Abstract
The study of elevational diversity gradients dates back to the foundation of biogeography. Although elevational patterns of plant and animal diversity have been studied for centuries, such patterns have not been reported for microorganisms and remain poorly understood. Here, in an effort to assess the generality of elevational diversity patterns, we examined soil bacterial and plant diversity along an elevation gradient. To gain insight into the forces that structure these patterns, we adopted a multifaceted approach to incorporate information about the structure, diversity, and spatial turnover of montane communities in a phylogenetic context. We found that observed patterns of plant and bacterial diversity were fundamentally different. While bacterial taxon richness and phylogenetic diversity decreased monotonically from the lowest to highest elevations, plants followed a unimodal pattern, with a peak in richness and phylogenetic diversity at mid-elevations. At all elevations bacterial communities had a tendency to be phylogenetically clustered, containing closely related taxa. In contrast, plant communities did not exhibit a uniform phylogenetic structure across the gradient: they became more overdispersed with increasing elevation, containing distantly related taxa. Finally, a metric of phylogenetic beta-diversity showed that bacterial lineages were not randomly distributed, but rather exhibited significant spatial structure across the gradient, whereas plant lineages did not exhibit a significant phylogenetic signal. Quantifying the influence of sample scale in intertaxonomic comparisons remains a challenge. Nevertheless, our findings suggest that the forces structuring microorganism and macroorganism communities along elevational gradients differ.
Keywords: elevation gradient, microbial ecology, phylogenetic diversity, macroecology, biogeography
Roughly 250 years ago, Carolus Linnaeus (1) documented how distinct plant and animal communities characterized the succession of climatic zones along the slopes of mountains. Such elevational gradients are characterized by dramatic changes in climate and biotic turnover over short geographic distances. The patterns observed by Linnaeus and his contemporaries played a foundational role in the development of ecology and biogeography (2). Studies of how individual taxa and community composition respond to elevational gradients have led to a search for generalized elevational patterns of biodiversity (3–5). These studies have documented elevational patterns of diversity across a wide variety of taxonomic groups, including trees, mammals, birds, reptiles, insects, and amphibians. In sum, this work has shown that taxa generally exhibit either monotonically decreasing or hump-shaped richness patterns with elevation (6, 7). However, despite a large number of proposed hypotheses to explain elevation patterns of diversity, their causes remain poorly understood. Improved knowledge of elevation gradients is fundamental to advancing basic ecology and predicting the potential consequences of climate change. Species in montane regions are often cited as being very sensitive to the impacts of warming (8–10).
Although elevational patterns of diversity for plants and animals are well established, we know very little about how microbial diversity varies across elevational gradients. This is a serious gap in our general understanding of biodiversity, given that microbes are abundant and diverse, play a central role in ecosystem functioning, and will likely be an important component of ecosystem response to global warming (11–13). Elevational diversity studies that consider empirical patterns of macroorganisms and microorganisms in parallel are needed to provide a more unified framework for understanding diversity patterns in Earth's major environmental gradients and predicting systemwide ecological responses to climatic change.
Traditional elevational diversity studies have focused on how patterns of species richness, abundance, and range size change with altitude. These analyses have used a nomenclatural approach by focusing on species identities. However, the increasing availability of molecular phylogenies has renewed interest in using phylogenetic approaches to study the forces that influence patterns of biodiversity and biogeography (e.g., ref. 14). Because many species traits are generally conserved during the evolution of a lineage, one would expect a positive relationship between a measure of the phylogenetic relatedness of two species and a measure of their overall ecological similarity (phylogenetic niche conservatism) (15). As a result, analysis of the degree of phylogenetic relatedness of taxa found within and across communities should provide insight into the ecological and evolutionary processes that organize these communities.
Here, in an effort to assess the generality of elevational diversity patterns and the forces that structure these patterns, we quantified both plant and soil bacterial diversity patterns along an elevational gradient in the Colorado Rocky Mountains. A parsimonious hypothesis is that if the forces structuring biodiversity across the gradient are the same for bacteria and plants, then the resulting taxon and phylogenetic biogeographic patterns will be similar for both groups. Alternatively, if ecological and evolutionary processes along elevational gradients differ between the two groups (e.g., the taxa differ in their dispersal ability, response to environmental heterogeneity, interspecific interactions, or speciation rates), then we would expect them to be characterized by distinct patterns of diversity. To test these hypotheses, we adopted a multifaceted approach that examines diversity in the context of both ecological and evolutionary patterns. Therefore, in addition to the established convention of quantifying patterns of taxon richness and taxon turnover along the gradient (e.g., refs. 16 and 17), we examined several biodiversity measures that incorporate information about the phylogenetic structure, phylogenetic diversity, and phylogenetic turnover of plant and bacterial communities.
Elevational Diversity in a Phylogenetic Context
While the sampling methods and taxonomy used to quantify plant diversity are well established and standardized, microbial surveys vary greatly in their approach to characterizing diversity (18). We determined the bacterial community composition of our soil samples by analyzing a PCR-amplified region of 16S ribosomal DNA, the most commonly used indicator of microbial biodiversity. Because bacteria are overwhelmingly diverse in soils we chose PCR primers that narrowed our focal group to the phylum Acidobacteria. This subgroup of bacteria is diverse and ubiquitous in soils (19) and thought to play an important role in biogeochemical cycling (20).
We followed the classic approach to intertaxonomic diversity analysis by comparing patterns of species richness and phylotype richness of plants and bacteria, respectively, along the gradient. We also quantified the phylogenetic diversity of every sampled community by calculating the sum of the branch length in a phylogeny that connects all species in a community and the root (21). Phylogenetic diversity is more inclusive than a simple count of species or types, in that it quantifies the evolutionary history of a group of taxa (22). Conservation biologists are interested in preserving phylogenetic diversity, as this is fundamental to maximizing evolutionary options for the future (23–26). Phylogenetic diversity is also believed to correspond to “feature diversity,” meaning the number of evolutionarily derived traits within a biological community (21).
