Abstract
Rationale: Inhaled β-agonists are one of the most widely used classes of drugs for the treatment of asthma. However, a substantial proportion of patients with asthma do not have a favorable response to these drugs, and identifying genetic determinants of drug response may aid in tailoring treatment for individual patients.
Objectives: To screen variants in candidate genes in the steroid and β-adrenergic pathways for association with response to inhaled β-agonists.
Methods: We genotyped 844 single nucleotide polymorphisms (SNPs) in 111 candidate genes in 209 children and their parents participating in the Childhood Asthma Management Program. We screened the association of these SNPs with acute response to inhaled β-agonists (bronchodilator response [BDR]) using a novel algorithm implemented in a family-based association test that ranked SNPs in order of statistical power. Genes that had SNPs with median power in the highest quartile were then taken for replication analyses in three other asthma cohorts.
Measurements and Main Results: We identified 17 genes from the screening algorithm and genotyped 99 SNPs from these genes in a second population of patients with asthma. We then genotyped 63 SNPs from four genes with significant associations with BDR, for replication in a third and fourth population of patients with asthma. Evidence for association from the four asthma cohorts was combined, and SNPs from ARG1 were significantly associated with BDR. SNP rs2781659 survived Bonferroni correction for multiple testing (combined P value = 0.00048, adjusted P value = 0.047).
Conclusions: These findings identify ARG1 as a novel gene for acute BDR in both children and adults with asthma.
Keywords: pharmacogenetics, asthma, bronchodilator agents
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Investigations on asthma pharmacogenetics of the β-agonist response to date have mostly studied only one or a few single nucleotide polymorphisms in the β2-adrenergic receptor gene (ADRB2). Because response to inhaled β-agonists in asthma is a complex phenotype, it is likely that other genes are involved.
What This Study Adds to the Field
This study identifies the arginase 1 gene (ARG1) as a potential β-agonist response gene, using a family-based method to screen variants in genes in the steroid and β-adrenergic pathways.
Asthma is a complex genetic disorder that currently affects about 300 million people worldwide (1). Asthma remains the most common chronic disease of childhood in the developed world (2, 3), and incurs a significant health care cost (4). β-Agonists form one of the oldest classes of drugs in medicine (5). They are the most effective medications for the treatment of acute asthma and remain one of the cornerstones of chronic asthma therapy. However, variability in the response to inhaled β-agonists exists (6), and it has been estimated that a substantial proportion of that response is genetic in nature.
To date, asthma pharmacogenetic studies in general (7), and response to inhaled bronchodilators in particular (8), have been based on one or more polymorphisms in a single gene. Recent studies from the Asthma Clinical Research Network, for instance, have reported adverse effects of regular albuterol treatment among patients with asthma who were homozygous for the +49 A allele (Arg16) of the ADRB2 gene (9, 10). However, because asthma is a complex disorder and response to inhaled β-agonist drugs is a complex phenotype, it is likely that other genes also impact on this phenotype. When more than one or a few polymorphisms are tested, the chances of obtaining false positives increases—the multiple testing issue—and methods to adjust for the total number of tests need to be applied. In addition, the ideal design for these studies would use samples with large numbers (i.e., in the thousands) of subjects that would help to offset the more stringent statistical criteria. Unfortunately, most existing datasets available for asthma pharmacogenetics are of modest size, and thus screening methods to limit the number of tests in the first stages of the analysis can help alleviate the multiple testing problem.
We conducted an analysis to screen 844 single nucleotide polymorphisms (SNPs) from 111 candidate genes for association with bronchodilator response (BDR) to inhaled β-agonist in an asthma clinical trial cohort. Because of the issue of multiple testing, we used an algorithm in a family-based association testing (FBAT) framework that allowed us to screen SNPs based on power for replication (11). This screening methodology allows for the identification of the most promising SNPs for testing without biasing the nominal significance level of the test statistic, and recently has led to the identification of disease-susceptibility genes (12–14). In addition, this algorithm allowed us to screen and test in the same population. After identifying the most promising SNPs, we then attempted replication in three additional asthma clinical trial cohorts. Some of the results from these analyses have been previously reported in abstract form (15).
METHODS
Study Populations
We used DNA samples from four clinical trials. All patients or their legal guardians consented to each trial study protocol and ancillary genetic testing. The population we used for the screening algorithm was the Childhood Asthma Management Program (CAMP). Trial design and methodology have been published (16, 17). A total of 209 white probands (randomized to the placebo group) and their parents were included as part of parent–child trios for the screening analyses. Only subjects randomized to the placebo group were used for the screening analyses to avoid confounding effects of medications (corticosteroids and nedocromil) other than inhaled β-agonist.
