Abstract
Galectin-1 is an anti-inflammatory lectin with pleiotropic regulatory functions at the crossroads of innate and adaptive immunity. It is expressed in immune privileged sites and is implicated in establishing maternal–fetal immune tolerance, which is essential for successful pregnancy in eutherian mammals. Here, we show conserved placental localization of galectin-1 in primates and its predominant expression in maternal decidua. Phylogenetic footprinting and shadowing unveil conserved cis motifs, including an estrogen responsive element in the 5′ promoter of LGALS1, that were gained during the emergence of placental mammals and could account for sex steroid regulation of LGALS1 expression, thus providing additional evidence for the role of galectin-1 in immune–endocrine cross-talk. Maximum parsimony and maximum likelihood analyses of 27 publicly available vertebrate and seven newly sequenced primate LGALS1 coding sequences reveal that intense purifying selection has been acting on residues in the carbohydrate recognition domain and dimerization interface that are involved in immune functions. Parsimony- and codon model-based phylogenetic analysis of coding sequences show that amino acid replacements occurred in early mammalian evolution on key residues, including gain of cysteines, which regulate immune functions by redox status-mediated conformational changes that disable sugar binding and dimerization, and that the acquired immunoregulatory functions of galectin-1 then became highly conserved in eutherian lineages, suggesting the emergence of hormonal and redox regulation of galectin-1 in placental mammals may be implicated in maternal–fetal immune tolerance.
Keywords: decidua, estrogen, glycocode, immune–endocrine cross-talk, pregnancy
The success of mammalian pregnancy, in which the developing fetus and mother exchange nutrients, gases, and other molecules via the chorioallantoic placenta, requires maternal immune tolerance to fetal allo-antigens (1–4). This tolerance presumably prevents the occurrence of exaggerated inflammation at the implantation site and reduces the danger of destructive immune attacks on the fetus, a danger that the first mammals with an invasive placenta would have faced (5, 6). It seems likely that mechanisms for immune tolerance to invasive placentation were already functioning in the early placental mammals and that immunoregulatory molecules, which had existed before the mammalian placenta evolved, were incorporated in this tolerance and have undergone evolutionary modifications coincident with the emergence of the mammalian placenta. Here, we present evidence that such modifications occurred in a key immunoregulatory molecule, galectin-1.
Molecules that have been implicated in conferring maternal–fetal immune tolerance include galectin-1, B7 proteins, Crry, Fas ligand, HLA-G, indoleamine 2,3-dioxygenase, and killer cell immunoglobulin-like receptors (2–4, 7–11). These proteins are involved in pathways regulating adaptive or innate immune responses at the maternal–fetal interface, and their disruption may lead to pregnancy complications, such as miscarriage, fetal loss, and preeclampsia (2, 8–12). Galectin-1 is a member of a conserved lectin family that regulates immune responses by deciphering the high-density glycocode of cell surface glycans (13–16). Galectin-1 triggers apoptosis of activated T cells and suppresses T cell-mediated autoimmune diseases, such as collagen-induced arthritis in mice (17, 18). Its up-regulation in tumors correlates with poor clinical outcome, possibly because galectin-1 reduces the survival of tumor resident T cells and promotes tumor immune escape (19). In addition, galectin-1 has been implicated in transplantation tolerance because it diminishes morbidity and mortality of graft-versus-host disease in a mouse allogeneic bone marrow transplantation model (20).
Galectin-1 is up-regulated in uterine natural killer cells and is a key moderator of regulatory T cell functions (21, 22). A recent study reported that galectin-1 induces the generation of tolerogenic dendritic cells and regulatory T cells in mice, and the knockout of LGALS1 led to a higher rate of fetal loss in allogeneic mating (9). Additionally, this study found that galectin-1 is involved in immune–endocrine cross-talk, maternal decidual expression of galectin-1 is regulated by progesterone, and galectin-1 increases progesterone concentrations, suggesting its central role in the maintenance of pregnancy. Moreover, estrogen also regulates uterine LGALS1 expression in mice and humans (23, 24).
In light of these findings, we examined the molecular evolution of LGALS1 coding sequences and regulatory elements in a wide range of mammals and outgroup vertebrates. We also determined LGALS1's placental expression pattern in the main clades of primates. The analysis of the data focused on testing the hypothesis that galectin-1 would show functionally significant evolutionary modifications early in placental mammalian phylogeny and then would be highly conserved in the descendent mammalian species. The experimental and inferential evidence presented herein indicate that indeed functionally important changes likely involved in the emergence of maternal–fetal immune tolerance were gained and then conserved in placental mammals.
Results
Galectin-1 Expression in Placentas and Membranes.
