Abstract
Although it is generally accepted that plant community composition is key for predicting rates of ecosystem processes in the face of global change, microbial community composition is often ignored in ecosystem modeling. To address this issue, we review recent experiments and assess whether microbial community composition is resistant, resilient, or functionally redundant in response to four different disturbances. We find that the composition of most microbial groups is sensitive and not immediately resilient to disturbance, regardless of taxonomic breadth of the group or the type of disturbance. Other studies demonstrate that changes in composition are often associated with changes in ecosystem process rates. Thus, changes in microbial communities due to disturbance may directly affect ecosystem processes. Based on these relationships, we propose a simple framework to incorporate microbial community composition into ecosystem process models. We conclude that this effort would benefit from more empirical data on the links among microbial phylogeny, physiological traits, and disturbance responses. These relationships will determine how readily microbial community composition can be used to predict the responses of ecosystem processes to global change.
Keywords: functional redundancy, microbial diversity, modeling
Recent rates of plant and animal species' extinctions have spurred ecologists to consider the consequences of biodiversity loss. Beyond the ethical and aesthetic reasons for conserving it, biodiversity supplies economically valuable ecosystem goods and services on which human society depends (1, 2). Although most biodiversity and conservation research has focused on the value and importance of large organisms, the sheer abundance of microorganisms confers on them a principal role in providing ecosystem services, such as water purification and soil fertility. Bacteria and Archaea alone contain most of the total nitrogen (N) and phosphorus (P) and up to half of the carbon (C) stored in living organisms (3), and the metabolic machinery of microorganisms drives a variety of ecosystem processes. Indeed, microbes carry out the bulk of decomposition and catalyze important transformations in the C, N, sulfur, and P cycles.
Despite their importance to the functioning of ecosystems, microorganisms are rarely explicitly considered in individual ecosystem or global process models. In addition to methodological hurdles, a primary reason for this gap is their overwhelming diversity. Estimates of soil microbial diversity range from thousands to a million microbial “species” in a few grams of soil (4, 5), and how this diversity is related to ecosystem processes is generally unknown (6–8). Moreover, it is infeasible to assess and track each microbial taxon in an ecosystem, let alone include even a small fraction of these taxa in ecosystem models.
Because of these obstacles, ecosystem models often “black box” microbiology. In other words, microorganisms are buried within equation structure as kinetic constants and response functions and are “simplified beyond recognition” (9). As a result, the abundance, diversity, and interactions of microorganisms are often assumed to be unimportant to ecosystem processes, particularly in terrestrial ecosystem models. [A number of ocean ecosystem models include various phytoplankton groups (e.g., refs. 10 and 11)].
In contrast to microorganisms, it is generally accepted that plant biodiversity (both richness and composition) affects terrestrial ecosystem processes (12–14) and influences ecosystem responses to disturbances such as CO2 and N addition (e.g., ref. 15). Plant community composition is often incorporated into large-scale models through the use of functional groups, which are based on plant traits (14, 16). Global change models—whether of ecosystems (17, 18), the terrestrial biosphere (19), or global climate (20, 21)—routinely incorporate 5–10 plant functional groups to improve model predictions. Nonetheless, researchers are calling for a better understanding of the functional traits of plant species to help predict ecosystem processes under changing environmental conditions (22–24).
Given the central role of microbes in ecosystem processes, we contend that microbial composition might be at least as important as plant composition for building predictive ecosystem models. Here we discuss two major hurdles to including microorganisms in ecosystem models. The first is a general idea that microbial composition does not matter to ecosystem processes. The second is that microbial composition may be too diverse to model. To address the first hurdle, we outline three conditions that would need to be true for changes in microbial composition to matter to ecosystem processes. We then review recent studies to assess whether particular microbial groups may be more or less subject to particular disturbances. We address the second hurdle by proposing a simple model of microbial process rates that incorporates information on community composition. The model is used to illustrate how empirical data could be used to predict microbial process rates under disturbance, even for relatively diverse communities. Because it is not feasible to add a parameter for each microbial population in a community, we consider when coarse information about microbial composition, such as the relative abundance of a few key clades within a functional group, could help decrease uncertainty about predictions of ecosystem processes.
A Framework Within the Black Box
Schimel (9) points out that black box ecosystem models make two implicit assumptions: that microbial processes can be represented across a range of environmental conditions with one mathematical function, and that microbial processes are never limited by the abundance of any microorganism. These assumptions are implicit because no major ecosystem models include parameters that explicitly represent components of microbial community composition. At best, some models include total microbial biomass as a parameter (e.g., ref. 25), but many widely used models such as CENTURY (26), MEL (multiple element limitation), (27), and TEM (terrestrial ecosystem model) (28) contain parameters related to microbial processes but not the microbial communities themselves. Ocean biogeochemistry models have only just begun to incorporate explicit parameters that capture plankton community composition (10, 11, 29).
These models implicitly assume that changes in community composition will not affect ecosystem processes, because there is no mechanism for such changes to influence model predictions. This assumption may be valid if microbial composition is resistant, resilient, and/or functionally redundant (these terms and others are defined in Table 1.) Microbial composition is resistant if it is similar across a variety of environmental conditions—in other words, it is difficult to perturb from an original state. It is resilient if, when composition does change, it recovers quickly, whether by growth or by physiological or genetic adaptation. Finally, microbial composition may change in response to new environmental conditions but contain functionally redundant taxa such that ecosystem process rates are not altered. If these conditions are not generally true for microbial communities, then many current ecosystem models may fail at predicting the rates of microbe-driven processes under disturbance.
