Abstract
Rationale: Polymorphisms affecting Toll-like receptor (TLR)–mediated responses could predispose to excessive inflammation during an infection and contribute to an increased risk for poor outcomes in patients with sepsis.
Objectives: To identify hypermorphic polymorphisms causing elevated TLR-mediated innate immune cytokine and chemokine responses and to test whether these polymorphisms are associated with increased susceptibility to death, organ dysfunction, and infections in patients with sepsis.
Methods: We screened single-nucleotide polymorphisms (SNPs) in 43 TLR-related genes to identify variants affecting TLR-mediated inflammatory responses in blood from healthy volunteers ex vivo. The SNP associated most strongly with hypermorphic responses was tested for associations with death, organ dysfunction, and type of infection in two studies: a nested case–control study in a cohort of intensive care unit patients with sepsis, and a case–control study using patients with sepsis, patients with sepsis-related acute lung injury, and healthy control subjects.
Measurements and Main Results: The SNP demonstrating the most hypermorphic effect was the G allele of TLR1−7202A/G (rs5743551), which associated with elevated TLR1-mediated cytokine production (P < 2 × 10−20). TLR1−7202G marked a coding SNP that causes higher TLR1-induced NF-κB activation and higher cell surface TLR1 expression. In the cohort of patients with sepsis TLR1−7202G predicted worse organ dysfunction and death (odds ratio, 1.82; 95% confidence interval, 1.07–3.09). In the case-control study TLR1−7202G was associated with sepsis-related acute lung injury (odds ratio, 3.40; 95% confidence interval, 1.59–7.27). TLR1−7202G also associated with a higher prevalence of gram-positive cultures in both clinical studies.
Conclusions: Hypermorphic genetic variation in TLR1 is associated with increased susceptibility to organ dysfunction, death, and gram-positive infection in sepsis.
Keywords: innate immunity, genetic variation, genetic predisposition
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Toll-like receptor (TLR)–related innate immune pathways have been shown to be important in animal models of sepsis. However, the importance of TLR-mediated responses in human sepsis is less clear.
What This Study Adds to the Field
We identify the polymorphisms in the TLR pathway that most strongly affect cellular responses to TLR agonists and show that one of these in the gene for TLR1 associates with susceptibility to organ dysfunction and death in patients with sepsis.
The sepsis syndrome and its more extreme manifestation, septic shock, are a major cause of mortality in the intensive care unit (ICU). More than 750,000 cases of sepsis occur in the United States each year (1), resulting in more than 210,000 deaths per year (2). The pathophysiology of sepsis involves highly complex interactions between invading microorganisms, the innate and adaptive immune systems of the host, and multiple downstream events leading to organ dysfunction (3). There has been considerable interest in the identification of genetic risk factors for clinical outcomes in sepsis. Common sequence variations within genes involved in innate immune responses have received particular attention (4).
The innate immune system provides host defense against microbial pathogens and involves the recognition of highly conserved pathogen-associated molecular patterns (PAMPs) by cell surface proteins of the Toll-like receptor (TLR) family (5, 6). Animal models of sepsis have shown that responses mediated by TLRs and their related intracellular signaling molecules are important determinants of host survival (6). In humans, purified PAMPs such as LPS, a TLR4 agonist, are capable of inducing vigorous inflammatory responses and reproducing the clinical manifestations of sepsis (7). These data implicate the TLR pathway in the pathophysiology of sepsis and suggest that excessive inflammation mediated by the TLR pathway might contribute to morbidity and mortality in this setting.
We have previously shown that peripheral leukocyte inflammatory responses to PAMPs show high interindividual variation in healthy volunteers (8). In this study, we sought to quantify the relative effects of functional genetic variation in the TLR pathway on leukocyte inflammatory responses to PAMPs in whole blood ex vivo and then to determine if genetic variants showing strong hyperinflammatory effects ex vivo might also increase the risk for poor outcomes, such as organ dysfunction or death, in patients with sepsis. Using this approach we identified several genetic variants with highly significant effects on TLR-mediated responses in healthy volunteers. We then identified a strong association between a linkage disequilibrium bin in TLR1 and a large increase in TLR1-mediated inflammatory cytokine production, providing functional validation of in vitro findings (9, 10). We report that the tag single-nucleotide polymorphism (SNP) for this bin, TLR1−7202A/G (rs5743551), is strongly associated with increased mortality and organ dysfunction in a large prospective cohort of patients with sepsis and septic shock. In a separate case–control study, TLR1−7202G showed a strong association with sepsis-induced acute lung injury (ALI), the predominant form of organ dysfunction in sepsis. Finally, we show that coding variants tagged by this SNP are functionally active, mediating higher TLR1 agonist–induced nuclear factor (NF)-κB activation and increased TLR1 cell surface expression. Taken together, these data provide strong genetic evidence that a predisposition to hyperinflammatory innate immune responses is associated with poorer clinical outcomes in patients with sepsis. Some of the results of these studies have been reported in abstract form (11).
METHODS
Subjects
Healthy cohort.
Nonsmoking individuals between 18 and 65 years of age were recruited from the metropolitan Seattle area. Exclusions to enrollment included recent antibiotic use, symptoms consistent with infection, a history of autoimmune disease, immunodeficiency, use of immunosuppressive medications, cancer, or pregnancy. Total and differential leukocyte counts were performed on each blood sample in the clinical laboratory at Harborview Medical Center (Seattle, WA). The study was approved by the Division of Human Subjects Research, University of Washington (Seattle, WA).
Sepsis cohort.
This prospectively collected cohort of patients with sepsis admitted to an ICU has been described (12, 13). Briefly, we screened 1,183 consecutive patients admitted to a mixed medical–surgical ICU for the presence of sepsis or septic shock (14). Patients were monitored for the primary outcome variable of 28-day in-hospital mortality. Secondary outcome variables included (1) the days alive and free of organ dysfunction (cardiovascular, respiratory, hepatic, coagulation, and neurologic) as defined by the Brussels criteria (15), (2) days alive and free of vasopressor use or mechanical ventilation, and (3) the prevalence of positive bacterial cultures. This study was approved by the University of British Columbia (Vancouver, BC, Canada) Human Subjects Research Committee.
Patients in Consortium to Evaluate Lung Edema Genetics.
We used white American participants in the Consortium to Evaluate Lung Edema Genetics (CELEG) (16), which enrolled patients with severe sepsis (17) with (n = 138) or without (n = 107) ALI (18) and healthy control subjects (n = 167). This study was approved by human subjects research committees at Johns Hopkins University (Baltimore, MD) and the University of Maryland (Baltimore, MD), Emory University (Atlanta, GA), the Medical College of Wisconsin (Milwaukee, WI), and the University of Colorado (Denver, CO).
Functional Assays
Whole blood assay of TLR-mediated responses.
We performed measurements of cellular responses to PAMPs in whole blood ex vivo according to a previously described protocol (19). Eight different TLR agonists were tested: (1) pam3CSK4 (N-palmitoyl-S-[2,3-bis(palmitoyl-oxy)-(2RS)-propyl]-[R]-cysteinyl-[S]-seryl-[S]-lysyl-[S]-lysyl-[S]-lysyl-[S]-lysine) (10 ng/ml) (EMC Microcollections GmbH, Tübingen, Germany); (2) ultrapure Salmonella minnesota Re595 LPS (1 ng/ml) (List Biological Laboratories, Inc., Campbell, CA), (3) Yersinia pestis LPS (10 ng/ml) (gift of R. K. Ernst, University of Washington), (4) ultrapure Escherichia coli 0111:B4 LPS (1 ng/ml) (List Biological Laboratories, Inc.), (5) zymosan derived from Saccharomyces cerevisiae (10 μg/ml) (Sigma-Aldrich, St. Louis, MO), (6) flagellin (100 ng/ml) from Salmonella typhimurium (gift of T. Hawn, University of Washington), (7) muramyl dipeptide (10 μg/ml; Sigma-Aldrich), and (8) peptidoglycan (10 μg/ml) from Staphylococcus aureus (Sigma-Aldrich) (20). All compounds were used at concentrations that elicited cytokine/chemokine responses that fell within the logarithmic portion of the dose–response curve in human whole blood. Blood was drawn from fasting subjects before 10:00 a.m. and was anticoagulated with pyrogen-free citrate (0.1 M, pH 7.2), diluted 1:1 with RPMI 1640 medium, and then added to each well of a 96-well plate containing each of the TLR agonists. After incubation for 6 hours at 37°C in 5% CO2, supernatants were harvested and concentrations of the mediators granulocyte colony-stimulating factor, IL-1ra, IL-1β, IL-6, IL-8, IL-10, and tumor necrosis factor-α were measured with a cytometric bead-based system (Luminex, Austin, TX) as previously described (21, 22).