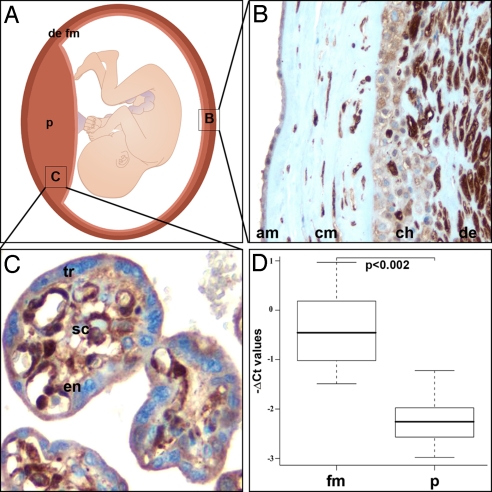
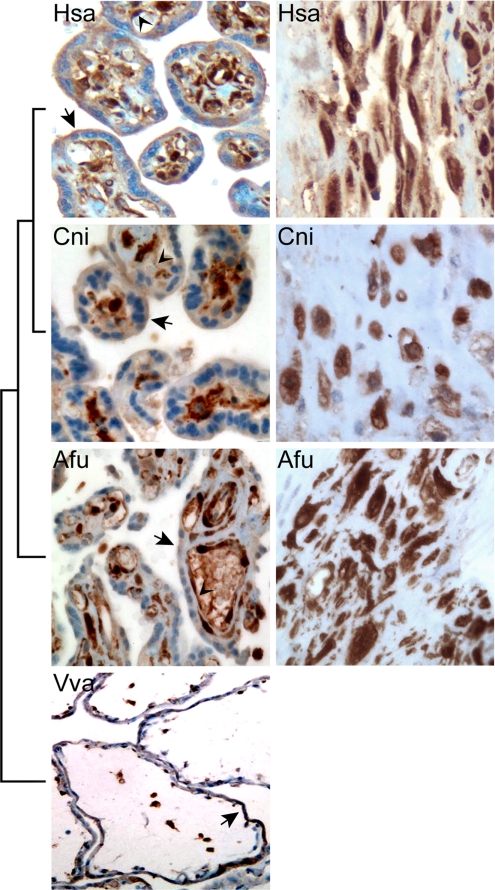
Galectin-1 is abundantly expressed by the placenta and the extraembryonic membranes (Fig. 1 A–C); however, its highest expression was detected in maternal decidual stromal cells (Fig. 1C). Indeed, LGALS1 expression was 3.6 fold higher in membranes containing decidua than in placentas of the same patients (P < 0.002; Fig. 1D). We detected a similar immunostaining pattern of galectin-1 in a Cercopithecus nictitans (Old World monkey) and an Ateles fusciceps (New World monkey) placenta [taxon names are listed in supporting information (SI) Dataset S1]. Trophoblasts and stromal cells were also galectin-1-immunopositive in a Varecia variegata (strepsirrhine primate) placenta. The strongest galectin-1 staining was detected in decidual cells in the basal plates of these placentas (Fig. 2). In agreement, in silico analysis of 298 human tissues and cell lines represented in the SAGE Genie database revealed that LGALS1 is most abundantly expressed in the endometrium (Dataset S2).
Fig. 1.
Galectin-1 immunostaining and LGALS1 expression in normal-term placenta and extraembryonic membranes. (A) Galectin-1 expression, shown with shades of brown, is strong in the placenta (p) and fetal membranes (fm), and the most intense in maternal decidua (de). (B) All layers of the fetal membranes [amnion (am); chorioamniotic mesenchyma (cm); chorion (ch)] stained for galectin-1; maternal decidua (de) is the richest source of galectin-1. (C) All placental cell types (trophoblasts [tr], stromal cells [sc], villous endothelium [en]) stained for galectin-1. Immunohistochemistry, hematoxylin counterstain, magnification ×200 (B) and ×400 (C). (D) LGALS1 expression was 3.6-fold higher in the membranes including maternal decidua (n = 6) than in placentas (n = 6) of the same patients. Boxes represent medians and interquartile ranges, whereas whiskers represent the most extreme data points.
Fig. 2.
Galectin-1 immunostaining in term primate placentas. Left: Phylogenetic tree depicting the evolutionary relationships among sampled primates. Middle: In Homo sapiens (Hsa), C. nictitans (Cni), and A. fusciceps (Afu) placentas, galectin-1 was detected in trophoblasts, stromal cells, and villous endothelium (arrowheads). The syncytiotrophoblast apical membrane (arrows) and the villous stroma was also galectin-1-positive. Trophoblasts (arrow) and stromal cells were galectin-1 positive in a V. variegata (Vva) placenta. Right: Maternal decidual cells in the basal plates of these placentas were strongly galectin-1 positive. [Immunohistochemistry with hematoxylin counterstain, magnification ×200 (Left) and ×400 (Right)].
Conservation of the LGALS1 Gene.
