Abstract
Background
Aeromonas salmonicida subsp. salmonicida is a Gram-negative bacterium that is the causative agent of furunculosis, a bacterial septicaemia of salmonid fish. While other species of Aeromonas are opportunistic pathogens or are found in commensal or symbiotic relationships with animal hosts, A. salmonicida subsp. salmonicida causes disease in healthy fish. The genome sequence of A. salmonicida was determined to provide a better understanding of the virulence factors used by this pathogen to infect fish.
Results
The nucleotide sequences of the A. salmonicida subsp. salmonicida A449 chromosome and two large plasmids are characterized. The chromosome is 4,702,402 bp and encodes 4388 genes, while the two large plasmids are 166,749 and 155,098 bp with 178 and 164 genes, respectively. Notable features are a large inversion in the chromosome and, in one of the large plasmids, the presence of a Tn21 composite transposon containing mercury resistance genes and an In2 integron encoding genes for resistance to streptomycin/spectinomycin, quaternary ammonia compounds, sulphonamides and chloramphenicol. A large number of genes encoding potential virulence factors were identified; however, many appear to be pseudogenes since they contain insertion sequences, frameshifts or in-frame stop codons. A total of 170 pseudogenes and 88 insertion sequences (of ten different types) are found in the A. salmonicida genome. Comparison with the A. hydrophila ATCC 7966T genome reveals multiple large inversions in the chromosome as well as an approximately 9% difference in gene content indicating instances of single gene or operon loss or gain.
A limited number of the pseudogenes found in A. salmonicida A449 were investigated in other Aeromonas strains and species. While nearly all the pseudogenes tested are present in A. salmonicida subsp. salmonicida strains, only about 25% were found in other A. salmonicida subspecies and none were detected in other Aeromonas species.
Conclusion
Relative to the A. hydrophila ATCC 7966T genome, the A. salmonicida subsp. salmonicida genome has acquired multiple mobile genetic elements, undergone substantial rearrangement and developed a significant number of pseudogenes. These changes appear to be a consequence of adaptation to a specific host, salmonid fish, and provide insights into the mechanisms used by the bacterium for infection and avoidance of host defence systems.
Background
The genus Aeromonas comprises a collection of Gram-negative bacteria that are widespread in aquatic environments and that have been implicated as causative agents of a number of human and animal diseases. A. hydrophila, A. veronii biovar sobria, A. caviae, A. jandaei, A. veronii biovar veronii, A. schubertii and A. trota have been associated with various human infections including gastroenteritis, wound infections and septicaemia [1]. Aeromonas salmonicida, a non-motile aeromonad, is the aetiological agent of a bacterial septicaemia in fish, called furunculosis [2-4]. Furunculosis is an important disease in wild and cultured stocks of salmonid and other fish species and can have significant negative economic impacts on aquaculture operations. Motile Aeromonas species have also been implicated as the causative agents of various fish septicemias [5]. A. hydrophila is also associated with red leg disease in amphibians and infections in turtles [6] and birds [7].
In addition to their role as disease agents, Aeromonas species can be found in non-pathogenic association with a variety of animals [8-10]. Most Aeromonas species are opportunistic pathogens, entering through wounds or affecting only stressed or otherwise immunocompromised hosts [1]. A. salmonicida subsp. salmonicida, however, is a specific pathogen of salmonid fish and is capable of causing disease in healthy fish at very low levels of infection (LD50 < 10 cfu by intraperitoneal injection [11]). Although Bergey's Manual of Systematic Bacteriology [12] recognizes five subspecies of A. salmonicida: salmonicida, achromogenes, masoucida, smithia, and pectinolytica, many laboratories currently classify A. salmonicida subsp. salmonicida as "typical" and any isolate deviating phenotypically as "atypical". Hosts for atypical strains include a wide variety of non-salmonid fish, as well as salmonids [4]. On the basis of DNA relatedness, A. salmonicida also includes a group of mesophilic, motile strains isolated from humans [12]. Morphological and biochemical differences such as pigment production, colony size and growth rate, haemolysis, and sucrose fermentation [4,13-15] are used to distinguish typical and atypical isolates. A. salmonicida subsp. salmonicida (i.e. typical) isolates grow well on blood agar with large colonies, produce a brown diffusible pigment, are haemolytic and do not ferment sucrose [12]. Historically, typical strains are thought to be extremely homogenous [16,17], and therefore any deviation in any of these characteristics has been considered enough evidence to classify a strain as "atypical" [13]. Phylogenetic analyses based on gene sequences [18,19] or biochemical analyses based on carbohydrates [20] appear to be better able to sort out the complex taxonomy and classification of A. salmonicida subspecies and related species.
A. salmonicida subsp. salmonicida appears to be an example of the evolution of pathogen specificity for a particular host from within a group of mainly opportunistic pathogens or commensal bacteria. It thus provides opportunities to identify genes involved in host invasion and virulence and to investigate the evolution of host specificity. In this communication, the genome, including both the chromosome and large plasmids, of an isolate of A. salmonicida subsp. salmonicida is characterized. The three small plasmids of this strain have been described previously [21]. Genes associated with virulence are identified and comparisons with the genome of A. hydrophila ATCC 7966T [22] provide insights into the changes in the genome that may be associated with adaptation to fish hosts. The genome sequence is an essential tool for the understanding of the infection process of A. salmonicida.
Results and discussion
Genome features
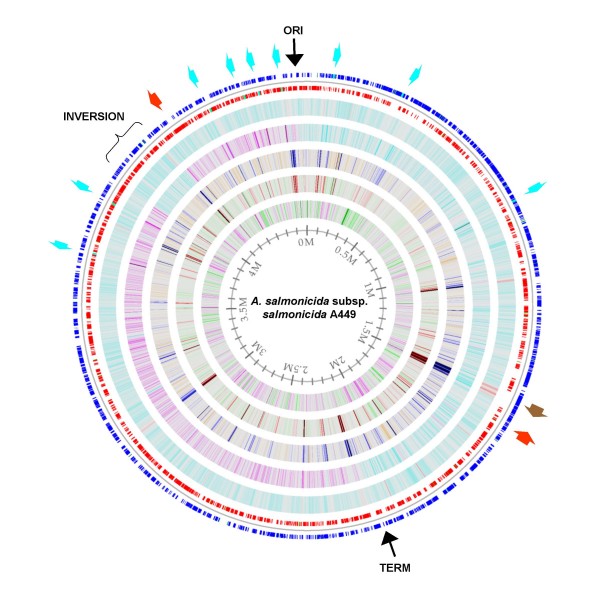
The genome of A. salmonicida subsp. salmonicida A449 (hereon A449) consists of a single circular chromosome, two large plasmids and three small plasmids. The 4,702,402 bp chromosome has a G+C content of 58.5% and contains 4388 genes, with 4086 encoding proteins (Table 1). Generally, the chromosome matches the restriction map previously constructed for this strain [23], although there are differences in the placement of some genes. The chromosome has a number of major structural features. The origin of replication (oriC), as inferred from the presence of multiple DnaA binding sites and GC skew, occurs at 4666400 – 4666750, which is approximately 35,700 bp from dnaA (Fig. 1). Replication terminates near 2134850 as judged by GC skew and the presence of a dif-like sequence, which has been recently implicated as the DNA replication terminus [24]. GC skew also detected the presence of a large inversion (3963279 – 4158772) that appears to have occurred between two identical insertion sequences. PCR analysis confirmed that this inversion was not due to misassembly of the sequence (not shown). In addition, two prophages have been detected by similarity to phage genes (Fig. 1, red arrows), but these regions of the chromosome do not show any obvious alteration in G+C content.
Table 1.
A. salmonicida genome characteristics
| chromosome | pAsa1 | pAsa2 | pAsa3 | pAsa4 | pAsa5 | |
| length (bp) | 4702402 | 5424 | 5247 | 5616 | 166749 | 155098 |
| # genes | 4388 | 9 | 9 | 10 | 178 | 164 |
| # CDS | 4086 | 8 | 7 | 9 | 173 | 154 |
| # pseudogenes | 155 | 5 | 10 | |||
| # rRNA | 28 | |||||
| # tRNA | 110 | |||||
| # sRNA | 9 | |||||
| # misc. RNA | 1 | 2 | 1 | |||
| # riboswitches | 11 | |||||
| # IS elements | 71 | 7 | 10 | |||
| # Tn21/In2 | 1 | |||||
| # prophage | 2 |
Figure 1.
