Abstract
After activation, CD4+ helper T (TH) cells differentiate into distinct effector lineages. Although CXCR5+ follicular TH (TFH) cells are important in humoral immunity, their developmental regulation is unclear. Here we show that TFH cells have a distinct gene expression profile from other effector T cells and develop in vivo independent of the TH1 or TH2 lineages. TFH cell generation is regulated by B7h expressed on B cells and, similar to TH17 cell development, is dependent on IL-21, IL-6 and STAT3. However, differentiation of TFH cells, unlike TH17 cells, does not require TGFβ signaling or TH17-specific orphan nuclear receptors RORα and RORγ in vivo. Finally, naïve T cells activated in vitro in the presence of IL-21 but not TGFβ signaling preferentially acquire TFH gene expression and function to promote germinal center reactions in vivo. This study thus demonstrates TFH as a distinct lineage of effector TH differentiation.
Introduction
Naïve CD4+ helper T (TH) cells, upon encountering their cognate antigens presented on professional antigen-presenting cells (APC), differentiate into effector cells that are characterized by their distinct cytokine production profiles and immune regulatory functions (Dong and Flavell, 2000; Glimcher and Murphy, 2000). TH1 cells produce IFNγ and regulate antigen presentation and immunity against intracellular pathogens, whereas TH2 cells produce IL-4, IL-5 and IL-13, and mediate humoral responses and immunity against parasites. IFNγ and IL-4 are not only the key effector cytokines but also mediate the differentiation of TH1 and TH2 cells, respectively.
Recently, TH17, a third subset of TH cells, has been identified, which produce IL-17, IL-17F and IL-22 and regulate inflammatory responses by tissue cells (Bettelli et al., 2007; Dong, 2006; Reiner, 2007; Weaver et al., 2006). TH17 differentiation, at least in mouse, is initiated by TGFβ and IL-6 (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006a), possibly via regulating the chromatin remodeling of the Il17-Il17f locus (Akimzhanov et al., 2007). While IL-6 is necessary for TH17 differentiation (Korn et al., 2007; Yang et al., 2007), IL-21 was recently reported as an autocrine factor induced by IL-6 to regulate TH17 differentiation (Korn et al., 2007; Nurieva et al., 2007a; Zhou et al., 2007). On the other hand, TGFβ signaling has also been clearly demonstrated to mediate TH17 differentiation in vivo (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006b), and activated T cells may serve as an important sources of TGFβ for this regulation (Li et al., 2007). TH17 development is dependent on STAT3 (Laurence et al., 2007; Yang et al., 2007), which functions to upregulate the expression of two TH17-specific orphan nuclear receptors RORγt and RORα that ultimately determines TH17 terminal differentiation (Ivanov et al., 2006; Yang et al.).
A fundamental function of TH cells is to provide “help” to B cells and regulate their proliferation and immunoglobulin class-switching, especially in the germinal center structures. TH1 and TH2 cells have been shown to regulate B cell responses to some extents. For example, IFNγ regulates IgG2a production while IL-4 is critical in IgE class-switching. However, an additional TH subset called follicular helper T (TFH) cells are recently found to be present in germinal centers and are characterized by their expression of CXCR5 (Vinuesa et al., 2005b). Although activated T cells may transiently express CXCR5, TFH cells appear to have more stable expression of this chemokine receptor. These cells are thought to regulate humoral immunity, especially germinal center reactions. Consistent with this notion, CXCR5 has been shown to be important for proper T and B cell localization in immune responses and antibody production (Haynes et al., 2007; Junt et al., 2005). In addition to CXCR5, other markers have been also reported for TFH cells, such as ICOS costimulatory receptor, IL-21 cytokine and Bcl-6 transcription factor. ICOS was found essential for generation of TFH cells in vivo (Akiba et al., 2005). In addition to TH17 cells, IL-21 is also expressed in TFH cells and may serve as an important regulator of humoral responses by TFH cells. IL-21 directly regulates B cell proliferation and class-switching; IL-21R deficiency results in defective antibody responses and impaired germinal center formation (Spolski and Leonard, 2008). On the other hand, sanroque mice, which have a mutation in a RING-type E3 ubiquitin ligase, Roquin, developed spontaneous autoantibody production and lupus-like autoimmunity, associated with greatly increased numbers of CXCR5+ CD4 T cells and enhanced expression of IL-21 and ICOS (Vinuesa et al., 2005a).
