Abstract
The serine and arginine-rich protein family (SR proteins) are highly conserved regulators of pre-mRNA splicing. SF2/ASF, a prototype member of the SR protein family, is a multifunctional RNA binding protein with roles in pre-mRNA splicing, mRNA export and mRNA translation. These observations suggest the intriguing hypothesis that SF2/ASF may couple splicing and translation of specific mRNA targets in vivo. Unfortunately the paucity of endogenous mRNA targets for SF2/ASF has hindered testing of this hypothesis. Here, we identify endogenous mRNAs directly cross-linked to SF2/ASF in different sub-cellular compartments. Cross-Linking Immunoprecipitation (CLIP) captures the in situ specificity of protein-RNA interaction and allows for the simultaneous identification of endogenous RNA targets as well as the locations of binding sites within the RNA transcript. Using the CLIP method we identified 326 binding sites for SF2/ASF in RNA transcripts from 180 protein coding genes. A purine-rich consensus motif was identified in binding sites located within exon sequences but not introns. Furthermore, 72 binding sites were occupied by SF2/ASF in different sub-cellular fractions suggesting that these binding sites may influence the splicing or translational control of endogenous mRNA targets. We demonstrate that ectopic expression of SF2/ASF regulates the splicing and polysome association of transcripts derived from the SFRS1, PABC1, NETO2 and ENSA genes. Taken together the data presented here indicate that SF2/ASF has the capacity to co-regulate the nuclear and cytoplasmic processing of specific mRNAs and provide further evidence that the nuclear history of an mRNA may influence its cytoplasmic fate.
Introduction
Eukaryotic messenger RNA (mRNA) must be processed prior to programming protein synthesis. The minimal modifications for most mRNAs include capping, pre-mRNA splicing and polyadenylation [1]. These reactions occur in the nucleus and must be completed prior to nuclear export of the mRNA to the cytoplasm. The cytoplasmic fate of mRNA is also subject to regulation at the level of localization, stability and translational efficiency [2]. RNA processing reactions have been extensively studied using biochemical systems; however, these are functionally linked in living cells providing increased efficiency and regulatory potential for gene expression [3]. The molecular mechanisms responsible for the coupling of post-transcriptional regulatory networks are poorly understood. A subset of multi-functional mRNA binding proteins operating at the interface of distinct RNA processing machineries may contribute to coupling of post-transcriptional gene expression.
The Serine and Arginine-rich (SR) protein family consists of eight phylogenetically conserved proteins ranging in molecular weight from 20–75 kDa. SR proteins have well-characterized roles in pre-mRNA splicing including exon definition and assembly of the mature spliceosome [4], [5]. A subset of the SR protein family also function outside pre-mRNA splicing [6]. Indeed, we recently established a role of the shuttling SR protein SF2/ASF in mRNA translation [7], [8]. SF2/ASF enhances translation initiation by promoting phosphorylation of the translational repressor protein 4E-BP1 by the mammalian target of rapamycin (mTOR) [9]. Further roles for SRp20 and 9G8 in internal ribosome entry site (IRES)-mediated translation and intronless mRNA export have been documented [10], [11]. These observations suggest the hypothesis that shuttling SR proteins may couple nuclear and cytoplasmic steps of pre-mRNA processing. Elucidating endogenous mRNA targets that are regulated by SF2/ASF in both the nucleus and cytoplasm is critical to testing the validity of this hypothesis.
The cross-linking immunoprecipitation (CLIP) method is a powerful method for elucidating the target specificity of RNA binding proteins in vivo [12]. In this protocol, living cells are exposed to UV irradiation to induce covalent cross-links between RNA binding proteins and their in situ RNA targets. Prior to immunoprecipitation of a specific RNA binding protein lysates are treated with RNase in order to generate 40–60 nt cross-linked RNA tags. Co-purifying RNA tags are then cloned and sequenced revealing genomic locus of the RNA as well as the position of the RNA-protein interaction within the RNA molecule. CLIP analysis of the neural specific splicing factor Nova identified a post-transcriptional regulatory network related to synaptic function. Mechanistic studies determined the relationship between positions of Nova binding sites and effects on splice site selection [13], [14]. These data were used to build an RNA map capable of predicting Nova-dependent splice site selection [15]. Here, we use CLIP to identify RNA targets for the shuttling SR protein, SF2/ASF, in order to test the hypothesis that this SR family protein member regulates nuclear and cytoplasmic expression of specific mRNAs in living cells. This sampling of nuclear, cytoplasmic and polyribosome-associated mRNA targets suggests that SF2/ASF may remain associated with a subset of specific transcripts as they are trafficked from the nucleus to the cytoplasm. Furthermore we provide evidence that SF2/ASF couples alternative splicing with enhanced translation for several endogenous mRNAs. These data provide insights to the targets and roles of an essential shuttling RNA binding protein in coupling nuclear and cytoplasmic steps of post-transcriptional gene expression.
Results
Cross-linking immunoprecipitation of SF2/ASF
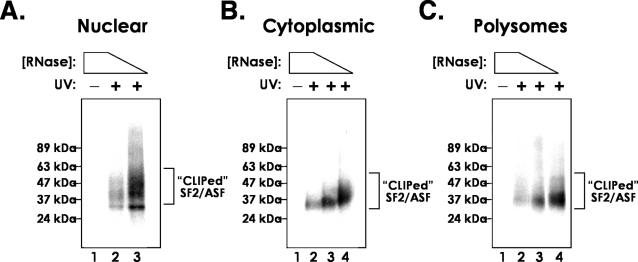
Previously, we have shown that SF2/ASF binds directly to cytoplasmic mRNA in vivo and enhances the translation of reporter mRNAs both in vitro and in vivo [8], suggesting that SF2/ASF may regulate the nuclear and cytoplasmic fate of specific endogenous mRNAs. Despite nearly two decades of study, few endogenous targets of SR proteins have been identified. In order to determine if SF2/ASF can regulate nuclear and cytoplasmic processing of endogenous mRNAs we used the CLIP protocol to identify binding sites for SF2/ASF in mRNAs from different subcellular fractions. Nuclear, cytoplasmic and polyribosome-containing sub-cellular fractions were prepared as previously described [7] from control cells or those exposed to UV irradiation and cell extracts were treated with decreasing amounts of RNase A/T1 to partially degrade cross-linked RNA molecules. SF2/ASF was immunoprecipitated from each extract under stringent conditions. Purified SF2/ASF-RNA complexes were modified on beads by the addition of a 3′ RNA linker and 32P at the 5′ end. Complexes were resolved on 10% Novex Nupage gels (Invitrogen, USA) and transferred to nitrocellulose. Fig. 1 shows UV-dependent, RNase-sensitive complexes immunoprecipitated by the anti-SF2/ASF monoclonal antibody. Note the increased molecular weight of the complex as RNase concentrations are decreased. The heterogeneous migration of these complexes is due to increasing length of captured RNA fragments. Complexes between 37 and 42 kDa were purified from nitrocellulose membranes. RNA was extracted, ligated to a 5′ RNA linker and amplified by RT-PCR (not shown). Amplicons were then concatamerized, ligated in pcDNA3.1, cloned and sequenced.