A. salmonicida subsp. salmonicida A449 chromosome. A genome atlas representation of the A. salmonicida chromosome. Indicated outside the circular chromosome are the origin of replication (ORI), replication termination site (TERM), a large inversion in the genome (INVERSION), nine rRNA operons (light blue arrows), two prophages (red arrows) and the surface layer protein operons (brown arrow). Moving inward are circles representing annotations (blue: + strand CDS, red: – strand CDS, light blue: rRNA, green: tRNA), percent AT (blue: 20% to red: 80%), GC skew (pink: -0.10 to blue: 0.10), intrinsic curvature (yellow: 0.11 to blue: 0.21), stacking energy (green: -9.21 to red: -7.78) and position preference (green: 0.13 to pink: 0.16).
Twenty-eight ribosomal RNA genes are encoded on the chromosome, arranged in nine operons, with one operon containing an extra copy of the 5S rRNA gene (Table 1; Fig. 1, light blue arrows). The nine operons are arranged around the origin of replication so as to be transcribed in the same direction as replication proceeds. Small variations (1 – 3 bp) in sequence occur between the copies of the rRNA genes, with only the "extra" 5S rRNA gene (rrfG1) having 6 bp that vary when compared to the other copies. A total of 110 tRNA genes are encoded on the A449 chromosome, most of which are present in at least two copies, and some of which occur in clusters of multiple tandem copies, similar to the A. hydrophila genome [22]. There are single genes for tryptophan (trnW) and selenocysteine tRNAs as well as a suppressor tRNA that translates TAG codons as tryptophan. This suppressor tRNA differs from the trnW sequence at only two bases, one of which is in the anticodon. Twenty-one protein coding genes appear to use the suppressor tRNA to allow the translation of the encoded protein. Analysis of the genome to identify small non-coding RNA features that regulate gene expression by binding to RNA or proteins [25] revealed the presence of nine small RNAs. In addition, 11 riboswitches, which regulate translation through the detection of small molecules [26], were detected near the 5' ends of genes they presumably regulate.
Two striking aspects of the A449 genome are the presence of large numbers of insertion sequences (IS) (n = 88) and pseudogenes (n = 170) on the chromosome and two large plasmids. Ten different types of IS are found in multiple copies in the A449 genome (Table 2) with ISAs7 present in 37 complete copies. One IS previously identified in Aeromonas species (ISAs4) [27] is not present in the A449 genome. In addition to the 88 complete IS elements, 14 partial IS sequences are present. This observation along with the finding that some IS are located within other IS, suggests that these dynamic elements have undergone recent transposition. Insertion sequences have also contributed to the apparent formation of pseudogenes, with more than 20 genes being interrupted by IS elements. Most pseudogenes, however, are created by small (1–37 bp) deletions or sequence duplications, although several genes have larger deletions. Additional pseudogenes appear to have arisen through mutations that introduce in-frame stop codons (TAA or TGA, but not TAG, due to the suppressor tRNA). These observations are in marked contrast to the A. hydrophila genome [22], which has no IS elements and only seven pseudogenes.
Table 2.
A. salmonicida Insertion Elements
| Name | Length (bp) | # copies | IS family | Reference |
| ISAs1 | 1223 | 2 | ISAs1 | [73] |
| ISAs2 | 1084 | 5 | IS30 | [73] |
| ISAs3 | 1326 | 4 (+2 partial) | IS256 | Genbank NC_004338 |
| ISAs4 | 1062 | 0 | IS5 | [27] |
| ISAs5 | 1233 | 12 | IS3 | [45] |
| ISAs6 | 1240 | 7 (+5 partial) | IS3 | this work |
| ISAs7 | 1165 | 37 (+3 partial) | IS630 | this work |
| ISAs8 | 754 | 3 | IS1 | this work |
| ISAs9 | 1624 | 4 | IS3 | this work |
| ISAs10 | 1229 | 2 (+1 partial) | IS30 | this work |
| ISAs11 | 2614 | 12 (+3 partial) | IS21 | this work |
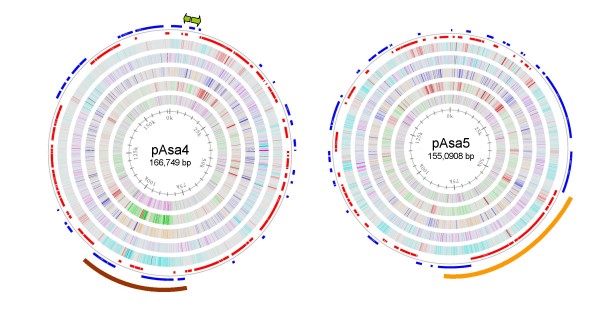
Both large plasmids contain genes involved in replication, plasmid partition and conjugative transfer. Plasmid 4 (pAsa4) carries an origin of replication that can be propagated in E. coli, since this plasmid was isolated by transformation of E. coli with a plasmid DNA extract from A449. This plasmid also contains a Tn21 composite transposon (bases 78182 – 101330) [28] that carries genes for resistance to mercury as well as an In2 integron encoding resistance to streptomycin/spectinomycin, quaternary ammonia compounds, sulphonamides and chloramphenicol (Fig. 2, brown bar). The Tn21 sequence has a considerably higher G+C content (61.43%) than the remainder of the plasmid (52.18%), as well as noticeable differences in stacking energy and position preference, as expected for a transposon.
Figure 2.
A. salmonicida subsp. salmonicida A449 large plasmids. Genome atlas representations of the two A. salmonicida large plasmids. Green arrows outside the pAsa4 circle indicate tetracycline resistance genes and the brown bar indicates the position of the Tn21 containing the In2 integron. The orange bar on the pAsa5 circle indicates the position of the type III secretion apparatus operons. The interior circles are as described in Figure 1.
Virulence genes
Secretion systems
The most notable aspect of the other large plasmid, pAsa5, is the presence of genes for a type III secretion system (T3SS) (Fig. 2, orange bar; Table 3) that has been shown to be required for virulence in A. salmonicida [29,30]. The 36 genes encoding the T3SS needle apparatus and regulatory proteins are organized identically to those described previously for the partial sequence of A. salmonicida [31] and the complete T3SS sequences of A. hydrophila: AH-3 [32], AH-1 [33]and SSU [34]. As well, three presumptive effector proteins (AopH, AopO, Ati2) and their associated chaperones (SycH, SycO, Ati1) are located on pAsa5 (Table 3). Two of the effector proteins AopH and AopO (ASA_P5G009 and ASA_P5G098) show significant similarity to Yersinia YopH and YopO, and thus are expected to encode a protein tyrosine phosphatase and a protein serine/threonine kinase, respectively [30]. A third effector, Ati2 (ASA_P5G045), was identified by the presence of its chaperone (Ati1 (ASA_P5G046)) [35] and its similarity to hypothetical proteins in the T3SS operons of Photorhabdus luminescens and Vibrio parahaemolyticus. On the basis of conserved domain structure, this effector appears to have inositol polyphosphate phosphatase activity. A fourth, well-characterized effector, AexT (ASA_4266) [36,37], is encoded on the chromosome. Two pseudogenes also appear to encode T3SS effectors: ASA_0010 is located on the chromosome and is disrupted by an in-frame TAA stop codon; ASA_P5G084 is located on pAsa5 and is disrupted by a 20 bp duplication that results in a frameshift. Genes encoding presumptive T3SS chaperones are adjacent to both pseudogenes. The A449 genome does not include the AopP effector found in several A. salmonicida subsp. salmonicida strains including JF2267and ATCC 33658T [38].
Table 3.