Despite their potential importance in humoral immunity and immunopathology, the developmental regulation of TFH cells and their relationship with other TH subsets is unclear, particularly considering the fact that IL-21 is expressed by both TH17 and TFH cells. A previous study revealed that human TFH cells express distinct genes from TH1 or TH2 cells (Chtanova et al., 2004). Here we show that mouse TFH cells have a divergent gene expression profile from TH1, TH2 and TH17 cells, and develop in vivo independent of these lineages. IL-21, IL-6 and STAT3 are critical in the generation of TFH cells. Moreover, T cells activated in vitro in the presence of IL-21 but without TGFβ signaling preferentially acquire TFH gene expression and function to promote humoral immunity in vivo. Thus, our data indicate that TFH cells represent a distinct TH lineage and suggest a reciprocal relationship between the TFH and TH17 lineages.
Results
Distinct gene expression profile of TFH cells
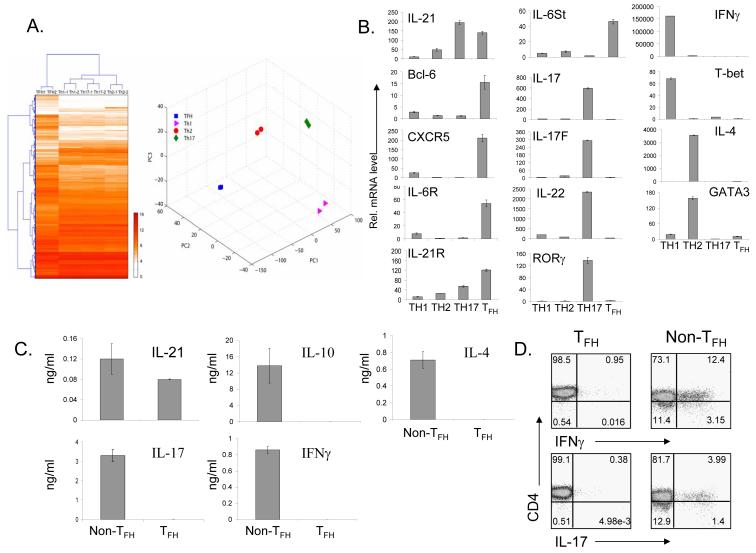
As a first step toward understanding TFH cell regulation, we compared the gene expression profiles of TH1, TH2 and TH17 cells differentiated in vitro with in-vivo generated TFH cells. Taking advantage of the co-expression of BTLA by CXCR5+ TFH cells (Supplementary Figure 1), CD4+CD44hiCXCR5+BTLA+ were FACS sorted from splenocytes of C57BL/6 mice 7 days after immunization with KLH. These cells as well as TH1, TH2 and TH17 cells were restimulated with anti-CD3 for 4 hours and subject to gene profiling analysis in duplicates using Affimetrix gene chips. The microarray data were normalized using GCRMA and the genes whose expression levels were changed across the TH1, TH2, TH17 and TFH cells were then selected using a False Discovery Rate (FDR) estimation method. Then, the expression levels of 8350 probe-sets showing differential expressions among the four types of cells were used for hierarchical clustering, which revealed that TFH cells have a very distinct gene expression profile (Figure 1A).
Figure1. TFH cells express distinct genes from TH1, TH2 and TH17 cells.