Genotyping
Selection and genotyping of tagging SNPs.
We selected polymorphisms previously identified through gene resequencing by the Program for Genomic Applications (PGA) (http://pga.gs.washington.edu/) (Table 1). A full list of candidate genes and informative SNPs is included in Table E1 (see the online supplement). We identified “tag SNPs” for genotyping that marked groups of highly correlated SNPs within each candidate gene as measured by the linkage disequilibrium statistic r2 (23). Tag SNPs were selected by a modification of the LD-select algorithm (23, 24) (minor allele frequencies ≥ 5%, linkage disequilibrium [r2] ≥ 0.65). Genotypes in the healthy cohort were determined for each of the selected tag SNPs, using a BeadStation laboratory system (Illumina, Inc., San Diego, CA) paramagnetic microbead array as previously described (25). We replicated 2% of the samples for quality control.
TABLE 1.
CANDIDATE GENES SCREENED ORGANIZED BY FUNCTIONAL CLASS
| Cell Surface Receptors | Signaling | Transcription | Cytokines and Chemokines |
|---|---|---|---|
| CD14 | IRAK1 | FOS | IL1A |
| LBP | IRAK2 | JUN | IL1B |
| TLR1 | IRAK3 (IRAKM) | NFKB1 | IL1RN |
| TLR2 | IRAK4 | NFKBIA (IKBa) | IL6 |
| TLR4 | JAK3 | NFKBIB (IKBb) | IL8 |
| TLR5 | MYD88 | IKBKA | IL10 |
| TLR6 | TIRAP | IKBKB | IL8RA |
| LY96 (MD2) | TOLLIP | RELA (NF-κB p65) | IL8RB |
| TRAF6 | STAT1 | CSF3 (G-CSF) | |
| TICAM1 (TRIF) | STAT2 | CSF3R (G-CSFR) | |
| TTRAP | STAT3 | TNFA | |
| STAT4 | MCP1 |
Human Genome Organization gene symbol in italics. Common gene aliases presented in parentheses.
Genotypes for TLR1−7202A/G (rs5743551), TLR11517G/A (rs5743614), and TLR11804G/T (rs5743618) in the sepsis cohort and CELEG patients were determined by TaqMan-based real-time polymerase chain reaction (RT-PCR) as previously described (26). Primer–probe sets were obtained from the Applied Biosystems (Foster City, CA) repository (TLR1−7202A/G) or synthesized (TLR11517G/A: forward primer, AGCAGCCTTTCTGTATTGATCATTGA; reverse primer, CATCTTCTGGCAGCTCTGGAA; probe, CACCCATC[G/A]GCTGAT; TLR11804G/T: forward primer, GGTGTTGGCTGTGACTGTGA; reverse primer, GCACACCATCCTGAGATACCA; probe, CCTCTGCA[G/T]CTACTT) and run according to the recommended guidelines on an ABI PRISM 7900HT (Applied Biosystems).
Functional Characterization
Cloning, mutagenesis, and expression of TLR1.
cDNA for TLR1 was isolated from lymphocyte cell lines homozygotic for either the A (wild-type) or G (variant) allele of TLR1−7202A/G (rs5743551). The cDNA was amplified and cloned into a pcDNA3.1 TOPO cloning vector (Invitrogen, Carlsbad, CA). Site-directed mutagenesis of the cloned wild-type TLR1 cDNA was performed with a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). Endotoxin-free plasmids were prepared for transfection with an EndoFree plasmid purification kit (Qiagen, Valencia, CA). Transfections were performed as previously described (27) with the following plasmids: NF-κB–reporter construct (ELAM-1–firefly luciferase), a transfection control (β-actin–Renilla luciferase), a human TLR2 expression construct (28), and the TLR1 constructs detailed above. After 3 hours, the medium was replaced with fresh medium and, the next day, the cells were stimulated with medium alone, pam3CSK4 (100 ng/ml), or Re595 LPS (10 ng/ml) for 4 hours and then lysed. Luciferase activity was measured with a Dual-Luciferase reporter assay system (Promega, Madison, WI). Quantitation of TLR1 gene expression in transfected HEK293 cells was performed by reverse transcription followed by quantitative real-time PCR, using commercially available primer–probe sets (Applied Biosystems), reference numbers Hs00413978_m1 (human TLR1) and Hs99999905_m1 (human GAPDH; glyceraldehyde-3-phosphate dehydrogenase) and standard reaction conditions (50°C for 10 min followed by 55 cycles of 95°C for 15 s and 60°C for 45 s). Ct values observed for each sample were normalized to the level of GAPDH mRNA detected in each sample and are presented as fold change from measurements in mock-transfected cells.
Flow cytometry.
Peripheral blood mononuclear cells were isolated in CPT density gradient tubes (Becton Dickinson, Franklin Lakes, NJ) and stained with the anti-huTLR1 monoclonal antibody GD2 or an isotype (IgG1) control (BD Biosciences, San Jose, CA) and fluorescence was measured with an Easycyte system (Guava Technologies, Hayward, CA) with forward and side scatter gating for the monocyte population. TLR1 staining was determined as the percentage of cells showing fluorescence greater than 95% of all cells stained with the isotype-control antibody. To determine intracellular staining we treated the cells with 4% paraformaldehyde followed by FACS permeabilizing solution 2 (BD Biosciences) before antibody staining.
Statistical Analysis
Observed genotype frequencies were compared with expected frequencies to test for deviations from Hardy-Weinberg equilibrium as described (29). The 56 PAMP-induced phenotypes (production of 7 cytokines/chemokines in response to 8 different PAMPs) for each subject were normalized to their blood monocyte count and these values were logarithmically (log10) transformed. The relationship between the copy number of each tag SNP (655 total) and the 56 PAMP-induced phenotypes was analyzed assuming a codominant effect, using linear regression. We limited the false discovery rate to 10%, using the QVALUE program (http://cran.r-project.org/web/packages/qvalue/) (30), and resultant findings were ranked by the proportion of variance accounted for by each SNP (r2). The analyses were performed for subjects of white and African American background separately to minimize confounding due to racial differences in polymorphism frequency.
We tested for associations between TLR1 genotypes and 28-day mortality in the sepsis cohort, using recessive logistic regression models adjusted for confounding variables including age, sex, the presence of serious preexisting comorbidities, and medical versus surgical diagnosis. The 28-day survival was compared between the two groups of patients, using Kaplan-Meier plots. To minimize the risk of confounding due to racial differences in SNP frequency (31), analyses were limited to white patients (n = 724) successfully genotyped for at least one tagging SNP. Differences in secondary outcome measures by genotype were analyzed by chi-square test for dichotomous outcomes (culture prevalence) and the Mann-Whitney U test for continuous variables (days alive and free of organ failure). Statistical significance was set at P < 0.05.
We tested for associations between TLR1 genotypes and case–control status (control, severe sepsis, or severe sepsis with ALI) among the CELEG subjects by logistic regression models, including age and sex as covariates. Age was recorded as a categorical variable with three categories: age not more than 40 years, 41–59 years, and 60 years or more. Departures from Hardy-Weinberg equilibrium proportions at each SNP were tested among cases and control subjects separately. Analyses were performed with Stata 9.0 (Stata Corp., College Station, TX).
Online Supplemental Material
Table E1 provides detailed information about each of the SNPs genotyped in the healthy volunteer population, including Single-Nucleotide Polymorphism Database (dbSNP) reference number, allele nucleotides, chromosome number, and flanking sequence.