Potential Virulence Genes of Aeromonas salmonicida A449
|
Virulence function |
ASA Locus |
A. hydrophila ATCC 7966T orthologue1 |
Description |
Reference or Comments2 |
| Secretion | 0514–0515 | 3785–3786 | T2SS (ExeAB) | [55] |
| 3774–3785 | 0568–0579 | T2SS (ExeC-N) | [56] | |
| P5G048 – 083 | NP3 | T3SS structural & regulatory proteins | [31] | |
| P5G008 & 009 | NP | AopH effector & chaperone | [30] | |
| P5G097 & 098 | NP | AopO effector & chaperone | [30] | |
| 4266 & 4267 | NP | AexT effector & chaperone | [36] | |
| P5G045 & 046 | NP | Ati2 putative effector & chaperone | [35] | |
| P5G084 & 085 | NP | putative T3SS effector & chaperone | frameshift | |
| 0010 & 0011 | NP | putative T3SS effector & chaperone | in frame TAA, frameshift | |
| 2455 – 2470 | 1847 – 1832 | T6SS operon | 2455 & 2458 disrupted | |
| P4G080 – 082 | 1118 – 1119 1826 – 1827 1847 – 1848 |
T6SS | 082 interrupted by Tn21 [39] | |
| Adhesion | 1422 – 1459 | NP | surface layer & assoc. secretion system | [40,44] |
| 0346 – 0386 | NP | Lateral flagella | 0365, 0376 [45] disrupted | |
| 1336 – 1360 | 1364 – 1388 | Polar flagella | ||
| 1484 – 1499 | 2847 – 2832 | Polar flagella | 1499 frameshift | |
| 1505 – 1507 | 2826 – 2824 | Polar flagella | 1505 frameshift | |
| 2656 – 2662 | 1698 – 1703 | Polar flagella | 2656 frameshift | |
| 3725 – 3730 | 0519 – 0524 | Type I pilus | ||
| 0411 – 0414 | 3868 – 3671 | Tap type IV pilus | 0412 frameshift [93] | |
| 2903 – 2915 | 1462 – 1450 | Flp type IV pilus | 2906, 2908 & 2913 disrupted [47] | |
| 3938 – 3947 | 0383 – 0399 | Msh type IV pilus | multiple gene deletion [47] | |
| Toxins | 3906 | 0438 | aerolysin | [48] |
| 2854 | 1512 | hemolysin | ||
| 0826 | 3491 | RTX toxin | ||
| 2128 | NP | cytolytic δ-endotoxin | ||
| 2003 | NP | zona occludens toxin | frameshift | |
| 2015 | NP | zona occludens toxin | IS insertion | |
| Secreted enzymes | 2540 | 2687 | serine protease Ahe2 | [51] |
| 3321 | 0978 | zinc metalloprotease TagA | [52] | |
| 3440 | 0851 | elastase AhpB | ||
| 1723 | NP | metalloprotease | ||
| 3723 | 0517 | collagenase | ||
| 1660 | 2713 | AsaP1 protease | frameshift [53] | |
| 0509 | 3791 | glycerophospholipid cholesterol acyltransferase | [51] | |
| 4288 | 0104 | phospholipase A1 | [54] | |
| 0635 | 0635 | phospholipase C | [54] | |
| 1199 | 3126 | extracellular nuclease | ||
| 2206 | 2180 | extracellular nuclease NucH | ||
| 1286 | 1304 | amylase Amy1 | ||
| 3455 | 0837 | amylase AmyA | ||
| 0873 | 3440 | chitinase CdxA | ||
| 2142 | 2363 | chitinase Chi2 | ||
| 3320 | 0979 | chitinase ChiB | ||
| 0628 | 0628 | pullulanase PulA | ||
| Antibiotic resistance | P4G087 – 105 | NP | Tn21/In2 | |
| P4G004 – 005 | NP | tetracycline resist. | ||
| 1191 | 3135 | β-lactamase: ampC | [57] | |
| 4346 | 4258 | β- lactamase: ampS | [57] | |
| 3612 | 0740 | β- lactamase: cphA | [57] | |
| Iron acquisition | 1838 – 1851 | 2479 – 2473 1964 – 1970 |
amonabactin synthesis & uptake | [59,60] |
| 4368 – 4380 | NP | anguibactin synthesis & uptake | [59,62] | |
| 4363 – 4367 | 4275 – 4279 | hydroxymate siderophore receptor | ||
| 3328 | 0972 | putative heme receptor | [59] | |
| 3332 – 3336 | 0968 – 0964 | heme uptake | [63] | |
| Quorum sensing | 3762 | 0556 | N-acyl homoserine lactone synthase | [65] |
| 3763 | 0557 | Quorum sensing regulon activator | [65] | |
| 0697 | 0700 | AI-2 synthase | ||
| 2781 | 1576 | Quorum sensing phosphorelay protein | ||
| 3295 | 1004 | Quorum sensing response regulator |
1indicates AHA_ number
2Disrupted genes are indicated
3NP – not present in A. hydrophila genome
Genes for a type VI secretion system (T6SS), which is also involved in the transfer of bacterial proteins into host cells, are encoded on the A449 chromosome (ASA_2455 – ASA_2470) (Table 3). These 16 proteins show high similarity to T6SS proteins from A. hydrophila, P. aeruginosa and other Gram-negative bacteria. Three additional genes usually associated with this operon are encoded on pAsa4 (ASA_P4G080 – ASA_P4G082). However, a key T6SS gene is interrupted in A449: the gene encoding IcmF (ASA_2458) contains a 5 bp deletion and is fused to the upstream coding sequence in the operon. In addition, two proteins transported by the T6SS are disrupted: a partial VgrG homolog is fused to a transposon subunit (ASA_2455), although a complete vgrG gene is encoded on pAsa4 (ASA_P4G080), and Hcp, which is encoded on pAsa4 (ASA_P4G082), is interrupted by an insertion sequence into which the Tn21 element has inserted. These gene disruptions are in contrast to A. hydrophila where the T6SS genes are uninterrupted and a functional T6SS has been demonstrated [39]. Since deletion of the A. hydrophila icmF homolog (vasK) blocks T6SS secretion [39], the A449 T6SS is unlikely to be functional. The presence of defects in two T3SS effectors and the T6SS suggests that A449 could be considerably more virulent, since functional versions of these genes would provide additional means to manipulate the response of the host.
A notable aspect of the T3SS and T6SS is the location of genes for these systems on both the chromosome and the large plasmids. For the T3SS, most of the genes are encoded on pAsa5, although one functional effector gene, aexT, and a putative effector pseudogene (ASA_0010), as well as their T3SS chaperone protein genes (ASA_4267, ASA_0011), are encoded on the chromosome. In A. hydrophila ATCC 7966T, T3SS genes are absent, while other A. hydrophila strains carry T3SS operons on the chromosome [32-34]. The ancestral state of the T3SS in the genus Aeromonas is thus unclear, making it difficult to surmise how it ended up in two locations in A449. The T6SS situation is somewhat reversed, with the majority of genes located on the chromosome, but with three genes located on pAsa4. Since A. hydrophila has a complete, intact T6SS on the chromosome, one might infer that these genes were transferred to pAsa4 following the acquisition of that plasmid, but prior to the capture of the Tn21 element.
Adhesins
Genes for several types of adhesins (e.g., surface layer, flagella, pili), which are important in host cell attachment and entry, are present in the A449 genome (Table 3). The abundant surface layer protein VapA (ASA_1438) [40], which has been implicated as an important virulence factor in several studies [41-43], is located downstream from an operon for a VapA-specific type II secretion system (ASA_1427 – ASA_1437). The identification of these genes as a VapA secretion system is based the observation that disruption of spsE (ASA_1427) blocks VapA secretion [44] and that many of the genes in this operon show some similarity to genes of the general secretion pathway (exeA-N). In the same region of the genome are multiple carbohydrate synthesis and modification genes (ASA_1422 – ASA_1426, ASA_1441 – ASA_1459) that appear to be involved in the synthesis of lipopolysaccharide, which anchors the surface layer to the cell. The genes involved in VapA synthesis and secretion have an unusually low G+C content that can be seen in Fig. 1 at approximately base 1500000 (Fig. 1, brown arrow).