C57BL/6 mice were immunized with KLH in CFA. 7 days later CD4+CD44hiCXCR5+ BTLA+ (TFH) cells were sorted and restimulated with anti-CD3 for 4 hours for gene profiling analysis (A) and for real-time PCR analysis (B). OT-II T cells differentiated under various conditions were restimulated with anti-CD3 for 4 hours and were analyzed together with TFH cells. (A). A distinct gene expression profile of TFH cells from TH1, TH2, and TH17 cells. Hierarchical clustering and PCA were applied to the expression levels of 8350 probe-sets showing differential expressions among the four types of cells (FDR = 0.1, corresponding to an unadjusted p-value = 0.033 from one-way ANOVA). (B). Real-time RT-PCR analysis of TH specific genes. (C-D). CD4+CD44hiCXCR5+ BTLA+ and CD4+CD44hiCXCR5- cells were restimulated with anti-CD3/anti-CD28 for 48 hours for cytokine measurement by ELISA (C) and with PMA/Ion for 6 hours for detection of IL-17 and IFNγ expression by intracellular cytokine staining (D). The data represent one of two independent experiments with similar results.
To confirm the above results, we performed real-time RT-PCR analysis on multiple subset-specific genes. The data indicate that TFH cells did not express the typical markers for TH1 (IFNγ and T-bet) or TH2 (IL-4 and GATA3) cells (Figure 1B), consistent with a previous report on human TFH cells (Chtanova et al., 2004). Although TFH cells shared IL-21 expression with TH17 cells, they did not express IL-17, IL-17F, IL-22 or RORγt (Figure 1B). Instead, similar to their human counterparts, (Chtanova et al., 2004; Kim et al., 2004), mouse TFH cells express mRNAs for CXCR5 as well as Bcl-6 (Figure 1B). In addition, TFH cells preferentially expressed mRNAs for IL-6R and IL-6st (gp130) and also upregulated the expression of IL-21R (Figure 1B), suggesting possible regulation of TFH cells by IL-6 and IL-21. Moreover, consistent with human TFH cells (Chtanova et al., 2004) and a recent report indicating that TFH cells express PD-1 protein (Haynes et al., 2007), PD-1 mRNA was highly upregulated in TFH cells compare to other TH subsets (data not shown).
To substantiate the above results, we also measured cytokine secretion of purified TFH cells after they were activated ex vivo with anti-CD3 and anti-CD28 for 24 hours. High levels of IL-21 but not IL-4, IL-10, IFNγ or IL-17 were observed in TFH cells (Figure 1C). In addition, purified TFH cells were activated with KLH and irradiated splenic APC. Consistent with above results, TFH cells preferentially produced IL-21, but not TH1, TH2 and TH17 cytokines (Supplementary Figure 3). Furthermore, intracellular analysis on CXCR5+ and CXCR5- cells following PMA and ionomycin restimulation also revealed that TFH cells did not express IFNγ or IL-17 (Figure 1D). These results indicate that TFH cells are distinct from TH1, TH2 and TH17 cells in their gene expression and cytokine production.
Naïve T cell differentiation into TFH cells is independent of TH1 and TH2 lineages
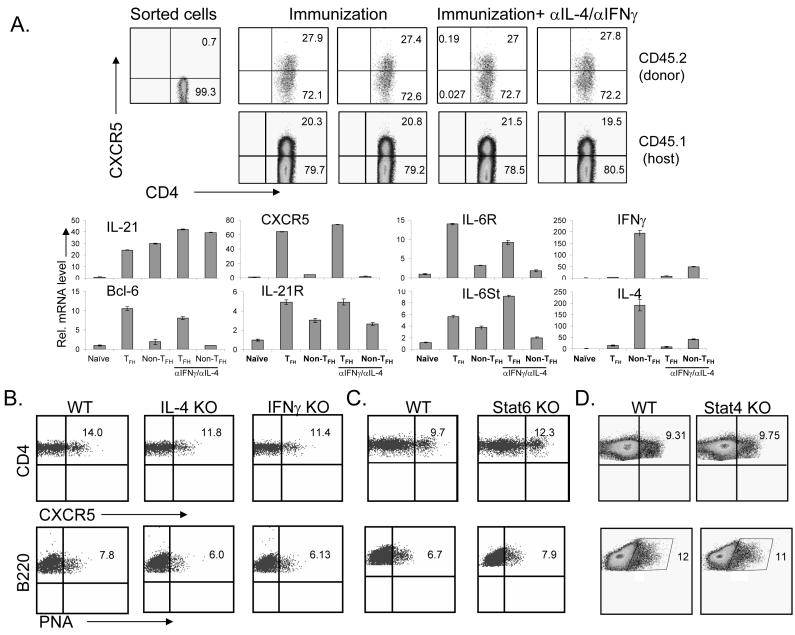