Figure 1. Cross-linking immunoprecipitation of SF2/ASF from different HEK293T cellular fractions.
SF2/ASF-RNA complexes, indicated by the bracket, were visualized by autoradiography. Positions of molecular weight standards are given at the left of each panel and exposure of cells to ultra violet radiation (UV) is indicated by + or −.
Mapping of SF2/ASF mRNA targets
We obtained sequences from more than 3,500 amplicons corresponding to putative SF2/ASF RNA targets. We used BLAT to align sequences to the human genome allowing for two mis-matches or gaps within the alignment [16]. For this analysis we focused on RNA fragments corresponding to protein coding genes, any other sites were ignored. 1,237 amplicons could be mapped to a total of 326 positions in 180 protein coding genes; 43% of the binding sites were represented by multiple sequences. A majority of binding sites (68%) fell within exonic sequences (Fig. 2A). We also detected binding sites for SF2/ASF within intronic sequences. However, as shown below, we were unable to detect a consensus motif within the pool of intronic binding sites, therefore we focused the remaining analysis on the exonic pool of sequences. We compared the binding specificity of SF2/ASF RNA targets identified in different sub-cellular compartments in order to determine if any RNA transcripts were in common. We found that 72 exonic binding sites are engaged by SF2/ASF in both the nucleus and the cytoplasm or polysome fraction. Interestingly, 15 binding sites in 12 different genes are engaged by SF2/ASF in all three compartments (Fig. 2B). These data provide strong evidence that SF2/ASF associates with a subset of messenger RNAs in the nucleus and that the same cis-acting element is recognized in the cytoplasm. We hypothesize that SF2/ASF remains bound to the mRNA as the mRNA is trafficked from the nucleus to the cytoplasm [7].
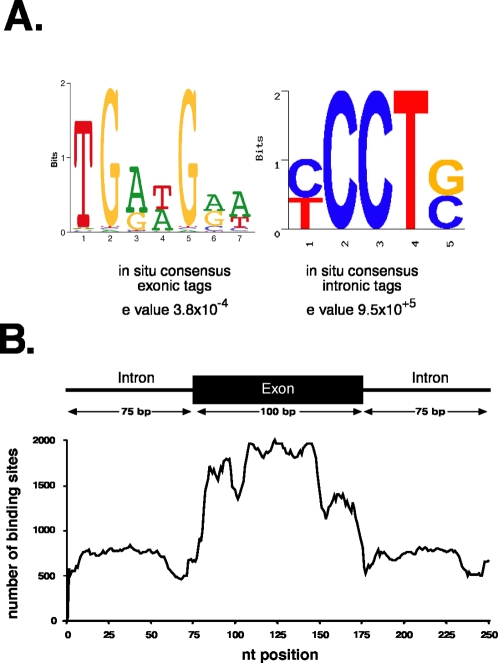
Figure 2. Distribution of SF2/ASF binding sites in relation to gene structure and sub-cellular localization.
A) The “coding” SF2/ASF CLIP tags from nuclear, cytoplasmic and polysomal sets were analyzed in relation to their relative position within genes. This showed that the majority map to intronic or to coding exonic sequences and only a small proportion mapped to UTRs. B) Venn diagram of exonic SF2/ASF binding sites from Nuclear, cytoplasmic and polysomal sub-cellular fractions. The number of unique binding sites identified in each fraction is designated as “n”.
Identification of a consensus binding site for SF2/ASF
The in vitro RNA binding specificity of many SR proteins including SF2/ASF, have been well-characterized using approaches such as selected evolution of ligands by exponential enrichment (SELEX) [17]. Different consensus sites for SF2/ASF have been obtained using different SELEX strategies [18], [19]. We used the motif finding algorithm MEME, to search exonic or intronic RNA fragments for over represented sequences [20], [21]. Fig. 3A shows that a purine-rich consensus motif is present within exonic sequences targeted by SF2/ASF with a highly significant expectation value (e value) of 3.8×10−4. No other statistically significant motifs were identified within the exonic sequences. By contrast MEME was unable to detect an enriched sequence motif within the intronic pool of binding sites identified by CLIP. These data suggest that intronic binding sites may prove to be a nonspecific contaminant. To test the validity of the consensus model, we asked if the consensus sequence is enriched in exonic or intronic sequences across the genome using composite exons [18]. We then used the position weight matrix calculated from the consensus motif to determine the number of binding sites at each position across all intron/exon/intron composite sequences. Fig. 3B demonstrates that the consensus motif for SF2/ASF is highly enriched at the edges of exons relative to intronic sequences or the interior of exon sequences. These data support the hypothesis that SF2/ASF plays a role in the establishment of exon identity.
Figure 3. Identification of the SF2/ASF consensus binding site and its frequency distribution in coding exons and flanking introns.
A) The exonic or intronic clip tags were analyzed using MEME to identify sequence patterns that are over-represented. MEME was unable to identify a consensus motif within the intronic binding sites. B) This pattern was used to analyze composite sequences made for all exons with more than 100 nucleotides, from protein coding genes. This analysis shows that the pattern is over-represented within exonic sequences and in particular within the exon 5′ prime and middle of the exons.