Table 1.
Summary of definitions used in this article
| Term | Definition |
|---|---|
| Functional group | All organisms that directly contribute to the rate of a particular functional process in an ecosystem |
| Functional redundancy | The ability of one microbial taxon to carry out a process at the same rate as another under the same environmental conditions |
| Functional similarity | The ability of two microbial communities to carry out a functional process at a similar rate, regardless of differences in composition |
| Microbial composition | The richness, relative abundance, and phylogenetic structure of taxa in an assemblage |
| Microbial taxon | A group of phylogenetically related microbes |
| Resilience | The rate at which microbial composition returns to its original composition after being disturbed |
| Resistance | The degree to which microbial composition remains unchanged in the face of a disturbance |
| Physiological trait | A physiological characteristic that determines the contribution of a microbial taxon to a functional process |
| Physiological response curve | The function describing the relationship between the process contribution of a microbial taxon and disturbance intensity |
The idea that microorganisms are resistant, resilient, and functionally redundant is pervasive in ecology. Some microbial groups show a high degree of metabolic flexibility and physiological tolerance to changing environmental conditions (e.g., ref. 30), which could result in microbial communities that are resistant to change. These traits and others associated with microbes—such as high abundances, widespread dispersal, and the potential for rapid growth rates—have also led to the suggestion that microbial communities will be resilient to change (31, 32). Furthermore, rapid evolutionary adaptation through horizontal gene transfer could allow sensitive microorganisms to adapt to new environmental conditions and quickly return the community to its original composition. The extremely high abundance and diversity of microorganisms are used as an argument for functional redundancy, because it is difficult to imagine that biogeochemical cycling is limited by microbial abundance (32) or genetic diversity (33).
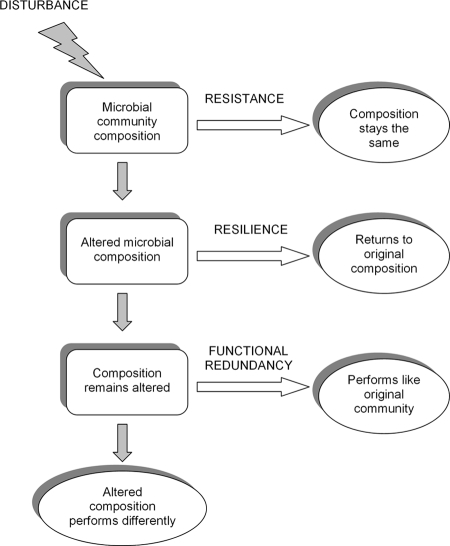
Fig. 1 illustrates the potential impacts of a disturbance on microbial composition and/or ecosystem processes. Consider a disturbance applied to an ecosystem and the microbial communities within it. Microbial composition might be resistant to the disturbance, and not change. Alternatively, if the community is sensitive and does change, it could be resilient and quickly recover to its initial composition. Finally, a community whose composition is sensitive and not resilient might produce process rates similar to the original community if the members of the community are functionally redundant.
Fig. 1.
A schematic of how disturbance can change microbial composition and thereby affect ecosystem processes versus when disturbance would not have this effect (when the microbial community is resistant, resilient, or functionally redundant).
Only if community composition is sensitive to a disturbance, not resilient, and functionally dissimilar to the original community do changes in community composition matter for predicting ecosystem process rates. In addition, the degree to which a community is resistant, resilient, and functionally similar will influence the degree to which community composition matters to a particular process. For instance, even if microbial composition is highly sensitive and not resilient to a disturbance, if all of the taxa perform a process at very similar rates (that is, they are nearly functionally redundant), then predictions of ecosystem process rates will not be improved much by including information about microbial composition.
This conceptual framework does not exclude the possibility that there is little change in microbial composition but large changes in ecosystem process rates. This scenario would suggest that the changes are a direct effect of environmental changes, the result of compositional changes in other organisms such as plants, or immediate physiological responses of microorganisms that are not accompanied by compositional changes. An example of the latter is that some transformations such as decomposition occur faster at higher temperatures. This is seen immediately, before changes in composition could be responsible (34).
Resistance of Microbial Composition
To assess whether microbial composition is often resistant to disturbance, we reviewed studies that experimentally exposed microbial communities to various disturbances. We searched Web of Science for papers including “microb*” and “community composition” in their titles, abstract, or subject words. In addition, we specifically searched the journal Global Change Biology for global change experiments that assessed microbial composition. We did not limit papers by study system, but the majority of studies returned by these search parameters focused on terrestrial soils. We acknowledge that there are many manipulative marine studies that we did not pick up in our search. The disturbances we examined were limited to CO2 enrichment, temperature, fertilization with mineral nutrients, and enrichment with C substrates (including complex organics such as manure and potential toxins such as pesticides). We reason that these four disturbances are typical of those that ecosystems experience under human-driven global change. Finally, we did not intend the literature search to be exhaustive, but to be representative of these types of studies.
We hypothesized that differences in focus and methodologies among the studies would influence the likelihood of detecting a compositional response. Therefore, for each study we recorded the breadth of the taxonomic group targeted (e.g., bacteria and fungi; methanotrophic bacteria), the habitat, the method used to examine composition, and the disturbance applied, as well as whether composition of the target group changed in response to the disturbance. We also recorded the time after the disturbance was first applied at which composition was assessed, which is relevant for the discussion of resilience below.
