serpin genes with multiple reactive centre-containing exon cassettes in insects and nematodes. Gene. 2002;293:97–105. doi: 10.1016/S0378-1119(02)00697-2. [DOI] [PubMed] [Google Scholar]
- Oley M, Letzel MC, Ragg H. Inhibition of furin by serpin Spn4A from Drosophila melanogaster. FEBS Lett. 2004;577:165–169. doi: 10.1016/j.febslet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Kuhnen A, Leiserson WM, Kim YS, Keshishian H. Drosophila serpin 4 functions as a neuroserpin-like inhibitor of subtilisin-like proprotein convertases. J Neurosci. 2004;24:5482–5491. doi: 10.1523/JNEUROSCI.5577-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer MJ, Keays CA, Waterhouse J, Minhas J, Hashimoto C, Jean F. The Spn4 gene of Drosophila encodes a potent furin-directed secretory pathway serpin. Proc Natl Acad Sci. 2004;101:10560–10565. doi: 10.1073/pnas.0401406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving JA, Cabrita LD, Kaiserman D, Worrall MW, Whisstock JC. In: Molecular and cellular aspects of the serpinopathies and disorders in serpin activity. Silverman GA, Lomas DA, editor. Singapore: World Scientific Publishing; 2007. Evolution and classification of the serpin superfamily; pp. 1–33. [Google Scholar]
- Barbour KW, Wei F, Brannan C, Flotte TR, Baumann H, Berger FG. The murine α1-proteinase inhibitor gene family: polymorphism, chromosomal location, and structure. Genomics. 2002;80:515–522. doi: 10.1016/S0888-7543(02)96864-3. [DOI] [PubMed] [Google Scholar]
- Forsyth S, Horvath A, Coughlin P. A review and comparison of the murine α1-antitrypsin and α1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003;81:336–345. doi: 10.1016/S0888-7543(02)00041-1. [DOI] [PubMed] [Google Scholar]
- Börner S, Ragg H. Functional diversification of a protease inhibitor gene in the genus Drosophila and its molecular basis. Gene. 2008;415:23–31. doi: 10.1016/j.gene.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Ragg H, Lokot T, Kamp PB, Atchley WR, Dress A. Vertebrate serpins: construction of a conflict-free phylogeny by combining exon-intron and diagnostic site analyses. Mol Biol Evol. 2001;18(4):577–84. doi: 10.1093/oxfordjournals.molbev.a003838. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Lokot T, Wollenberg K, Dress A, Ragg H. Phylogenetic analyses of amino acid variation in the serpin proteins. Mol Biol Evol. 2001;18:1502–1511. doi: 10.1093/oxfordjournals.molbev.a003936. [DOI] [PubMed] [Google Scholar]
- Benarafa C, Remold-O'Donnell E. The ovalbumin serpins revisited: Perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc Natl Acad Sci USA. 2005;102:11367–11372. doi: 10.1073/pnas.0502934102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserman D, Bird PI. Analysis of vertebrate genomes suggests a new model for clade B serpin evolution. BMC Genomics. 2005;6:167. doi: 10.1186/1471-2164-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelissier P, Delourme D, Germot A, Blanchet X, Becila S, Maftah A, Leveziel H, Ouali A, Bremaud L. An original SERPINA3 gene cluster: elucidation of genomic organization and gene expression in the Bos taurus 21q24 region. BMC Genomics. 2008;9:151. doi: 10.1186/1471-2164-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss K, Stahl S, Schleider E, Ullrich S, Nickel J, Mueller TD, Felbor U. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8:249–256. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M, Tournier-Lasserve E. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachert C, Lee TH, Linstedt AD. Lumenal endosomal and Golgi-retrieval determinants involved in pH-sensitive targeting of an early Golgi protein. Mol Biol Cell. 2001;12:3152–3160. doi: 10.1091/mbc.12.10.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan R, Linstedt AD. A cycling cis-Golgi protein mediates endosome-to-Golgi traffic. Mol Biol Cell. 2004;15:4798–4806. doi: 10.1091/mbc.E04-05-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T, Forsten-Williams K, Storrie B. Both post-Golgi and intra-Golgi cycling affect the distribution of the Golgi phosphoprotein GPP130. Traffic. 2007;8:1265–1279. doi: 10.1111/j.1600-0854.2007.00607.x. [DOI] [PubMed] [Google Scholar]
- Bentele C, Krüger O, Tödtmann U, Oley M, Ragg H. A proprotein convertase-inhibiting serpin with an ER targeting signal from Branchiostoma lanceolatum, a close relative of vertebrates. Biochem J. 2006;395:449–456. doi: 10.1042/BJ20051947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, Ruddock L. A molecular specificity code for the three mammalian KDEL receptors. J Cell Biol. 2007;179:1193–1204. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair Hedges S, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Raff RA. Written in stone: fossils, genes and evo-devo. Nat Rev Genet. 2007;8:911–920. doi: 10.1038/nrg2225. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Chen PY, Chang WS, Chou RH, Lai YK, Lin SC, Chi CY, Wu CW. Two non-homologous brain diseases-related genes, SERPINI1 and PDCD10, are tightly linked by an asymmetric bidirectional promoter in an evolutionarily conserved manner. BMC Mol Biol. 2007;8:2. doi: 10.1186/1471-2199-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krem MM, Di Cera E. Conserved Ser residues, the shutter region, and speciation in serpin evolution. J Biol Chem. 2003;278:37810–37814. doi: 10.1074/jbc.M305088200. [DOI] [PubMed] [Google Scholar]
- Danielli A, Kafatos FC, Loukeris TG. Cloning and characterization of four Anopheles gambiae serpin isoforms, differentially induced in the midgut by Plasmodium berghei invasion. J Biol Chem. 2003;278:4184–4193. doi: 10.1074/jbc.M208187200. [DOI] [PubMed] [Google Scholar]
- Raible F, Tessmar-Raible K, Osoegawa K, Wincker P, Jubin C, Balavoine G, Ferrier D, Benes V, de Jong P, Weissenbach J, Bork P, Arendt D. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310:1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- Zhuo D, Madden R, Elela SA, Chabot B. Modern origin of numerous alternatively spliced human introns from tandem arrays. Proc Natl Acad Sci USA. 2007;104:882–886. doi: 10.1073/pnas.0604777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babenko VN, Rogozin IB, Mekhedov SL, Koonin EV. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 2004;32:3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, Rapp BA. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2001;29:11–16. doi: 10.1093/nar/29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36:D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetraodon genome browser at the French National Sequencing Center – Genoscope. http://www.genoscope.cns.fr/externe/tetranew/
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Kober KM, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium. Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Bellé R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Genevière AM, Goel M, Kelkar H, Morales J, Mulner-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stolc V, Tongprasit W, Tu Q, Bergeron KF, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallböök F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su YH, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Röttinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu SY, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu LL, Thorn R, Wright R. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II. GeneDoc: analysis and visualization of genetic variation. EMBNEWNEWS. 1997;4:14. [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes flanking PDCD10 orthologs in D. melanogaster and C. elegans. Genes flanking PDCD10 orthologs in D. melanogaster and C. elegans. Neighbouring genes of PDCD10
Sources of data for genomes investigated in this study. Sources of data for genomes investigated in this study. Web adresses of genomes