Annotation of alternatively spliced pre-mRNA targets of SF2/ASF
SR proteins have well characterized roles in both constitutive and alternative pre-mRNA splicing. Resolving these dual functions of SR proteins solely by the use of heterologous substrates and mini-gene reporters has proven difficult. In order to determine if the endogenous binding sites for SF2/ASF may reveal any clues to the roles of SR proteins in alternative splicing we manually curated annotated ENSEMBL transcripts targeted by SF2/ASF for examples of alternative splicing using two distinct databases: AceView and FAST-DB [22], [23]. Of the 234 exonic binding sites identified by CLIP, only 72 were subject to some form of alternative processing (Table 1– 4 for details on binding sites associated with specific types of alternative splicing). We also identified 9 binding sites within constitutive exons located downstream and adjacent to alternative cassette exons (See Table 5). This type of arrangement suggests that SF2/ASF may promote skipping of the upstream cassette exon, as recently demonstrated for alternative splicing of the receptor tyrosine kinase RON during breast cancer metastasis [24]. Within this list of alternative exons targeted by SF2/ASF there is a modest enrichment for genes encoding RNA binding proteins (SFRS1, PABPC1, hnRNPDL, hnRNPAB, SERBP1, RPL5, SON, RPL0, RPS10, RPL12, RPS24, RPL19, PKRA) and genes involved in regulation of biological processes such as cell division, proliferation and apoptosis (CDK4, MAPK3, ACIN1). The ENSA gene is also of interest as it is bound by SF2/ASF in each sub-cellular compartment and is a Type 2 diabetes candidate gene. ENSA is an endogenous ligand for the sulfonylurea receptor which plays an important role in insulin release from pancreatic beta cells [25], [26]. Several of the alternative splicing events involving exons targeted by SF2/ASF lead to premature termination codons and potentially NMD (SFRS1, CDK4, MAPK3, for example) however the majority of alternative events alter the primary structure of the encoded polypeptide (not shown). All of these splicing events can be visualized using the Friendly Alternative Splicing and Transcript Diversity database (http://www.fast-db.com; [22]).
Table 1. Annotation of alternative cassette exons bound by SF2/ASF.
| Partial list of alternative cassette exons bound by SF2/ASF | |||||
| Gene ID | Exon Position | Gene Description | Localization | Gene Family | Function |
| ACIN1 | 12 | apoptotic chromatin condensation inducer 1 | Nucleus | enzyme | apoptotic chromosome condensation |
| BECN1 | 11 | beclin 1 (coiled-coil, myosin-like BCL2 interacting protein) | Nucleus; Polysome | other | anti-apoptosis;cellular defense response |
| CALM2 | 4 | calmodulin 2 (phosphorylase kinase, delta) | Cytoplasm | other | Calcium Binding;G-protein coupled receptor protein signaling pathway |
| DHX15 | 5 | DEAH (Asp-Glu-Ala-His) box polypeptide 15 | Cytoplasm | enzyme | mRNA processing |
| ERCC6 | 2 | excision repair cross-complementing rodent repair deficiency, complementation group 6 | Cytoplasm | transcription regulator | transcription-coupled nucleotide-excision repair |
| MAPK3 | 6 | mitogen-activated protein kinase 3 | Cytoplasm | kinase | intracellular signaling cascade |
| NUP43 | 4 | nucleoporin 43kDa | Nucleus; Cytoplasm; Polysome | transporter | nuclear envelope |
| PIP5K2A | 7 | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | Nucleus | kinase | glycerophospholipid metabolic process |
| SON | 3 | SON DNA binding protein | Nucleus | other | anti-apoptosis |
| TCF12 | 9 | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | Cytoplasm | transcription regulator | regulation of transcription from RNA polymerase II promoter |
| WDR42A | 11 | WD repeat domain 42A | Nucleus; Cytoplasm | other | Unknown |
| Y13871 | 1 | hypothetical protein LOC202181 | Nucleus; Cytoplasm; Polysome | other | Unknown |
| ZHX1 | 3 | zinc fingers and homeoboxes 1 | Nucleus | transcription regulator | negative regulation of transcription, DNA-dependent |
Exons were annotated manually according to AceView and FAST-DB databases. Column Headers: Gene ID, official gene symbol; Exon position, number of exon within the primary transcript; Gene Description; Localization, the sub-cellular fraction used for CLIP experiment; Gene Family; Function, the primary encoded function of gene. This table is a partial list of cassette exons bound by SF2/ASF. The complete list can be found in Table S1.
Table 2. Annotated as described in Table 1.
| SF2/ASF target exons containing alternative 3′ splice sites | |||||
| Gene ID | Exon Position | Gene Description | Localization | Gene Family | Function |
| PSAP | 7 | prosaposin | Nucleus | other | glycosphingolipid metabolic process; lipid transport |
| HN1 | 3 | hematological and neurological expressed 1 | Nucleus | other | Not Determined |
| IGF1R | 21 | insulin-like growth factor 1 receptor | Nucleus | transmembrane receptor | insulin receptor signaling pathway |
| PRELID1 | 5 | PRELI domain containing 1 | Nucleus | other | multicellular organismal development |
| CALM2 | 3 | calmodulin 2 (phosphorylase kinase, delta) | Cytoplasm | other | Calcium Binding; G-protein coupled receptor protein signaling pathway |
| AHCY | 10 | S-adenosylhomocysteine hydrolase | Cytoplasm | enzyme | one-carbon compound metabolic process |
| ZFP91 | 11 | zinc finger protein 91 homolog (mouse) | Cytoplasm | transcription regulator | |
| UBE3B | 28 | ubiquitin protein ligase E3B | Cytoplasm | enzyme | Protein degredation |
| RPL10A | 6 | ribosomal protein L10a | Cytoplasm | other | Ribosome Component |
| GK | 17 | glycerol kinase | Cytoplasm | kinase | |
| RPS10 | 3 | ribosomal protein S10 | Nucleus; Cytoplasm | other | Ribosome Component |
| MGST3 | 2 | microsomal glutathione S-transferase 3 | Nucleus; Cytoplasm | enzyme | lipid metabolic process; signal transduction |
| RPL19 | 6 | ribosomal protein L19 | Nucleus; Cytoplasm | other | Ribosome Component |
| NAGPA | 5 | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucos-aminidase | Nucleus; Cytoplasm | enzyme | carbohydrate metabolic process; lysosome organization and biogenesis |
| B2M | 2 | beta-2-microglobulin | Nucleus; Cytoplasm | transmembrane receptor | immune response |
| ETNK1 | 2 | ethanolamine kinase 1 | Cytoplasm; Polysome | kinase | Phosphatidylethanol-amine biosynthetic process |
| EEF1A1 | 3,6 | eukaryotic translation elongation factor 1 alpha 1 | Nucleus; Cytoplasm; Polysome | translation | translational elongation |
| FLJ00313 | 7 | Not Determined | Nucleus | Unknown | Unknown |
| NR2C1 | 12 | nuclear receptor subfamily 2, group C, member 1 | Nucleus | transcription regulator | Unknown |
| PWP1 | 13 | PWP1 homolog (S. cerevisiae) | Nucleus | other | Unknown |
| CENPO | 5 | centromere protein O | Nucleus | other | role in cell growth and/or transcription |
| TCF12 | 9 | transcription factor 12 | Cytoplasm | transcription regulator | Regulation of transcription by RNA Polymerase II |
Table 3. Annotated as described in Table 1.