Papers from this search included studies that targeted composition broadly such as with phospholipid fatty acid analysis, which targets bacteria and fungi. In contrast, other studies examine narrower functional guilds within the Bacteria by PCR-amplifying a functional gene followed by a fingerprinting technique (such as denaturing gradient gel electrophoresis or terminal restriction fragment length polymorphism). Thus, the breadth of the taxa studied varied widely and is related to the methodology used to evaluate community composition. The methodology itself might also have an influence on whether a change in composition is detected. For instance, sequencing of PCR-amplified clone libraries will reveal finer-scale genetic changes than fingerprinting methods that target the same gene. Finally, because we were interested in compositional changes, our search excluded many studies that measure only total microbial biomass (e.g., those reviewed in ref. 35).
Table 2summarizes the results by the four disturbance types. The majority of these studies demonstrate that composition is sensitive to disturbance. More than 80% of the mineral fertilization (N/P/K), temperature, and C amendment studies found significant effects of disturbance on microbial composition. In contrast, the effect of elevated CO2 was found less often, in only 60% of the studies.
Table 2.
Summary of studies, including the percentage of studies in which microbial community composition was sensitive to the disturbance, and the average length of studies that found resistant versus sensitive composition
| Disturbance | No. of studies | Percentage of studies sensitive | Average length of study, years ± SD |
P value | |
|---|---|---|---|---|---|
| Resistant | Sensitive | ||||
| CO2 increase | 20 | 60 | 3.3 ± 1.9 | 3.6 ± 1.9 | 0.78 |
| N/P/K fertilization | 38 | 84 | 4.8 ± 4.5 | 8.2 ± 8.7 | 0.17 |
| Temperature | 11 | 82 | 1.1 ± 1.3 | 3.0 ± 5.0 | 0.35 |
| C amendments | 41 | 83 | 0.15 ± 0.09 | 4.9 ± 12.6 | 0.03 |
The P value reported is the result of a t test (assuming unequal variances) between the study times for the resistant versus sensitive studies.
These studies did not suggest that broad taxonomic groups are more or less sensitive to disturbances than narrow taxonomic groups. Supporting information (SI) Tables S1–S4 list the studies from the most taxonomically broad (e.g., Bacteria and Fungi) to the most taxonomically narrow (e.g., ammonia-oxidizing bacteria or Pseudomonads). The studies that did observe compositional shifts are scattered throughout this list. This pattern suggests that taxonomic breadth is not related to whether a compositional shift was detected. Perhaps more surprisingly, there are no patterns suggesting that methodology influences whether a compositional change was detected. In addition, we were not able to discern whether particular taxonomic or functional groups are more or less sensitive to particular disturbance types. Overall, the low number of studies observing a resistant microbial composition hinders our ability to recognize any patterns among these studies. However, we can conclude that microbial composition is generally sensitive to disturbance.
Resilience of Microbial Composition
Even if microbial composition is sensitive to a disturbance, the community might still be resilient and quickly return to its predisturbance composition. A number of features of microorganisms, and in particular Bacteria and Archaea, suggest that resilience could be common. First, many microorganisms have fast growth rates; thus, if their abundance is suppressed by a disturbance, they have the potential to recover quickly. Second, many microbes have a high degree of physiological flexibility. This is famously the case for the purple nonsulfur bacteria, which can be phototrophs under anoxic conditions and heterotrophs under aerobic conditions. Thus, even if the relative abundance of some taxa decreased initially, these taxa might physiologically acclimate to the new abiotic conditions over time and return to their original abundance. Finally, if physiological adaptation is not possible, then the rapid evolution (through mutations or horizontal gene exchange) could allow microbial taxa to adapt to new environmental conditions and recover from disturbance. All of these arguments assume that abundance is reduced by a disturbance, but some microbial taxa may benefit from the new conditions and increase in abundance. Thus, in order for some taxa to recover in abundance, those that responded positively to the disturbance would also need to decrease in abundance to return the community to its original composition.
Few studies explicitly focus on the time course of microbial composition after a disturbance; instead, most focus solely on the sensitivity of composition. Consequently, we recorded the length of time between the application of the disturbance and when microbial composition was assessed for the studies in Tables S1–S4. If composition is highly resilient, then one should be less likely to detect a compositional change as time from disturbance increases.
We compared the time from initial disturbance for those studies that found composition to be sensitive versus resistant. Generally, the timing of compositional assessment varied widely, from just a few hours to decades. For C amendments, the studies in which microbial community composition changed were significantly longer than studies that did not detect a change (Table 2). This result implies that there is a lag in the response of microbial communities to C additions and does not support the idea that these communities are resilient. For elevated CO2, mineral fertilization, and temperature, all studies were equally likely to find shifts in community composition, regardless of time since disturbance. On average, the reviewed studies examined composition after several years of the disturbance application. Thus, as a conservative boundary, microbial composition is often not resilient within a few years.
Certainly, the strength of the disturbance and how often it is applied will have an effect on the resilience of microbial composition. Most of the studies we reviewed continued to apply the disturbance throughout the study (as occurs for most global change disturbances), rather than a one-time application at the beginning of the experiment. For instance, Enwall et al. (36) compared fertilized and unfertilized soil plots that have been maintained since 1956. The composition of the general bacteria and ammonia-oxidizing bacteria still differs between the plot types. In contrast, Stark et al. (37) applied organic and inorganic forms of N to soil samples and compared the composition of Actinomycetes, alpha-Proteobacteria, and Pseudomonads. After 10 days composition differed between the soil treatments, but after 91 days composition differed only among the Pseudomonads. Conversely, some of the studies that found no effect of disturbance on composition might have found an effect if the study was carried out longer.