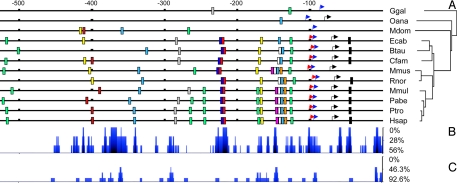
Multiple sequence alignment of 27 publicly available vertebrate LGALS1 sequences plus primate sequences determined by the present study (Dataset S1) showed that LGALS1 has a conserved 4-exon structure in all investigated taxa. The coding sequences including indels spans 408 bp; the lengths of the coding regions in the aligned exons were 9 bp, 80 bp, 172 bp, and 147 bp, respectively. The Pan troglodytes, Pongo abelii, and Rattus norvegicus promoter sequences and the Callithrix jacchus, Microcebus murinus, Tupaia belangeri, Sorex araneus, Felis catus, Monodelphis domestica, Xenopus tropicalis, and Ambystoma mexicanum coding sequences were briefly disrupted by Ns, probably as a result of assembly gaps and/or sequencing errors. The individual Macaca mulatta sequence we obtained differs from the draft genome sequence by a synonymous and non-synonymous difference in nucleotide positions 78 and 79, resulting in a codon for Asp instead of Tyr. Pair-wise p-distance comparison and phylogenetic shadowing of the 2-kb 5′ promoters and first exons revealed the highest conservation in the proximal 500 bp in 12 investigated vertebrates (Fig. 3 and Dataset S3).
Fig. 3.
Phylogenetic footprinting and shadowing of LGALS1 5′ promoter and exon 1. (A) A conserved, non-canonical TATA box (blue arrow) is present in all vertebrates, an overlapping, consensus Inr (red arrow), an adjacent SP1 (green boxes) motif, and an alternative transcriptional start site (black arrow) in all placental mammals. LGALS1 directs the transcription of two distinct-length mRNAs encoding for the same protein in humans and mice, and the transcription initiation for both transcripts is mediated by the Inr, TATA box, and SP1 binding site, the latter of which is crucial for the basal LGALS1 expression (35–38). From the conserved cis elements (AP-2, yellow boxes; AP-4, black boxes, CAAT box-binding protein, white boxes; CAC-binding protein, gray boxes; C/EBP-α, pink boxes; c-ETS-2, brown boxes; ERE, red boxes; HSF-1, orange boxes; NF-1, light blue boxes, NF-Y, dark blue boxes; and SP1, green boxes), 10 have been gained on the stem of placental mammals. Positions relative to the translational start codon are shown at the top. (B) Phylogenetic shadowing in nine placental mammals and (C) in 12 vertebrates demonstrates the extent of conservation of these cis elements. The x-axis shows the positions along the human promoter in the same scale as in A and the y-axis shows the percentage of difference in blue based on a 5-bp sliding window.
Conserved cis Elements in the 5′ Promoter.
Phylogenetic footprinting identifies a non-canonical TATA box in 12 investigated vertebrates as well as an overlapping consensus Inr, an adjacent SP1 motif, and an alternative transcriptional start site in nine placental mammals (Fig. 3). Phylogenetic analysis of 12 vertebrates reveals 32 gains and five losses of cis elements (Fig. S1). The stem of placental mammals gained 10 cis elements, including a highly conserved activator protein (AP)-4, a half estrogen response element (ERE), an overlapping nuclear transcription factor (NF)-Y, and an adjacent AP-2 binding motif. There were six gains and one loss of cis elements on the primate stem (Fig. S1 and Dataset S4). There were significantly more cis element gains and losses per million years of evolutionary time on the placental versus the therian stem (Fisher's exact test, P = 0.009).
Phylogenetic History of Galectin-1 Coding Sequences.
The phylogenetic tree from 34 coding nucleotide sequences was inconsistent with vertebrate phylogeny. The eight most parsimonious trees each have a length of 1,061 steps (Fig. S2). In contrast, the presumed phylogenetic species tree has a length of 1,090 steps and is a significantly worse fit for the data (Kishino-Hasagawa test, P = 0.005; Templeton test, P = 0.013). These differences suggest a complex pattern of LGALS1 evolution with the possibility of independent gene gains and losses on different lineages. Therefore, we used both gene and species trees for reconstructing character evolution and for examining the effects of natural selection.
In the 3ω model, ω varies among placental mammals, the placental stem lineage, and outgroups. Codeml reconstructed 19 non-synonymous and 70.5 synonymous substitutions on the placental stem lineage. However, this model is not significantly better (χ12 test, P = 0.868) than the 2ω model, in which non-placental mammals (including the placental stem) have a different ratio than placental mammals as the crown group. Thus, the protein as a whole was under intense purifying selection throughout tetrapod evolution (Dataset S5). Comparison of chicken and human LGALS1 coding sequences demonstrate stronger purifying selection (kA/kS = 0.0042) than human–chicken orthologous genes as a whole on micro- and macrochromosomes (median kA/kS = 0.052 and 0.073, respectively) (25).