| SF2/ASF target exons containing alternative 5′ splice sites | |||||
| Gene ID | Exon Position | Gene Description | Localization | Gene Family | Function |
| NAGPA | 5 | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | Nucleus; Cytoplasm | enzyme | carbohydrate metabolic process; lysosome organization and biogenesis |
| SERBP1 | 6 | SERPINE1 mRNA binding protein 1 | Nucleus | other | mRNA processing |
| CDK4 | 2 | cyclin-dependent kinase 4 | Nucleus | kinase | Cell Cycle Regulation |
| CDC14A | 15 | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | Nucleus | phosphatase | dual specificity protein tyrosine phosphatase |
| BCOR | 4 | BCL6 co-repressor | Cytoplasm | transcription regulator | Transcriptional corepressor |
| RPL5 | 1 | ribosomal protein L5 | Cytoplasm | other | Ribosome Component |
| NOL1 | 16 | nucleolar protein 1, 120kDa | Polysome | other | Cell Cycle Regulation; RNA Methyltransferase |
| PKM2 | 2 | pyruvate kinase, muscle | Cytoplasm; Polysome | kinase | Glycolytic enzyme |
| ETNK1 | 2 | ethanolamine kinase 1 | Cytoplasm; Polysome | kinase | phosphatidylethanolamine biosynthetic process |
| MAPK3 | 6 | mitogen-activated protein kinase 3 | Cytoplasm | kinase | intracellular signaling cascade |
| EEF1A1 | 2,3,6 | eukaryotic translation elongation factor 1 alpha 1 | Nucleus; Cytoplasm; Polysome | translation | translational elongation |
Table 4. Annotated as described in Table 1.
| SF2/ASF target exons containing a retained intron | |||||
| Gene ID | Exon Position | Gene Description | Localization | Gene Family | Function |
| ATP5B | 2 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | Nucleus; Cytoplasm | transporter | generation of precursor metabolites and energy |
| ATP7B | 2 | ATPase, Cu++ transporting, beta polypeptide | Nucleus | transporter | cellular copper ion homeostasis |
| EEF1A1 | 1 | eukaryotic translation elongation factor 1 alpha 1 | Nucleus; Cytoplasm; Polysome | translation regulator | translational elongation |
| GNL3 | 13 | guanine nucleotide binding protein-like 3 (nucleolar) | Cytoplasm | other | Stem Cell Proliferation |
| NAGPA | 5 | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | Nucleus; Cytoplasm | enzyme | carbohydrate metabolic process; lysosome organization and biogenesis |
| RAN | 7 | RAN, member RAS oncogene family | Nucleus | enzyme | GTP-binding protein involved in nucleocytoplasmic transport |
| RPL12 | 7 | ribosomal protein L12 | Nucleus | other | Ribosome Component |
| RPL23 | 3 | ribosomal protein L23 | Polysome | other | Ribosome Component |
| RPLP0 | 5 | ribosomal protein, large, P0 | Nucleus | other | Ribosome Component |
| SFRS1 | 4 | splicing factor, arginine/serine-rich 1 (splicing factor 2, alternate splicing factor) | Cytoplasm | other | mRNA processing |
| TH1L | 15 | TH1-like (Drosophila) | Cytoplasm | other | Transcription regulator |
| TSSC4 | 4 | tumor suppressing subtransferable candidate 4 | Nucleus | other | Unknown |
| WDR59 | 12 | WD repeat domain 59 | Nucleus | transporter | Not Determined |
| ZMYM3 | 7 | zinc finger, MYM-type 3 | Cytoplasm | other | multicellular organismal development |
Table 5. Annotated as described in Table 1.