Functional Redundancy of Microbial Composition
Our literature survey clearly indicates that microbial communities are sensitive to disturbance and often do not rapidly recover to their original state. These responses beg the question of whether compositional shifts will affect ecosystem processes—will the disturbed community be functionally similar to the original community? There are two reasons why changes in microbial composition might not affect ecosystem process rates. First, the new community might contain taxa that are functionally redundant with the taxa in the old community. Second, taxa in the new community may function differently but result in the same process rate when combined at the community level.
What do we know about functional redundancy in microbial communities? Few studies compare the degree of redundancy within different microbial functional groups (but see for example refs. 38 and 39). Functional redundancy is difficult to establish because it requires detailed knowledge about the microbial populations that perform a specific process. Furthermore, organisms that are functionally redundant under one set of conditions may not be under different conditions. In general, we know little about the distribution of functional traits across microbial taxa despite years of recognition of this need (9).
Nonetheless, a growing body of literature has begun to address the more basic question of whether microbial composition matters to ecosystem processes to any degree versus whether the members of a functional group are completely redundant. To address this question, one needs to manipulate microbial composition while controlling for the abiotic environment. This is because any differences in process rates can then be attributed directly to compositional differences and not simply to physiological responses of the original community under new environmental conditions. In plant communities, composition and diversity can be manipulated directly by sowing and weeding plots in the field. Although this is not an easy task, it is much more feasible than manipulating microbial community composition in the field.
A number of approaches can be used to disentangle compositional versus environmental effects on ecosystem processes. In one approach, process rates are measured before and after a disturbance manipulation but before microbial composition changes. This measurement gives some idea about the direct effect of the disturbance on process rates independent of community composition. Schimel and Gulledge (40) review studies that subject microbial communities from different habitats to parallel short-term, environmental treatments and compare the communities' process rates. For instance, Gulledge et al. (41) found that rates of methane consumption by methanotroph communities in response to ammonium fertilization varied depending on whether the communities were associated with paper birch or white spruce taiga forests. This result suggests that differences in methanotroph composition are responsible for differences in the physiological responses.
Although they may not mimic natural communities, bioreactors are useful model systems for linking microbial functional groups with process rates. Community composition can be manipulated directly to test the functional contribution of different microbial taxa. In addition, the communities can be perturbed and monitored for function and composition over time. For example, Fernandez et al. (42) constructed two different methanogenic bioreactor communities that functioned similarly before disturbance with a glucose pulse. After the pulse, the communities metabolized glucose using different pathways and regained prepulse function at different rates. The authors were able to attribute these differences to specific groups of bacteria that showed different metabolic functions.
Transplant experiments can also be used to separate environmental versus compositional effects on process rates (43). If different microbial communities produce different process rates in a common environment, then it can be inferred that the compositional differences are responsible for the functional differences. Balser and Firestone (44) provide a good example of how the transplant approach can also be used to make linkages between microbial taxa and process rates under disturbance. They transplanted soil microbial communities across a climate gradient and demonstrated that community composition affected process rates independent of climate. Furthermore, they used phospholipid fatty acid data to correlate process rates with specific members of the microbial community and concluded that nitrification potential and N2O flux were likely driven by Gram-negative bacteria.
Although not often possible, direct manipulations of microbial composition can provide useful information about the functional status of microbial groups, especially when coupled with process rate measurements. For example, specific taxa can be targeted for elimination from a community via chemical or physical means and process rates compared in communities with and without the taxa (45–47). Wertz et al. (48) manipulated soil microbial composition by serial dilution and reinnoculation of sterile microcosms; they found no effect of composition on functioning in the microcosms. Alternatively, communities can be artificially constructed to contain specific taxa and to establish links between composition and process rates (49). For instance, Bell et al. (50) showed that the diversity and composition of bacteria influenced respiration rates in aquatic microcosms.
The literature reviewed in the sections above suggests that microbial composition is often altered by disturbances and does not recover over some time. Furthermore, these changes often impact the rates of ecosystem processes, suggesting that at least some microbial taxa are functionally dissimilar. In light of these observations, we propose a broad framework in the next section for integrating information about microbial composition into predictive models of ecosystem processes.
Incorporating Microbes into Models: Physiological Traits and Process Response Curves
As more data are collected on the relationship between microbial composition and ecosystem functioning, explicitly incorporating microbes into process models will become increasingly tractable. Indeed, analogous efforts have been successful with plant functional groups and ecosystem models. However, there are some gaps to bridge between microbial ecologists and ecosystem modelers. Modelers are uncertain about how to aggregate extremely diverse microbial communities into a manageable number of functional units. Conversely, microbial ecologists often have a poor understanding of the types of studies that would be useful to modelers in carrying out this aggregation step. In this section we outline a simple microbial process model to frame some of the results above and consider how experimentalists might further inform predictive models.
Consider a number of taxa within a functional group that all contribute to an ecosystem process. The functional group has n taxa with abundances ai (in units of biomass) and biomass-specific physiological rates ri. The community process rate R is the sum of the products of the abundances and the rates:
If taxon 1 and taxon 2 have the same physiological rates ri, then they are functionally redundant, and their abundances can be aggregated together:
If communities 1 and 2 have different compositions but the same process rates (R1 = R2), then we can define these communities as functionally similar, although they may contain taxa that are not functionally redundant.