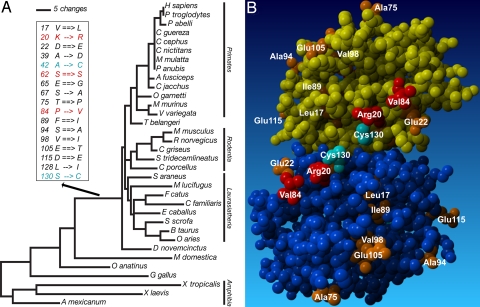
In contrast to purifying selection, there is evidence for adaptive evolution on the stem of placental mammals. Using the gene tree, the likelihood ratio in the branch-site test was significant (χ12 test, P = 0.035); when the species tree was used, the χ12 had a P value of 0.05 (Dataset S6). On the placental stem, 85% of the codons experienced purifying selection (ω = 0.069), 11% belonged to the neutral class, and 4% showed evidence for positive selection (ω = 29.6). Parsimony reconstructions of ancestral states of inferred amino acids resulted in 407 inferred amino acid replacements, from which those on the stem placental lineage are indicated in Fig. 4A. Residues and their Bayes–empirical Bayes posterior probabilities, detected as being positively selected on the stem placental lineage, are as follows: Arg-20, Ser-62, and Val-84 (Fig. 4 A and B, and Dataset S6).
Fig. 4.
(A) Phylogenetic tree of galectin-1. DELTRAN inferred amino acid replacements on the species tree are drawn to scale. Parsimony analysis resulted in a tree length of 407 steps; inferred amino acid replacements on the placental stem lineage are presented. Positively selected sites are highlighted in red. The S->S change at position 62 is considered non-synonymous because of the switch between disjunct codon classes. Derived cysteines (cyan blue) can disable sugar binding and dimerization upon oxidation as a result of the formation of disulfide bridges. (B) Residues replaced on the placental stem lineage are shown on the space-fill model of human dimeric galectin-1. Replaced residues are orange, positively selected residues are red, Cys-130 is cyan blue. Two positively selected residues (Arg-20, Val-84), along with Cys-130 and Glu-22, form a subdomain next to the dimerization interface. X-ray crystallographic data: 1GZW (43).
Conserved Functional Domains.
In all 34 investigated taxa, the translation start site and the stop codon are in the same positions in exons 1 and 4, respectively. Eighteen percent of the residues (24 of 134) are strictly conserved in the 34 vertebrates studied, 56% (75 of 134) in 28 mammals, and 81% (108 of 134) in 14 primates. Six of eight residues involved in carbohydrate binding are conserved in 100% of the analyzed taxa, the exceptions being conservative replacements for Val-59 in X. tropicalis, Xenopus laevis, and Otolemur garnetti, and for Arg-73 in X. tropicalis. Of the five residues in the growth-inhibitory domain, two are identical in all investigated species, and the remaining are at least 87% identical. Seven of the nine residues at the N- and C-termini that create the hydrophobic core for the dimerization are 100% conserved in placental mammals, with 96% and 78% conservation of the two additional residues, respectively (Fig. S3).
Substitutions with Impact on Functional Regulation.
Among the parsimony reconstructions (i.e., ACCTRAN and DELTRAN) and maximum likelihood (ML) reconstructions of ancestral states on the species tree, several functionally important replacements occurred in which derived cysteines would have resulted in the formation of disulfide bridges. According to all ancestral character state reconstruction methods, Cys-16, Cys-60, and Cys-88 were present at the time of the last common ancestor of tetrapods and have been conserved in nearly all descendant lineages. Cys-42 and Cys-132 emerged on either the stem mammalian lineage (ACCTRAN and ML) or the stem placental mammal lineage (DELTRAN). Finally, Cys-2 emerged on either the stem lineage leading to birds and mammals (parsimony) or was present at the time of the last common ancestor of tetrapods (ML). The emergence of Cys-42 and Cys-132 would have enabled the formation of disulfide bridges with Cys-2 and Cys-60, respectively, early in mammalian evolution (Fig. S3). Of additional importance, amino acid positions involved in forming a subdomain juxtaposed with the dimerization interface (i.e., Arg-20, Glu-22, Val-84, Cys-130) were replaced on the stem lineage leading to placental mammals according to at least one method of reconstruction, and two of these residues (20 and 84) showed evidence of positive selection according to the conservative Bayes empirical Bayes procedure as applied to ML analysis. Additional residues that were replaced on this stem are shown in Fig. 4.
Discussion
Galectin-1 is highly conserved throughout evolution (Dataset S5) and has a similar expression pattern in placentas and extraembryonic membranes in the investigated taxa. We identified phylogenetic footprints, including a half ERE, in highly conserved regions of the LGALS1 5′ promoter that emerged on the stem of placental mammals and may be involved in the sex steroid regulation of its expression. Functional domains in galectin-1 are highly conserved among the investigated vertebrates, especially among extant placental mammals. Inferred amino acid replacements on the stem of placental mammals confer redox regulatory changes that have been maintained in all but one descendent species included in the study (Fig. S3).
Galectin-1 and Maternal–Fetal Immune Tolerance.
Mechanisms of vertebrate immune tolerance were presumably necessary to prevent self-attack during the emergence of the adaptive immune system (1, 26). The presence of fetal allo-antigens at the maternal–fetal interface poses a challenge to the maternal immune system that generates the need for a specialized version of tolerance. Accumulating evidence suggests that several immunoregulatory molecules, including galectin-1, may have been incorporated by the placenta and uterine mucosa during mammalian evolution to confer this tolerance (2–4, 7–9, 11). Indeed, galectin-1 is abundantly expressed in human third trimester placentas and extraembryonic membranes (23, 24, 27–33), and its up-regulation in the placenta in preeclampsia as well as in the extraembryonic membranes in chorioamnionitis (31–33) suggests a fetal contribution to anti-inflammatory responses.