RESULTS
Associations between Tag SNPs in TLR Pathway Genes and PAMP-induced Responses
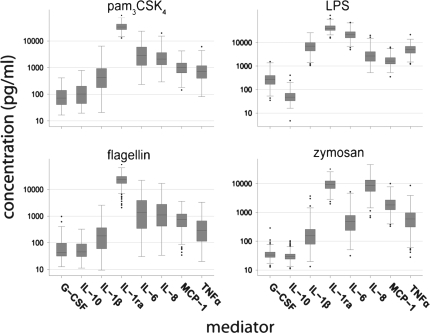
We tested PAMP-induced responses in whole blood samples obtained from 337 healthy volunteers. Figure 1 shows the distribution of cytokine and chemokine responses, in whole blood from 296 white volunteers, to four TLR agonists representing the four major classes of TLRs tested: pam3CSK4, an agonist for TLR2/1; zymosan, an agonist for TLR2/6; LPS, an agonist for TLR4; and flagellin, an agonist for TLR5. Cytokine and chemokine responses to all PAMPs showed high interindividual variability in the normal population. For example, LPS-induced responses varied over 2 log10 units (Figure 1), consistent with our previous findings (8). For all of our subsequent analyses, the PAMP-induced cytokine and chemokine values were normalized to each subject's peripheral blood monocyte count to isolate genetic effects related to differences in TLR-related cell signaling processes as opposed to effects related to peripheral leukocyte concentrations. We then genotyped 309 of these subjects (white Americans, n = 275; African American, n = 34) across 43 candidate genes involved in the TLR pathway and several inflammatory mediators induced by this pathway (Table 1). We genotyped 255 linkage disequilibrium (LD) bin-tagging SNPs (tag SNPs) (see Table E1), selected from the candidate genes to mark groups of common SNPs (minor allele frequency ≥ 5%) in high LD (r2 ≥ 0.65). We limited our initial analysis to white Americans (n = 275). All tag SNP genotypes were found to be in Hardy-Weinberg equilibrium and the genotype calls for all replicated samples were in agreement. Our analysis identified 18 tag SNPs in 12 different candidate genes that were significantly associated with PAMP-induced responses after correction for multiple comparisons (Table 2).
Figure 1.
Pathogen-associated molecular pattern (PAMP)–induced responses show high interindividual variation in whole blood. Healthy white American volunteers (n = 296) provided whole blood samples that were stimulated as described in methods with pam3CSK4 (N-palmitoyl-S-[2,3-bis(palmitoyl-oxy)-(2RS)-propyl]-[R]-cysteinyl-[S]-seryl-[S]-lysyl-[S]-lysyl-[S]-lysyl-[S]-lysine; 10 ng/ml), LPS (from Escherichia coli; 1 ng/ml), flagellin (from Salmonella typhimurium; 10 ng/ml), and zymosan (from Saccharomyces cerevisiae; 10 μg/ml) and cytokine and chemokine production (expressed as picograms per milliliter) was measured by immunoassay. Data are represented as box plots showing median, intraquartile range (IQR) (box), and range (up to 1.5 times the upper and lower quartiles) (whiskers), and outliers (dots). G-CSF = granulocyte colony-stimulating factor; MCP-1 = monocyte chemotactic protein-1; TNF-α = tumor necrosis factor-α.
TABLE 2.
GENETIC VARIANTS ASSOCIATED WITH WHOLE BLOOD INNATE IMMUNE RESPONSES
| Genotypes
|
Phenotypes Associated
|
Linear Regression*
|
||||
|---|---|---|---|---|---|---|
| Gene | dbSNP | PAMP | Mediator | Effect† | r2 (range) | −log10(P) (range) |
| TLR1 | rs5743551 | pam3CSK4 | All | + | 0.06–0.50 | 6.9–20.0 |
| Zymosan | IL-6, G-CSF | + | 0.06 | 4.5–4.6 | ||
| rs5743594 | pam3CSK4 | IL-1β, IL-6 | − | 0.05–0.07 | 4.1–5.5 | |
| rs4624663 | pam3CSK4 | IL-10 | + | 0.04–0.05 | 4.4 | |
| TLR6 | rs5743795 | pam3CSK4 | All | + | 0.05–0.35 | 6.0–20.0 |
| rs1039560 | pam3CSK4 | IL-6, IL-8, IL-10, G-CSF | − | 0.05–0.10 | 4.0–7.3 | |
| rs3775073 | pam3CSK4 | IL-6 | − | 0.05–0.06 | 4.7 | |
| TLR4 | rs7864330 | LPS (Y. pestis) | IL-1β, IL-6, TNF-α | − | 0.04–0.10 | 5.6–7.8 |
| rs1927906 | LPS (Y. pestis) | IL-1β, IL-6 | − | 0.05–0.06 | 4.0–4.8 | |
| TLR5 | rs5744168 | Flagellin | IL-6, IL-8 | − | 0.05–0.08 | 5.0–5.5 |
| TIRAP | rs8177352 | Peptidoglycan | IL-6, TNF-α, G-CSF | − | 0.06–0.07 | 4.6–5.5 |
| rs611953 | Peptidoglycan | IL-6, G-CSF | + | 0.06 | 4.7–4.8 | |
| IL1RN | rs4251961 | Peptidoglycan | IL-1RA | − | 0.05 | 4.2 |
| Zymosan | IL-1RA | − | 0.07 | 5.4 | ||
| TOLLIP | rs5743856 | MDP | G-CSF | + | 0.06 | 4.8 |
| STAT1 | rs2280233 | LPS (Y. pestis) | IL-6, IL-1RA | − | 0.05–0.06 | 4.0–4.8 |
| CSF3 | rs2227319 | Zymosan | G-CSF | − | 0.06 | 4.6 |
| LY96 | rs11466004 | LPS (Y. pestis) | IL-1β | − | 0.06 | 4.5 |
| IL10 | rs3024493 | Peptidoglycan | IL-10 | + | 0.05 | 4.3 |
| NFKBIA | rs8904 | LPS (S. minnesota) | G-CSF | + | 0.05 | 4.0 |
Definition of abbreviations: dbSNP = Single-Nucleotide Polymorphism Database; G-CSF = granulocyte colony-stimulating factor; IL-1RA = interleukin-1-receptor antagonist; MDP = muramyl dipeptide; PAMP = pathogen-associated molecular pattern; pam3CSK4 = N-palmitoyl-S-[2,3-bis(palmitoyl-oxy)-(2RS)-propyl]-[R]-cysteinyl-[S]-seryl-[S]-lysyl-[S]-lysyl-[S]-lysyl-[S]-lysine; S. minnesota = Salmonella minnesota; Y. pestis = Yersinia pestis.
r2 and P values were determined for relationships between genotypes and PAMP-induced mediator phenotypes by linear regression in white Americans. Resulting P values were analyzed with QVALUE (30), limiting the false discovery rate to 0.1. Only associations meeting this threshold are presented.
Effect of the rare allele relative to the common allele is represented as the sign (+ or –) of the regression coefficient (β).
Our approach identified tag SNPs in the genes for TLR4 and TLR5 that had previously been found to be associated with reduced inflammatory responses to LPS (32) and flagellin (33). We found that the T allele at TLR47260T/G (rs7864330) was associated with lower cytokine responses to LPS from Yersinia pestis (Table 2). This SNP shows perfect linkage disequilibrium with TLR4Asp299Gly (rs4986790) (r2 = 1.0; see http://innateimmunity.net/IIPGA2/index_html), an SNP previously reported to confer hyporesponsiveness to LPS (32). We also found a strong association between the stop codon–inducing polymorphism TLR5Arg392Ter (rs5744168) and lower responses to flagellin, consistent with a previous report (33). Finally, we found highly significant associations between the production of IL-1 receptor antagonist (IL-1RA) and IL-10 and genetic variants in the IL1RN and IL-10 genes, respectively (Table 2), also consistent with prior reports of functional variants in these genes (34–36).
Another notable finding was an association between reduced whole blood responses to peptidoglycan, a TLR2 agonist, and a variant in TIR domain–containing adaptor protein (TIRAP) (rs8177352; Table 2). This variant is in high LD with the Ser180Leu variant (see http://gvs.gs.washington.edu/GVS/) that has been shown to be associated with protection from invasive pneumococcal disease, pneumococcal bacteremia, severe malaria, and tuberculosis (37). In addition, we identified several novel associations with genetic variants in TOLLIP, STAT1, CSF3, and NFKBIA and PAMP-induced responses (Table 2). These findings provide evidence that this screening approach can identify true functional polymorphisms in the TLR pathways that are of clinical relevance. However, it is notable that, overall, most of the SNPs identified had only a small effect on the phenotype, and explained only a small portion of the variance in the data. For example, SNPs in TLR4 identified a maximum of 10% of the variance in LPS-induced responses (r2 = 0.1; Table 2) and SNPs in TLR5 explained a maximum of 8% of the variance in flagellin-induced responses (r2 = 0.1; Table 2). SNPs in TIRAP conferred similar effect sizes.