The population we used for the first replication study (hereafter called the Asthma Trial) was composed of 432 white subjects with asthma (18, 19) who were part of an asthma medication trial conducted by Sepracor, Inc., in the United States. Two completed trials conducted by the American Lung Association Asthma Clinical Research Centers, the Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol trial (LOCCS) (20), and the Effectiveness of Low Dose Theophylline as Add-on Treatment in Asthma (LODO) trial (21) were used as the second and third replication samples. The 166 white subjects from the LOCCS trial and the 155 white subjects from the LODO trial for whom DNA was available were used for this analysis. Detailed information on these subjects has been previously published and is included in the online supplement.
Selection of Genes and SNPs: Genotyping
We genotyped 844 SNPs in 111 candidate genes: 42 genes involved in β-adrenergic signaling and regulation; 28 genes involved in innate glucocorticoid synthesis and metabolism, cellular receptors, and transcriptional regulators; and 41 genes from prior asthma association studies that had been previously conducted in the CAMP dataset (Table E1 in the online supplement). Candidate genes in the β-adrenergic and corticosteroid pathways were selected based on prior studies in the literature, their known involvement in metabolic pathways (22, 23), and on expert opinion (S.B.L. and K.G.T.). Corticosteroid pathway candidate genes were included in this analysis because of the known interactions between β2-agonists and corticosteroids (24, 25). Finally, we included the candidate genes that our group had previously genotyped and studied in CAMP, because these were already available and so as to appropriately adjust our current analyses for all prior tests conducted with these genes. SNPs were primarily selected using public databases, although resequencing of several core genes was performed. We oversampled exonic and promoter regions and attempted coverage of at least one SNP every 10 kb. We emphasized golden-gate validated and linkage disequilibrium tag SNPs, where available.
SNPs were genotyped via an Illumina BeadStation 500G (Illumima, Inc., San Diego, CA) and via a Sequenom MassArray matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Sequenom, San Diego, CA). Further details are included in the online supplement. SNPs were also checked for mendelian inconsistencies and for Hardy-Weinberg equilibrium.
Statistical Methodology
The primary outcome measure of the association analyses was acute response to inhaled bronchodilator (BDR), and calculated as the percentage difference between the pre- and post-bronchodilator FEV1 value (BDR = 100 × [post-FEV1 − pre-FEV1]/pre-FEV1). In all analyses, both screening and replication, BDR was treated as a continuous variable. We initially screened the genotypic association with BDR in CAMP using a modified version of the screening algorithm as detailed by Van Steen and associates (11). Further details are included in the online supplement. The rationale for using CAMP data as the screening set is that the screening methodology was designed for family data. No screening methodology has yet been published for population-based data. We used the 11 repeated measures of BDR over the 4 years of the trial in the placebo group using the FBAT-PC statistic (26) to maximize the heritability of a given marker and thereby maximize power for the screening stage. Only additive genetic models were evaluated and all analyses in the screening stage were adjusted for age, sex, height, and baseline FEV1. We selected the most powerful candidate genes by first ranking the individual SNPs based on conditional power and then evaluating the median rank of all of the SNPs within a given candidate gene. We selected genes whose median SNP ranks for power were within the top 25% of all SNPs genotyped to be taken forward for genotyping in the Asthma Trial population. For those selected SNPs, we evaluated the FBAT-PC statistics for directionality of the association and P value for each SNP, and this allowed us to conduct one-sided tests in the replication analyses. We selected 99 SNPs from 17 genes for replication in the Asthma Trial population.
The analyses of the replicate populations were performed using generalized linear models as incorporated into PROC GLM of the SAS statistical analysis software (version 9.0; SAS Institute, Cary, NC), and SNP genotypes were coded for additive models. Only BDR calculated from spirometric measurements at all baseline visits (before randomization) for all the trials were used. All analyses in the replication populations were adjusted for age, sex, height, and baseline FEV1. Genes with at least one SNP that was at least marginally associated with BDR (one-sided P value < 0.05) were then genotyped in the final two replicate populations. For each SNP in which the direction of the association was in the same direction in each of the four populations, we then combined the P values (two-sided) from the original family-based analysis and the one-sided P values from the replication cohorts using Fisher's method (27) to increase statistical efficiency (28). No evidence for population stratification was found in any of the three populations. Further details on analytic issues are included in the online supplement.
RESULTS
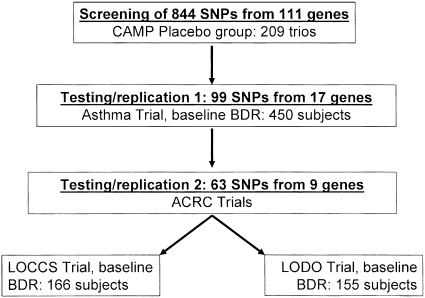
The baseline characteristics of participants of the four asthma cohorts are shown in Table 1. The CAMP subjects on whom we performed the initial screen were composed of children, whereas the three replication cohorts were primarily adults with asthma. In the CAMP subjects, we screened 844 SNPs from 111 candidate genes (Table E1 and Figure 1). In the screening analysis, we ranked the individual SNPs based on the highest to lowest power estimates from the FBAT screening analysis. From these, we identified 19 genes whose median SNP ranks for power were within the top 25% of all SNPs genotyped (Table E1). Because the family-based analysis suggested directionality of the association, we conducted one-sided tests in the replication datasets. The first replication analysis was conducted on 432 white adults with asthma who had participated in a clinical trial of an asthma medication (Asthma Trial). We successfully genotyped 99 SNPs (11.7% of all the SNPs that were screened) from 17 genes in the Asthma Trial. In this first replication analysis, nine genes contained at least one SNP that was at least marginally (one-sided P ≤ 0.05) associated with BDR. We then genotyped 63 SNPs from these nine genes and tested them in the final two asthma clinical trial populations from the American Lung Association Asthma Clinical Research Network: the LOCCS trial and the LODO trial.