The placental immunostaining pattern of galectin-1 is similar in all major primate groups. All investigated taxa had detectable immunoreactive galectin-1 in the syncytiotrophoblast on the fetal side of the interface, and it was strongly expressed in the decidua on the maternal side. These results are of importance in light of recent phylogenetic studies that demonstrated that the hemochorial placenta is the ancestral state in placental mammals (5, 6), and that this invasive type of placentation would have placed great demands on the maternal immune system at early stages of mammalian evolution (2). We found that the depth of the maternal–fetal interface (34) did not affect the selective constraint on galectin-1 and its placental and decidual expression pattern. This finding suggests that either the challenge to the maternal immune system by fetal allo-antigens may be similar in different types of placentation or that the functional constraint on LGALS1 results from its role in other tissues.
ERE May Participate in Regulation of Uterine/Placental LGALS1 Expression.
The finding that galectin-1 is most strongly expressed in decidua agrees that, among human tissues and cell lines, LGALS1 expression is highest in non-pregnant endometrium (Dataset S2), as well as in uterine tissues of mice (23). There is evidence that sex steroids regulate LGALS1 expression in human endometrium and in mouse uterine tissues during the menstrual and estrus cycles, as well as during decidualization in pregnancy (23, 24).
Uterine LGALS1 expression increases as early as 6 h after treatment with 17β-estradiol in ovariectomized mice and 12 h after treatment with progesterone; treatment with 17β-estradiol rapidly increases uterine LGALS1 expression in progesterone-primed endometrium (23). These effects can be blocked by estrogen and progesterone receptor antagonists (9, 23), suggesting the involvement of these nuclear receptors in LGALS1 expression.
To examine the sex steroid regulation of LGALS1 expression, we attempted to find conserved cis elements in the LGALS1 promoter that could control this effect. Indeed, the stem of placental mammals underwent a gain of 10 cis elements, including a highly conserved half ERE, an overlapping NF-Y, and a nearby AP-2 motif, which were identified in humans and mice but functionally not yet characterized (35–37). Estrogens act through estrogen receptors that are involved in the stimulation of uterine growth, progesterone receptor expression, and immunoregulation (39). Only 71% of the functional EREs bear a consensus palindromic motif, whereas 25% have half-ERE; estrogen receptors can also be tethered to other nuclear proteins found in the LGALS1 promoter, such as SP1 (40). Of note, NF-Y is enriched in promoters of genes induced by estrogen (41), and AP-2 is also involved in estrogen-dependent transcriptional regulation (42). Our data suggest that these cis motifs are candidate sites for the sex steroid regulation of LGALS1 expression at the maternal–fetal interface. We propose that the sex steroid regulation of galectin-1 function in immune tolerance (9) may have been gained via cis-regulatory evolution on the stem of placental mammals and that these elements have been preserved in all descendent placental lineages.
Highly Conserved Structure and Functional Domains of Galectin-1.
The “jelly-roll” structure of galectin-1 is highly conserved among vertebrates (13–16, 43). Residues crucial for sugar binding, growth inhibition, and dimerization were subject to strict purifying selection during vertebrate evolution: (i) eight residues in the carbohydrate recognition domain are identical in 31 taxa with only conservative replacements for Val-59 and Arg-73 in three species; thus, both the ability and specificity of galectin-1 to bind sugars have been maintained; (ii) site-directed mutagenesis have shown that Asp-27-Phe-31 are crucial for the growth inhibitory function (44). These five residues showed 87 to 100% conservation among the studied taxa, suggesting their functional importance; and (iii) seven of the nine residues comprising the hydrophobic core for the dimerization interface are 100% conserved among the studied placental mammals. As the integrity of the galectin-1 dimer depends on β-sheet interactions across the monomers (45), it is not surprising that the chicken CG-14, which has four radical replacements, is a monomer (46). Based on its highly conserved dimerization and sugar-binding properties, we propose that the immunoregulatory functions of galectin-1 are conserved across vertebrates, in accord with our observation that intense purifying selection has been acting on galectin-1 (Dataset S5). When this finding is viewed in the light of embryonic, placental, and chorioamniotic expression of LGALS1 during mammalian development (23, 24, 27–33), it provides support for a previous prediction (26) that genes expressed early in fetal development are under more selective constraint than those expressed only later in life.
Residues Under Selective Constraint Are Involved in Redox Regulation.
Despite the purifying selection acting on galectin-1, evidence for adaptive evolution for key residues was found on the stem of placental mammals. The disjunct codon S->S change at position 62 emphasizes the importance of this residue in sugar binding, as Ser-62 forms H-bonds with Arg-111 to maintain the architecture of the carbohydrate recognition domain (43). The conservative L->I change at position 128 suggests constraint on the hydrophobic nature of the dimerization interface (45). Importantly, Cys-42 and Cys-130 emerged during mammalian evolution. It has been shown that substitution of cysteines with serines in galectin-1 moderately affects sugar binding activity under non-reducing conditions, whereas WT galectin-1 loses its sugar-binding capacity shortly under oxidating conditions (47). Oxidation of these cysteines causes conformational changes that hinder lectin activity and dimerization of galectin-1 in humans, cows, and rats (43, 48–50).