TLR1−7202G Is Highly Associated with Hyperinflammatory Responses to TLR1 Ligands
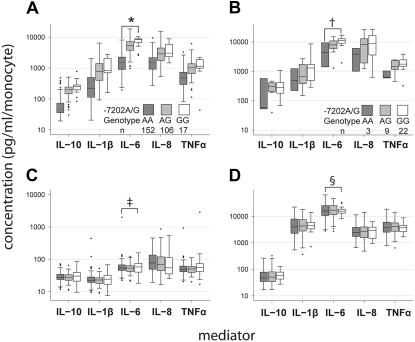
Among the associations we identified, tag SNPs in the TLR1 gene were, by far, the most strongly associated with PAMP-induced responses (Table 2). The G allele of a tag SNP in TLR1 at position −7202 relative to the start codon (TLR1−7202G, rs5743551) demonstrated a hypermorphic effect as it was strongly associated with higher cytokine responses to pam3CSK4 (Figure 2), a synthetic agonist for TLR1/TLR2 heterodimers (38). Notably, in contrast with the other TLR-related alleles described previously, this allele explained a large proportion of the variance in pam3CSK4-induced responses. For example, linear regression of pam3CSK4-induced IL-6 on the copy number of TLR1−7202G indicated that more than 40% of the variance in this response was accounted for by this genotype (r2 = 0.43, P < 1 × 10−20). The magnitude of the difference in IL-6 production associated with TLR1−7202G was large, with more than eight times as much pam3CSK4-induced IL-6 production in homozygotes for TLR1−7202G as in homozygotes for TLR1−7202A (Figure 2). Similar effects were observed for all pam3CSK4-induced cytokine and chemokine responses tested. These effects were specific to pam3CSK4-induced responses and were not seen for baseline IL-6 responses (r2 = 0.02, P > 0.05) or LPS-induced IL-6 responses, which should involve signaling through TLR4 (r2 = 0.02, P > 0.05). Notably, the association seen with the TLR1−7202G allele was also observed in subjects of African descent (n = 34) (Figure 2). This provides strong support for the association between elevated pam3CSK4-induced responses and the SNPs tagged by the TLR1−7202G allele.
Figure 2.
Genetic variation in Toll-like receptor 1 (TLR1) explains a large portion of interindividual variation in whole blood responses to pam3CSK4. Cytokine and chemokine responses to (A) pam3CSK4 (10 ng/ml), (C) medium, and (D) LPS (E. coli, 1 ng/ml) in whole blood from white Americans, and (B) pam3CSK4 (10 ng/ml) in whole blood from African Americans by TLR1−7202A/G genotype. Associations determined by linear regression of cytokine and chemokine production on the copy number of TLR1–7202G. *r2 = 0.43, P < 1 × 10−20; †r2 = 0.15, P = 0.013; ‡P > 0.05; §P > 0.05.
We also identified tag SNPs in TLR6 that had strong associations with responses to pam3CSK4. The TLR1 gene resides on chromosome 4 in close proximity to and in tandem with TLR6 and likely arose from a duplication event (39). Given that pam3CSK4 is a specific ligand for TLR1/TLR2, as opposed to TLR6/TLR2 heterodimers (38), it is likely that this association with rs5743795 in TLR6 reflects the high LD between this SNP and causative SNPs in TLR1.
Coding SNPs in LD with TLR1−7202G and TLR1-mediated Responses
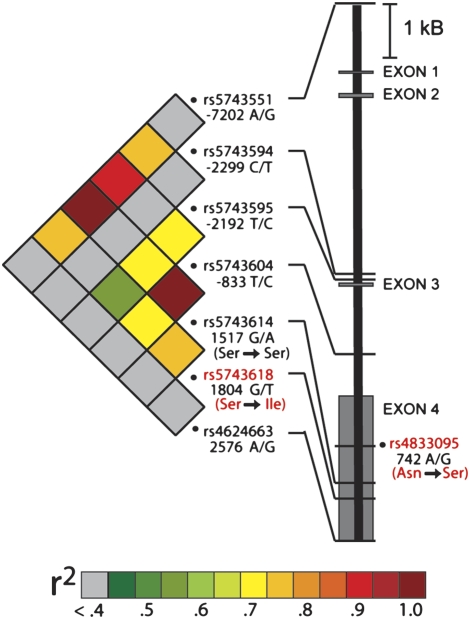
Genotyping information from a reference population revealed that two nonsynonymous SNPs [TLR1742A/G(Asn248Ser), rs4833095; and TLR11804G/T(Ser602Ile), rs5743618] and one synonymous SNP [TLR11517G/A(Ser506Ser), rs5743614] were in LD with TLR1−7202A/G (http://innateimmunity.net/IIPGA2/index_html). TLR1742A/G(Asn248Ser), which falls within the extracellular domain, and TLR11804G/T(Ser602Ile), located within the transmembrane domain of TLR1, are predicted to alter protein function by the PolyPhen program (40). We genotyped TLR11804G/T(Ser602Ile) in the population of normal volunteers and confirmed a high level of LD between this SNP and TLR1−7202A/G (r2 = 0.76) (Figure 3). We were unable to design an accurate genotyping assay for TLR1742A/G(Asn248Ser) because it is located in a region of high sequence homology to the TLR6 gene. Therefore, we used TLR11517G/A(Ser506Ser) as a surrogate for TLR1742A/G(Asn248Ser), given that the two SNPs are proximal. We observed perfect LD (r2 = 1) between TLR11517G/A(Ser506Ser) and TLR1−7202A/G (Figure 3). These data show that both common nonsynonymous SNPs [TLR1742A/G(Asn248Ser) and TLR11804G/T(Ser602Ile)] in TLR1 are in high LD with the tag SNP TLR1−7202A/G.
Figure 3.
Linkage disequilibrium (LD) structure and gene model of TLR1. Genotyped single-nucleotide polymorphisms (SNPs) in TLR1 are presented as Single-Nucleotide Polymorphism Database (dbSNP) reference numbers with corresponding gene position relative to the ATG start codon. The two nonsynonymous coding variants are highlighted in red. The amino acids corresponding to coding nucleotide changes are also shown in red. LD between each of the SNPs in the population of healthy white volunteers was calculated with LD-select (23) and the LD is expressed as the r2 value presented as colors as per the legend.
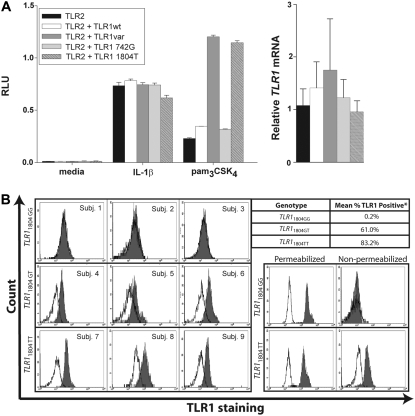
To determine the functional importance of these two nonsynonymous SNPs [TLR1−7202A/G(Asn248Ser) and TLR11804G/T(Ser602Ile)], we cloned the cDNA for TLR1 from lymphocyte cell lines obtained from subjects homozygous for either the wild-type (TLR1−7202A) or variant (TLR1−7202G) allele, and then cotransfected the wild-type or variant TLR1 cDNA constructs into HEK293 cells with an NF-κB–dependent luciferase construct. We found that the variant cDNA promoted greater pam3CSK4-induced activation of NF-κB relative to the wild-type cDNA (Figure 4A). This effect was due to the TLR11804T(602Ile) allele, because reversion of TLR1742G(248Ser) to TLR1742A(Asn248) by site-directed mutagenesis did not alter pam3CSK4-induced responses, whereas reversion of TLR11804T(602Ile) to TLR11804G(Ser602) reduced responses to that observed with the wild-type clone. This effect was not mediated by differences in gene expression, because we observed no significant differences in the levels of TLR1 transcripts induced by all of the TLR1 cDNA constructs (Figure 4A). Furthermore, there was not a general suppression of cell signaling, because all of the transfectants responded similarly to IL-1β (Figure 4A). In primary peripheral monocytes we found that carriers of the TLR11804T(602Ile) allele showed high surface expression of TLR1, whereas the subjects homozygous for TLR11804G(Ser602) had almost no detectable surface expression of TLR1 (Figure 4B). In contrast, permeabilized monocytes showed similar levels of expression between the two groups (Figure 4B). These data provide evidence that coding polymorphisms in high LD with TLR1−7202G contribute to the hypermorphic effect associated with this allele in healthy volunteers and suggest that enhanced cell surface trafficking of TLR1 is one mechanism responsible for this finding.