In addition to measuring phylogenetic diversity, we quantified community phylogenetic structure along the gradient by using two commonly used metrics: a mean pairwise distance metric sensitive to phylogeny-wide patterns [net relatedness index (NRI)] and a nearest-taxon-based measure sensitive to patterns at the “tips” of the phylogeny [nearest taxon index (NTI)] (27). The degree of phylogenetic relatedness quantified by these metrics provides insight into drivers of community assembly. Assuming phylogenetic niche conservatism, phylogenetic clustering within a local assemblage is considered consistent with the hypothesis that selective filters (e.g., environmental conditions) cause local assemblages to comprise closely related taxa (27). Phylogenetic overdispersion, on the other hand, can be explained by two possible biotic interactions: competition (27) or facilitation (28, 29). In the case of competition, more closely related species are hypothesized to compete more strongly with one another. This results in competitive exclusion, which leads to a community of distantly related species. In the case of facilitation, facilitator species are hypothesized to create microhabitats that permit distantly related species adapted to different environments to persist within a local assemblage.
In addition to considering patterns in the diversity and phylogenetic structure within communities along the elevation gradient (alpha-diversity), we investigated how community composition changes across a landscape (beta-diversity). Ecologists have long recognized that beta-diversity is important for understanding the biodiversity of montane ecosystems (16, 30–33). We examined beta-diversity in terms of compositional similarity, defined as the fraction of taxa shared between two samples (Sørensen index), and phylogenetic similarity, defined as the fraction of branch lengths shared between two samples. By analogy with the well established distance–decay relationship, which describes the decrease in compositional similarity between two communities with increasing geographic distance (or equivalently elevational separation) between them (34), we described the decrease in phylogenetic similarity with distance (phylogenetic distance–decay). Our objective in exploring both measures of beta-diversity is to understand not only if there are shifts in compositional similarity with increasing elevational distance, as expected along an environmental gradient, but to quantify the phylogenetic nature of the shifts.
Phylogenetic similarity reflects the combined additive influence of: (i) lineages that are shared between two communities that lead to shared taxa, and (ii) lineages that are shared but ultimately lead to unshared taxa. One can test whether the phylogenetic similarity between two communities is solely a consequence of compositional similarity, or if it is also caused by a nonrandom structure of shared and unshared lineages. A significant phylogenetic distance–decay pattern (i.e., one that differs from that expected by taxa turnover alone; see Materials and Methods) reflects significant spatial variability in lineage composition across a landscape. Based on the assumption of phylogenetic niche conservatism described above, changes in lineage composition should correspond to changes in the traits of species. Under this model, a significant phylogenetic distance–decay relationship should reflect strong variability in the ecologically relevant traits of biological communities across a landscape.
Results and Discussion
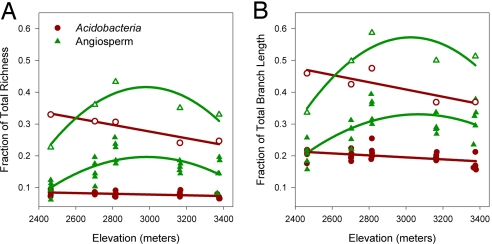
Whereas bacterial richness decreased monotonically from the lowest to highest elevations, plant richness followed a unimodal pattern with a peak in species richness at midelevations (Fig. 1A). These contrasting diversity patterns emerged when richness values were calculated for bacterial and plant samples individually and also when the samples for each respective group were pooled together at every elevational band [following the protocol suggested by Whittaker (16)]. To our knowledge, an altitudinal richness pattern has never been reported for microorganisms. The patterns observed here for microbes and plants are consistent, respectively, with the classical monotonically decreasing and hump-shaped patterns observed across most macroorganism groups (4, 6, 7). It has been argued that these two contrasting richness patterns may emerge as a result of inconsistent sampling approaches among different studies, rather than an underlying ecological mechanism (3, 7). By implementing a parallel sample design for the bacteria and plants, we controlled for two potential biases: variation in sampling extent (i.e., the geographic distance between the furthest sampled elevations), which often occurs among studies, and sampling intensity (or effort) along the gradient within a taxonomic group, which often occurs within studies. As a result, the disparity in elevational richness patterns observed between bacteria and plants is likely caused by differences in how ecological and evolutionary processes have operated across the gradient (although see below for a discussion on the potential influence of scaling effects).
Fig. 1.
Variation in taxon richness (A) and phylogenetic diversity (B) across the elevation gradient. Data are presented as the fraction of total richness and phylogenetic diversity across the gradient. Solid symbols indicate sample richness (core or quadrat), and open symbols indicate the pooled richness at each elevational site (n = 5 per site). At the sample level, Acidobacteria richness and phylogenetic diversity linearly decrease with elevation (regression analysis, r2 = 0.22, P < 0.05; r2 = 0.23, P < 0.05, respectively), whereas angiosperm richness and phylogenetic diversity patterns are hump-shaped (regression analysis, r2 = 0.53, P < 0.0005; r2 = 0.47, P < 0.005, respectively). Model choice was based on Akaike information criteria.