Cysteines are normally protected against oxidation by the reducing intracellular environment (51). Oxidation of galectin-1 occurs upon cell surface externalization, and this causes galectin-1 to lose lectin activity (48–50). Of note, Cys-130 is among the residues that emerged during mammalian evolution and is in a subdomain near the dimerization interface. It is possible that residues in this subdomain may be involved in the redox regulation of the dimerization of galectin-1; however, further functional experiments are needed to support this view.
Concluding Remarks.
Our results support the hypothesis that molecules implicated in immune tolerance to invasive placentation adapted their immunoregulatory functions at the maternal–fetal interface during early eutherian evolution (1), and these adaptations have been conserved regardless of the degree of placental invasion. These results further support the proposal that the emergence of cis elements on the eutherian stem enabled the establishment of galectin-1 mediated immune tolerance at the maternal–fetal interface. Redox status mediated conformational changes in galectin-1 appear to be involved in the regulation of this tolerance. These features remain present in all placental mammals, suggesting their importance in the immunoregulation of pregnancy.
Materials and Methods
Placental specimens of six pregnant women were retrieved from the bank of biological samples of the Perinatology Research Branch. Fresh-frozen tissues were used for RNA isolation, quantification, or sequence analysis; formalin-fixed tissues were applied for immunohistochemistry. Written informed consent was obtained from all women before the collection of samples, and the research was approved by the institutional review boards of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and Wayne State University.
RNAlater-preserved placentas taken from Colobus guereza, C. nictitans, M. mulatta, Papio anubis, A. fusciceps, and V. variegata were used for RNA isolation, cDNA synthesis, and sequence analysis (Dataset S7). Formalin-fixed placentas from the same animals were used for immunohistochemistry.
Eight newly generated primate LGALS1 coding sequences were deposited into GenBank (Dataset S1). The evolutionary conservation of the 2-kb promoter regions and first exons of LGALS1 in 12 vertebrate species was examined by pair-wise p-distance comparison and phylogenetic shadowing. Phylogenetic footprinting and transcriptional element searches were applied to predict conserved cis motifs in these 12 promoters. Maximum parsimony and ML methods were used for phylogenetic analyses of 34 publicly available vertebrate or newly sequenced primate LGALS1 coding sequences. Parsimony and codon-model based methods were used to infer the amino acid replacements on the phylogenetic tree.
LGALS1 expression was quantified in H. sapiens placentas (n = 6) and fetal membranes (n = 6) containing maternal decidua by quantitative RT-PCR. In silico LGALS1 expression analysis was performed using publicly available data within the SAGE Genie database. Immunostaining was performed on H. sapiens, C. nictitans, A. fusciceps, and V. variegata placentas. Further details for all methods are described in the SI Text.
Supplementary Material
Acknowledgments.
We acknowledge the valuable contributions of Dr. Chong Jai Kim, Sandy Field, Nancy Hauff, Gerardo Rodriguez, and the nursing staff of the Perinatology Research Branch. The authors thank Dr. Susan Land and Daniel Lott at the Applied Genomics Technology Center of Wayne State University for performing the qRT-PCR reactions, and Sara Tipton for critical reading of the manuscript. The following kindly provided DNA and/or tissue samples: Dr. Caro-Beth Stewart (SUNY, Albany, NY), Dr. Kathy Neiswanger (University of Pittsburgh, Pittsburgh, PA), the New England Regional Primate Center (Southborough, MA), the Duke University Primate Center (Durham, NC), and the San Diego Zoo (San Diego, CA). This research was supported by the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health/Department of Health and Human Services.
Footnotes
The authors declare no conflict of interest.
Data deposition: The new sequences reported in this paper have been deposited in the GenBank database (accession nos. EU152915–EU152920, EU363769, and EU363770).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807606105/DCSupplemental.