TABLE 1.
BASELINE CHARACTERISTICS OF PARTICIPANTS IN THE FOUR ASTHMA TRIALS
| CAMP | Asthma Trial | LOCCS | LODO | |
|---|---|---|---|---|
| (n = 209) | (n = 432) | (n = 166) | (n = 155) | |
| Age, mean yr (SD) | 8.8 (2.1) | 32.5 (13.7) | 34.5 (15.3) | 42.9 (14.7) |
| Range | 5.2–13.2 | 12–80 | 7–71 | 15–76 |
| Sex, n (%) | ||||
| Male | 125 (59.8) | 216 (50.0) | 58 (34.9) | 39 (25.2) |
| Female | 84 (40.2) | 216 (50.0) | 108 (65.1) | 116 (74.8) |
| FEV1, mean %predicted (SD) | 95.0 (13.1) | 61.4 (6.9) | 90.8 (9.7) | 78.2 (16.5) |
| BDR, mean % (SD) | 10.4 (9.4) | 39.9 (20.4) | 6.4 (6.1) | 9.7 (11.1) |
Definition of abbreviations: BDR = bronchodilator response; CAMP = Childhood Asthma Management Program; LOCCS = Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol trial; LODO = Effectiveness of Low Dose Theophylline as Add-on Treatment in Asthma trial.
Figure 1.
Overall strategy of screening and replication. ACRC = Asthma Clinical Research Centers; BDR = bronchodilator response; CAMP = Childhood Asthma Management Program; LOCCS = Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol; LODO = Effectiveness of Low Dose Theophylline as Add-on Treatment in Asthma.
Table 2 summarizes the results of the replication analyses. Several SNPs were individually associated with BDR in the four populations. The P values from each of these populations were combined, and four SNPs from ARG1 (rs2781659, rs2781663, rs2781665, and rs2749935) showed the strongest evidence for association with BDR. After applying Bonferroni correction for the 99 tests in the initial replication analysis, SNP rs22781659 remained significantly associated with BDR (combined P value = 0.00048; Bonferroni-corrected P value = 0.047). Evidence for association of SNPs rs2781663 and rs2781665 was borderline significant after adjustment for multiple testing (Bonferroni-corrected P values of 0.075 and 0.085, respectively).
TABLE 2.
SUMMARY OF RESULTS OF TESTING AND REPLICATION IN THE FOUR ASTHMA CLINICAL TRIALS*
| Gene Name | rs Number | CAMP P Value | Asthma Trial P Value | LODO P Value | LOCCS P Value | Combined P Value |
|---|---|---|---|---|---|---|
| ARG1 | rs2781659 | 0.310 | 0.030 | 0.029 | 0.003 | 0.00048 |
| ARG1 | rs2781663 | 0.428 | 0.022 | 0.060 | 0.003 | 0.00076 |
| ARG1 | rs2781665 | 0.559 | 0.031 | 0.036 | 0.003 | 0.00086 |
| ARG1 | rs2749935 | 0.918 | 0.072 | 0.011 | 0.029 | 0.00596 |
| CRHR2 | rs1003929 | 0.159 | 0.003 | 0.484 | 0.222 | 0.01244 |
| CRHR2 | rs2190242 | 0.022 | 0.071 | 0.176 | 0.281 | 0.01509 |
| CPM | rs1144961 | 0.401 | 0.265 | 0.140 | 0.009 | 0.02189 |
| CRHR2 | rs2240403 | 0.053 | 0.270 | 0.040 | 0.342 | 0.02917 |
| CRHR2 | rs2284220 | 0.209 | 0.016 | 0.263 | 0.315 | 0.03747 |
| CRHR2 | rs917195 | 0.097 | 0.106 | 0.107 | 0.270 | 0.03906 |
| CRHR2 | rs2284217 | 0.472 | 0.068 | 0.154 | 0.084 | 0.04869 |
| CREBL2 | rs4555 | 0.860 | 0.077 | 0.280 | 0.023 | 0.05003 |
| CREM | rs10827492 | 0.827 | 0.009 | 0.465 | 0.136 | 0.05331 |
| CREM | rs4934736 | 0.884 | 0.009 | 0.260 | 0.242 | 0.05694 |
| CREM | rs1148247 | 0.610 | 0.016 | 0.397 | 0.140 | 0.05758 |
| CRHR2 | rs929377 | 0.652 | 0.180 | 0.076 | 0.066 | 0.06155 |
| CRHR2 | rs7793837 | 0.873 | 0.096 | 0.015 | 0.480 | 0.06336 |
| CRHR2 | rs2267716 | 0.342 | 0.366 | 0.026 | 0.244 | 0.07444 |
| CREM | rs7077242 | 0.835 | 0.024 | 0.232 | 0.174 | 0.07590 |
| CREM | rs10827493 | 0.956 | 0.010 | 0.322 | 0.273 | 0.07867 |
Definition of abbreviations: CAMP = Childhood Asthma Management Program; LOCCS = Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol trial; LODO = Effectiveness of Low Dose Theophylline as Add-on Treatment in Asthma trial.