To predict how microbial processes will respond to disturbance, we also need to know the physiological responses of each taxon to disturbance. Assume that the physiological response curve r(I) is a linear function of disturbance intensity I:
where m is the slope and r0 is the physiological rate under undisturbed conditions (Fig. 2). For the microbial community as a whole, the process rate as a function of disturbance intensity I would be
Because it is not feasible to model all taxa and their responses individually, the challenge is to determine properties of the functional group that will help predict the responses of ecosystem processes to disturbance. A worthwhile goal for future experimental studies would be to identify the level of redundancy in physiological traits and disturbance responses within microbial functional groups. Then one could estimate the parameters m and r0 without having to measure them for every taxon. For example, if there is correspondence between phylogeny and a physiological trait (r0), or between phylogeny and functional responses (m), then we could use phylogenetic information to estimate the parameters. Given that microbial composition is usually assessed with phylogenetic markers, this information could be used to predict how members of a functional group influence ecosystem process rates.
Fig. 2.
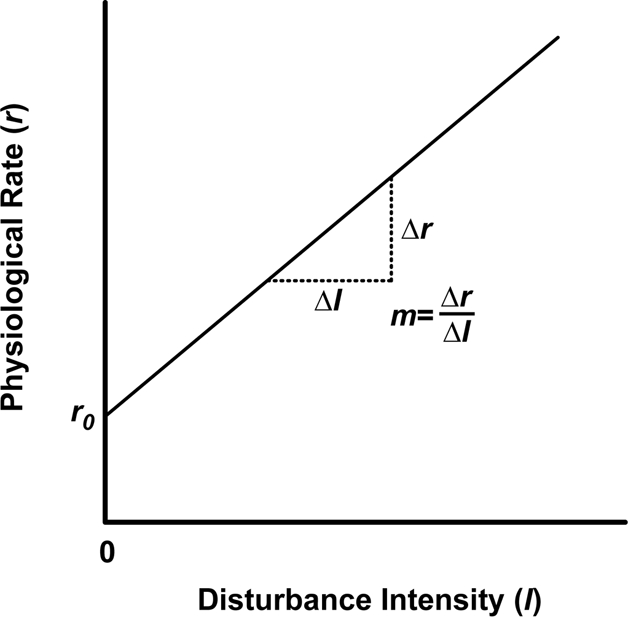
A physiological response curve for a microbial taxon. The curve illustrates the rate at which the taxon contributes to an ecosystem process as a function of disturbance intensity. For simplicity, this function is assumed to be linear, although other forms are likely for microbial taxa. The slope m of the line indicates how quickly the physiological rate changes with I, and r0 is the physiological rate in the absence of disturbance.
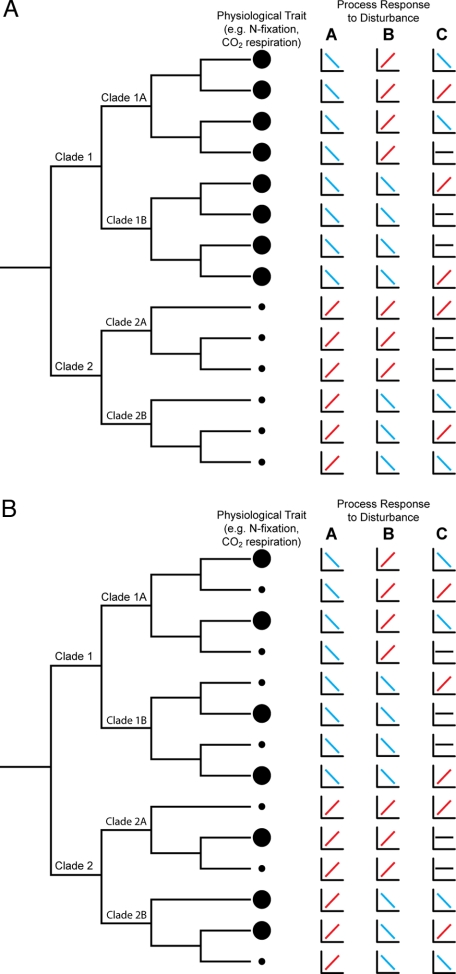
As an illustration of this approach, consider the functional group in Fig. 3A. All of the taxa in the top clade perform the functional process at a high rate (denoted by the large circles), and those in the bottom clade perform the process at a lower rate (denoted by the small circles). If the total abundance of these two clades stays the same, then shifts in composition within the two clades are not functionally important, as taxa within the clades perform the function at the same rate. In the model more generally, these clades define the level of aggregation for an undisturbed community contributing to process R, such that
where ac1 and ac2 are the summed abundances of all taxa in clades 1 and 2 and r0c1 and r0c2 are the physiological rates for the taxa in clades 1 and 2, respectively.
Fig. 3.
A stylized illustration comparing the phylogenetic relationship of physiological traits versus process responses to disturbance among taxa. Different rates of the physiological traits are represented by differently sized circles (see text for further explanation). In A physiological trait values are correlated with phylogenetic similarity, whereas in B physiological trait values are randomly distributed among taxa. Three different disturbances (A, B, and C) produce process responses from the taxa that are also either phylogenetically related (disturbances A and B) or randomly distributed among taxa (disturbance C).
As with the physiological trait in Fig. 3A, the disturbance responses may also be related to phylogeny. For instance, the taxa within clade 1 all respond similarly to disturbance A, as do the taxa within clade 2. Therefore, only the abundances of these two clades must be considered to predict the overall process rate R under disturbance:
where mc1 and mc2 represent the slopes of the physiological response curves for clades 1 and 2, respectively. We note that the abundances under disturbance must be determined before the calculation of the new process rate unless they are known to be stable with disturbance.