References
- 1.Medawar PB. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol. 1953;44:320–338. [Google Scholar]
- 2.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 3.Niederkorn JY. See no evil, hear no evil, do no evil: the lessons of immune privilege. Nat Immunol. 2006;7:354–359. doi: 10.1038/ni1328. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J, Betz AG. Mother's little helpers: mechanisms of maternal-fetal tolerance. Nat Immunol. 2006;7:241–246. doi: 10.1038/ni1317. [DOI] [PubMed] [Google Scholar]
- 5.Wildman DE, et al. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc Natl Acad Sci USA. 2006;103:3203–3208. doi: 10.1073/pnas.0511344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliot MG, Crespi BJ. Placental invasiveness mediates the evolution of hybrid inviability in mammals. Am Nat. 2006;168:114–120. doi: 10.1086/505162. [DOI] [PubMed] [Google Scholar]
- 7.Petroff MG, Chen L, Phillips TA, Hunt JS. B7 family molecules: novel immunomodulators at the maternal-fetal interface. Placenta. 2002;23(Suppl A):S95–101. doi: 10.1053/plac.2002.0813. [DOI] [PubMed] [Google Scholar]
- 8.Terness P, et al. Tolerance signaling molecules and pregnancy: IDO, galectins, and the renaissance of regulatory T cells. Am J Reprod Immunol. 2007;58:238–254. doi: 10.1111/j.1600-0897.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- 9.Blois SM, et al. A pivotal role for galectin-1 in fetomaternal tolerance. Nat Med. 2007;13:1450–1457. doi: 10.1038/nm1680. [DOI] [PubMed] [Google Scholar]
- 10.Saito S, et al . The role of the immune system in preeclampsia. Mol Aspects Med. 2007;28:192–209. doi: 10.1016/j.mam.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 12.Aluvihare VR, Kallikourdis M, Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. 2004;5:266–271. doi: 10.1038/ni1037. [DOI] [PubMed] [Google Scholar]
- 13.Barondes SH, et al. Galectins: a family of animal beta-galactoside-binding lectins. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 14.Cooper DN. Galectinomics: finding themes in complexity. Biochim Biophys Acta. 2002;1572:209–231. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 15.Gabius HJ, Andre S, Kaltner H, Siebert HC. The sugar code: functional lectinomics. Biochim Biophys Acta. 2002;1572:165–177. doi: 10.1016/s0304-4165(02)00306-9. [DOI] [PubMed] [Google Scholar]
- 16.Houzelstein D, et al. Phylogenetic analysis of the vertebrate galectin family. Mol Biol Evol. 2004;21:1177–1187. doi: 10.1093/molbev/msh082. [DOI] [PubMed] [Google Scholar]
- 17.He J, Baum LG. Presentation of galectin-1 by extracellular matrix triggers T cell death. J Biol Chem. 2004;279:4705–4712. doi: 10.1074/jbc.M311183200. [DOI] [PubMed] [Google Scholar]
- 18.Rabinovich GA, et al. Recombinant galectin-1 and its genetic delivery suppress collagen-induced arthritis via T cell apoptosis. J Exp Med. 1999;190:385–398. doi: 10.1084/jem.190.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubinstein N, et al. Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection: a potential mechanism of tumor-immune privilege. Cancer Cell. 2004;5:241–251. doi: 10.1016/s1535-6108(04)00024-8. [DOI] [PubMed] [Google Scholar]
- 20.Baum LG, et al . Amelioration of graft versus host disease by galectin-1. Clin Immunol. 2003;109:295–307. doi: 10.1016/j.clim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Koopman LA, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garin MI, et al. Galectin-1: a key effector of regulation mediated by CD4+CD25+ T cells. Blood. 2007;109:2058–2065. doi: 10.1182/blood-2006-04-016451. [DOI] [PubMed] [Google Scholar]
- 23.Choe YS, et al. Expression of galectin-1 mRNA in the mouse uterus is under the control of ovarian steroids during blastocyst implantation. Mol Reprod Dev. 1997;48:261–266. doi: 10.1002/(SICI)1098-2795(199710)48:2<261::AID-MRD14>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 24.von Wolff M, Wang X, Gabius HJ, Strowitzki T. Galectin fingerprinting in human endometrium and decidua during the menstrual cycle and in early gestation. Mol Hum Reprod. 2005;11:189–194. doi: 10.1093/molehr/gah144. [DOI] [PubMed] [Google Scholar]
- 25.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 26.Goodman M. In: Viking Fund publications in anthropology. Number thirty-seven. Classification and human evolution. Washburn SL, editor. New York: Wenner-Gren Foundation for Anthropological Research Inc.; 1963. pp. 204–234. [Google Scholar]
- 27.Bevan BH, et al. Immunohistochemical localization of a beta-D-galactoside-binding lectin at the human maternofetal interface. Histochem J. 1994;26:582–586. doi: 10.1007/BF00158592. [DOI] [PubMed] [Google Scholar]
- 28.Walzel H, et al. Immunohistochemical and glycohistochemical localization of the beta-galactoside-binding S-type lectin in human placenta. Acta Histochem. 1995;97:33–42. doi: 10.1016/s0065-1281(11)80204-7. [DOI] [PubMed] [Google Scholar]
- 29.Maquoi E, van den Brule FA, Castronovo V, Foidart JM. Changes in the distribution pattern of galectin-1 and galectin-3 in human placenta correlates with the differentiation pathways of trophoblasts. Placenta. 1997;18:433–439. doi: 10.1016/s0143-4004(97)80044-6. [DOI] [PubMed] [Google Scholar]