As expected, for both bacteria and plants (26) we found that the patterns of phylogenetic diversity mirrored those of taxon richness (Fig. 1B). However, a more detailed look at the phylogenetic structure of the bacterial and plant communities revealed another significant difference. At all elevations bacterial communities had a tendency to be more phylogenetically clustered than expected by chance (Fig. 2). This observation is consistent with results reported by Horner-Devine and Bohannan (35) who found that bacterial communities in a wide range of environments tended to be phylogenetically more closely related than expected by chance. Given the parsimonious hypothesis that closely related taxa are more ecologically similar (i.e., phylogenetic niche conservatism), our results suggest that abiotic filtering tends to be a more prominent force in the structuring of bacterial communities along the gradient. Several studies have suggested that for most macroorganisms, ecological traits are phylogenetically conserved (36–38). It is important to emphasize that although this statement may be correct for macrorganisms, the generality of niche conservatism for microorganisms and in particular bacteria is unknown. Observed phylogenetic clustering in microbial communities could also be the result of radiation events combined with dispersal limitation (35). As we discuss below, alternative explanations for the patterns we observe relate to the phylogenetic and spatial scale of our analyses (39, 40). Scaling issues are relevant to all of the biodiversity patterns we examined.
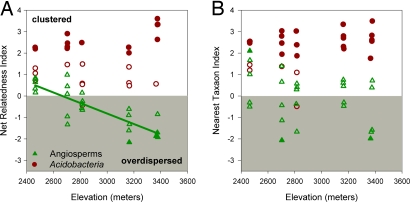
Fig. 2.
Variation in community phylogenetic relatedness along the elevation gradient as measured with the NRI (A) and NTI (B). Positive index values indicate phylogenetic clustering, and negative values indicate phylogenetic overdispersion. Observed community phylogenetic structures unlikely to arise by chance (P < 0.05) are depicted by solid symbols. All microbial communities are clustered, with >50% being significantly clustered. Angiosperm communities are not uniformly clustered or dispersed across the gradient, but rather become increasingly overdispersed with increasing elevation. This trend in increased overdispersion with elevation is significant when measuring relatedness with the NRI (solid line; r2 = 0.70, P < 0.001).
In contrast to bacteria, plant communities did not show a uniform phylogenetic structure across the gradient. Plant communities tended to exhibit either random phylogenetic structure or phylogenetic overdispersion. Surprisingly, our analyses indicated that plant communities also tended to become increasingly overdispersed at higher elevations (Fig. 2). Given niche conservatism, phylogenetic overdispersion is consistent with the importance of biotic forces (competitive exclusion or facilitation) structuring community diversity. Recent experimental evidence suggests that both of these forces are important drivers in alpine plant community assembly, with a shift from competition at lower elevations, where conditions are less physically stressful, to facilitation at higher elevations where abiotic stress is high (41). Increased overdispersion at high elevations suggests that the influence of facilitation on high-elevation communities is stronger than the influence of competition at low elevations. An alternative explanation is that the evolution of traits necessary to cope with environmental conditions at high elevations has occurred independently in distantly related lineages (i.e., convergent evolution in high alpine plants) (27). This explanation goes against the assumption of phylogenetic niche conservatism.
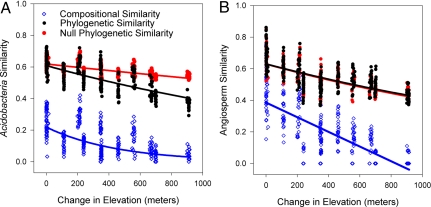
We observed that both plant and bacterial compositional similarity significantly decreased with elevational distance (Fig. 3). Plant and bacterial communities differed, however, in their phylogenetic distance–decay patterns. The bacterial phylogenetic distance–decay curve was significantly steeper than expected from the observed bacterial taxa turnover alone (Fig. 3A). In contrast, the plant phylogenetic distance–decay curve was not significantly different from expected from the observed decay in plant compositional similarity. These results are consistent with those reported above for the NRI and NTI measures of community phylogenetic structure, indicating that, bacteria lineages were not randomly distributed across the elevation gradient. Rather, bacterial lineages exhibited a spatially structured pattern across the gradient. Given the parsimonious hypothesis that closely related taxa are more ecologically (or functionally) similar, our observations suggest that bacterial lineages harbor increasingly disparate ecological features (or functions) at increased elevational distances as a probable consequence of abiotic filtering. These findings highlight the utility of gathering information on phylogenetic relationships between communities in montane regions as a means to quantify the potential consequences of selectively trimming evolutionary lineages under the scenario of mountaintop extinctions in response to global warming.
Fig. 3.
Compositional and phylogenetic similarity of Acidobacteria communities (A) and angiosperm communities (B), as a function of the elevation separating the communities. The compositional (blue) and phylogenetic (black) similarity for both angiosperm and Acidobacteria communities significantly decrease with increasing elevational separation (Mantel test, P < 0.001). Lines represent best-fit regressions of similarity versus change in elevation (see Materials and Methods). The slope of the decay of phylogenetic similarity between Acidobacteria communities is significantly steeper than predicted by a null model constrained by the decrease in taxon turnover (red) (P < 0.05). The slope of the decay in phylogenetic similarity across the angiosperm communities is not significantly different from the null prediction given species turnover. Fig. S7 illustrates within-site similarity as a function of elevational distance.