Only the top 20 SNPs with the smallest P values are included in the table. All analyses investigated additive genetic models. All analyses were adjusted for age, sex, height, and baseline FEV1. P values for CAMP were obtained from FBAT (two-sided P values); P values for the replication analyses were obtained from linear regression models (one-sided P values) based on the direction of association obtained from the FBAT analysis. SNPs are ranked based on the smallest to the largest combined P value.
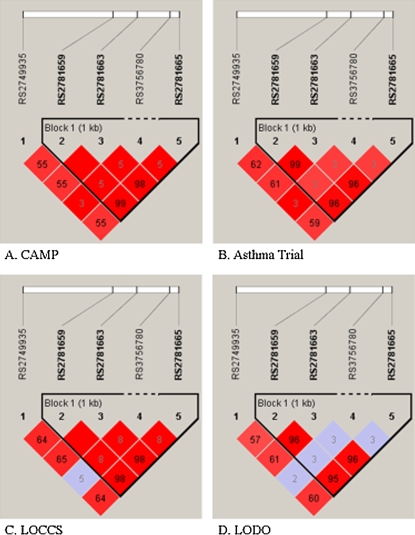
We examined the effect of each of these three SNPs on the magnitude of BDR in each of the populations (Table 3). In each case, the presence of the minor allele was associated with lower adjusted BDR compared with the homozygous major allele, consistent with the initial FBAT-PC results. Figure 2 compares the LD patterns of the five ARG1 SNPs in the four asthma populations by plotting pairwise r2 in physical order. The pairwise r2 values are similar in each of the populations. The three SNPs that were associated with BDR (rs2781659, rs2781663, and rs2781665) were in strong LD with each other (r2 values ranging from 95 to 100%, depending on the population). For example, in the CAMP population, r2 between SNPs rs2781659 and rs2781663 was 100%, whereas the r2 between SNPs rs2781659 and rs2781665 was 99%. In contrast, the other SNP that was more weakly associated with BDR, rs2749935, was not as tightly linked with the other three SNPs (r2 was 55% with rs2781659, rs2781663, and rs2781665).
TABLE 3.
EFFECTS OF ARG1 SINGLE NUCLEOTIDE POLYMORPHISMSS ON BRONCHODILATOR RESPONSE*
| Asthma Trial
|
LOCCS
|
LODO
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | n (%) | BDR | SE | n (%) | BDR | SE | n (%) | BDR | SE | ||||||||
| ARG1_RS2781665 | |||||||||||||||||
| AA | 189 (46.55) | 41.08 | 1.33 | 71 (44.38) | 7.53 | 0.69 | 70 (47.30) | 11.53 | 1.17 | ||||||||
| AT | 169 (41.63) | 39.44 | 1.41 | 68 (42.50) | 6.42 | 0.72 | 62 (41.89) | 7.95 | 1.25 | ||||||||
| TT | 48 (11.82) | 35.33 | 2.65 | 21 (13.13) | 3.14 | 1.29 | 16 (10.81) | 8.59 | 2.44 | ||||||||
| ARG1_RS2781663 | |||||||||||||||||
| TT | 190 (44.92) | 41.71 | 1.32 | 71 (44.38) | 7.43 | 0.68 | 67 (46.85) | 10.78 | 1.12 | ||||||||
| AT | 184 (43.50) | 39.14 | 1.34 | 67 (41.88) | 6.68 | 0.70 | 61 (42.66) | 7.85 | 1.18 | ||||||||
| AA | 49 (11.58) | 36.28 | 2.62 | 22 (13.75) | 2.99 | 1.22 | 15 (10.49) | 8.40 | 2.38 | ||||||||
| ARG1_RS2781659 | |||||||||||||||||
| AA | 193 (45.43) | 41.39 | 1.31 | 74 (47.13) | 7.35 | 0.69 | 71 (47.65) | 11.53 | 1.15 | ||||||||
| AG | 183 (42.86) | 39.50 | 1.35 | 62 (33.49) | 6.39 | 0.76 | 61 (40.94) | 7.51 | 1.24 | ||||||||
| GG | 50 (11.71) | 35.93 | 2.59 | 21 (13.38) | 2.90 | 1.30 | 17 (11.41) | 8.73 | 2.35 | ||||||||
Definition of abbreviations: BDR = bronchodilator response; LOCCS = Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol trial; LODO = Effectiveness of Low Dose Theophylline as Add-on Treatment in Asthma trial.