Complete sets of genes for two types of flagella, lateral and polar, are also encoded in the A449 genome. The genes for lateral flagella are found in a single cluster (ASA_0346 – ASA_0386) but include two disrupted genes: lafA, encoding the lateral flagellin, which has been shown previously to be interrupted by an insertion sequence [45], and lfgD, encoding the lateral flagellar hook-capping protein, which has a 1 bp deletion. The genes for the polar flagella are dispersed around the genome in multiple operons (ASA_1336 – ASA_1360, ASA_1484 – ASA_1499, ASA_1505 – ASA_1507, ASA_2656 – ASA_2662), but also include interrupted genes: flgL (ASA_1499), encoding a flagellar hook-associated protein, has a 5 bp duplication; flrA (ASA_1505), encoding a transcriptional activator, contains a 13 bp deletion; and, maf1 (ASA_2656), encoding a motility accessory factor [46] has a 1 bp deletion. The disruption of genes involved in the production of both types of flagella suggests that neither structure can be synthesized, which is consistent with the characterization of A. salmonicida as non-motile.
An additional class of adhesins, the pili, is well-represented in the A449 genome with genes for four different pili (three type IV, one type I) distributed throughout the genome. The type I pilus operon (ASA_3725 – ASA_3730) appears to be complete and intact. However, for each of three types of type IV pili [47], there are frameshifted genes encoding proteins involved in pilin assembly (tap, flp) or a multiple gene deletion (msh) (Table 3). Nevertheless, a mutant deleted for tapA showed reduced virulence when delivered by immersion, but not by intraperitoneal injection, suggesting a role for the Tap pilus in host invasion [47].
Toxins
Another class of putative virulence factors are pore-forming toxins that create channels in host membranes resulting in cell lysis (Table 3). Aerolysin, one of the earliest virulence factors to be discovered among Aeromonas spp. [48], is represented by two genes in the A449 genome: ASA_3906, which encodes the classical aerolysin [48], and ASA_2854, which encodes a conserved hemolysin found in other Aeromonas species, Vibrio species and Listonella anguillarum. A large (9588 bp) gene, asx (ASA_0826), encodes an RTX (repeats in toxin) protein, homologs of which are important virulence determinants in a range of Gram-negative bacteria [49]. The A449 genome also encodes a cytolytic delta-endotoxin (ASA_2128) that is 61% similar to the Bacillus thuringiensis insecticidal toxin CryET29 (Genbank accession AAK50455), which may be an indication of interaction with invertebrates. Two additional genes (ASA_2003 and ASA_2015) encode proteins that are 53 and 44% similar to the zonula occludens toxin (Zot) of Colwellia psychrerythraea (Genbank accession YP_267119). Zot was first described in Vibrio cholerae as a toxin that transiently loosens intracellular tight junctions in the intestinal mucosa [50]. In V. cholerae, Zot is associated with cholera toxin A and B and is also encoded on the CTXΦ plasmid [50]. In A449, however, both genes are interrupted, either by an IS (ASA_2015) or by a single bp insertion (ASA_2003), suggesting that functional proteins can not be synthesized from them.
Secreted enzymes
An additional class of potential virulence factors in A. salmonicida are extracellular enzymes, some of which have been previously investigated (Table 3). Among secreted proteases are a serine protease previously tested for its contribution to virulence (ASA_2540) [51], a zinc metalloprotease (TagA, ASA_3321) implicated in complement inhibition [52], another secreted metalloprotease (ASA_1723) and a microbial collagenase (ASA_3723). A gene encoding an extracellular endopeptidase (AsaP1, ASA_1660) contributing to virulence in atypical A. salmonicida strains [53] is present, but interrupted by a single bp insertion. The phospholipases encoded by satA (glycerophospholipid cholesterol acyltransferase, ASA_0509), pla (phospholipase A1, ASA_4288) and plc (phospholipase C, ASA_0635) have been investigated for their role in virulence in A. salmonicida [51] and A. hydrophila [54]. Extracellular nucleases (ASA_1199, ASA_2206), amylases (ASA_1286, ASA_3455), pullulanase (ASA_0628) and chitinases (ASA_0873, ASA_2142, ASA_3320) may also contribute to A449 virulence. All of these enzymes have a predicted Sec-dependent signal sequence and are expected to be secreted via the type II secretion system (exeA-N, ASA_0514–0515, ASA_3777–3785) [55,56].
Antibiotic resistance
In addition to the antibiotic resistance genes encoded in the Tn21 element, pAsa4 also carries genes for tetracycline resistance: tetA(E) (ASA_P4G005) encodes a class E tetracycline efflux pump that is presumably regulated by the adjacent class E tetracycline repressor protein (tetR(E), ASA_P4G005). Three β-lactamase genes, ampC (ASA_1191), ampS (ASA_4346) and cphA (ASA_3612), previously described in A. sobria (as cepS, ampS and imiS, respectively) [57] are carried on the A449 chromosome (Table 3). The presence of more than 25 genes for multidrug resistance and major facilitator efflux family proteins indicates that A449 carries an array of genes to counteract antimicrobials.
Iron acquisition
Iron acquisition is an important virulence factor for many bacterial pathogens and for A. salmonicida, it may also be a key process for survival in aquatic environments. Mesophilic Aeromonas species have been found to produce two types of catecholate siderophores, amonabactin and enterobactin [58]. When A449 is grown under low iron conditions, either in vivo or in the presence of 2,2'-dipyridyl, three outer membrane proteins are induced that appear to be ferric siderophore or heme receptors [59]. On the A449 chromosome, both of the ferric siderophore receptors are located adjacent to clusters of genes encoding ABC-type ferric transporter subunits as well as non-ribosomal peptide synthetase modules, suggesting complete systems for siderophore synthesis and uptake. The gene for the FstC receptor is located within a cluster (ASA_1838 – ASA_1851) that includes the amonabactin synthesis gene [60], indicating that these genes are likely involved in the synthesis and uptake of amonabactin. The gene for the FstB receptor is encoded in a gene cluster (ASA_4368 – ASA_4380) that is similar to the Listonella anguillarum anguibactin and the Acinetobacter baumannii acinetobactin synthesis genes [61], suggesting that A449 has the ability to synthesize and recapture an anguibactin-like siderophore. Some of the genes in this cluster have been recently characterized in A. salmonicida and shown to be required for siderophore synthesis [62]. Adjacent to this gene cluster are five genes (ASA_4363 – ASA_4367) encoding a hydroxamate-type ferric siderophore receptor and an ABC transporter system, indicating that A449 may also use a hydroxamate siderophore for iron acquisition. The gene for a presumptive heme receptor, hupA (ASA_3328), that is induced by low iron conditions [59], is located near hutZXBCD (ASA_3332 – ASA_ 3336), which encode proteins involved in heme uptake and utilization [63]. Genes for several additional Ton-B dependent outer membrane receptors that may be involved in heme or hemoprotein transport are also present in the A449 genome, but require further characterization to establish their function.
Quorum sensing
Another bacterial process implicated in virulence is quorum sensing [64]. The A449 chromosome contains the luxI and luxR homologs, asaI (ASA_3762) and asaR (ASA_3763), which encode proteins for the synthesis of the acylhomoserine lactone quorum sensing molecule and the transcriptional regulator that responds to it, respectively [65]. In addition, genes for a second quorum sensing pathway that uses autoinducer-2 [66] are present: luxS (ASA_0697) encodes the autoinducer-2 synthase, luxU (ASA_2781) is a putative phosphorelay protein involved in transduction of the signal and luxO (ASA_3295) is a transcriptional response regulator (Table 3). Other unidentified genes in the A449 genome may also participate in these systems since in Vibrio spp. receptor proteins and multiple small RNAs are involved in the complete signal transduction pathway [67].