Although our study was not designed to directly examine the environmental drivers of elevational diversity patterns, our results do illuminate their potential role in shaping biodiversity patterns across the gradient. The contrasting phylogenetic diversity patterns we observed in plants and microbes suggest a differing role in how abiotic forces structure communities across the gradient. Soil temperature and pH were consistently correlated with diversity in both plants and bacteria, and bacterial diversity was also strongly correlated with slope in a univariate analysis [supporting information (SI) Table S1]. A multivariate analysis suggested that soil temperature was the major explanatory variable of taxon richness and phylogenetic diversity for both plants and bacteria (P < 0.001 in six of eight multivariate models). Turnover in taxon and phylogenetic composition of plant and bacterial communities was significantly correlated with changes in the majority of our measured environmental parameters (Table S2); however, the combined influence of soil temperature, pH, and total nitrogen was the most important predictor for both groups. After controlling for these environmental parameters, geographic distance between samples significantly correlated with all turnover patterns (partial Mantel test, P < 0.001). These results imply that dispersal limitation could be occurring, but given the small geographic range of our gradient, they are more likely caused by effects of environmental heterogeneity that we did not characterize. The correlation of richness and turnover with temperature and pH is consistent with the findings of other studies of plant (42–44) and microbial (45) diversity.
It is well documented that the scale over which biodiversity is sampled will strongly influence observed patterns. For example, recent empirical studies have shown that decreasing the spatial grain at which organisms are sampled shifts their diversity patterns (7, 39, 40, 46, 47). Although the spatial extent of our study was the same for bacteria and plants, the grain of our sample observations was different between these two groups. The spatial scales over which bacteria interact with each other are likely to be several orders of magnitude smaller than the scale at which they were sampled. Therefore, relative to plants, bacteria were likely sampled at a coarser grain, and thus we may have included a greater amount of environmental heterogeneity within a bacterial sample. Sampling bacteria at a spatial scale that more closely approaches the “ecologically equivalent” grain of plants may result in convergent biodiversity patterns between these two groups.
Taxonomic scale also influences biodiversity patterns. For example, taxonomic breadth, which defines how broadly or narrowly a target community is defined from a phylogenetic perspective (e.g., bacteria versus Acidobacteria), can shift the degree of observed overdispersion or clustering in that community (39). Species are a natural taxonomic unit by which to measure plants (48). Such an intuitive unit does not exist for prokaryotes. In this study we classified partial Acidobacteria 16S ribosomal DNA sequences into taxonomic units based on the commonly used 99% sequence similarity designation (see Materials and Methods). It is unknown how taxonomic resolution, defined as the threshold at which individuals are binned into taxonomic units, should influence phylogenetic patterns, although it has been shown to impact taxonomic patterns such as the taxa–area relationship (49). We found that binning bacteria into increasingly broader taxonomic units (i.e., 97%, 94% and 90% sequence similarity) tended to dampen the strength of all observed elevational diversity patterns. However, general trends did not qualitatively change (Figs. S1–S3), suggesting that taxonomic resolution is not the cause of disparate bacterial and plant biodiversity patterns in this study. Alternative approaches to defining bacterial taxonomic units such as “ecotypes” (50) could significantly change the results and lead to plant and microbial diversity patterns that more resemble one another.
Differences in the approach to building the Acidobacteria and Angiosperm phylogenetic trees should also be considered when comparing phylogenetic patterns between these two groups. The Acidobacteria phylogeny was estimated solely from molecular data identified in this study, whereas the Angiosperm phylogenic tree topology was constructed by using the widely accepted super tree approach (51), and branch lengths were assigned based on estimates of the minimum age of internal nodes (see Materials and Methods). Comparative analyses using molecular approaches alone for both plants and microbes would improve our confidence in such phylogenetic comparisons. Such approaches will be facilitated in the future by increased accessibility to molecular data.
Microorganisms (especially prokaryotes) are very diverse in soils (19, 52). On par with most microbial diversity studies, it is likely that we sampled the most abundant taxa in each soil core along the elevational gradient. Sampling effort (i.e., the proportion of a community that is sampled) is known to significantly influence taxonomic biodiversity patterns (53–55, 75). To our knowledge the influence of sampling effort on phylogenetic biodiversity patterns has not been explored. For example, estimators are available to predict the taxon richness (56) and taxon similarity (57) of ecological communities from sample data, but there are currently no estimators to predict phylogenetic richness, phylogenetic structure, or phylogenetic turnover from sample data. A new generation of estimators is needed for future comprehensive studies that examine taxonomic and phylogenetic diversity patterns in parallel.
As discussed by others, a promising approach to understanding elevational diversity patterns (7), and more generally biodiversity patterns (58), is to conduct intertaxonomic comparisons to elucidate the spatial and taxonomic scales and degree of sampling effort over which microbial biodiversity relationships approach those of macroorganisms. Such an approach is ambitious, but increasingly tractable as molecular approaches advance our ability to comprehensively characterize biodiversity. Here, we have shown that across an elevation gradient, plant and microbial communities exhibit different patterns of diversity. Phylogenetic-based analyses suggest that the evolutionary and ecological processes driving the biogeographic patterns may differ significantly between these two domains of life. Further work is needed to link the phylogenetic patterns to functional differences among plant and bacterial taxa. Such comparative analyses are needed to provide the empirical foundation for a truly inclusive and predictive theory linking patterns of biodiversity to ecosystem function.
Materials and Methods
Study Site and Sampling.
We sampled angiosperm and Acidobacteria communities at five sites along an elevational transect located near the Rocky Mountain Biological Laboratory, Gunnison County, Colorado. The sites extend from 2,460 to 3380 m above sea level and spanned a geographic distance of 39 km. Within each study site, we placed five 1-m by 1-m quadrats in a subtransect running down the slope. Three soil samples, separated by 1 cm, were collected adjacent to the middle, highest, and lowest quadrats at each site (nine total). All soil samples were collected from the B-horizon by using sterile glass collection jars. After collection, the soil samples were homogenized and stored at −80°C until analysis.