BDR is expressed as the mean for each genotype category. BDR means are obtained from multiple linear regression models adjusted for age, height, sex, and baseline FEV1, using the lsmeans (least squares means) option in the GLM procedure in SAS (SAS Institute, Cary, NC).
Figure 2.
Linkage disequilibrium patterns of ARG1 single nucleotide polymorphisms (SNPs) in the four asthma cohorts. Numbers in the individual blocks represent r2 values for each pair of SNPs (blank values = 100%), with the colors corresponding to D′ values. Plots were created using the program Haploview (http://www.broad.mit.edu/mpg/haploview/).
DISCUSSION
Prior investigations into the pharmacogenetics of asthma have generally been limited to one or a few SNPs from one gene. We investigated 844 SNPs from 111 candidate genes selected from asthma β-agonist and corticosteroid pathways, and from our prior candidate gene studies, and screened these SNPs for association with BDR using a family-based screening algorithm that allowed us to rank the SNPs based on estimated power for replication. We then genotyped 99 SNPs from 17 genes in a population-based cohort of patients with asthma, who participated in an asthma clinical trial. Finally, we genotyped 83 SNPs in seven genes in two separate cohorts of subjects with asthma. We found SNPs in the ARG1 gene to be associated with BDR in these three asthma populations, after adjusting for multiple comparisons.
ARG1 has recently been implicated in asthma. Zimmermann and colleagues (29) reported increased expression of ARG1 and ARG2 in murine lung, and also found increased arginase 1 protein expression from human asthma bronchoalveolar lavage cells. Variants in ARG1 were associated with atopy in a cohort of Mexicans with asthma (30). ARG1 maps to chromosome 6q23 and encodes one isoform of the enzyme arginase, which metabolizes l-arginine. l-Arginine homeostasis is involved in the regulation of airway function, because the availability of this amino acid to nitric oxide synthase (NOS) determines the production of the endogenous bronchodilator nitric oxide (NO) (31). Changes in l-arginine homeostasis may contribute to many of the features of asthma, such as airway hyperresponsiveness, airway inflammation, and airway remodeling (32). Intracellular l-arginine levels are regulated by at least three distinct mechanisms (reviewed by Maarsingh and colleagues [32]): (1) cellular uptake by cationic amino acid transporters, (2) recycling from l-citrulline, and (3) metabolism by NOS and arginase. Arginase is postulated to be involved in asthma by depleting stores of l-arginine, an NOS substrate, which leads to decreased production of NO, a potent bronchial smooth muscle relaxer (33, 34), and it has been shown to inhibit airway smooth muscle relaxation (35, 36). Finally, RNA interference of arginase 1 in the lungs resulted in complete loss of airway hyperresponsiveness to methacholine due to IL-13 treatment (37). This correlated with arginase 1 expression, which suggests that the polymorphisms involved with the current findings in human asthma may cause a loss of expression or function of arginase 1.
We used a gene-based strategy to select SNPs to take forward for replication. In this method, after ranking SNPs from 1 (most power) to 844 (least power), we grouped all SNPs for each gene and calculated the median SNP rank for that gene. Thus, whereas some genes had one or two SNPs that were assigned high ranks, these genes may not be taken forward because the median SNP rank did not meet the predetermined cutoff. We adopted this strategy because we were not sure that LD patterns across the four asthma populations would be similar. It is interesting to note that ADRB2, a gene that has been widely studied in asthma pharmacogenetics (38), was not one of the genes that was selected using this strategy, despite including 18 SNPs from this gene in the screening analysis. It is possible that there was insufficient power in the screening stage, because we only analyzed the 209 trios in the placebo group in CAMP. However, it should be noted that a prior analysis using all 400 trios also did not find an association with any of the ADRB2 SNPs and BDR (39). Furthermore, the phenotype that we investigated is different from that reported in other studies reporting on the pharmacogenetic effect of ADRB2 (9, 10). We are currently performing additional genotyping and analyses using an SNP-based strategy for replication, rather than the gene-based strategy that we used here, to see if we identify important SNPs in this gene and others for association with BDR.
Our analysis used the phenotype of acute response to a short-acting β2-agonist, albuterol, in part because this was the phenotype that was common to all asthma cohorts. In the screening algorithm, we used the information from repeated measures of BDR among the 209 white children randomized to the placebo group in the CAMP study over the 4 years of the trial. This was done to increase the power for the screening method. In contrast, for the replication cohorts, we only used the information on BDR response on entry into the respective studies, to standardize the phenotype. Thus, our results may not be applicable to patients with asthma who are on regular β2-agonist treatment (either short- or long-acting). We also did not address interactions with any other class of asthma medication, because baseline medication was different for all the populations: BDR was performed in both CAMP and the Adult Trial populations after several weeks of being off all asthma medications; LOCCS subjects were receiving inhaled corticosteroids for 4–6 weeks before BDR testing; and drug regimens for LODO subjects were not changed before entry into the trial.