Comparison to the A. hydrophila genome
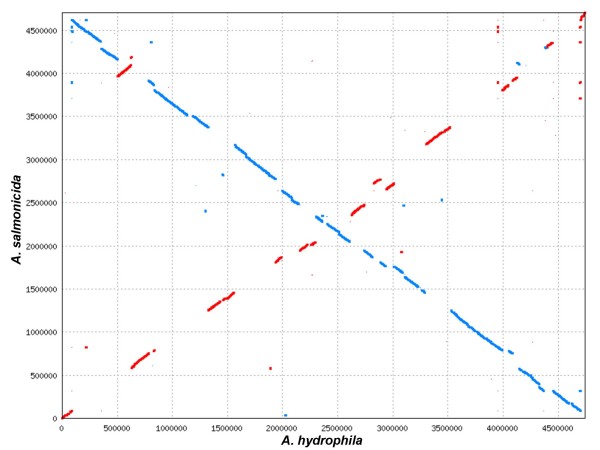
The genome sequence of A. hydrophila ATCC 7966T [22] provides an excellent basis for comparative sequence analysis leading to enhanced understanding of genome evolution within the genus Aeromonas. A comparative analysis of the two chromosomes using Mummer [68] is shown in Fig. 3. Due to an inversion around the origin of replication, the A449 sequence primarily aligns with the A. hydrophila sequence on the reverse strand (blue line in Fig. 3). As expected for two chromosomes of nearly the same size, there are no large gap regions indicative of significant insertions or deletions. Nearly all regions of sequence similarity fall along one of the diagonals, indicating generally similar gene and sequence order. Approximately 15 large sequence inversions (red lines in Fig. 3) around the origin of replication have occurred in the A449 chromosome relative to the A. hydrophila chromosome, accounting for the regions of forward strand alignment. The large inversion already noted in the A449 chromosome, which appears to be an evolutionarily recent change since it is bounded by transposons and is absent in A. hydrophila, stands out as a red line along the blue (reverse strand) diagonal at 500,000 bp in the A. hydrophila sequence.
Figure 3.
Comparison of A. salmonicida and A. hydrophila chromosomes. Mummer comparison of the A. salmonicida and A. hydrophila chromosomes. Red lines/dots indicate forward sequence matches. Blue lines/dots indicate reverse matches.
On a global scale, the A449 and A. hydrophila chromosomes appear generally similar and encode similar numbers of proteins (4086 in A449, 4128 in A. hydrophila). However, there are multiple instances of single gene or operon loss and gain between the two genomes, leading to a 9% difference in gene content. There are 477 coding sequences (CDS) present in the A. salmonicida chromosome that are not found on the A. hydrophila chromosome. Many of these are transposon (101 CDS) or phage related (69 CDS) and 122 represent CDS unique to A. salmonicida. However, there are also 97 conserved hypothetical CDS found in other bacterial species and 88 known CDS that are present in the A. salmonicida genome, but not in that of A. hydrophila. Conversely, the A. hydrophila genome contains 278 CDS not present in A. salmonicida (72 unique CDS, 67 conserved hypothetical CDS and 139 known CDS). Clearly, significant changes in gene content have occurred following the separation of these two species.
Pseudogenes
An additional obvious difference between the A. salmonicida and A. hydrophila genomes is the number of pseudogenes present. The A. hydrophila genome [22] has only 7 pseudogenes: 2 in tRNA genes, 2 protein CDS with in-frame stop codons and 3 frameshifted protein CDS. Only one of these CDS (AHA_2264) is present in A. salmonicida (ASA_2042) and both genes contain the same frameshift. To investigate the frequency and occurrence of frameshifts in the genus Aeromonas, we attempted to amplify and sequence 16 A. salmonicida pseudogenes (Additional file 1) having a variety of lesions from five A. salmonicida strains (two strains of subspecies salmonicida, one each of subspecies masoucida, achromogenes and smithia) and from one strain each of five other Aeromonas species (hydrophila, veronii, caviae, sobria and bestiarum) (Table 4). In addition, these sequences were amplified from A449 cDNA to determine whether transcriptional frameshifting corrected any of them. All the cDNA sequences were identical to the genomic sequence (Table 4 and Additional file 1). While most of the genes could be amplified from the A. salmonicida strains and subspecies, the amplification of genes from the other Aeromonas species was considerably less successful (Table 4 and Additional file 1), presumably due to sequence changes at the primer sites. However, it is clear (Table 4) that in the single species of A. salmonicida subsp. masoucida, achromogenes and smithia that were tested, only 3 or 4 of the pseudogenes are present and that none of the amplified sequences from the other Aeromonas species showed disruptions. While this analysis tests less than 10% of the A449 pseudogenes and although the data are incomplete for many of the non-A. salmonicida species, these pseudogenes appear to be limited to A. salmonicida with the majority present only in A. salmonicida subsp. salmonicida.
Table 4.
Summary of disrupted genes in Aeromonas species
| Species | # genes amplified1 | # disrupted genes | % disrupted |
| A449 | 162 | 16 | 100 |
| A449 cDNA | 16 | 16 | 100 |
| A. salmonicida subsp. salmonicida ATCC 33658T | 15 | 15 | 100 |
| A. salmonicida subsp. salmonicida ATCC 51413 (non-pigmented) | 16 | 16 | 100 |
| A. salmonicida subsp. masoucida ATCC 27013T | 13 | 3 | 23 |
| A. salmonicida subsp. achromogenes ATCC 33659T | 14 | 3 | 21 |
| A. salmonicida subsp. smithia ATCC 49393T | 15 | 4 | 27 |
| A. bestarium ATCC 51108T | 7 | 0 | 0 |
| A. veronii bv. sobria ATCC 9071 | 4 | 0 | 0 |
| A. sobria ATCC 43979T | 2 | 0 | 0 |
| A. caviae ATCC 15468T | 2 | 0 | 0 |
| A. hydrophila ATCC 7966T | 132 | 0 | 0 |
Genomic evidence for pathogen speciation
Analyses of the genomes of bacterial pathogens provide evidence that three key processes, the acquisition of mobile genetic elements, genome rearrangements and gene loss in the process of adapting to a specific host, result in substantial changes in the genomes of pathogens (see [69] for a recent review). Since its separation from the last common ancestor with A. hydrophila, A449 appears to have acquired multiple plasmids, two prophages, a variety of IS elements and a number of individual genes and operons, presumably through horizontal gene transfer. While mechanisms for the acquisition of prophage, plasmids and insertion sequences are understood, mechanisms for the gain of individual genes, such as the B. thuringiensis toxin, or operons, such as VapA and its secretion system, are less obvious. The acquisition of foreign DNA appears to be an ongoing process in A. salmonicida based on the diversity of plasmids identified in various strains [70-72]. IS transposition also continues to be active in A449 since mutants can be generated by IS transposition ([73], JMB unpublished results), usually by growth under stressful conditions such as elevated temperature (30°C).
Substantial genetic rearrangements have occurred in the A449 genome, relative to the A. hydrophila genome. Many of these rearrangements have been assisted by the presence of IS elements, such as the apparent transfer of T6SS genes to pAsa4 and the large inversion. Most of the other inversions, relative to A. hydrophila, do not appear to be associated with IS elements, indicating that other mechanisms are also generating genetic rearrangements. While these large scale rearrangements do not obviously affect gene sequences, reorientation of large regions relative to the origin of replication may impact the regulation of gene expression.
The third trait of recently evolved pathogen genomes, gene loss or decay, has also occurred frequently in the A449 genome. The number of A449 pseudogenes, 170, is comparable to that seen in Yersinia pestis (~150) [74], but less than other recently evolved human pathogens such as Salmonella enterica serovar Typhi CT18 (>200) [75] or Mycobacterium leprae (>1100) [76]. Several of the A449 pseudogenes prevent the expression of cell surface structures such as flagella and pili. Loss of flagellar motility is common among recently emerged pathogens [69], perhaps as a means to evade the host innate immune system. Loss of genes for type IV pili is also associated with pathogen speciation [77] and may also help pathogens avoid innate immune responses [78]. Significant accumulations of pseudogenes are also found among genes for transcriptional regulators (17 pseudogenes), genes encoding carbohydrate synthesis and modification enzymes (12 pseudogenes) and genes for basic metabolic enzymes (e.g., sulfite reductase, α and β galactosidase, acetolactate synthase, etc.) (10 pseudogenes). Compared to A. hydrophila ATCC 7966T, the "jack of all trades" [22], the accumulation of pseudogenes in A449 has considerably reduced its capacity to produce some organelles (e.g., pili or flagella) and to synthesize some enzymes and their products.
The A449 genome thus carries all the hallmarks of an organism that has undergone adaptation to a specific host. Clearly, substantial horizontal gene transfer, genome rearrangements and gene decay have occurred in A449 relative to A. hydrophila ATCC 7966T. The small survey of pseudogenes in other members of the genus Aeromonas suggests that pseudogene accumulation coincided with the speciation of A. salmonicida but increased substantially during the evolution of the subspecies salmonicida. Further analysis of Aeromonas sequences and genomes should provide insights into the process and timing of the evolution of host specialization as well as a better understanding of the genes and proteins involved in virulence.