Average soil temperature at each site was measured by placing Hobo Temperature Data Loggers (OnSet) at 10-cm depth in relatively open patches and recording soil temperature every hour for the month of July in 2007. Total carbon and nitrogen in the soil samples were measured by using a Costech ECS 4010 CHNS-O system. Soil pH was measured after shaking a soil water (1:3 wt/vol) suspension for 30 min. Soil moisture was measured gravimetrically. ArcGIS data and area photos were used to calculate slope and aspect of the sites. These data had a 15.24-cm resolution per pixel and a horizontal accuracy of 60–90 cm. The environmental data along with a description of how environmental parameters correlate with one another is provided in Table S3, and Table S4, respectively.
Characterization of Acidobacteria Communities with 16S Clone Libraries.
At each site, the bacterial communities within the three soil samples collected adjacent to the middle quadrat and one soil sample adjacent to the lower and upper quadrats were characterized by using sequence analysis of clone libraries (five total). DNA was extracted by using Mobio Power Soil DNA Isolation kits (MoBio Laboratories). Triplicate PCRs were carried out on each soil extraction by using the Acidobacteria-specific PCR primer set Acid31/Eub518 (59). The 25-μl PCR mixtures were composed of 10 μl of 5 Prime MasterMix (5 Prime, Inc.), 14 μl of water, 1 μmol of each primer, and 1 μl of DNA extract. The PCR conditions used were as follows: 3 min at 94°C 25 cycles of 30 s at 94°C, 30 s at 50°C (60), 60 s at 72°C, and a final extension for 5 min at 72°C.
Triplicate PCRs were pooled and then gel-purified by using a MinElute PCR Purification kit (Qiagen). Amplicons were ligated into pCR4-TOPO vectors and cloned by using a TOPO-TA cloning kit (Invitrogen). Ninety-six clones from each soil sample were selected for sequencing. Plasmid purification and sequencing of cloned PCR products was done at the Qiagen Genomic Services/Sequencing facility with an ABI 377 or 377xl sequencer (Applied Biosystems).
A total of 2,239 cloned 16s sequences were aligned with the NAST alignment tool (61), and the alignments were manually edited based on conserved primary sequence and secondary structure information in the ARB software package (62). The phylum affiliation of each sequence was checked by using the BLAST tool within the National Center for Biotechology Information (64). Potentially chimeric sequences were identified by using the Bellerophon server (65). Putative chimeric sequences were manually assessed by building trees in ARB that contained a set of reference sequences obtained from the Greengenes database (63) and the 5′ and 3′ sides of the putative chimeras. Sequences were removed from the analysis that had 5′ and 3′ ends affiliating with different groups of reference sequences in the tree (66).
There is no standard definition of microbial species. Therefore we grouped our 2,196 nonchimeric Acidobacteria sequences into phylotypes with a <99% sequence similarity cutoff by using the programs PHYLIP (67) and DOTUR (68). This is a commonly used phylotype designation (69), which provides high phylogenetic resolution. One sequence was randomly chosen to represent each phylotype. The representative sequences were used to build a phylogenetic tree by maximum-likelihood methods using the program phyML (70). We used Jukes–Cantor and gamma substitution models where the gamma distribution parameter was estimated from the data. Only informative base positions were used to bin sequences into phylotypes and build the microbial phylogenetic tree. A newick format and image of the Acidobacteria phylogenetic tree along with a list of phylotypes identified in each soil core are available in SI Text, Fig. S4, and Table S3, respectively. All diversity analyses were later repeated by using 97%, 94% and 90% sequence similarity cutoffs (Figs. S1–S3).
Characterization of Plant Communities.
Angiosperms within each quadrat were identified to species level and checked against Rocky Mountain Biological Laboratory (RMBL) Herbarium specimens jointly by B.J.E., A.J.K., and C.L. Vouchers are being prepared for deposition in the RMBL Herbarium and the University of Arizona Herbarium. All plants were identified in 2005, except for the plants at the lowest site, which were sampled in 2006. Plots were sampled near the peak of the growing season, and thus some individuals with later phenologies could not be identified to species. We staggered the plant sampling dates with the aim of sampling each community at the same relative phenological time point. Any individuals that could not be identified to species or differentiated from known species were excluded from the analysis. This affected between 10% and 15% of the possible species at each site. We used version R20031202 of phylomatic to construct a tree topology consisting of all of the angiosperms identified in all our quadrats, based on the Angiosperm Phylogeny Group (APG) II backbone (51, 71) and used results from recent plant cladistics studies to resolve polytomies (72, 73). The final tree we used for our analyses was almost completely resolved to the family level. We assigned branch lengths to the tree by using the phylocom module BLADJ to constrain the internal nodes with available age estimates (74) and interpolated the other nodes for which direct age estimates are not available. A newick format and image of the angiosperm phylogenetic tree along with a list of plants identified in each quadrat are available in SI Text, Fig. S5, and Table S3, respectively.
Diversity Analyses.
Taxon richness and phylogenetic diversity.
We define the term community as all phylotypes originating from a single soil core (bacteria) or species identified in a single quadrat (plants). Taxon richness within each community was quantified as the total number of species or phylotypes within that community. Phylogenetic diversity within each community was quantified as the minimum total branch length connecting all species within the community to the root of the phylogenetic tree (21). Phylogenetic diversity was calculated by using the pd module within Phylocom-3.40 (by C. O. Webb, D. D. Ackerly, and S. W. Kemble; available at http://phylodiversity.net/phylocom/). We used a rarefaction sampling approach to account for the unequal sample sizes of each microbial community (number of clones) by calculating the mean of the taxon richness and phylogenetic diversity of 1,000 randomized subsamples of each community. Each community was subsampled by the number of clones in the smallest library (75 clones).
Phylogentic community structure.