For modeling purposes, it would be useful to know the relationships among physiological rates, disturbance responses, and phylogenetic position among taxa. Then, predictions of process rates could be estimated based on the abundances of relatively broad phylogenetic groups. However, we currently lack the empirical evidence necessary to establish which microbial processes show a strong correspondence between phylogeny and physiology. Although this topic has received much attention in the microbial ecology literature (51–53), few studies have tested the linkage convincingly.
Disturbance B in Fig. 3 illustrates a case where the physiological response curves are phylogenetically grouped at the level of subclades (denoted 1A, 1B, 2A, and 2B). In the general model, we would calculate the process rate as
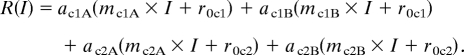 |
This model is considerably more complex than the prior model because of the heterogeneity in disturbance responses at the subclade level. In a complex microbial community this situation is likely to occur but is still tractable for modeling if the heterogeneity is relatively low and well characterized. For instance, Horz et al. (54) found that two different clades of methanotrophs responded differently to simulated global change. The worst-case scenario is that the responses of the taxa to disturbance are randomly distributed across the phylogeny as illustrated by disturbance C in Fig. 3. In this case, calculation of the overall process rate requires the abundances of each individual taxon. A similar challenge would arise if the functional trait itself is not phylogenetically determined (Fig. 3B).
One promising set of tools for overcoming the challenges of these “worst-case” scenarios are metagenomic or metatranscriptomic approaches. For example, environmental gene tags (EGTs) could potentially be used as a proxy for physiological rates or disturbance responses across the whole community, even if these traits are unrelated to phylogeny (55). With this technique, the abundances of genes specific to particular environmental processes (such as phosphate metabolism) could be extracted from community metagenomic data and used in modeling process rates. Another advantage of this approach is that multiple EGTs and processes can be examined in a single sample, rather than constructing separate clone libraries for each different functional gene of interest.
In our model, when does changing composition matter to ecosystem processes? Changing the abundance of a taxon will affect the process rate unless the abundances of other taxa also change to compensate. In undisturbed communities, exchanging one taxon for another (with similar biomass) can affect the community process rate if the two groups have different r0 (physiological rate) values. With disturbance, composition matters if taxa with different m (disturbance response) values change in abundance (even if they had the same r0 values). Although our simple model considers only the total abundances (i.e., biomasses) of different taxa, we note that changes in body size distributions within or across taxonomic groups could also affect ecosystem process rates as suggested by metabolic scaling theory (56).
In communities with a large number of taxa, a “portfolio effect” may prevent the community process rate from changing with disturbance, even if the taxa change in abundance and are not functionally redundant (57). The portfolio effect can occur when positive responses of some taxa are averaged with negative responses of other taxa, resulting in no net change in function. Thus, the greater the number of taxa that perform a process, the more buffered the process is to environmental perturbations (58). This portfolio mechanism (in addition to functional redundancy) could lead to similarity in community function despite changes in microbial composition.
Conclusions
There are three ways in which microbial composition might not matter to ecosystem functioning in the face of disturbance. First, microbial communities might be resistant to change. However, we find that microbial composition is, in the majority of cases that we reviewed, sensitive to elevated CO2, mineral fertilization, temperature changes, and C amendments. Second, microbial composition might be resilient and quickly return to its original state. The literature suggests that, at least over the time scale of a few years, microbial composition usually still differs from that of undisturbed communities. Third, even if microbial composition changes, the new community might be functionally similar to the original. Although this hypothesis is currently difficult to test, recent studies suggest that the taxa in many microbial communities are probably not functionally redundant and different communities are not functionally similar.
How can the information that microbial composition affects ecosystem functioning be used to improve predictions of ecosystem process rates under disturbance? The simple model we presented in the previous section highlights two lacking pieces of information. First, more data are needed on the responses of microbial taxa to disturbance, in addition to knowledge about physiological traits. Microbial taxa may vary in their responses to different disturbances, and these taxa may not correspond to functional groups defined by physiological traits.
Second, it would be useful to know the relationship among microbial phylogeny, physiological traits, and response curves. Although it is clear that phylogenetic relationships of taxa are not perfect predictors of microbial physiology (59, 60), there are phylogenetic signals of physiological traits (e.g., ref. 61). The genetic scale at which these traits are clustered would guide modelers in aggregating microbial taxa for their models. Similarly, we know of no studies that address the relationship between phylogeny and microbial responses to disturbance. For instance, perhaps the response of microbial taxa to particular C amendments are predictable at very fine phylogenetic scales (e.g., >99% 16S rDNA similarity), whereas the responses of taxa to temperature changes can be aggregated at a broader scale (such as at >95% similarity).
In sum, there has been increasing recognition that microbes are relevant to ecosystem processes and enormous progress in characterizing the response of microbial composition to disturbance, particularly in soils. Despite these advances, the field of microbial ecology lacks a strong predictive framework to interpret the functional consequences of changes in microbial composition. Much more empirical work is needed to define microbial functional groups and their responses to various disturbances. Greater efforts toward improving culture techniques and assessing the physiological responses of microbial populations under controlled conditions would be especially useful. Once explicitly incorporated into models, this information could greatly enhance our ability to predict ecosystem responses to global change.
Acknowledgments.