- 30.Vicovac L, Jankovic M, Cuperlovic M. Galectin-1 and −3 in cells of the first trimester placental bed. Hum Reprod. 1998;13:730–735. doi: 10.1093/humrep/13.3.730. [DOI] [PubMed] [Google Scholar]
- 31.Jeschke U, et al. Expression of galectin-1, -3 (gal-1, gal-3) and the Thomsen-Friedenreich (TF) antigen in normal, IUGR, preeclamptic and HELLP placentas. Placenta. 2007;28:1165–1173. doi: 10.1016/j.placenta.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Than NG, et al. Severe preeclampsia is characterized by increased placental expression of galectin-1. J Matern Fetal Neonatal Med. 2008;21:429–442. doi: 10.1080/14767050802041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Than NG, et al. Chorioamnionitis and increased galectin-1 expression in PPROM–an anti-inflammatory response in the fetal membranes? Am J Reprod Immunol. 2008 doi: 10.1111/j.1600-0897.2008.00624.x. doi: 10.1111/j.1600-0897.2008.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mossman HW. Vertebrate fetal membranes. New Brunswick, NJ: Rutgers Univ Press; 1987. [Google Scholar]
- 35.Gitt MA, Barondes SH. Genomic sequence and organization of two members of a human lectin gene family. Biochemistry. 1991;30:82–89. doi: 10.1021/bi00215a013. [DOI] [PubMed] [Google Scholar]
- 36.Salvatore P, et al. Characterization and functional dissection of the galectin-1 gene promoter. FEBS Lett. 1995;373:159–163. doi: 10.1016/0014-5793(95)01032-a. [DOI] [PubMed] [Google Scholar]
- 37.Lu Y, Lotan R. Transcriptional regulation by butyrate of mouse galectin-1 gene in embryonal carcinoma cells. Biochim Biophys Acta. 1999;1444:85–91. doi: 10.1016/s0167-4781(98)00257-7. [DOI] [PubMed] [Google Scholar]
- 38.De Gregorio E, Chiariotti L, Di Nocera PP. The overlap of Inr and TATA elements sets the use of alternative transcriptional start sites in the mouse galectin-1 gene promoter. Gene. 2001;268:215–223. doi: 10.1016/s0378-1119(01)00437-1. [DOI] [PubMed] [Google Scholar]
- 39.Dahlman-Wright K, et al. International Union of Pharmacology LXIV Estrogen receptors. Pharmacol Rev. 2006;58:773–781. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- 40.Lin CY, et al . Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scafoglio C, et al. Comparative gene expression profiling reveals partially overlapping but distinct genomic actions of different antiestrogens in human breast cancer cells. J Cell Biochem. 2006;98:1163–1184. doi: 10.1002/jcb.20820. [DOI] [PubMed] [Google Scholar]
- 42.Orso F, et al. Activator protein-2gamma (AP-2gamma) expression is specifically induced by oestrogens through binding of the oestrogen receptor to a canonical element within the 5′-untranslated region. Biochem J. 2004;377:429–438. doi: 10.1042/BJ20031133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Lucendo MF, et al. Growth-regulatory human galectin-1: crystallographic characterisation of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J Mol Biol. 2004;343:957–970. doi: 10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 44.Scott K, Zhang J. Partial identification by site-directed mutagenesis of a cell growth inhibitory site on the human galectin-1 molecule. BMC Cell Biol. 2002;3:3. doi: 10.1186/1471-2121-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho M, Cummings RD. Characterization of monomeric forms of galectin-1 generated by site-directed mutagenesis. Biochemistry. 1996;35:13081–13088. doi: 10.1021/bi961181d. [DOI] [PubMed] [Google Scholar]
- 46.Varela PF, et al. The 2.15 Å crystal structure of CG-16, the developmentally regulated homodimeric chicken galectin. J Mol Biol. 1999;294:537–549. doi: 10.1006/jmbi.1999.3273. [DOI] [PubMed] [Google Scholar]
- 47.Hirabayashi J, Kasai K. Effect of amino acid substitution by sited-directed mutagenesis on the carbohydrate recognition and stability of human 14-kDa beta-galactoside-binding lectin. J Biol Chem. 1991;266:23648–23653. [PubMed] [Google Scholar]
- 48.Tracey BM, et al. Subunit molecular mass assignment of 14,654 Da to the soluble beta-galactoside-binding lectin from bovine heart muscle and demonstration of intramolecular disulfide bonding associated with oxidative inactivation. J Biol Chem. 1992;267:10342–10347. [PubMed] [Google Scholar]
- 49.Yamaoka K, et al. Structural and functional characterization of a novel tumor-derived rat galectin-1 having transforming growth factor (TGF) activity: the relationship between intramolecular disulfide bridges and TGF activity. J Biochem. 1996;119:878–886. doi: 10.1093/oxfordjournals.jbchem.a021325. [DOI] [PubMed] [Google Scholar]
- 50.Inagaki Y, et al. Oxidized galectin-1 promotes axonal regeneration in peripheral nerves but does not possess lectin properties. Eur J Biochem. 2000;267:2955–2964. doi: 10.1046/j.1432-1033.2000.01311.x. [DOI] [PubMed] [Google Scholar]
- 51.Sen CK. Redox signaling and the emerging therapeutic potential of thiol antioxidants. Biochem Pharmacol. 1998;55:1747–1758. doi: 10.1016/s0006-2952(97)00672-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.