Using the classical NRI and NTI (27, 76), we measured the extent to which co-occurring species in a community are phylogenetically related compared with what is expected by chance. With both indices, the phylogenetic structure of the observed community was compared to a null expectation obtained by randomly sampling the pool of all of the species identified in the study 1,000 times, while constraining both the number of taxa in the community and species occurrence across communities (77). Observed values smaller or larger than 975 of the randomizations were considered significantly structured (P < 0.05).
Compositional and phylogenetic similarity.
Compositional similarity between all pairwise comparisons of communities was quantified with the Sørensen Index:
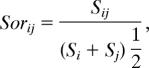 |
where Sij is the number of taxa common to both communities i and j, and Si and Sj are the total number of species found in community i and j, respectively (78). By analogy, phylogenetic similarity between two communities was quantified by using an index, coined PhyloSor:
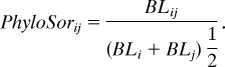 |
Here, BLij is the branch length common to both communities i and j, and BLi and BLj are the total branch lengths of community i and j, respectively.
The PhyloSor index ranges from indefinably close to 0 (two communities only share a very small root) to 1 (both communities are composed of the same taxa). Similar approaches have been carried out by Lozupone and Knight (79) and Ferrier et al. (80) when considering the closely related Jaccard and Bray-Curtis similarity indices. Using PhyloSor, one can test whether two communities are phylogenetically more or less similar than what is expected given their taxa similarity. This is done by comparing the phylogenetic similarity of the observed communities to a null expectation obtained by randomly sampling the pool of all of the species identified in the study while constraining the number of taxa in each community and the number of taxa shared by the two communities.
By analogy with the well established distance–decay relationship, which describes the decrease in compositional similarity between two communities with increasing geographic distance (or equivalently elevational separation) between them (34), we used PhyloSor to quantify the decrease in phylogenetic similarity with distance (phylogenetic distance–decay). We tested whether the slope in the decay of phylogenetic similarity was greater or less than what was expected given the taxonomic decay in similarity by comparing the observed slope with a distribution of distance–decay slopes obtained by randomizing the location of taxa at the tips of the community phylogenetic tree 1,000 times. This is equivalent to randomly sampling the taxa while constraining the number of taxa in each community, the number of taxa shared by any two communities, and taxa occurrence across all communities. The observed phylogenetic slope was assumed significantly different from the null if it was greater than or less than 975 of the slopes of the randomizations (two-tailed test, P < 0.05). To address the unequal sampling of microbial communities, we repeatedly calculated and tested the significance of distance–decay slope on subsampled communities, where communities were subsampled by the number of clones in the smallest library. The significance of results did not differ between repeated subsamples.
Linking Diversity Measurements to Environmental Parameters.
To determine the correlation between elevation and potential environmental drivers with the observed diversity patterns, we used polynomial regression analysis. For each environmental variable, we fit a linear and a quadratic regression model. The best model was determined based on Akaikes Information Criterion differences (81). Using a stepwise regression to select variables and interactions, a multivariate model was constructed for each alpha diversity measurement.
Mantel tests (999 permutations) were used to determine whether compositional and phylogenetic similarity decayed significantly with elevational distance (82). Similarity values between pairwise comparisons of microbial communities were the averages of 1,000 rarefaction samples, as described above. The best fit and the most homoscedastic residuals were found in models that used the log transformation of similarity against elevational distance, with the exception of angiosperm taxa similarity, which was best described by a linear–linear distance–decay model. We used Mantel tests to examine correlations between community similarity and environmental similarity (for a discussion of these methods see refs. 83 and 84). We chose the combination of environmental variables that best explained the changes in angiosperm and Acidobacteria community composition between samples with BIO-ENV (85) and tested the importance of these variables after controlling for geographic distance and vice versa by using partial mantel tests. For all analyses, moisture, carbon, and nitrogen were arcsin(sqrt(y))-transformed and aspect was 1/y-transformed (82).
Acknowledgments.
We thank our field crew from the University of Arizona (Laura Crumbacher, Peter Gaube, Melissa Wilson, James Stegen, Jason Pither, and Robin Sleith) for their help collecting the plant data and soil cores; the staff and researchers of the Rocky Mountain Biological Laboratory, in particular John Harte and Ian Billick for their support; the staff and faculty at the University of California, Merced, for their support; Brendan Bohannan, Kathryn Docherty, Michael Donoghue, Rebecca Mueller, James O'Dwyer, Nathan Swenson, Ian Wright, and an anonymous reviewer for useful comments; and John Avise, Francisco Ayala, and Stephen Hubbell for organizing the Sackler Colloquium. Logistical support was provided by National Science Foundation Grant DBI 0420919 (to Ian Billick). J.A.B., H.M., and J.L.G. were supported by Natinal Science Foundation Grant MCB 0500124 (to J.L.G.). B.J.E., A.J.K., and C.L. were supported by a National Science Foundation CAREER award, the University of Arizona, and a Los Alamos (Department of Energy) Laboratory Directed Research and Development award (to B.J.E.).
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution II: Biodiversity and Extinction,” held December 6–8, 2007, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at www.nasonline.org/Sackler_biodiversity.
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU424348–EU426543).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801920105/DCSupplemental.