| SF2/ASF target exons located adjacent to alternative cassette exons | |||||
| Gene ID | Exon Position | Gene Description | Localization | Gene Family | Function |
| NETO2 | 9 | neuropilin (NRP) and tolloid (TLL)-like 2 | Cytoplasm | Transmembrane Protein | development; neuronal function |
| PRKRA | 4 | protein kinase, interferon-inducible double stranded RNA dependent activator | Cytoplasm | Kinase | Pro-apoptosis |
| RPS24 | 6 | ribosomal protein S24 | Cytoplasm | Ribosome Component | Translation factor |
| PABPC1 | 12 | poly(A) binding protein, cytoplasmic 1 | Nucleus; Polysome | Other | translation regulator |
| BRD2 | 13 | bromodomain containing 2 | Nucleus | kinase | generation of spermatozoa |
| YWHAH | 2 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | Nucleus; Cytoplasm | Other | transcription regulator |
| HNRPDL | 9 | heterogeneous nuclear ribonucleoprotein D-like | Nucleus | Other | mRNA processing |
| HNRPAB | 8 | heterogeneous nuclear ribonucleoprotein A/B | Nucleus | Other | mRNA processing |
| MPZL1 | 6 | myelin protein zero-like 1 | Nucleus; Cytoplasm | Transmembrane Protein | Receptor |
Regulation of endogenous nuclear and cytoplasmic mRNA processing by SF2/ASF
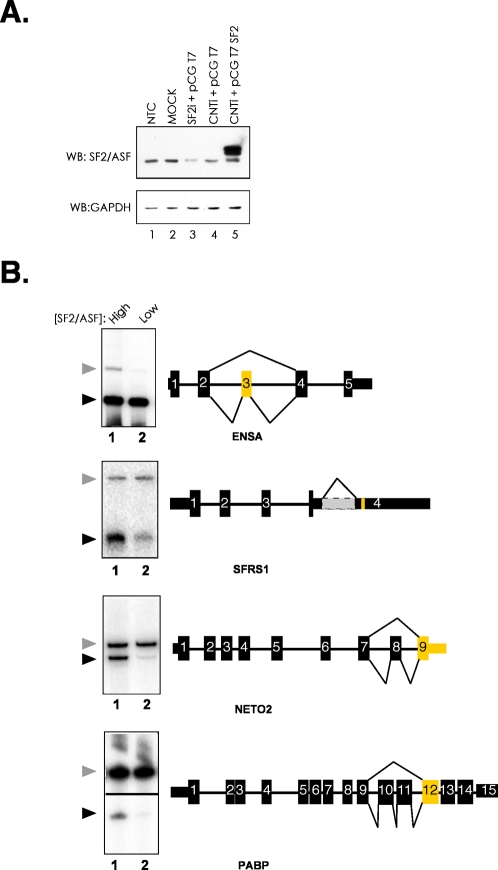
A majority of RNA fragments that co-purified with SF2/ASF contain a purine-rich consensus motif, reminiscent of previous SELEX binding sites [19]. Interestingly, a subset of mRNA targets was bound by SF2/ASF in both the nucleus and the cytoplasm. Three of these targets, ENSA (endosulfine α), PABPC1 (poly A binding protein 1) and NETO2 (neuropillin-like tolloid receptor 2) contained binding sites for SF2/ASF within or flanking alternative cassette exons, whereas the SFRS1 (pre-mRNA encoding SF2/ASF) contains a binding site adjacent to a retained intron. We then asked whether SF2/ASF modulates the alternative splicing of these pre-mRNAs in the nucleus and enhance their translation in the cytoplasm. We manipulated the levels of SF2/ASF in HEK293T cells by transfecting cells with siRNA targeting endogenous SF2/ASF, scrambled control siRNA or plasmid to increase levels of SF2/ASF by transient ectopic expression and assayed alternative splicing of endogenous ENSA, SF2/ASF, PABP and NETO2. Fig. 4A shows a Western blot from control cells (lanes 1,2, and 4, respectively) or cells depleted or over expressing SF2/ASF (lanes 3 and 5, respectively). In the context of the ENSA pre-mRNA, the observed binding site is located within an alternative cassette exon. As expected ectopic expression of SF2/ASF increases inclusion of the alternative exon, relative to the cells depleted for SF2/ASF (Fig. 4B). Binding sites for SF2/ASF within the PABP and Neto2 pre-mRNAs are in exons proximal to alternative cassette exons. We found that ectopic expression of SF2/ASF causes increased skipping of the alternative cassette exon in both cases (Fig. 4B). Finally, the observed binding site for SF2/ASF within its' own pre-mRNA (transcribed from the SFRS1 gene) is located near an ultra-conserved element adjacent to a retained intron within the 3′UTR [27], [28]. This intron is believed to attenuate expression of the SFRS1 gene through regulated unproductive splicing and translation (RUST) mechanism [27]. We observe that ectopic expression of SF2/ASF increase splicing of this intron and provides supporting evidence for the RUST hypothesis (Fig. 4B).
Figure 4. Alternative splicing of endogenous RNA transcripts targeted by SF2/ASF.
A) Western blot analysis of HEK293T cells co-transfected with siRNA duplexes and expression plasmids. The upper panel is probed with anti-SF2/ASF monoclonal antibody, the lower panel is probed with anti-GAPDH monoclonal antibody. Lane 1, non transfected control (NTC); Lane 2, Mock (no siRNA or plasmid); Lane 3, siRNA targeting SF2/ASF and empty vector (pCGT7); Lane 4, scrambled siRNA and empty vector; Lane 5, scrambled siRNA and T7-tagged SF2/ASF expression plasmid (pCGT7 SF2). Total RNA was purified for RT-PCR analysis from samples corresponding to lanes 3 and 5. B) RT-PCR analysis of alternative splicing. Radiolabeled products were visualized by phosphorimaging. Schematic diagrams show exon-intron structure of target genes and patterns of alternative splicing. Thin rectangles indicate untranslated regions, thick rectangles indicate coding regions, thick lines indicate introns and thin lines indicate the alternative splicing event. Exons identified by CLIP are yellow. The grey region in the SFRS1 3′UTR indicates a retained intron. Arrowheads to the left of each panel indicate the included (gray arrowhead) and skipped (black arrowhead) alternative mRNA isoform. Lane 1 and 2 contain RNA purified from SF2/ASF overexpressing and depleted cells, for example see panel A, lanes 5 and 3, respectively. The sub-cellular fraction corresponding to the initial CLIP experiment is indicated below each gene symbol.
The splicing of each of these endogenous pre-mRNA targets is effected by increasing the intracellular concentrations of SF2/ASF. It is known that SF2/ASF can remain associated with mRNA following the splicing reaction and that SF2/ASF can enhance the translation of reporter mRNAs [7], [8]. The CLIP method revealed that a subset of SF2/ASF mRNA targets, including PABP, NETO2, ENSA and SFRS1, were bound by SF2/ASF in the cytoplasm of HEK293T cells. This observation suggests that SF2/ASF may also enhance the translation of these particular mRNA targets. To test this hypothesis we asked whether a specific mRNA target of SF2/ASF exhibited increased association with polyribosomes, the actively translating pool of ribosomes, when SF2/ASF is overexpressed. Cytoplasmic extracts were prepared from HEK293T cells transfected with empty expression vector or epitope tagged SF2/ASF (Fig. 5A). Cytoplasmic extracts were resolved across 10–45% sucrose gradients. Following fractionation, total RNA was extracted from every other fraction and cDNA was synthesized using oligo dT primer. Upon ectopic expression of SF2/ASF all four endogenous mRNA targets showed increased association with polyribosomes (Fig. 5A, lanes 5–8). By contrast, levels of GAPDH remain constant across the gradient when SF2/ASF is overexpressed. We also examined the distribution of several other endogenous targets, but found no difference when SF2/ASF is over expressed, suggesting that this effect is specific for a subset of transcripts rather than a global phenomenon (data not shown). Additionally, there are no obvious differences between the distributions of total RNA across the gradient. These data are in good agreement with our previous findings that SF2/ASF can enhance the polysome association and translation of luciferase reporter mRNAs both in vitro and in vivo [7]–[9]. Finally these data provide clear evidence that SF2/ASF can drive enhaced polysome association of endogenous mRNAs in vivo.