Figure 4.
Coding polymorphisms tagged by TLR1–7202G confer increased pam3CSK4-induced nuclear factor (NF)-κB activation and are associated with increased cell surface expression of TLR1. (A) Pam3CSK4-induced NF-κB activation in transiently transfected HEK293 cells. cDNA clones for wild-type TLR1 isolated from an individual with the TLR1–7202AA genotype (TLR1wt) or a clone for variant TLR1 isolated from an individual with the TLR1–7202GG genotype (TLR1var) or clones containing only TLR1742G(248Ser) (TLR1742G) or TLR11804T(602Ile) (TLR11804T), generated by site-directed mutagenesis, were cotransfected into HEK293 cells with a human TLR2 cDNA construct, an NF-κB–report construct (ELAM-1–firefly luciferase), and a transfection control (β-actin–Renilla luciferase) followed by incubation with medium alone, IL-1β (10 ng/ml), or pam3CSK4 (100 ng/ml). Each column represents the mean firefly/Renilla luciferase activity for three replicate wells ± the standard error of the mean and is representative of an experiment repeated three times. Right: TLR1 mRNA gene expression in transfected cells by quantitative real-time polymerase chain reaction (PCR). Data are presented as the TLR1/GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression ratio normalized to the mock-transfected control and is the mean of three wells ± the standard error of the mean. (B) Cell surface and intracellular expression of TLR1 by TLR11804G/T. Peripheral blood mononuclear cells from nine volunteers with different TLR11804G/T genotypes were stained with an anti-TLR1 monoclonal antibody (shaded histogram) or an isotype control (open histogram). The proportion of TLR1-positive cells (mean percentage) represents the percentage of cells with fluorescence ≥95% of that seen with the isotype-control antibody. Values for each genotype are shown in the accompanying table. *P < 0.05 using a nonparametric test (Kruskal-Wallis). Monocytes from TLR11804GG and TLR11804TT individuals were also stained for TLR1 with and without permeabilization (bottom right, representative of experiment repeated three times).
TLR1−7202G Is Associated with Higher Risk of Death in Sepsis
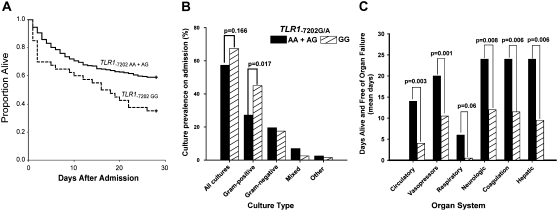
Having shown that individuals carrying TLR1−7202G are predisposed to hypermorphic TLR1-mediated responses, we sought to determine whether this genetic predisposition to high innate immune inflammation was associated with an increased risk for poor outcomes in patients with sepsis or septic shock. We performed a nested case-control study in a cohort of 724 patients admitted to an ICU and who met clinical criteria for sepsis (14) and were prospectively monitored for infection, organ dysfunction, and survival. We also analyzed the subset of patients who were hemodynamically unstable and who met criteria for septic shock (14). Subjects with sepsis enrolled in this cohort had a mean age of 57.3 years (SD, ±16.2 yr), were predominantly male (64.1%), and were admitted primarily for medical (vs. surgical) diagnoses (74.9%). TLR1−7202A/G was successfully genotyped in 711 subjects and the genotype distribution was consistent with Hardy-Weinberg equilibrium (Table 3). We identified an association between the tag SNP TLR1−7202G and death at 28 days in patients with sepsis (Table 3) that was strongest when analyzed assuming a recessive effect of the allele (adjusted odds ratio [OR], 1.82; P = 0.028). This effect was more pronounced in the subset of patients with septic shock (adjusted OR, 2.56; P = 0.008). The recessive effect of TLR1−7202G on mortality contrasts with the codominant effect seen with ex vivo inflammatory responses and may reflect the need for an extreme proinflammatory predisposition before the risk of death is altered in patients. Kaplan-Meier survival curves showed that the difference in mortality observed in TLR1−7202G homozygotes was established early in the hospital stay, consistent with the hypothesis that the hyperfunctioning TLR1 variant contributes to the induction of lethal inflammation during the initial host response to infection (Figure 5A). We also genotyped TLR11804G/T(Ser602Ile) in the sepsis cohort. The genotype frequencies deviated significantly from Hardy-Weinberg equilibrium with an excess of rare homozygotes [TLR11804TT(602Ile/Ile)]. Genotyping assay error is an unlikely cause for this deviation because our assays correctly identified control samples and duplicates. We cannot exclude population admixture as a potential cause for the excess of TLR11804T(602Ile) homozygotes, but given that all other genotypes measured in this population were in Hardy-Weinberg equilibrium this possibility is also unlikely. With this caveat, we found that TLR11804T(602Ile) was associated with higher mortality in patients with sepsis, although this relationship was weaker than that observed with TLR1−7202G and achieved statistical significance only in the subset of patients with septic shock (Table 3). This weaker association for TLR11804T(602Ile) implies that, although this SNP is sufficient to alter TLR1-mediated responses ex vivo, additional functional SNPs in LD with TLR1−7202G are necessary to express an effect on mortality in sepsis and septic shock in vivo. In subsequent analyses of this cohort we focused on TLR1−7202A/G and assumed a recessive effect of the variant on phenotype.
TABLE 3.
TLR1 GENOTYPES AND ASSOCIATIONS WITH 28-DAY MORTALITY IN SEPSIS AND SEPTIC SHOCK
|
−7202A/G
|
1804G/T
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | OR (95% CI) | P Value* | GG | GT | TT | OR (95% CI) | P Value | |
| Healthy volunteers, count (%) | 152 (55.3) | 106 (38.5) | 17 (6.2) | NA | NA | 137 (49.8) | 108 (39.3) | 30 (10.9) | NA | NA |
| HWE† | 0.966 | HWE | 0.468 | |||||||
| Sepsis | ||||||||||
| All (n = 724) | ||||||||||
| Count, % | 374 (52.6) | 273 (38.4) | 64 (9.0) | 333 (48.5) | 253 (36.8) | 101 (14.7) | ||||
| HWE | 0.385 | HWE | 0.0001 | |||||||
| Mortality, % | 35.0 | 33.7 | 50.0 | 1.82 (1.07–3.09) | 0.028 | 34.8 | 34.4 | 39.6 | 1.39 (0.88–2.20) | 0.154 |
| Shock (n = 493) | ||||||||||
| Count, % | 261 (53.8) | 184 (37.9) | 40 (8.2) | 230 (49.4) | 172 (36.9) | 64 (13.7) | ||||
| Mortality | 41.0 | 40.8 | 65.0 | 2.56 (1.28–5.15) | 0.008 | 40.9 | 41.9 | 53.1 | 1.79 (1.02–3.13) | 0.042 |
Definition of abbreviations: CI = confidence interval; HWE = Hardy-Weinberg equilibrium; NA = not applicable; OR = odds ratio.
P values for association with mortality were determined by logistic regression, adjusted for age, sex, medical/surgical admitting diagnosis, and chronic health score.
P value calculated from χ2 value with 1 degree of freedom.
Figure 5.
TLR1–7202G is associated with higher mortality, organ failure, and gram-positive infection in patients with septic shock. (A) Patients with septic shock (n = 493) were genotyped for TLR1−7202A/G and Kaplan-Meier survival curves were generated to compare A/A and A/G individuals (n = 445) (solid line) against G/G (n = 40) (dashed line). (B) Prevalence of bacterial cultures within 24 hours of enrollment in patients with septic shock are presented by TLR1−7202A/G genotype. Prevalence across genotypes was compared by Fisher exact test, assuming a recessive model. (C) The number of days alive and free of organ failure by TLR1−7202A/G genotype in patients with septic shock. Data are expressed as median days alive and free of organ failure over the initial 28 days after enrollment and P values were determined by Mann-Whitney U test.