References
- 1.Linnaeus C. On the increase of the habitable earth. Amoenitates Academicae. 1781;2:17–27. [Google Scholar]
- 2.Briggs J, Humphries C. In: Foundations of Biogeography. Lomolino MV, Sax DF, Brown JH, editors. Chicago: Chicago Univ Press; 2004. pp. 5–266. [Google Scholar]
- 3.Lomolino MV. Elevation gradients of species density: Historical and prospective views. Glob Ecol Biogeogr. 2001;10:3–13. [Google Scholar]
- 4.McCain CM. Elevation gradients in diversity of small mammals. Ecology. 2005;86:366–372. [Google Scholar]
- 5.Brown JH. Mammals on mountainsides: Elevational patterns of diversity. Glob Ecol Biogeogr. 2001;10:101–109. [Google Scholar]
- 6.Stevens GC. The elevational gradient in altitudinal range: An extension of Rapoport's latitudinal rule to altitude. Am Nat. 1992;140:893–911. doi: 10.1086/285447. [DOI] [PubMed] [Google Scholar]
- 7.Rahbek C. The role of spatial scale and the perception of large-scale species-richness patterns. Ecol Lett. 2005;8:224–239. [Google Scholar]
- 8.Thuiller W. Biodiversity: Climate change and the ecologist. Nature. 2007;448:550–552. doi: 10.1038/448550a. [DOI] [PubMed] [Google Scholar]
- 9.McDonald KA, Brown JH. Using motane mammals to model extinctions due to global change. Conserv Biol. 1992;6:409–415. [Google Scholar]
- 10.Parmesan C. Ecologial and evolutionary responses to recent climate change. Annu Rev Ecol Syst. 2006;37:637–639. [Google Scholar]
- 11.Carney KM, Hungate BA, Drake BG, Megonigal JP. Altered soil microbial community at elevated CO2 leads to loss of soil carbon. Proc Natl Acad Sci USA. 2007;104:4990–4995. doi: 10.1073/pnas.0610045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rillig MC, Wright SF, Shaw MR, Field CB. Artificial climate warming positively affects arbuscular mycorrhizae but decreases soil aggregate water stability in an annual grassland. Oikos. 2002;97:52–58. [Google Scholar]
- 13.Monson RK, et al. Winter forest soil respiration controlled by climate and microbial community composition. Nature. 2006;439:711–714. doi: 10.1038/nature04555. [DOI] [PubMed] [Google Scholar]
- 14.Chave J, Chust G, Thebaud C. In: Scaling Biodiversity. Storch D, Marquet P, Brown JH, editors. Cambridge, UK: Cambridge Univ Press; 2007. pp. 151–167. [Google Scholar]
- 15.Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford: Oxford Univ Press; 1991. [Google Scholar]
- 16.Whittaker RH. Vegetation of the Siskiyou Mountains, Oregon, and California. Ecol Monogr. 1960;30:279–338. [Google Scholar]
- 17.Whittaker RH. Gradient analysis of vegetation. Biol Rev. 1967;42:207–264. doi: 10.1111/j.1469-185x.1967.tb01419.x. [DOI] [PubMed] [Google Scholar]
- 18.Eisen JA. Environmental shotgun sequencing: Its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:0384–0388. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichorst SA, Breznak JA, Schmidt TM. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl Environ Microbiol. 2007;73:2708–2717. doi: 10.1128/AEM.02140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faith DP. Systematics and conservation: On predicting the feature diversity of subsets of taxa. Cladistics. 1992;8:361–373. doi: 10.1111/j.1096-0031.1992.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 22.Vane-Wright RI, Humphries CJ, Williams PH. What to protect? systematics and the agony of choice. Biol Conserv. 1991;55:235–254. [Google Scholar]
- 23.Faith DP. Phylogenetic pattern and the quantification of organismal biodiversity. Philos Trans R Soc London Ser B. 1994;345:45–58. doi: 10.1098/rstb.1994.0085. [DOI] [PubMed] [Google Scholar]
- 24.Myers N, Knoll AH. The biotic crisis and the future of evolution. Proc Natl Acad Sci USA. 2001;98:5389–5392. doi: 10.1073/pnas.091092498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sechrest W, et al. Hotspots and the conservation of evolutionary history. Proc Natl Acad Sci USA. 2002;99:2067–2071. doi: 10.1073/pnas.251680798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forest F, et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- 27.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 28.Lortie CJ. An ecological tardis: The implications of facilitation through evolutionary time. Trends Ecol Evol. 2007;22:627–630. doi: 10.1016/j.tree.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Valiente-Banuet A, Verdu M. Facilitation can increase the phylogenetic diversity of plant communities. Ecol Lett. 2007;10:1029–1036. doi: 10.1111/j.1461-0248.2007.01100.x. [DOI] [PubMed] [Google Scholar]
- 30.Jaccard P. The distribution of the flora in the Alpine zone. New Phytol. 1912;11:37–50. [Google Scholar]
- 31.Brehm G, Homeir J, Fiedler K. Beta diversity of geometrid moths,(Lepidoptera: Geometridae) in an Andean montane rainforest. Divers Distrib. 2003;9:351–366. [Google Scholar]
- 32.Harte J, et al. Estimating species-area relationships from plot to landscape scale using species spatial-turnover data. Oikos. 1999;86:45–54. [Google Scholar]
- 33.Mena JL, Vazequez-Dominguez E. Species turnover on elevational gradients in small rodents. Glob Ecol Biogeogr. 2005;14:539–547. [Google Scholar]
- 34.Soininen J, McDonald R, Hillebrand H. The distance decay of similarity in ecological communities. Ecography. 2007;30:3–12. [Google Scholar]
- 35.Horner-Devine MC, Bohannan BJM. Phylogenetic clustering and overdispersion in bacterial communities. Ecology. 2006;87:S100–S108. doi: 10.1890/0012-9658(2006)87[100:pcaoib]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Prinzing A, Durka W, Klotz S, Brandl R. The niche of higher plants: Evidence for phylogenetic conservatism. Proc R Soc London Ser B. 2001;268:2383–2389. doi: 10.1098/rspb.2001.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution (Lawrence, Kans.) 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 38.Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. Phylogenetic overdispersion in Floridian oak communities. Am Nat. 2004;163:823–843. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- 39.Swenson NG, et al. The problem and promise of scale dependency in community phylogenetics. Ecology. 2006;87:2418–2424. doi: 10.1890/0012-9658(2006)87[2418:tpapos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Swenson NG, Enquist BJ, Thompson J, Zimmerman JK. The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology. 2007;88:1770–1780. doi: 10.1890/06-1499.1. [DOI] [PubMed] [Google Scholar]