Figure 5. Polyribosome association of endogenous SF2/ASF mRNA targets.

A) Western blot analysis of cytoplasmic extracts prepared from cells transfected with pCGT7 of pCGT7 SF2/ASF (lanes 1 and 2, respectively). Blots were probed with anti-SF2/ASF and anti-GAPDH (left and right panel, respectively). B) Polyribosome association of endogenous mRNAs. Cytoplasmic extracts from control or pCGT7 SF2/ASF cells were resolved by sucrose gradient fractionation. The upper panel shows the absorbance of rRNA across the gradient, the positions of ribosomal subunits and complexes are indicated. Lower panels: RT-PCR analysis of RNA targets. Products were resolved by agarose gel electrophoresis. The sub-cellular fraction corresponding to the initial CLIP experiment is indicated below each gene symbol.
Discussion
SF2/ASF is a nucleo-cytoplasmic shuttling RNA binding protein with clearly defined activities in spliceosome assembly. Our previous work established a role for SF2/ASF in mRNA translation [7], [8]. These findings suggested that SF2/ASF may co-ordinate the nuclear and cytoplasmic steps of post-transcriptional gene expression for a subset of pre-mRNAs. This hypothesis is further supported by the observation that extrinsic signals leading to activation of the AKT protein kinase enhance the alternative splicing and translation of reporter RNAs containing a consensus SF2/ASF binding site [29]. We used the CLIP protocol to extend our findings to endogenous cellular mRNAs. CLIP allowed us to sample direct mRNA targets of SF2/ASF under conditions that preserve the in situ specificity of protein-RNA interactions. By coupling CLIP with sub-cellular fractionation we were able to identify mRNAs associated with SF2/ASF in the nucleus, cytoplasm and in the actively translating pool of ribosomes (Fig. 2 and Table S1). These data revealed a set of mRNAs whose splicing and translation may be coordinated by SF2/ASF. Collectively, our data do not support the view that SF2/ASF plays a general role in mRNA translation, but suggests that the biological importance of this aspect of SF2/ASF function may be at transcript and cell types-specific events. Our recent findings that mTOR is critical to translational control by SF2/ASF suggest that SF2/ASF may connect cellular signal transduction pathways with post-transcriptional control of specific target mRNAs [9].
Using the CLIP approach we were able to not only identify potential RNA targets for SF2/ASF but also define a consensus binding motif present in exonic binding sites (Fig. 3). This purine-rich motif closely resembles the well-characterized exonic splicing enhancers (ESE) present in the fibronectin EDA alternative cassette exon as well as exon 5 of the cardiac troponin T (cTNT) gene [30], [31]. The binding specificity of SR proteins has also been extensively studied using SELEX strategies. Our data are in good agreement from results obtained using only the RRM domains of SF2/ASF for in vitro evolution of binding sites [19]. By contrast, functional SELEX experiments, in which bona fide ESEs within reporter constructs are replaced by randomized sequence and in vitro splicing is driven by a single recombinant SR protein in S100 complementation assays, yielded different consensus motifs for SF2/ASF [18]. The functional SELEX motifs are more heterogeneous in nature but may represent binding sites for SF2/ASF that can lead to productive splicing. The differences between motifs identified by functional SELEX and CLIP may reflect inherent differences in the experimental conditions. For example, photo cross-linking of protein-RNA interactions is believed to be fairly inefficient, but it allows for recovery of sequences under stringent conditions. It remains possible that sequences identified by CLIP represent only a subset of possible binding sites that are both abundant and suitable for cross-linking. Data presented in Fig. 4B demonstrates that SF2/ASF can regulate the alternative splicing of a subset of pre-mRNAs suggesting that the observed binding sites are functionally relevant. Indeed, 26 out of 44 endogenous pre-mRNAs containing binding sites for SF2/ASF within or proximal to cassette exons appear to be functionally relevant when tested by RT-PCR (Fig. 4B and data not shown). Thus we feel the CLIP method has captured a subset of biologically relevant SF2/ASF binding sites.
Cataloguing the collection of cis-acting elements directing post-transcriptional gene regulation on a global scale is only at the initial stages of investigation. In silico studies have successfully elucidated cis-acting elements that are enriched or co-evolve with constitutive and alternative exons. Several recent computational screens have defined thousands of hexa- or octamers as splicing regulatory sequences [32]–[34]. If a combinatorial code governing splice site selection exists, the solution will likely emerge through interdisciplinary approaches merging computational methods with biochemical and functional genomics approaches. Methods such as SELEX, CLIP and RIP-Chip have the potential to illuminate the specificity and functions of RNA binding proteins by making connections between trans-acting RNA binding proteins and the biological processes they regulate [13], [35]–[37]. CLIP also provides the added value of determining potential mechanisms of RNA binding protein action by revealing the positional context of cis-acting RNA elements within a transcript [13]. RNA binding protein target specificity can also be evaluated using platforms to profile alternative splicing such as exon tiling arrays, spliced junction array platforms and RNA-Seq [38]–[40]. Discrimination of direct versus indirect effects is a limitation of these experiments however this may be overcome by applying computational models of consensus binding sites generated by CLIP or SELEX to classify global changes in alternative splicing [39], [41]. Our initial CLIP experiment with SF2/ASF revealed just a small snap shot of putative RNA targets. One clear limitation of this study is the inefficiency of the Sanger sequencing. Merging CLIP with high throughput genome wide sequencing platforms such as Solexa and 454 (Illumina, USA and Roche Diagnostics, USA, respectively), will vastly expand our ability to define the RNA binding specificity. These data will allow for a high resolution map of SF2/ASF binding sites that not only illuminate mechanisms of action in pre-mRNA splicing but also reveal connections to important biological processes such as genome stability, cellular transformation and non-coding RNA processing.