TLR1−7202G Is Associated with Greater Organ Dysfunction in Sepsis
Multiorgan dysfunction is within the causal pathway for death in sepsis and septic shock. Therefore we examined whether TLR1−7202G might increase the risk of death in patients with sepsis and septic shock through increased organ dysfunction. We found that TLR1−7202G was associated with greater organ dysfunction in patients with septic shock (Figure 5C). Relative to carriers of TLR1−7202A, patients homozygous for TLR1−7202G had less time alive and free of cardiovascular dysfunction (P = 0.003), greater need for vasopressors (P = 0.001), more neurologic dysfunction (P = 0.008), greater dysfunction of coagulation pathways (P = 0.006), worse liver dysfunction (P = 0.006), and a trend toward greater respiratory dysfunction (P = 0.06) (Figure 5C). Notably, when broadened to the patients with sepsis, all measures of organ dysfunction remained significantly greater in homozygotes for TLR1−7202G relative to carriers of TLR1−7202A (data not shown), including a markedly higher degree of respiratory dysfunction (1 vs. 12 d alive and free of mechanical ventilation in TLR1−7202GG vs. TLR1−7202AA and TLR1−7202A/G; P = 0.02). This broad association of TLR1−7202G with organ dysfunction in both sepsis and septic shock supports the assertion that TLR1-mediated inflammatory responses affect sepsis pathophysiology proximally, resulting in multiple downstream effects.
We sought to validate our findings of an association between TLR1−7202G and organ dysfunction in an independent clinical sample. We chose to focus on the development of sepsis-related respiratory dysfunction in the form of ALI, the most common form of organ dysfunction in sepsis that is characterized by diffuse alveolar damage, noncardiogenic edema, and hypoxemia (41). We performed a case-control study using three groups, healthy control subjects, patients with severe sepsis, and patients with severe sepsis ALI, collected by the CELEG as previously described (16). We successfully genotyped TLR1−7202A/G and TLR11804G/T(Ser602Ile) in white patients from CELEG and observed no deviation from Hardy-Weinberg equilibrium (Table 4). The mean ages of subjects with severe sepsis and severe sepsis–associated ALI from this cohort were 61.2 (SD, ±18.1) and 54.2 (SD, ±15.3) years, respectively; and subjects were predominantly male (55.6 and 62.2%, respectively). We found that TLR1−7202A/G was associated with the severity of disease, with increased frequency of the rare homozygote in patients with severe sepsis relative to control subjects, and an even higher frequency in the subjects with severe sepsis-induced ALI (Table 4). Carriers of the rare homozygous GG variant for TLR1−7202A/G had an approximately 3.4-fold increased risk of development of ALI (P = 0.002), and the result remained significant after adjusting for age and sex (P = 0.041; OR, 2.87; 95% confidence interval, 1.01–8.10). We conducted similar analyses for TLR11804G/T(Ser602Ile) in the CELEG subjects, but found no significant association with severe sepsis or severe sepsis-related ALI (Table 4). We did not observe a significant association with 60-day mortality in the CELEG subjects with either SNP, but this smaller sample size had insufficient statistical power to detect a modest effect size.
TABLE 4.
ASSOCIATIONS BETWEEN TLR1 GENOTYPES AND SEPSIS AND SEPSIS-ASSOCIATED ACUTE LUNG INJURY IN CELEG
|
−7202A/G
|
1804G/T
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | OR (95% CI) | P Value* | Padj† | GG | GT | TT | OR (95% CI) | P Value | |
| Control subjects (n = 167) | |||||||||||
| Count, % | 72 (43.9) | 79 (48.2) | 13 (7.9) | NA | NA | NA | 68 (40.7) | 84 (50.3) | 15 (8.9) | NA | NA |
| HWE | 0.17 | HWE | 0.13 | ||||||||
| Sepsis (n = 107) | |||||||||||
| Count, % | 41 (40.6) | 47 (46.5) | 13 (12.9) | 1.76 (0.74–4.15) | 0.199 | NA | 46 (43.0) | 49 (45.8) | 12 (11.2) | 1.18 (0.51–2.76) | 0.69 |
| HWE | 0.93 | HWE | 0.85 | ||||||||
| ALI (n = 138) | |||||||||||
| Count, % | 44 (31.9) | 67 (48.6) | 27 (19.6) | 3.40 (1.59–7.27) | 0.002 | 0.041 | 54 (42.5) | 62 (48.8) | 11 (8.7) | 0.92 (0.39–2.17) | 0.86 |
| HWE | 0.87 | HWE | 0.23 | ||||||||
Definition of abbreviations: CELEG = Consortium to Evaluate Lung Edema Genetics; HWE = Hardy-Weinberg equilibrium; NA = not applicable.
Unadjusted P values for association with susceptibility determined by logistic regression under the general genotype model.
P values adjusted for age and sex.
TLR1−7202G Is Associated with Higher Prevalence of Gram-positive Infection
Analysis of the relationship between TLR1−7202G and the prevalence of specific types of infection revealed other mechanisms through which variation in TLR1 function could alter outcomes in sepsis. Patients with septic shock who were homozygous for TLR1−7202G had a markedly higher prevalence of positive cultures for gram-positive organisms (27% [TLR1−7202A carriers] vs. 45% [TLR1−7202G homozygotes]; p = 0.017) (Figure 5B). In contrast, there was no significant difference in the overall prevalence of bacterial cultures, gram-negative cultures, or mixed and atypical cultures. Of all the gram-positive cultures, 64.0% came from blood, 26.6% came from sputum, 4.3% came from soft tissue, 2.2% came from urine, and 2.9% originating from other sources, and these rates were similar across all genotypes.
We also examined the prevalence of gram-positive cultures in the CELEG subjects and found that, similar to our initial cohort of sepsis patients, patients who were homozygous for TLR1−7202G with severe sepsis in the CELEG cohort were more likely to have had cultures positive for gram-positive organisms than patients homozygous for TLR1−7202A (33.3 vs. 12.5%; P = 0.022; OR, 3.45; 95% confidence interval, 1.19–10.0). Stratification by gram-positive species in the CELEG dataset did not reveal any significant species-specific associations with TLR1−7202A (data not shown) but this sample size is likely to lack power to detect such relationships.
DISCUSSION
Critical illness after severe infection, as manifested by patients with sepsis, is characterized by severe inflammation and can result in multiple organ dysfunction and death. Our findings provide genetic evidence that the innate immune system is an important source of this inflammation. Our data show that a genetic predisposition to hyperinflammatory innate immune responses is associated with a higher risk of death and organ dysfunction after development of sepsis or septic shock. Furthermore, our findings associating common genetic variation in the TLR1 gene with clinical outcomes in sepsis suggests an important role for TLR1-mediated responses in modulating the risk for common bacterial infections.
There exists considerable genetic variation, primarily in the form of SNPs, within the genes involved in TLR-mediated responses. Previous efforts to characterize the effect of these SNPs on TLR-mediated responses have focused on single genes and have not provided information concerning the relative effects of these variants. We have used a broad screen for functional polymorphisms controlling TLR-mediated responses in healthy volunteers and we have identified associations between several tag SNPs and the quantitative trait of PAMP-induced cytokine production in whole blood ex vivo. We identified previously described functional variants in several genes including TLR4, TLR5, and TIRAP. Notably, the variants from all three of these genes explained only a small portion of the variance observed in response to their cognate ligands. In contrast, we identified a strong association between several closely linked SNPs in TLR1 tagged with TLR1−7202G, and markedly increased responses to pam3CSK4, a specific agonist for TLR1/TLR2 heterodimers (38). The genotype for this SNP explained a large portion of the interindividual variability of pam3CSK4-induced IL-6 responses in the healthy volunteers (r2 > 0.4; Figure 2), substantially higher than the portion of variability explained by other SNPs for other phenotypes (Table 2). Cloning and transfection experiments showed that a coding polymorphism linked with TLR1−7202G, TLR11804G/T(Ser602Ile) can confer hyperresponsiveness to pam3CSK4. Furthermore, we found that cell surface expression of TLR1 is much higher on monocytes from individuals carrying the serine allele of TLR11804G/T(Ser602Ile) whereas total expression measured in permeabilized cells was similar between individuals carrying the serine and isoleucine alleles. This suggests that the effect observed with TLR1−7202G is mediated, in part, through altered trafficking of the TLR1 protein.