We used a novel method of screening a large number of SNPs for association analysis (11). This method has been successfully used to identify disease-susceptibility genes (12–14). Because this method has only been developed for family-based studies and not population-based studies, we used the CAMP population for screening the original 844 SNPs. The traditional method would have been to analyze all the SNPs in one population, determine which SNPs were associated with BDR at a predetermined level of significance, then test these SNPs in the replication populations. However, if we had used this usual method for gene finding, we would then have had to adjust our overall results for the 844 SNPs that were originally tested, and it is likely that no finding would have survived this adjustment for multiple testing, even if the association was real. In our method, because we screened on power and not P value, we only needed to adjust for the 99 tests in the first replication step. Thus, this screening method allows the use of modest-sized populations for gene discovery because it limits the number of tests that are actually being performed.
The population to which we applied our screening algorithm was a cohort of childhood asthmatics, whereas the three asthma replication cohorts were composed predominantly of adults with asthma. As we stated previously, the rationale for this is that the screening method was developed for the setting of family-based studies and not for population-based studies. There is no similar screening method that has yet been developed for population-based studies. Because our replication populations were of small to modest sizes, we applied the screening method as a means of minimizing the number of tests. Although there were only 209 parent–child trios included in the screening analysis, we maximized the power in the screening stage by using the 11 repeated measures of BDR over the 4 years of the trial. In addition, there were differences in the asthma severity and in the magnitude of the BDR between the populations as shown in Table 1. Despite these differences, we were able to detect associations between SNPs in ARG1 and BDR in each of the three replication populations. Although the association between these SNPs and BDR in CAMP was not statistically significant, the effect sizes of each SNP were of sufficient magnitude for them to be selected based on power in the screening analysis. We can only surmise at this point that there may be age-related effects associated with the SNPs in this gene. We believe, therefore, that the results of the association between ARG1 polymorphisms and BDR are robust and applicable to both children and adults with asthma in a variety of settings.
The three ARG1 SNPs that were associated with BDR were all in the promoter region of the gene and were in tight LD with each other. Genotyping of all known SNPs in the gene or resequencing of the gene will need to be performed to determine if these three promoter SNPs are in LD with the functional mutation. There is mounting evidence for “cross-talk” between pathways involved with the relaxation and constriction of airway smooth muscle (40, 41). It is thus not entirely unexpected that a gene involved with airway hyperresponsiveness in mouse studies is associated with a bronchodilator response in human asthma. However, arginase 1 has not been previously identified as one of the proteins involved in such cross-talk. This unexpected finding shows the potential value of whole-genome coverage to study drug response to uncover novel genetic determinants. The screening method that we used for this analysis would be easily applicable to the case of whole-genome association.
In summary, we have identified SNPs in ARG1 as novel BDR determinants. Further studies will need to identify the functional SNP or SNPs in this gene. Other pharmacogenetic studies using long-acting β2-agonists, either alone or in conjunction with corticosteroids, and investigation of other phenotypes (e.g., FEV1, peak flow) are needed to clarify the effects of variants in this gene. Our analysis shows the utility of a family-based algorithm to effectively screen SNPs for replication in other cohorts. This method is easily applicable to the case of whole-genome association.
Supplementary Material
Acknowledgments
The authors thank all families for their enthusiastic participation in the CAMP Genetics Ancillary Study. They thank the CAMP investigators and research team, for collection of CAMP Genetic Ancillary Study data. The authors also acknowledge the American Lung Association (ALA) and the ALA's Asthma Clinical Research Centers investigators and research teams for use of LOCCS and LoDo data, and Nemours Children's Clinic. The authors also acknowledge Sepracor, Inc., for use of the Asthma Trial data.
Supported by U01 HL65899: The Pharmacogenetics of Asthma Treatment from the NHLBI. The CAMP Genetics Ancillary Study was supported by the National Heart, Lung, and Blood Institute (NHLBI), NO1-HR16049. The CAMP investigators and research team received support from the NHLBI. Additional support for this research came from grants N01-HR16044, HR16045, HR16046, HR16047, HR16048, HR16049, HR16050, HR16051, and HR16052 from the NHLBI. Additional funding was received from HL071394 and HL074755 from the NHLBI. GlaxoSmithKline supported the conduct of the Leukotriene Modifier or Corticosteroid or Corticosteroid Salmeterol trial by an unrestricted grant to the American Lung Association.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200709-1363OC on July 10, 2008
Conflict of Interest Statement: A.A.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.L.-S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.G.T. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.J.L. is a coinvestigator on a grant from Merck to study montelukast transport. C.G.I. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.P.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.P.H. is a pulmonary physician employed full time by Sepracor, Inc., as the Executive Director for Pulmonary Clinical Research. Sepracor supported one of the clinical trials that provided data to this manuscript. S.B.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.A.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.A.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.R.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.T.W. received a grant for $900,065, Asthma Policy Modeling Study, from AstraZeneca for 1997–2003; has been a coinvestigator on a grant from Boehringer Ingelheim to investigate a COPD natural history model, which began in 2003 (he has received no funds for his involvement in this project); has been an advisor and chair of the advisory board to the TENOR Study for Genentech and has received $10,000 for 2005–2006; received a grant from Glaxo-Wellcome for $500,000 for genomic equipment for 2000–2003; was a consultant for Roche Pharmaceuticals in 2000 and received no financial renumeration for this consultancy; and also served as a consultant to Pfizer (2000–2003), Schering Plough (1999–2000), Variagenics (2002), Genome Therapeutics (2003), and Merck Frost (2002).