Conclusion
The genome of A. salmonicida subsp. salmonicida A449 consists of a circular chromosome and five plasmids that encode more than 4700 genes. A large number of genes encoding potential virulence factors have been identified, although a number of them have been disrupted to become pseudogenes. The acquisition of plasmids, insertion sequences and pseudogenes, along with large genome rearrangements is indicative of a genome that has decayed to adapt to the environment of a specific host.
Methods
Bacterial Strains
Aeromonas salmonicida subsp. salmonicida A449 was originally isolated from a brown trout in the Eure river, France by Christian Michel in 1975 [79]. Other Aeromonas strains were obtained from the American Type Culture Collection (ATCC): A. salmonicida subsp. salmonicida ATCC 33658T, A. salmonicida subsp. salmonicida ATCC 51413 (non-pigmented), A. salmonicida subsp. masoucida ATCC 27013T, A. salmonicida subsp. achromogenes ATCC 33659T, A. salmonicida subsp. smithia ATCC 49393T, A. bestarium ATCC 51108T, A. veronii bv. sobria ATCC 9071, A. sobria ATCC 43979T, A. caviae ATCC 15468T and A. hydrophila ATCC 7966T.
Genome Sequencing
A mixed strategy was employed for sequencing the genome of A449. A shotgun library was generated by cloning hydro-sheared and end-repaired 1–2 kb genomic inserts into the plasmid vector pUC19. Clones from this library were sequenced [80] from both direction on Li-Cor 4200 and MegaBace 1000 instruments. As well, a BAC library was constructed by partial digestion of genomic DNA with EcoRI and cloning in pBACe3.6 [81]. Twelve clones from this library were sequenced completely. All reads were assembled in gap4 [82] to produce ~2100 contigs with approximately 6× coverage. Contigs were joined using a read-pair approach as well as a two-step PCR-based approach involving two primers at the contig ends and a random primer. For contig closure, a fosmid library was made in the EpiFOS vector (Epicentre Biotechnologies) and clones were end-sequenced to locate their position in the assembly. Sequence from these clones was used for confirming assembly as well as to fill the remaining gaps. Finally, the sequence was completely disambiguated and polished by sequencing genomic PCR products generated with flanking primer pairs. Presumptive plasmid contigs were identified by similarity to common plasmid encoded genes, removed from the main assembly and joined by PCR experiments using primers at the contig ends. pAsa4 was cloned into E. coli DH5α by transformation with a plasmid DNA preparation from A449 and selection on chloramphenicol. This clone was used to identify pAsa4 contigs and to join and polish the sequence. The A449 chromosome and plasmid 4 and 5 sequences have been deposited in Genbank (NC_009348, NC_009439, NC_009350).
Annotation
Initial analysis of the genome sequences was done using a script written in Perl and relying heavily on the BioPerl modules [83]. The script initially searched for rRNA and transposon sequences using Blastn [84] followed by a tRNA search using tRNAscan-SE [85]. sRNA sequences were also identified with rfam_scan.pl which uses Blast and INFERNAL [86] searches of the Rfam database [87]. Open reading frames were identified with Glimmer2 [88] and searched for similarity using Blastp and for conserved domains with CDD [89]. Sequences between open reading frames with Blastp or CDD hits were extracted and further searched with Blastx and Blastn. All search results were assembled in an EMBL feature table file for editing in Artemis [90]. Final annotation was done by hand in Artemis. Chromosome and large plasmid representations were produced using the Genome Atlas website [91]. Comparisons between the A. salmonicida and A. hydrophila chromosomes used the Mummer package [68].
Frameshift Analysis
To investigate the presence of frameshifts in other Aeromonas species and subspecies, primers flanking frameshift sites (Additional file 2) were designed with Primer 3 [92]. Aeromonas species and subspecies were grown in tryptic soy broth and DNA was extracted for use as the template in standard PCR reactions. PCR products were gel purified and sequenced directly. Unsuccessful amplifications were attempted at least two more times using a lower annealing temperature. RNA extraction and cDNA synthesis were as described previously [35]. Sequences were deposited in Genbank under accession numbers FJ178190–FJ178298.
Authors' contributions
MER coordinated the project, edited and assembled sequence, annotated the sequence, conceived of the frameshift experiments, carried out the A. hydrophila genome comparison and wrote and edited the manuscript. RKS made the shotgun library, directed the DNA sequencing, designed and directed joining approaches and polished the sequence. BC edited and assembled the sequence and contributed bioinformatic analyses. JMB cloned pAsa4, contributed to annotation and edited the manuscript. AB performed and analyzed joining experiments. JK designed and implemented software for tracking clones and sequences. JM constructed and characterized the BAC library. CM performed and analyzed assembly PCRs and contributed to plasmid sequences. DS designed, performed and analyzed the frameshift PCR experiments. JW characterized and analyzed BAC clones and sequences. JHEN designed and directed joining approaches. SCJ and LLB co-led the conception and design of the host-pathogen interaction program and contributed to the writing and editing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Additional Table 1. Details of the analysis of disrupted genes in Aeromonas species.
Additional Table 2. Primers for pseudogene amplification.
Acknowledgments
Acknowledgements
We would like to thank the GHI Sequencing Platform (D. Barrington, J. Burbidge, R. Easy, J. Heal, B. Higgins, C. Kozera, A. Lewis, A. Maffey, B. Parsons, G. Simpson, C. Stone, H. Verheul) for sequencing services, The Atlantic Genome Centre for fosmid library construction and additional sequencing, E. Egbosimba for help with joining, K. Doody for help with frameshift PCRs, S. Penny and Y. Sebastian for bioinformatic assistance and K. Soanes for a critical review of the manuscript. Bioinformatics was carried out on the IMB Research Informatics infrastructure. This work was supported by the NRC Genomics and Health Initiative. This is NRCC publication 2007-42766.
Contributor Information
Michael E Reith, Email: michael.reith@nrc-cnrc.gc.ca.
Rama K Singh, Email: rama.singh@nrc-cnrc.gc.ca.
Bruce Curtis, Email: bcurtis@genomeatlantic.ca.
Jessica M Boyd, Email: jessica.boyd24@gmail.com.
Anne Bouevitch, Email: anne.bouevitch@dnagenotek.com.
Jennifer Kimball, Email: jennifer.kimball@nrc-cnrc.gc.ca.
Janet Munholland, Email: janet.munholland@nrc-cnrc.gc.ca.
Colleen Murphy, Email: colleen.murphy@nrc-cnrc.gc.ca.
Darren Sarty, Email: darren.sarty@nrc-cnrc.gc.ca.
Jason Williams, Email: jason.williams@nrc-cnrc.gc.ca.
John HE Nash, Email: john.nash@phac-aspc.gc.ca.
Stewart C Johnson, Email: stewart.johnson@dfo-mpo.gc.ca.
Laura L Brown, Email: laura.l.brown@dfo-mpo.gc.ca.