We thank Devon Bradley, Claire Horner-Devine, and Adam Martiny for comments on earlier versions of the manuscript and Francisco Ayala, John Avise, and Stephen Hubbell for organizing the Arthur M. Sackler Colloquium “In the Light of Evolution II: Biodiversity and Extinction.” Support to J.B.H.M. was provided by the National Science Foundation and the Gordon and Betty Moore Foundation.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution II: Biodiversity and Extinction,” held December 6–8, 2007, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at www.nasonline.org/Sackler_1020;biodiversity.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801925105/DCSupplemental.
References
- 1.Ehrlich PR, Ehrlich AH. The value of biodiversity. Ambio. 1992;21:219–226. [Google Scholar]
- 2.Daily GC. Nature's Services: Societal Dependence on Natural Ecosystems. Washington, DC: Island; 1997. [Google Scholar]
- 3.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 5.Torsvik V, Øvreås L. Microbial diversity and function in soil: From genes to ecosystems. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 6.Crawford JW, Harris JA, Ritz K, Young IM. Towards an evolutionary ecology of life in soil. Trends Ecol Evol. 2005;20:81–87. doi: 10.1016/j.tree.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 8.Torsvik V, Øvreås L, Thingstad TF. Prokaryotic diversity—Magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- 9.Schimel JP. In: Global Biogeochem Cycles in the Climate System. Schulze ED, et al., editors. San Diego, CA: Academic; 2001. pp. 239–254. [Google Scholar]
- 10.Moore JK, Doney SC, Glover DM, Fung IY. Iron cycling and nutrient limitation patterns in surface waters of the world ocean. Deep Sea Res II. 2002;49:463–508. [Google Scholar]
- 11.Salihoglu B, Hofmann EE. Simulations of phytoplankton species and carbon production in the equatorial Pacific Ocean 2. Effects of physical and biogeochemical processes. J Mar Res. 2007;65:275–300. [Google Scholar]
- 12.Spehn EM, et al. Ecosystem effects of biodiversity manipulations in European grasslands. Ecol Monogr. 2005;75:37–63. [Google Scholar]
- 13.Hector A, et al. Plant diversity and productivity experiments in European grasslands. Science. 1999;286:1123–1127. doi: 10.1126/science.286.5442.1123. [DOI] [PubMed] [Google Scholar]
- 14.Tilman D, et al. The influence of functional diversity and composition on ecosystem processes. Science. 1997;277:1300–1302. [Google Scholar]
- 15.Reich PB, et al. Plant diversity enhances ecosystem responses to elevated CO2 and nitrogen deposition. Nature. 2001;410:809–812. doi: 10.1038/35071062. [DOI] [PubMed] [Google Scholar]
- 16.Reich PB, Wright IJ, Lusk CH. Predicting leaf physiology from simple plant and climate attributes: A global GLOPNET analysis. Ecol Appl. 2007;17:1982–1988. doi: 10.1890/06-1803.1. [DOI] [PubMed] [Google Scholar]
- 17.Haxeltine A, Prentice IC. A general model for the light-use efficiency of primary production. Funct Ecol. 1996;10:551–561. [Google Scholar]
- 18.Moorcroft PR, Hurtt GC, Pacala SW. A method for scaling vegetation dynamics: The ecosystem demography model (ED) Ecol Monogr. 2001;71:557–585. [Google Scholar]
- 19.Foley JA, et al. An integrated biosphere model of land surface processes, terrestrial carbon balance, and vegetation dynamics. Global Biogeochem Cycles. 1996;10:603–628. [Google Scholar]
- 20.Cox PM, Betts RA, Jones CD, Spall SA, Totterdell IJ. Acceleration of global warming due to carbon-cycle feedbacks in a coupled climate model. Nature. 2000;408:184–187. doi: 10.1038/35041539. [DOI] [PubMed] [Google Scholar]
- 21.Higgins PAT, Schneider SH. Long-term potential ecosystem responses to greenhouse gas-induced thermohaline circulation collapse. Glob Change Biol. 2005;11:699–709. [Google Scholar]
- 22.Arndt SK. Integrated research of plant functional traits is important for the understanding of ecosystem processes. Plant Soil. 2006;285:1–3. [Google Scholar]
- 23.Diaz S, et al. The plant traits that drive ecosystems: Evidence from three continents. J Veg Sci. 2004;15:295–304. [Google Scholar]
- 24.McGill BJ, Enquist BJ, Weiher E, Westoby M. Rebuilding community ecology from functional traits. Trends Ecol Evol. 2006;21:178–185. doi: 10.1016/j.tree.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Harte J, Kinzig A. Mutualism and competition between plants and decomposers: Implications for nutrient allocation in ecosystems. Am Nat. 1993;141:829–846. doi: 10.1086/285511. [DOI] [PubMed] [Google Scholar]
- 26.Parton WJ, Schimel DS, Cole CV, Ojima DS. Analysis of factors controlling soil organic-matter levels in great-plains grasslands. Soil Sci Soc Am J. 1987;51:1173–1179. [Google Scholar]
- 27.Rastetter EB, Ågren GI, Shaver GR. Responses of N-limited ecosystems to increased CO2: A balanced-nutrition, coupled-element-cycles model. Ecol Appl. 1997;7:444–460. [Google Scholar]
- 28.McGuire AD, et al. Productivity response of climax temperate forests to elevated temperature and carbon dioxide: A North American comparison between two global models. Clim Change. 1993;24:287–310. [Google Scholar]
- 29.Le Quéré C, et al. Ecosystem dynamics based on plankton functional types for global ocean biogeochemistry models. Glob Change Biol. 2005;11:2016–2040. [Google Scholar]