References
- 1.Masoli M, Fabian D, Holt S, Beasley R; Global Initiative for Asthma (GINA) Program. Global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy 2004;469–478. [DOI] [PubMed]
- 2.Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma—United States, 1980–1999. MMWR Surveill Summ 2002;51:1–13. [PubMed] [Google Scholar]
- 3.Gergen PJ, Mullally DI, Evans R III. National survey of prevalence of asthma among children in the United States, 1976. to 1980. Pediatrics 1988;81:1–7. [PubMed] [Google Scholar]
- 4.Smith DH, Malone DC, Lawson KA, Okamoto LJ, Battista C, Saunders WB. A national estimate of the economic costs of asthma. Am J Respir Crit Care Med 1997;156:787–793. [DOI] [PubMed] [Google Scholar]
- 5.Sears MR, Lotvall J. Past, present and future: beta2-adrenoceptor agonists in asthma management. Respir Med 2005;99:152–170. [DOI] [PubMed] [Google Scholar]
- 6.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull 2000;56:1054–1070. [DOI] [PubMed] [Google Scholar]
- 7.Weiss ST, Litonjua AA, Lange C, Lazarus R, Liggett SB, Bleecker ER, Tantisira KG. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J 2006;6:311–326. [DOI] [PubMed] [Google Scholar]
- 8.Litonjua AA. The significance of beta2-adrenergic receptor polymorphisms in asthma. Curr Opin Pulm Med 2006;12:12–17. [DOI] [PubMed] [Google Scholar]
- 9.Israel E, Chinchilli VM, Ford JG, Boushey HA, Cherniack R, Craig TJ, Deykin A, Fagan JK, Fahy JV, Fish J, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet 2004;364:1505–1512. [DOI] [PubMed] [Google Scholar]
- 10.Israel E, Drazen JM, Liggett SB, Boushey HA, Cherniack RM, Chinchilli VM, Cooper DM, Fahy JV, Fish JE, Ford JG, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med 2000;162:75–80. [DOI] [PubMed] [Google Scholar]
- 11.Van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, Demeo DL, Murphy A, Su J, Datta S, Rosenow C, et al. Genomic screening and replication using the same data set in family-based association testing. Nat Genet 2005;37:683–691. [DOI] [PubMed] [Google Scholar]
- 12.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, et al. A common genetic variant is associated with adult and childhood obesity. Science 2006;312:279–283. [DOI] [PubMed] [Google Scholar]
- 13.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, Zhu X, Thorleifsson G, Gunnarsdottir S, Walters GB, Thorsteinsdottir U, et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet 2007;3:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA, Lazarus R, Klanderman B, Soto-Quiros ME, Avila L, et al. On the replication of genetic associations: timing can be everything! Am J Hum Genet 2008;82:849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litonjua AA, Tantisira KG, Su JA, Lazarus R, Klanderman B, Lange C, Weiss ST. Polymorphisms in ARG1 are determinants of bronchodilator response: screening and replication in 4 asthma cohorts [abstract]. Am J Respir Crit Care Med 2007;175:A968. [Google Scholar]
- 16.Childhood Asthma Management Program Research Group. The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 17.Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med 2000;343:1054–1063. [DOI] [PubMed] [Google Scholar]
- 18.Baron RM, Palmer LJ, Tantisira K, Gabriel S, Sonna LA, Le L, Hallock A, Libermann TA, Drazen JM, Weiss ST, et al. DNA sequence variants in epithelium-specific ETS-2 and ETS-3 are not associated with asthma. Am J Respir Crit Care Med 2002;166:927–932. [DOI] [PubMed] [Google Scholar]
- 19.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, Faffe DS, Shikanai T, Raby BA, Weiss ST, et al. Transforming growth factor-β1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med 2004;169:214–219. [DOI] [PubMed] [Google Scholar]
- 20.Peters SP, Anthonisen N, Castro M, Holbrook JT, Irvin CG, Smith LJ, Wise RA. Randomized comparison of strategies for reducing treatment in mild persistent asthma. N Engl J Med 2007;356:2027–2039. [DOI] [PubMed] [Google Scholar]
- 21.American Lung Association Asthma Clinical Research Centers. Clinical trial of low-dose theophylline and montelukast in patients with poorly controlled asthma. Am J Respir Crit Care Med 2007;175:235–242. [DOI] [PubMed] [Google Scholar]