References
- Figueras MJ. Clinical relevance of Aeromonas. Rev Med Microbiol. 2005;16:145–153. [Google Scholar]
- Bernoth EM. Furunculosis: the history of the disease and of disease research. In: Bernoth EM, Ellis AE, Midtlyng P, Olivier G, Smith P, editor. Furunculosis – Multidisciplinary Fish Disease Research. London: Academic Press; 1997. pp. 1–20. [Google Scholar]
- Hiney M, Olivier G. Furunculosis (Aeromonas salmonicida) In: Woo PTK, Bruno DW, editor. Fish Diseases and Disorders III: Viral, Bacterial and Fungal Infections. Oxford: CAB Publishing; 1999. pp. 341–425. [Google Scholar]
- Wiklund T, Dalsgaard I. Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis Aquat Organ. 1998;32:49–69. doi: 10.3354/dao032049. [DOI] [PubMed] [Google Scholar]
- Joseph SW, Carnahan A. The isolation, identification, and systematics of the motile Aeromonas species. Ann Rev Fish Dis. 1994;4:315–343. [Google Scholar]
- Pasquale V, Baloda SB, Dumontet S, Krovacek K. An outbreak of Aeromonas hydrophila infection in turtles (Pseudemis scripta) Appl Environ Microbiol. 1994;60:1678–1680. doi: 10.1128/aem.60.5.1678-1680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane SM, Gifford DH. Prevalence and pathogenicity of Aeromonas hydrophila. Avian Dis. 1985;29:681–689. [PubMed] [Google Scholar]
- Kueh CS, Chan KY. Bacteria in bivalve shellfish with special reference to the oyster. J Appl Bacteriol. 1985;59:41–47. doi: 10.1111/j.1365-2672.1985.tb01773.x. [DOI] [PubMed] [Google Scholar]
- Santavy DL, Willenz P, Colwell RR. Phenotypic study of bacteria associated with the Caribbean sclerosponge, Ceratoporella nicholsoni. Appl Environ Microbiol. 1990;56:1750–1762. doi: 10.1128/aem.56.6.1750-1762.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Brune A, Zimmer M. Bacterial symbionts in the hepatopancreas of isopods: diversity and environmental transmission. FEMS Microbiol Ecol. 2007;61:141–152. doi: 10.1111/j.1574-6941.2007.00329.x. [DOI] [PubMed] [Google Scholar]
- Daly JG, Kew AK, Moore AR, Olivier G. The cell surface of Aeromonas salmonicida determines in vitro survival in cultured brook trout (Salvelinus fontinalis) peritoneal macrophages. Microb Pathog. 1996;21:447–461. doi: 10.1006/mpat.1996.0075. [DOI] [PubMed] [Google Scholar]
- Martin-Carnahan A, Joseph SW. Aeromonadaceae. In: Garrity GM, editor. Bergey's manual of systematic bacteriology. 2. Vol. 2. New York, NY: Springer-Verlag; 2005. pp. 556–580. [Google Scholar]
- Austin B, Austin DA, Dalsgaard I, Gudmundsdottir BK, Hoie S, Thornton JM, Larsen JL, O'Hici B, Powell R. Characterization of atypical Aeromonas salmonicida by different methods. Syst Appl Microbiol. 1998;21:50–64. doi: 10.1016/s0723-2020(98)80008-8. [DOI] [PubMed] [Google Scholar]
- Bohm KH, Fuhrmann H, Schlotfeldt HJ, Korting W. Aeromonas salmonicida from salmonids and cyprinids – serological and cultural identification. Zentralbl Veterinarmed B. 1986;33:777–783. doi: 10.1111/j.1439-0450.1986.tb00099.x. [DOI] [PubMed] [Google Scholar]
- Dalsgaard I, Gudmundsdottir BK, Helgason S, Hoie S, Thoresen OF, Wichardt UP, Wiklund T. Identification of atypical Aeromonas salmonicida: inter-laboratory evaluation and harmonization of methods. J Appl Microbiol. 1998;84:999–1006. doi: 10.1046/j.1365-2672.1998.00435.x. [DOI] [PubMed] [Google Scholar]
- Dalsgaard I, Nielsen B, Larsen JL. Characterization of Aeromonas salmonicida subsp. salmonicida: a comparative study of strains of different geographic origin. J Appl Bacteriol. 1994;77:21–30. doi: 10.1111/j.1365-2672.1994.tb03039.x. [DOI] [PubMed] [Google Scholar]
- Garcia JA, Larsen JL, Dalsgaard I, Pedersen K. Pulsed-field gel electrophoresis analysis of Aeromonas salmonicida ssp. salmonicida. FEMS Microbiol Lett. 2000;190:163–166. doi: 10.1111/j.1574-6968.2000.tb09280.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Murcia AJ, Soler L, Saavedra MJ, Chacon MR, Guarro J, Stackebrandt E, Figueras MJ. Phenotypic, genotypic, and phylogenetic discrepancies to differentiate Aeromonas salmonicida from Aeromonas bestiarum. Int Microbiol. 2005;8:259–269. [PubMed] [Google Scholar]
- Nash JH, Findlay WA, Luebbert CC, Mykytczuk OL, Foote SJ, Taboada EN, Carrillo CD, Boyd JM, Colquhoun DJ, Reith ME, Brown LL. Comparative genomics profiling of clinical isolates of Aeromonas salmonicida using DNA microarrays. BMC Genomics. 2006;7:43. doi: 10.1186/1471-2164-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu X, Dacanay A, Harrison BA, Fast M, Colquhoun DJ, Lund V, Brown LL, Li J, Altman E. Carbohydrate analysis and serological classification of typical and atypical isolates of Aeromonas salmonicida: a rationale for the lipopolysaccharide-based classification of A. salmonicida. Fish Shellfish Immunol. 2007;23:1095–1106. doi: 10.1016/j.fsi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Boyd J, Williams J, Curtis B, Kozera C, Singh R, Reith M. Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid. 2003;50:131–144. doi: 10.1016/s0147-619x(03)00058-1. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol. 2006;188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umelo E, Trust TJ. Physical map of the chromosome of Aeromonas salmonicida and genomic comparisons between Aeromonas strains. Microbiology. 1998;144:2141–2149. doi: 10.1099/00221287-144-8-2141. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Lawrence JG. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol Microbiol. 2007;64:42–56. doi: 10.1111/j.1365-2958.2007.05596.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson WB, Gudkovs N, Strom MS. Atypical strains of Aeromonas salmonicida contain multiple copies of insertion element ISAsa4 useful as a genetic marker and a target for PCR assay. Dis Aquat Organ. 2006;70:209–217. doi: 10.3354/dao070209. [DOI] [PubMed] [Google Scholar]
- Liebert CA, Hall RM, Summers AO. Transposon Tn21, Flagship of the Floating Genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr SE, Pugovkin D, Wahli T, Segner H, Frey J. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology. 2005;151:2111–2118. doi: 10.1099/mic.0.27926-0. [DOI] [PubMed] [Google Scholar]
- Dacanay A, Knickle L, Solanky KS, Boyd JM, Walter JA, Brown LL, Johnson SC, Reith M. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology. 2006;152:1847–1856. doi: 10.1099/mic.0.28768-0. [DOI] [PubMed] [Google Scholar]
- Burr SE, Stuber K, Wahli T, Frey J. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J Bacteriol. 2002;184:5966–5970. doi: 10.1128/JB.184.21.5966-5970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches S, Urgell C, Merino S, Chacon MR, Soler L, Castro-Escarpulli G, Figueras MJ, Tomas JM. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl Environ Microbiol. 2004;70:6914–6919. doi: 10.1128/AEM.70.11.6914-6919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HB, Rao PS, Lee HC, Vilches S, Merino S, Tomas JM, Leung KY. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect Immun. 2004;72:1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J, Pillai L, Fadl AA, Galindo CL, Erova TE, Chopra AK. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect Immun. 2005;73:6446–6457. doi: 10.1128/IAI.73.10.6446-6457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebanks RO, Knickle LC, Goguen M, Boyd JM, Pinto DM, Reith M, Ross NW. Expression of and secretion through the Aeromonas salmonicida type III secretion system. Microbiology. 2006;152:1275–1286. doi: 10.1099/mic.0.28485-0. [DOI] [PubMed] [Google Scholar]
- Burr SE, Stuber K, Frey J. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J Bacteriol. 2003;185:6583–6591. doi: 10.1128/JB.185.22.6583-6591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr D, Burr SE, Gibert M, d'Alayer J, Frey J, Popoff MR. Aeromonas exoenzyme T of Aeromonas salmonicida is a bifunctional protein that targets the host cytoskeleton. J Biol Chem. 2007;282:28843–28852. doi: 10.1074/jbc.M704797200. [DOI] [PubMed] [Google Scholar]
- Fehr D, Casanova C, Liverman A, Blazkova H, Orth K, Dobbelaere D, Frey J, Burr SE. AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology. 2006;152:2809–2818. doi: 10.1099/mic.0.28889-0. [DOI] [PubMed] [Google Scholar]
- Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog. 2008;44:344–361. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay WW, Buckley JT, Ishiguro EE, Phipps BM, Monette JP, Trust TJ. Purification and disposition of a surface protein associated with virulence of Aeromonas salmonicida. J Bacteriol. 1981;147:1077–1084. doi: 10.1128/jb.147.3.1077-1084.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro EE, Kay WW, Ainsworth T, Chamberlain JB, Austen RA, Buckley JT, Trust TJ. Loss of virulence during culture of Aeromonas salmonicida at high temperature. J Bacteriol. 1981;148:333–340. doi: 10.1128/jb.148.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano RC, Bertolini J. Selection for virulence in the fish pathogen Aeromonas salmonicida, using Coomassie Brilliant Blue agar. J Wildl Dis. 1988;24:672–678. doi: 10.7589/0090-3558-24.4.672. [DOI] [PubMed] [Google Scholar]
- Thornton JC, Garduno RA, Newman SG, Kay WW. Surface-disorganized, attenuated mutants of Aeromonas salmonicida as furunculosis live vaccines. Microb Pathog. 1991;11:85–99. doi: 10.1016/0882-4010(91)90002-r. [DOI] [PubMed] [Google Scholar]
- Noonan B, Trust TJ. Molecular analysis of an A-protein secretion mutant of Aeromonas salmonicida reveals a surface layer-specific protein secretion pathway. J Mol Biol. 1995;248:316–327. doi: 10.1016/s0022-2836(95)80053-0. [DOI] [PubMed] [Google Scholar]
- Merino S, Gavin R, Vilches S, Shaw JG, Tomas JM. A colonization factor (production of lateral flagella) of mesophilic Aeromonas spp. is inactive in Aeromonas salmonicida strains. Appl Environ Microbiol. 2003;69:663–667. doi: 10.1128/AEM.69.1.663-667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals R, Ramirez S, Vilches S, Horsburgh G, Shaw JG, Tomas JM, Merino S. Polar flagellum biogenesis in Aeromonas hydrophila. J Bacteriol. 2006;188:542–555. doi: 10.1128/JB.188.2.542-555.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Dacanay A, Knickle LC, Touhami A, Brown LL, Jericho MH, Johnson SC, Reith M. Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.) Infect Immun. 2008;76:1445–1455. doi: 10.1128/IAI.01019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheimer AW, Avigad LS. Partial characterization of aerolysin, a lytic exotoxin from Aeromonas hydrophila. Infect Immun. 1974;9:1016–1021. doi: 10.1128/iai.9.6.1016-1021.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lally ET, Hill RB, Kieba IR, Korostoff J. The interaction between RTX toxins and target cells. Trends Microbiol. 1999;7:356–361. doi: 10.1016/s0966-842x(99)01530-9. [DOI] [PubMed] [Google Scholar]
- Baudry B, Fasano A, Ketley J, Kaper JB. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60:428–434. doi: 10.1128/iai.60.2.428-434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vipond R, Bricknell IR, Durant E, Bowden TJ, Ellis AE, Smith M, MacIntyre S. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect Immun. 1998;66:1990–1998. doi: 10.1128/iai.66.5.1990-1998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai L, Sha J, Erova TE, Fadl AA, Khajanchi BK, Chopra AK. Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect Immun. 2006;74:3742–3755. doi: 10.1128/IAI.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnlaugsdottir B, Gudmundsdottir BK. Pathogenicity of atypical Aeromonas salmonicida in Atlantic salmon compared with protease production. J Appl Microbiol. 1997;83:542–551. doi: 10.1046/j.1365-2672.1997.00247.x. [DOI] [PubMed] [Google Scholar]
- Merino S, Aguilar A, Nogueras MM, Regue M, Swift S, Tomas JM. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect Immun. 1999;67:4008–4013. doi: 10.1128/iai.67.8.4008-4013.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahagirdar R, Howard SP. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J Bacteriol. 1994;176:6819–6826. doi: 10.1128/jb.176.22.6819-6826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev AV, MacIntyre S. Cloning and study of the genetic organization of the exe gene cluster of Aeromonas salmonicida. Gene. 1995;158:77–82. doi: 10.1016/0378-1119(95)00139-w. [DOI] [PubMed] [Google Scholar]
- Walsh TR, Payne DJ, MacGowan AP, Bennett PM. A clinical isolate of Aeromonas sobria with three chromosomally mediated inducible {beta}-lactamases: a cephalosporinase, a penicillinase and a third enzyme, displaying carbapenemase activity. J Antimicrob Chemother. 1995;35:271–279. doi: 10.1093/jac/35.2.271. [DOI] [PubMed] [Google Scholar]
- Massad G, Arceneaux JE, Byers BR. Acquisition of iron from host sources by mesophilic Aeromonas species. J Gen Microbiol. 1991;137:237–241. doi: 10.1099/00221287-137-2-237. [DOI] [PubMed] [Google Scholar]
- Ebanks RO, Dacanay A, Goguen M, Pinto DM, Ross NW. Differential proteomic analysis of Aeromonas salmonicida outer membrane proteins in response to low iron and in vivo growth conditions. Proteomics. 2004;4:1074–1085. doi: 10.1002/pmic.200300664. [DOI] [PubMed] [Google Scholar]
- Barghouthi S, Payne SM, Arceneaux JE, Byers BR. Cloning, mutagenesis, and nucleotide sequence of a siderophore biosynthetic gene (amoA) from Aeromonas hydrophila. J Bacteriol. 1991;173:5121–5128. doi: 10.1128/jb.173.16.5121-5128.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey CW, Tomaras AP, Connerly PL, Tolmasky ME, Crosa JH, Actis LA. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology. 2004;150:3657–3667. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- Najimi M, Lemos ML, Osorio CR. Identification of siderophore biosynthesis genes essential for growth of Aeromonas salmonicida under iron limitation conditions. Appl Environ Microbiol. 2008;74:2341–2348. doi: 10.1128/AEM.02728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najimi M, Lemos ML, Osorio CR. Identification of heme uptake genes in the fish pathogen Aeromonas salmonicida subsp. salmonicida. Arch Microbiol. 2008 doi: 10.1007/s00203-008-0391-5. [DOI] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S, Karlyshev AV, Fish L, Durant EL, Winson MK, Chhabra SR, Williams P, Macintyre S, Stewart GS. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449:835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- Casas C, Anderson EC, Ojo KK, Keith I, Whelan D, Rainnie D, Roberts MC. Characterization of pRAS1-like plasmids from atypical North American psychrophilic Aeromonas salmonicida. FEMS Microbiol Lett. 2005;242:59–63. doi: 10.1016/j.femsle.2004.10.039. [DOI] [PubMed] [Google Scholar]
- Sorum H, L'Abee-Lund TM, Solberg A, Wold A. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob Agents Chemother. 2003;47:1285–1290. doi: 10.1128/AAC.47.4.1285-1290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Abee-Lund TM, Sorum H. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid. 2002;47:172–181. doi: 10.1016/s0147-619x(02)00001-x. [DOI] [PubMed] [Google Scholar]
- Gustafson CE, Chu S, Trust TJ. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–463. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Cummings CA, Brinig MM, Lepp PW, Pas S van de, Relman DA. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol. 2004;186:1484–1492. doi: 10.1128/JB.186.5.1484-1492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Yamamoto M, Akira S, Beutler B, Svanborg C. Mechanism of pathogen-specific TLR4 activation in the mucosa: Fimbriae, recognition receptors and adaptor protein selection. Eur J Immunol. 2006;36:267–277. doi: 10.1002/eji.200535149. [DOI] [PubMed] [Google Scholar]
- Michel C. Furunculosis of salmonids: vaccination attempts in rainbow trout (Salmo gairdneri) by formalin-killed germs. Ann Rech Vet. 1979;10:33–40. [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Oost J Van der. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics. 1999;58:250–253. doi: 10.1006/geno.1998.5693. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehvaslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Computational analysis of RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:117–128. doi: 10.1101/sqb.2006.71.003. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Hallin PF, Ussery DW. CBS Genome Atlas Database: a dynamic storage for bioinformatic results and sequence data. Bioinformatics. 2004;20:3682–3686. doi: 10.1093/bioinformatics/bth423. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Masada CL, LaPatra SE, Morton AW, Strom MS. An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss. Dis Aquat Organ. 2002;51:13–25. doi: 10.3354/dao051013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Table 1. Details of the analysis of disrupted genes in Aeromonas species.
Additional Table 2. Primers for pseudogene amplification.