Since completion of our studies, two other laboratories have reported functional effects associated with the TLR11804G/T(Ser602Ile) polymorphism. Similar to our studies, both Johnson and coworkers and Hawn and coworkers reported that, in primary cells, pam3CSK4-induced cytokine responses are increased in carriers of the isoleucine allele relative to the serine allele of TLR11804G/T(Ser602Ile) (9, 10). Johnson and coworkers also found that carriers of the isoleucine allele of TLR11804G/T(Ser602Ile) demonstrated markedly higher surface expression of TLR1 on peripheral monocytes (9). In cells transiently transfected with cDNA encoding TLR1 they found that the isoleucine allele conferred surface expression of TLR1 whereas the serine allele did not. However, similar to what we observed in monocytes, and consistent with the hypothesis that this SNP alters cell surface trafficking of TLR1, they observed that total TLR1 expression measured in permeabilized transfected cells was similar between the two alleles. In contrast, Hawn and coworkers observed no difference in TLR1 expression or localization by immunofluorescence microscopy in cells transfected cDNA containing either of the TLR11804G/T(Ser602Ile) alleles (10). This discrepancy may be due to differences in the sensitivity of these methods for detecting the magnitude of the difference in cell surface TLR1 expression observed in our report and that of Johnson and coworkers. Importantly, both reports showed that in cells transiently transfected with a cDNA encoding TLR1, the isoleucine allele conferred markedly higher NF-κB activation in response to pam3CSK4 relative to the serine allele. Taken together, these two studies clearly show that TLR11804G/T(Ser602Ile) is a functional variant and that differences in cell surface trafficking of TLR1 contribute to the mechanism accounting for its function.
Our screen identified TLR1−7202G as having the largest hypermorphic effect on innate immune inflammatory responses in human whole blood ex vivo. Thus, we tested the clinical relevance of these TLR1 variants in two separate groups of patients with sepsis; a disease state in which TLR-related pathways are thought to play an important role. In the first nested case–control study, we found that TLR1−7202G was associated with significantly higher mortality and organ dysfunction and higher susceptibility to gram-positive infection in patients with sepsis and septic shock. We verified the association between TLR1−7202A/G and organ dysfunction in a distinct group of control subjects and case subjects with either severe sepsis alone or in the presence of an important and highly prevalent form of sepsis-related organ dysfunction, ALI. We also verified the finding that TLR1−7202G is associated with higher rates of gram-positive infection. Notably, in both clinical study groups we found that the tag SNP TLR1−7202A/G, located in the 5′ untranslated region of the TLR1 gene, showed a stronger association with clinical outcomes than the linked, nonsynonymous SNP, TLR11804G/T(Ser602Ile), which causes an amino acid change in a region of the TLR1 protein predicted to be located in the transmembrane domain (42). Our genotyping data show that these two SNPs are in high (r2 = 0.76) but not perfect LD (Figure 3). These finding suggest that the variant allele at TLR11804G/T(Ser602Ile) requires other variants for expression of the clinical phenotype.
Notably, similar patterns of association with the various TLR1 genotypes have been observed by other investigators. Kesh and coworkers have reported on an association between TLR1743A/G(Asn248Ser) and invasive aspergillosis in patients who had undergone allogeneic stem cell transplantation (43). This SNP was not genotyped in our study but is predicted to be in high LD with TLR11517G/A(Ser506Ser) (http://innateimmunity.net/IIPGA2/PGAs/innateimmunity/TLR1), a SNP that we found to be in perfect LD with TLR1−7202A/G (Figure 2A). Analogous to our report, the authors found no significant association between TLR11804G/T(Ser602Ile) and invasive aspergillosis. Similarly, Ma and coworkers reported an association between susceptibility to tuberculosis and both TLR1743A/G(Asn248Ser) and TLR11804G/T(Ser602Ile) in a case–control study. However, only the association with TLR1743A/G(Asn248Ser) was replicated in a family-based study of tuberculosis cases and their parents (44). TLR11804A/T(Ser602Ile) was found to be associated with susceptibility to leprosy in a small case-control study but no other SNPs in TLR1 were tested (9). Taken together, these reports support the interpretation that, in some disease processes such as sepsis-related organ dysfunction and mortality, TLR11804A/T(Ser602Ile) requires additional variants to significantly elevate susceptibility. Notably, Hawn and coworkers showed that the isoleucine allele of TLR11804A/T(Ser602Ile) conferred higher diacyl lipoprotein-induced NF-κB activation only when the serine allele of TLR1248A/G(Asn248Ser) was also present in the transfected cDNA (10). This finding suggests that the subset of haplotypes containing both TLR11804A/T(Ser602Ile) and TLR1248A/G(Asn248Ser) might be the true “causative” genetic unit. However, it remains possible that other coding or noncoding SNPs fulfill this role. Future studies will need to more comprehensively examine genetic variation within TLR1 to clarify the haplotype responsible for the observed phenotypes. In addition, mouse knock-in models might be used to assess the role of the various haplotypes in TLR1 expression and function in vivo during experimentally induced sepsis.
How might hypermorphic TLR1 variants affect susceptibility to death and organ dysfunction in sepsis? It is possible that TLR1 variants simply amplify the initial innate immune inflammatory response to bacterial products beyond a threshold that is tolerated by the host organ systems. Serial sampling of serum and other relevant biologic fluids in sepsis patients genotyped for TLR1 variants will be needed to address this possibility. Another, more nuanced possibility involves the modulation of regulatory T-cell function. Levels and function of regulatory T cells have been associated with mortality in human sepsis and have been shown to play an important role in regulating the inflammatory and antimicrobial response in animal models of sepsis (45). Studies have shown that TLR2-mediated stimulation of regulatory T cells by bacterial lipoproteins such as pam3CSK4 causes expansion of regulatory T-cell populations but also transiently suppresses the “regulatory” function of these cells (46, 47). We, therefore, speculate that excessive TLR2/1 heterodimer–mediated stimulation of regulatory T cells conferred by TLR1 variants could lead to dysregulation of regulatory T-cell function in patients with sepsis and lead to organ dysfunction and death.
How might hyperfunctional responses to the triacyl lipoprotein pam3CSK4 relate to the higher incidence of gram-positive infection shown in Figure 5B? This finding was somewhat surprising given that biacyl, not triacyl, lipoproteins are believed to predominate in gram-positive cell walls (48, 49). An explanation for this apparent inconsistency is provided by results presented by Johnson and coworkers, who showed that the isoleucine allele of TLR11804G/T(Ser602Ile) confers higher NF-κB induction in transfected cells exposed to heat-killed S. aureus as well as the diacyl lipoprotein PamCysPam (9). Furthermore, we have now found that whole blood cytokine responses to the diacyl lipoprotein FSL-1 are increased in carriers of TLR1−7202G (data not shown). These data show that TLR1 variants are capable of affecting responses to components of gram-positive bacteria. We speculate that TLR1 agonists may predominate in gram-positive relative to gram-negative bacteria and, thus, lead to the differential effect of the TLR1 variants on the risk for gram-positive bacterial infections. Notably, our findings with TLR1 variants echo a report that a genetic variant in TLR-related adapter protein, TIRAP (Ser180Leu), causes higher TLR2-mediated signal transduction and is associated with increased risk of invasive pneumococcal disease and pneumococcal bacteremia (37). It remains to be seen whether this TIRAP allele has effects similar to the TLR1 variants in critically ill populations with sepsis.
In conclusion, through a broad screen of genetic variation in innate immune response genes in normal humans, we have identified a tag SNP (TLR1−7202G) in the TLR1 gene that marks a hypermorphic allele in both white and African Americans and have shown a strong association between this tag SNP and increased organ dysfunction and death in patients with sepsis. This effect is likely to be mediated, in part, by increased cell surface expression of TLR1 in carriers of the risk allele but additional effects, such as alterations in gene regulation in vivo, need to be studied. These data support the conclusion that TLR-mediated inflammatory responses play a clinically important role in host defense against bacterial infection in humans. Our studies provide the first association between variants in TLR1 and bacterial sepsis, and, more generally, one of the clearest associations between a genetic predisposition to elevated innate immune responses and poorer clinical outcomes in sepsis. Notably, this article used prospectively collected data from two different patient groups totaling more than 950 critically ill patients and, thus, should provide good estimates of effect size. Additional larger, multicenter studies will be needed to further establish the specific subgroups of patients, such as those with only gram-positive infection, for which the TLR1 variant has maximal predictive power and to determine the mechanisms underlying the effect of these variants. We also anticipate that pharmacologic inhibition of TLR2/1 activation in the setting of sepsis might represent a novel addition to the currently spare arsenal of specific therapeutic approaches.