- 22.Litonjua A, Thorn CF, Liggett SB. β-Agonist and β-blocker pathway [diagram on the Internet]. Palo Alto (CA): PharmGKB, Stanford University; c2008 [updated 2007 Aug 1; accessed 2008 May 11]. Available from: http://www.pharmgkb.org/do/serve?objId=PA2024
- 23.Weiss S, Litonjua A, Tantisira K, Wong M-L, Thorn C, Licinio J. Glucocorticoid and inflammatory genes pathway [diagram on the Internet]. Palo Alto (CA): PharmGKB, Stanford University; c2008 [updated 2007 Aug 1; accessed 2008 May 11]. Available from: http://www.pharmgkb.org/search/pathway/glucocorticoid/hpa-axis.jsp#
- 24.Barnes PJ. Scientific rationale for using a single inhaler for asthma control. Eur Respir J 2007;29:587–595. [DOI] [PubMed] [Google Scholar]
- 25.Johnson M. Interactions between corticosteroids and β2-agonists in asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc 2004;1:200–206. [DOI] [PubMed] [Google Scholar]
- 26.Lange C, van Steen K, Andrew T, Lyon H, DeMeo DL, Raby B, Murphy A, Silverman EK, MacGregor A, Weiss ST, et al. A family-based association test for repeatedly measured quantitative traits adjusting for unknown environmental and/or polygenic effects. Stat Appl Genet Mol Biol [serial on the Internet]. 2004 [accessed 2008 May 11];3:Article 17. Available from: http://www.bepress.com/sagmb/vol3/iss1/art17 [DOI] [PubMed]
- 27.Fisher RA. Statistical methods for research workers. New York: Hafner; 1950.
- 28.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006;38:209–213. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 2003;111:1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Romieu I, Sienra-Monge JJ, Ramirez-Aguilar M, Estela Del Rio-Navarro B, Kistner EO, Gjessing HK, Lara-Sanchez Idel C, Chiu GY, London SJ. Genetic polymorphisms in arginase i and ii and childhood asthma and atopy. J Allergy Clin Immunol 2006;117:119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricciardolo FL, Sterk PJ, Gaston B, Folkerts G. Nitric oxide in health and disease of the respiratory system. Physiol Rev 2004;84:731–765. [DOI] [PubMed] [Google Scholar]
- 32.Maarsingh H, Zaagsma J, Meurs H. Arginine homeostasis in allergic asthma. Eur J Pharmacol 2008;585:375–384. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann N, Rothenberg ME. The arginine-arginase balance in asthma and lung inflammation. Eur J Pharmacol 2006;533:253–262. [DOI] [PubMed] [Google Scholar]
- 34.Meurs H, Maarsingh H, Zaagsma J. Arginase and asthma: novel insights into nitric oxide homeostasis and airway hyperresponsiveness. Trends Pharmacol Sci 2003;24:450–455. [DOI] [PubMed] [Google Scholar]
- 35.Maarsingh H, Leusink J, Bos IS, Zaagsma J, Meurs H. Arginase strongly impairs neuronal nitric oxide-mediated airway smooth muscle relaxation in allergic asthma. Respir Res 2006;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maarsingh H, Tio MA, Zaagsma J, Meurs H. Arginase attenuates inhibitory nonadrenergic noncholinergic nerve-induced nitric oxide generation and airway smooth muscle relaxation. Respir Res 2005;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Rangasamy D, Matthaei KI, Frew AJ, Zimmmermann N, Mahalingam S, Webb DC, Tremethick DJ, Thompson PJ, Hogan SP, et al. Inhibition of arginase I activity by RNA interference attenuates IL-13-induced airways hyperresponsiveness. J Immunol 2006;177:5595–5603. [DOI] [PubMed] [Google Scholar]
- 38.Liggett SB, Hall IP. Beta2-adrenergic receptor polymorphisms and asthmatic phenotypes. In: Postma DS, Weiss ST, editors. Genetics of asthma and chronic obstructive pulmonary disease. New York: Informa Healthcare USA; 2007. pp. 299–316.
- 39.Silverman EK, Kwiatkowski DJ, Sylvia JS, Lazarus R, Drazen JM, Lange C, Laird NM, Weiss ST. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol 2003;112:870–876. [DOI] [PubMed] [Google Scholar]
- 40.McGraw DW, Almoosa KF, Paul RJ, Kobilka BK, Liggett SB. Antithetic regulation by beta-adrenergic receptors of GQ receptor signaling via phospholipase C underlies the airway beta-agonist paradox. J Clin Invest 2003;112:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGraw DW, Elwing JM, Fogel KM, Wang WC, Glinka CB, Mihlbachler KA, Rothenberg ME, Liggett SB. Crosstalk between GI and GQ/GS pathways in airway smooth muscle regulates bronchial contractility and relaxation. J Clin Invest 2007;117:1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.