- 30.Meyer AF, Lipson DA, Martin AP, Schadt CW, Schmidt SK. Molecular and metabolic characterization of cold-tolerant alpine soil Pseudomonas sensu stricto. Appl Environ Microbiol. 2004;70:483–489. doi: 10.1128/AEM.70.1.483-489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- 32.Meyer O. In: Biodiversity and Ecosystem Function. Schultze ED, Mooney HA, editors. Berlin: Springer; 1994. pp. 67–96. [Google Scholar]
- 33.Finlay BJ, Maberly SC, Cooper JI. Microbial diversity and ecosystem function. Oikos. 1997;80:209–213. [Google Scholar]
- 34.Fierer N, Craine JM, McLauchlan K, Schimel JP. Litter quality and the temperature sensitivity of decomposition. Ecology. 2005;86:320–326. [Google Scholar]
- 35.Wardle DA. A comparative assessment of factors which influence microbial biomass carbon and nitrogen levels in soil. Biol Rev Camb Philos Soc. 1992;67:321–358. [Google Scholar]
- 36.Enwall K, et al. Long-term impact of fertilization on activity and composition of bacterial communities and metabolic guilds in agricultural soil. Soil Biol Biochem. 2007;39:106–115. [Google Scholar]
- 37.Stark C, Condron LM, Stewart A, Di HJ, O'Callaghan M. Influence of organic and mineral amendments on microbial soil properties and processes. Appl Soil Ecol. 2007;35:79–93. [Google Scholar]
- 38.Wohl DL, Arora S, Gladstone JR. Functional redundancy supports biodiversity and ecosystem function in a closed and constant environment. Ecology. 2004;85:1534–1540. [Google Scholar]
- 39.Setälä H, McLean MA. Decomposition rate of organic substrates in relation to the species diversity of soil saprophytic fungi. Oecologia. 2004;139:98–107. doi: 10.1007/s00442-003-1478-y. [DOI] [PubMed] [Google Scholar]
- 40.Schimel JP, Gulledge J. Microbial community structure and global trace gases. Glob Change Biol. 1998;4:745–758. [Google Scholar]
- 41.Gulledge J, Doyle AP, Schimel JP. Low-concentration kinetics of atmoshperic CH4 oxidation in soil and mechanism of NH4+ inhibition. Appl Environ Microbiol. 1997;64:4291–4298. doi: 10.1128/aem.64.11.4291-4298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez AS, et al. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol. 2000;66:4058–4067. doi: 10.1128/aem.66.9.4058-4067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed HE, Martiny JBH. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol. 2007;62:161–170. doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 44.Balser TC, Firestone MK. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry. 2005;73:395–415. [Google Scholar]
- 45.Griffiths BS, et al. Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: An examination of the biodiversity-ecosystem function relationship. Oikos. 2000;90:279–294. [Google Scholar]
- 46.Austin AT, Sala OE, Jackson RB. Inhibition of nitrification alters carbon turnover in the Patagonian steppe. Ecosystems. 2006;9:1257–1265. [Google Scholar]
- 47.Santos PF, Whitford WG. The effects of microarthropods on litter decomposition in a Chihuahuan desert ecosystem. Ecology. 1981;62:654–663. [Google Scholar]
- 48.Wertz S, et al. Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ Microbiol. 2007;9:2211–2219. doi: 10.1111/j.1462-2920.2007.01335.x. [DOI] [PubMed] [Google Scholar]
- 49.Naeem S, Hahn DR, Schuurman G. Producer-decomposer co-dependency influences biodiversity effects. Nature. 2000;403:762–764. doi: 10.1038/35001568. [DOI] [PubMed] [Google Scholar]
- 50.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. The contribution of species richness and composition to bacterial services. Nature. 2005;436:1157–1160. doi: 10.1038/nature03891. [DOI] [PubMed] [Google Scholar]
- 51.Balser TC, Kinzig AP, Firestone MK. In: The Functional Consequence of Biodiversity: Empirical Progress and Theoretical Extensions. Kinzig A, Pacala S, Tilman D, editors. Princeton: Princeton Univ Press; 2002. pp. 265–293. [Google Scholar]
- 52.Andren O, Balandreau J. Biodiversity and soil functioning—from black box to can of worms? Appl Soil Ecol. 1999;13:105–108. [Google Scholar]
- 53.Nannipieri P, et al. Microbial diversity and soil functions. Eur J Soil Sci. 2003;54:655–670. [Google Scholar]
- 54.Horz HP, Rich V, Avrahami S, Bohannan BJM. Methane-oxidizing bacteria in a California upland grassland soil: Diversity and response to simulated global change. Appl Environ Microbiol. 2005;71:2642–2652. doi: 10.1128/AEM.71.5.2642-2652.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 56.Enquist BJ, et al. Scaling metabolism from organisms to ecosystems. Nature. 2003;423:639–643. doi: 10.1038/nature01671. [DOI] [PubMed] [Google Scholar]
- 57.Doak DF, et al. The statistical inevitability of stability-diversity relationships in community ecology. Am Nat. 1998;151:264–276. doi: 10.1086/286117. [DOI] [PubMed] [Google Scholar]
- 58.Schimel J. In: Arctic and Alpine Biodiversity: Patterns, Causes, and Ecosystem Consequences. Chapin FS, Koerner C, editors. New York: Springer; 1995. pp. 239–254. [DOI] [PubMed] [Google Scholar]
- 59.Achenbach LA, Coates JD. Disparity between bacterial phylogeny and physiology. ASM News. 2000;66:714–715. [Google Scholar]
- 60.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]