- 41.Callaway RM, et al. Positive interactions among alpine plants increase with stress. Nature. 2002;417:844–848. doi: 10.1038/nature00812. [DOI] [PubMed] [Google Scholar]
- 42.Allen AP, Brown JH, Gillooly JF. Global biodiversity, biochemical kinetics, and the energetic-equivalence rule. Science. 2002;297:1545–1548. doi: 10.1126/science.1072380. [DOI] [PubMed] [Google Scholar]
- 43.Currie DJ, et al. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol Lett. 2004;7:1121–1134. [Google Scholar]
- 44.Hawkins BA, et al. Energy, water, and broad-scale geographic patterns of species richness. Ecology. 2003;84:3105–3117. [Google Scholar]
- 45.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad of Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slingsby JA, Verboom GA. Phylogenetic relatedness limits co-occurrence at fine spatial scales: Evidence from the schoenoid sedges (Cyperaceae: Schoeneae) of the Cape Floristic Region, South Africa. Am Nat. 2006;168:14–27. doi: 10.1086/505158. [DOI] [PubMed] [Google Scholar]
- 47.Cavender-Bares J, Keen A, Miles B. Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology. 2006;87:109–122. doi: 10.1890/0012-9658(2006)87[109:psofpc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 48.Mayr E. Systematics and Origin of Species. New York: Columbia Univ Press; 1942. [Google Scholar]
- 49.Horner-Devine M, Lage M, Hughes J, Bohannan B. A taxa-area relationship for bacteria. Nature. 2004;432:750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- 50.Cohan F, Perry L. A systematics for discovering the fundamental units of bacterial diversity. Curr Biol. 2007;17:R373–R386. doi: 10.1016/j.cub.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 51.Webb CO, Donoghue MJ. Phylomatic: Tree assembly for applied phylogenetics. Mol Ecol Notes. 2005;5:181–183. [Google Scholar]
- 52.Torsvik V, Goksoyr J, Daae FL. High diversity of DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green JL, Plotkin JB. A statistical theory for sampling species abundances. Ecol Lett. 2007;10:1037–1045. doi: 10.1111/j.1461-0248.2007.01101.x. [DOI] [PubMed] [Google Scholar]
- 54.Plotkin JB, Muller-Landau HC. Sampling the species composition of a landscape. Ecology. 2002;83:3344–3356. [Google Scholar]
- 55.Woodcock S, et al. Taxa-area relationships for microbes: The unsampled and the unseen. Ecol Lett. 2006;9:805–812. doi: 10.1111/j.1461-0248.2006.00929.x. [DOI] [PubMed] [Google Scholar]
- 56.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chao A, Chazdon RL, Colwell RK, Shen T. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett. 2005;8:148–159. [Google Scholar]
- 58.Green JL, Bohannan BJM. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Barns SM, Takala SL, Kuske CR. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fierer N, Jackson JA, Vilgalys R, Jackson RB. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol. 2005;71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeSantis TZ, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludwig W, et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altschul SF, et al. Basic local aligment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 65.Huber T, Faulkner G, Hugenholtz P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 66.Horner-Devine MC, Carney KM, Bohannan BJM. An ecological perspective on bacterial biodiversity. Proc R Soc London Ser B. 2004;271:113–122. doi: 10.1098/rspb.2003.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 68.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad of Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 71.II A. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 72.Worberg A, et al. Phylogeny of basal eudicots: Insights from noncoding and rapidly evolving DNA. Org Divers Evol. 2007;7:55–77. [Google Scholar]
- 73.Winkworth R, Lundberg J, Donoghue MJ. Toward a resolution of campanulid phylogeny, with special reference to the placement of Dipsacales. Taxon. 2008 in press. [Google Scholar]
- 74.Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: Calibrating the family tree. Proc R Soc London Ser B. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morlon H, et al. A general framework for the distance-decay of similarity in ecological communities. Ecol Lett. 2008 doi: 10.1111/j.1461-0248.2008.01202.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webb CO. Exploring the phylogenetic structure of ecological communities: An examplfor rain forest trees. Am Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 77.Kembel SW, Hubbell SPH. The phylogenetic structure of a neotropical forest tree community. Ecology. 2006;87:S86–S99. doi: 10.1890/0012-9658(2006)87[86:tpsoan]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 78.Krebs CJ. Ecologial Methodology. New York: Harper Collins; 1998. [Google Scholar]
- 79.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferrier S, Manion G, Elith J, Richardson K. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers Distrib. 2007;13:252–264. [Google Scholar]
- 81.Burnham K, Anderson D. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. New York: Springer; 2002. [Google Scholar]
- 82.Legendre P, Legendre L. Numerical Ecology. Boston: Elsevier; 1998. [Google Scholar]
- 83.Legendre P, Borcard D, Peres-Neto PR. Analyzing beta diversity: Partitioning the spatial variation of community composition data. Ecol Monogr. 2005;75:435–450. [Google Scholar]
- 84.Toumisto H, Ruokolainen K. Analyzing or explaining beta diversity? Understanding the targets of different methods of analysis. Ecology. 2006;87:2697–2708. doi: 10.1890/0012-9658(2006)87[2697:aoebdu]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 85.Clark KR, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205–219. [Google Scholar]