Supplementary Material
Supported by HL629063 (M.M.W.), HL073996 and AI057141 (T.R.M.), HL73994 (R.B.), the Mary Beryl Patch Turnbull Scholar Program (K.C.B), and HL074366 (G.P.J).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200803-462OC on July 17, 2008
Conflict of Interest Statement: M.M.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.C.G. works as a consultant to Sirius Genomics, Inc., and owns $50,000 in stock options. T.D.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. F.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. O.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.T.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.A.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.P.J. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.M.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.A.N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.P.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.G.N.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.C.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.R.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.A.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. T.R.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 2003;348:1546–1554. [DOI] [PubMed] [Google Scholar]
- 2.Murphy SL. Deaths: final data for 1998. Natl Vital Stat Rep 2000;48:1–105. [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med 2003;348:138–150. [DOI] [PubMed] [Google Scholar]
- 4.Arcaroli J, Fessler MB, Abraham E. Genetic polymorphisms and sepsis. Shock 2005;24:300–312. [DOI] [PubMed] [Google Scholar]
- 5.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001;2:675–680. [DOI] [PubMed] [Google Scholar]
- 6.Beutler B, Hoebe K, Du X, Ulevitch RJ. How we detect microbes and respond to them: the Toll-like receptors and their transducers. J Leukoc Biol 2003;74:479–485. [DOI] [PubMed] [Google Scholar]
- 7.Taveira da Silva AM, Kaulbach HC, Chuidian FS, Lambert DR, Suffredini AF, Danner RL. Brief report: shock and multiple-organ dysfunction after self-administration of salmonella endotoxin. N Engl J Med 1993;328:1457–1460. [DOI] [PubMed] [Google Scholar]
- 8.Wurfel MM, Park WY, Radella F, Ruzinski J, Sandstrom A, Strout J, Bumgarner RE, Martin TR. Identification of high and low responders to lipopolysaccharide in normal subjects: an unbiased approach to identify modulators of innate immunity. J Immunol 2005;175:2570–2578. [DOI] [PubMed] [Google Scholar]
- 9.Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, Alpsoy E, Hamann L, Schumann RR, Tapping RI. Cutting edge: A common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol 2007;178:7520–7524. [DOI] [PubMed] [Google Scholar]
- 10.Hawn TR, Misch EA, Dunstan SJ, Thwaites GE, Lan NT, Quy HT, Chau TT, Rodrigues S, Nachman A, Janer M, et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur J Immunol 2007;37:2280–2289. [DOI] [PubMed] [Google Scholar]
- 11.Wurfel MM, Gordon AC, Holden TD, Radella F, Stratton S, Rona G, Strout J, Black RA, Nickerson DA, Walley KR, et al. Variation in the tlr1 gene affects TLR1 surface expression and is associated with increased mortality in septic shock [abstract]. Am J Respir Crit Care Med 2007;175:A898. [Google Scholar]
- 12.Wattanathum A, Manocha S, Groshaus H, Russell JA, Walley KR. Interleukin-10 haplotype associated with increased mortality in critically ill patients with sepsis from pneumonia but not in patients with extrapulmonary sepsis. Chest 2005;128:1690–1698. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland AM, Walley KR, Russell JA. Polymorphisms in CD14, mannose-binding lectin, and Toll-like receptor-2 are associated with increased prevalence of infection in critically ill adults. Crit Care Med 2005;33:638–644. [DOI] [PubMed] [Google Scholar]
- 14.American College of Chest Physicians; Society of Critical Care Medicine. American College of Chest Physicians/Society of Critical Care Medicine consensus conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med 1992;20:864–874. [PubMed] [Google Scholar]
- 15.Bernard GR, Wheeler AP, Arons MM, Morris PE, Paz HL, Russell JA, Wright PE; Antioxidant in ARDS Study Group. A trial of antioxidants N-acetylcysteine and procysteine in ARDS. Chest 1997;112:164–172. [DOI] [PubMed] [Google Scholar]
- 16.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol 2006;34:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. ACCP/SCCM Consensus Conference Committee; American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101:1644–1655. [DOI] [PubMed] [Google Scholar]
- 18.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American–European Consensus Conference on ARDS: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818–824. [DOI] [PubMed] [Google Scholar]
- 19.Lee JS, Wurfel MM, Matute-Bello G, Frevert CW, Rosengart MR, Ranganathan M, Wong VW, Holden T, Sutlief S, Richmond A, et al. The duffy antigen modifies systemic and local tissue chemokine responses following lipopolysaccharide stimulation. J Immunol 2006;177:8086–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol 2006;6:9–20. [DOI] [PubMed] [Google Scholar]
- 21.Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP Jr. Multiplex bead array assays for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers. Cytometry B Clin Cytom 2004;61:35–39. [DOI] [PubMed] [Google Scholar]
- 22.Prabhakar U, Eirikis E, Davis HM. Simultaneous quantification of proinflammatory cytokines in human plasma using the LabMAP assay. J Immunol Methods 2002;260:207–218. [DOI] [PubMed] [Google Scholar]
- 23.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 2004;74:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Carlson CS, Rieder MJ, Nickerson DA. Efficient selection of tagging single-nucleotide polymorphisms in multiple populations. Hum Genet 2006;120:58–68. [DOI] [PubMed] [Google Scholar]
- 25.Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, et al. Decoding randomly ordered DNA arrays. Genome Res 2004;14:870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 1999;14:143–149. [DOI] [PubMed] [Google Scholar]
- 27.Hajjar AM, O'Mahony DS, Ozinsky A, Underhill DM, Aderem A, Klebanoff SJ, Wilson CB. Cutting edge: functional interactions between Toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J Immunol 2001;166:15–19. [DOI] [PubMed] [Google Scholar]
- 28.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid a species that functionally interact with both Toll-like receptors 2 and 4. Infect Immun 2004;72:5041–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992;48:361–372. [PubMed] [Google Scholar]
- 30.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003;100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet 2001;2:91–99. [DOI] [PubMed] [Google Scholar]
- 32.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet 2000;25:187–191. [DOI] [PubMed] [Google Scholar]
- 33.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, Skerrett SJ, Beutler B, Schroeder L, Nachman A, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to Legionnaires' disease. J Exp Med 2003;198:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA 1998;95:9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schippers EF, van't Veer C, van Voorden S, Martina CA, Huizinga TW, le Cessie S, van Dissel JT. IL-10 and Toll-like receptor-4 polymorphisms and the in vivo and ex vivo response to endotoxin. Cytokine 2005;29:215–228. [DOI] [PubMed] [Google Scholar]
- 36.Rafiq S, Stevens K, Hurst AJ, Murray A, Henley W, Weedon MN, Bandinelli S, Corsi AM, Guralnik JM, Ferruci L, et al. Common genetic variation in the gene encoding interleukin-1-receptor antagonist (IL-1RA) is associated with altered circulating IL-1RA levels. Genes Immun 2007;8:344–351. [DOI] [PubMed] [Google Scholar]
- 37.Khor CC, Chapman SJ, Vannberg FO, Dunne A, Murphy C, Ling EY, Frodsham AJ, Walley AJ, Kyrieleis O, Khan A, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet 2007;39:523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol 2002;169:10–14. [DOI] [PubMed] [Google Scholar]
- 39.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol 2003;21:335–376. [DOI] [PubMed] [Google Scholar]
- 40.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002;30:3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 42.Seabury CM, Cargill EJ, Womack JE. Sequence variability and protein domain architectures for bovine Toll-like receptors 1, 5, and 10. Genomics 2007;90:502–515. [DOI] [PubMed] [Google Scholar]
- 43.Kesh S, Mensah NY, Peterlongo P, Jaffe D, Hsu K, van den Brink M, O'Reilly R, Pamer E, Satagopan J, Papanicolaou GA. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. Ann N Y Acad Sci 2005;1062:95–103. [DOI] [PubMed] [Google Scholar]
- 44.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals Toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS ONE 2007;2:e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venet F, Chung CS, Monneret G, Huang X, Horner B, Garber M, Ayala A. Regulatory T cell populations in sepsis and trauma. J Leukoc Biol 2008;83:523–535. [DOI] [PubMed] [Google Scholar]
- 46.Sutmuller RP, den Brok MH, Kramer M, Bennink EJ, Toonen LW, Kullberg BJ, Joosten LA, Akira S, Netea MG, Adema GJ. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest 2006;116:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Komai-Koma M, Xu D, Liew FY. Toll-like receptor 2 signaling modulates the functions of CD4+ CD25+ regulatory T cells. Proc Natl Acad Sci USA 2006;103:7048–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henneke P, Dramsi S, Mancuso G, Chraibi K, Pellegrini E, Theilacker C, Hubner J, Santos-Sierra S, Teti G, Golenbock DT, et al. Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J Immunol 2008;180:6149–6158. [DOI] [PubMed] [Google Scholar]
- 49.Stoll H, Dengjel J, Nerz C, Gotz F. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun 2005;73:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.