Materials and Methods
Cell culture and transient transfection
For CLIP analysis of SF2/ASF, HEK293T cells were grown to 75–80% confluence at 37°C, 5% CO2. HEK293T cells were cultured in DMEM containing 10% Fetal Calf Serum and antibiotics. Transient transfection of HEK293T cells with plasmid DNA was performed using Lipofectamine 2000 (Invitrogen) following the manufacturers instructions. Transient transfection of siRNA duplexes and plasmid DNA were performed using Duo-fect transfection reagent (Dharmacon) following the manufacturers specifications. Cells were harvested 48 hours following transfection. siRNA transfection efficiency was monitored using siGLO green siRNA duplexes (Dharmacon) and was found to be 80–95% efficient.
Plasmids and siRNA
T7 Epitope tagged SF2/ASF was expressed from the pCGT7 vector and has been described previously [42]. siRNA Smartpools targeting the endogenous SF2/ASF transcript, or scrambled nontargeting siRNA duplexes were purchased from Dharmacon.
Cross-linking Immunoprecipitation of SF2/ASF from HEK293T cells
CLIP analysis of SF2/ASF was performed as described by Ule et al.[32] with the following modifications relating to extract preparation and RNase treatment. Nuclear and cytoplasmic extracts were prepared from UV-treated or control cells as previously described [7]. The soluble extract was treated with 30 U RQ DNase 1 for 20 min at 37°C. The reactions were terminated by the addition of 20 mM EDTA. Subsequently, ribosomal subunits were cleared by centrifugation of the extract at 100,000× g using an Optima Max ultracentrifuge (Beckman Coulter, USA) in a TLA120.2 rotor for 20 min. Cleared extracts were then treated with a dilute cocktail of RNase A/T1 (Ambion, USA) at a final dilution range of 1∶1000–1∶10,000 for 20 min at 37°C. 200 U RNaseOut (Invitrogen, USA) was then added to the extract. Proteins were then partially denatured by addition of an equal volume of buffer A (2× PBS, 0.2% SDS, 1% NP-40). An aliquot of each UV-treated extract was used to prepare input RNA fragments. The remainder of the extract was used for immunoprecipitation with anti-SFRS1 monoclonal antibody.
Characterization of the CLIP tags
All Human exon and gene information was downloaded from Ensembl, using the BioMart tool. The sequence data was downloaded using Perl scripts to access the ensembl database through the Ensembl Core Perl API. The CLIPS sequences were mapped against unspliced gene, cDNA and coding sequences using Mega BLAST (Zhang et al. 2000). The unambiguous mapping of the CLIP tags allowed us to match the CLIPs to Ensembl gene ids. The characterization of type of splicing event that occurs on the alternatively spliced exons that were found to be bound by SF2/ASF was performed according to Ensembl exon information.
Identification of the SF2/ASF binding site
The distinct CLIP tags that match exons were used on the MEME (Bailey et al., 2006) web server (http://meme.sdsc.edu/meme/meme.html) to identify patterns that are probabilistically significant. The intronic set of CLIP tags was used in the same way but did not result in any meaningful pattern.
The genome wide distribution of the SF2/ASF binding site pattern
To identify the genome-wide distribution of SF2/ASF binding sites around exon splice sites, for each exon from a protein coding gene (greater than 100 bp), we generated a composite sequence made of the 75 bp upstream of the 3′ splice site, the 5′ exonic 25 bp, the 50 bp from the middle of the exon, the 25 bp 3′ exonic and 75 bp downstream of the 5′ splice site. There were in total 174,440 exons that satisfied this criteria. These were then analyzed using the binding sites in CLIP tags that were identified by MEME.
Parsing of Data
All parsing of data was performed using multiple Perl scripts.
in vivo splicing assay
HEK293T cells were transfected as described above. Cells were harvested 48 hours post-transfection. One aliquot was used for western blot analysis the other was used to prepare total RNA from the cytosolic fraction. RNA was extracted using TRI Reagent LS (Sigma) following the manufacturers instructions. cDNA was synthesized using oligo dT primer and Superscript RT III (Invitrogen). 50 ng of cDNA was used as a template for RT-PCR analysis. Primers were 5′ end labelled with 32PγATP and T4 PNK (NEW England Biolabs). Primers are available upon request. PCR was performed using the following conditions: 3 min at 94°C; 30 cycles of 94°C 30 sec, 59°C 30 sec, 72°C 60 sec; 72°C 5 min. Following PCR, the reactions were ethanol precipitated and resolved upon 10% polyacrylamide/7M urea gels. RT-PCR reactions were visualized by autoradiography or phosphorimager.
Polyribosome Profiles
Polyribosome profiles were obtained from HEK293T cells as previously described [7], [8]. Total RNA was extracted from each fraction using TRI Reagent LS. Every other fraction was analyzed by RT-PCR. Purification of polyribosome for CLIP analysis of SF2/ASF was accomplished by pelleting polyribosomes through 10–25% sucrose gradients as previously described [8]. Polysome pellets were washed gently with cold PBS, then resuspended in cold 0.5× buffer A (see above). The polysomes were then treated with RQ DNase, and RNase as described in the CLIP protocol [12].
Supporting Information
A supplementary excel spread sheet accompanies this manuscript (Table S1). It contains complete annotation of the CLIP data.
Supporting Information
A complete list of binding site annotation using the Ensembl, UCSC Known Gene and Rfam databases. The Excel file can be filtered in order to find binding sites identified by CLIP using nuclear, cytoplasmic or polysomal extracts. Headers for the table are as follows: Chromosome: Defines the specific chromosome from the human genome to which the seq-block mapped. Strand: CLIP preserves the orientation of the captured RNA marker, so that it is possible to determine the strandedness of the locus. Id: Generic description given to each binding site during our annotation work flow. Region start/end: The precise chromosomal coordinates defining the unique or overlapping sequence blocks. Regions are define by at least 2 partially overlapping binding sites mapping to the locus. # of fragments in the Cytoplasm/Nucleus/Polysome indicates if the binding site was identified in each compartment. The number specifies whether a sequence block was absent, present in a single assay or multiple assays. # of sample targets is useful for finding binding sites that are in 1, 2 or all three cellular fractions. Gene Annotation: This column describes the relationship of the seq block to annotated protein coding genes based on the UCSC Known Gene Database. Exon Style: This column describes the relationship of the seq block to annotated parts of protein coding genes (exon, intron etc). The strategy is presented in Supplementary Figure 2. USCS Known Gene database ID: This column refers to the name of a specific gene cluster by the UCSC Known Gene database. Gene Symbol: This column contains information pertaining to the approved HUGO Gene Nomenclature Committee symbol for each protein coding gene. Exon Position: This column describes the position of the exon within the protein coding gene. First/Last exon columns: Designation of “1” in either column indicates 5′ or 3′ terminal exon. “1” in both columns denotes that the sequence block is in a single exon gene. Upstream/Downstream Exon Position: These columns are useful for determining the position of introns within a protein coding gene. ncRNA Annotation: Describes the relationship of a sequence block to annotated non coding RNA (ncRNA). Annotation is based on the Rfam database. ncRNA Name: This column describes the gene symbol for each ncRNA containing a sequence block. UTR type: Describes the relationship between sequence blocks and untranslated regions of protein coding genes. Splicing Event: Gives alternative splicing annotation for exonic binding sites based upon AceVIEW, ALT Events and Fast-db, databases.
(0.13 MB XLS)
Acknowledgments
We wish to thank Dr. Veronica Vanheyningen (Medical Research Council Human Genetics Unit) for facilitating collaboration between J.R.S., J.C.F. and P.C.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by the Medical Research Council (J.F.C., P.C. and J.A.H) plus additional funds from EURASNET (European Alternative splicing Network) to J.F.C.. J.R.S. was initially supported by a postdoctoral fellowship from the Caledonian Research Foundation and is currently funded by American Heart Association Scientist Development Grant #0830206N. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 4.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13:302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 5.Ram O, Ast G. SR proteins: a foot on the exon before the transition from intron to exon definition. Trends Genet. 2007;23:5–7. doi: 10.1016/j.tig.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci U S A. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. Embo J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 12.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 14.Ule J, Ule A, Spencer J, Williams A, Hu JS, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 15.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006 doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 16.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 18.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, et al. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 19.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. Embo J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 21.Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 22.de la Grange P, Dutertre M, Martin N, Auboeuf D. FAST DB: a website resource for the study of the expression regulation of human gene products. Nucleic Acids Res. 2005;33:4276–4284. doi: 10.1093/nar/gki738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Drummond-Barbosa D, Spradling AC. Alpha-endosulfine, a potential regulator of insulin secretion, is required for adult tissue growth control in Drosophila. Dev Biol. 2004;266:310–321. doi: 10.1016/j.ydbio.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Craig RL, Schay J, Chu W, Das SK, et al. Alpha-endosulfine, a positional and functional candidate gene for type 2 diabetes: molecular screening, association studies, and role in reduced insulin secretion. Mol Genet Metab. 2004;81:9–15. doi: 10.1016/j.ymgme.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 28.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 30.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, et al. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- 36.Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 2006;7:R113. doi: 10.1186/gb-2006-7-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang ZD, Paccanaro A, Fu Y, Weissman S, Weng Z, et al. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 2007;17:787–797. doi: 10.1101/gr.5573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 39.Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, et al. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science. 2008 doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson S, Blanchette M, Park J, Savva Y, Yeo GW, et al. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cazalla D, Sanford JR, Caceres JF. A rapid and efficient protocol to purify biologically active recombinant proteins from mammalian cells. Protein Expr Purif. 2005;42:54–58. doi: 10.1016/j.pep.2005.03.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A complete list of binding site annotation using the Ensembl, UCSC Known Gene and Rfam databases. The Excel file can be filtered in order to find binding sites identified by CLIP using nuclear, cytoplasmic or polysomal extracts. Headers for the table are as follows: Chromosome: Defines the specific chromosome from the human genome to which the seq-block mapped. Strand: CLIP preserves the orientation of the captured RNA marker, so that it is possible to determine the strandedness of the locus. Id: Generic description given to each binding site during our annotation work flow. Region start/end: The precise chromosomal coordinates defining the unique or overlapping sequence blocks. Regions are define by at least 2 partially overlapping binding sites mapping to the locus. # of fragments in the Cytoplasm/Nucleus/Polysome indicates if the binding site was identified in each compartment. The number specifies whether a sequence block was absent, present in a single assay or multiple assays. # of sample targets is useful for finding binding sites that are in 1, 2 or all three cellular fractions. Gene Annotation: This column describes the relationship of the seq block to annotated protein coding genes based on the UCSC Known Gene Database. Exon Style: This column describes the relationship of the seq block to annotated parts of protein coding genes (exon, intron etc). The strategy is presented in Supplementary Figure 2. USCS Known Gene database ID: This column refers to the name of a specific gene cluster by the UCSC Known Gene database. Gene Symbol: This column contains information pertaining to the approved HUGO Gene Nomenclature Committee symbol for each protein coding gene. Exon Position: This column describes the position of the exon within the protein coding gene. First/Last exon columns: Designation of “1” in either column indicates 5′ or 3′ terminal exon. “1” in both columns denotes that the sequence block is in a single exon gene. Upstream/Downstream Exon Position: These columns are useful for determining the position of introns within a protein coding gene. ncRNA Annotation: Describes the relationship of a sequence block to annotated non coding RNA (ncRNA). Annotation is based on the Rfam database. ncRNA Name: This column describes the gene symbol for each ncRNA containing a sequence block. UTR type: Describes the relationship between sequence blocks and untranslated regions of protein coding genes. Splicing Event: Gives alternative splicing annotation for exonic binding sites based upon AceVIEW, ALT Events and Fast-db, databases.
(0.13 MB XLS)