Previous (Chtanova et al., 2004) and our current gene expression analysis on human and mouse TFH cells, respectively, suggested that they are distinct from TH1 and TH2 cells. However, it was not clear if generation of TFH cells is distinct from TH1 or TH2 differentiation. To address this question, we transferred FACS sorted naïve CD4+ T cells (CXCR5-) from OT-II TcR transgenic mice (CD45.2+, Figure 2A) into C57BL/6 recipients (CD45.1+) followed by immunization with Ova protein in CFA. 7 days later, we examined splenic CD45.1+ and CD45.2+ CD4 cells, and found a substantial increase in the frequency of CXCR5+CD4+ cells (Figure 2A). Interestingly, treatment of recipient mice with antibodies to IL-4 and IFNγ did not reduce the proportion of CXCR5+CD4+ cells (Figure 2A), suggesting that TFH cell development is independent of TH1 or TH2 development. In addition, donor CD44hiCXCR5+ and CD44hiCXCR5- OT-II cells from immunized mice were purified and real-time RT-PCR analysis of TFH specific genes were performed. CXCR5+ T cells were generated in the presence of blocking antibodies to IL-4 and IFNγ exhibited gene expression patterns comparable to those from control group (Figure 2A), supporting that these CXCR5+ cells were indeed TFH cells.
Figure 2. Generation of TFH cells is independent of TH1 and TH2 lineages.
(A). CD4+ T cells from CD45.2/OT-II mice were transferred into C57BL/6 (CD45.1) mice which were subsequently divided into 2 groups (3 mice per group). Mice were immunized subcutaneously with Ova protein emulsified in CFA and treated with a 300 μg of control rat Ig or anti-IFNγand anti-IL-4 mAbs. Seven days after the immunization, experimental mice were sacrificed and splenic CD45.1+ and CD45.2+ CD4 cells were stained with biotinylated anti-CXCR5 mAb, followed by APC-labeled streptavidin. Numbers in dot plot quadrants represent the percentages. CD44hiCXCR5+ and CD44hiCXCR5- cells from immunized mice were purified and real-time RT-PCR analysis of TFH specific genes were performed. (B-C). IL-4 KO, IFNγ KO (B), STAT6 KO (C) and STAT4 KO (D) and their appropriate controls (WT, 3 mice per group) were immunized with KLH emulsified in CFA. Seven days after the immunization, experimental mice were sacrificed and the germinal center B cells were determined by staining with FITC-labeled PNA and PerCP-labeled anti-B220 mAb. The TFH cells were analyzed by staining with PerCP-labeled anti-CD4 mAb and biotinylated anti-CXCR5 mAb, followed by APC-labeled streptavidin. Numbers in dot plot quadrants represent the percentages. The experiments were repeated three times with consistent results.
To substantiate the above finding, we also immunized mice deficient in IL-4, IFNγ STAT6 or STAT4 and their appropriate controls with KLH. 7 days after immunization, we did not detect any defect in TFH cells or PNA+ germinal center B cells in the knockout animals compared to wild-type controls (Figure 2B-D, Supplementary Figure 4A-C). To confirm the above results, we also measured cytokine secretion of purified CXCR5+ cells from IL-4- and IFNγ-deficient mice and their controls after they were activated ex vivo with anti-CD3 and anti-CD28 for 24 hours. TFH cells IL-4- or IFNγ-deficient mice produced similar level of IL-21, but did not express IL-4, IL-10, IFNγ or IL-17 (Supplementary Figure 4D), supporting that they were TFH cells. Thus, we conclude that CXCR5+CD4+ TFH cells develop independent of the TH1 and TH2 lineages.
B7h expressed on B cells regulates the generation of TFH cells
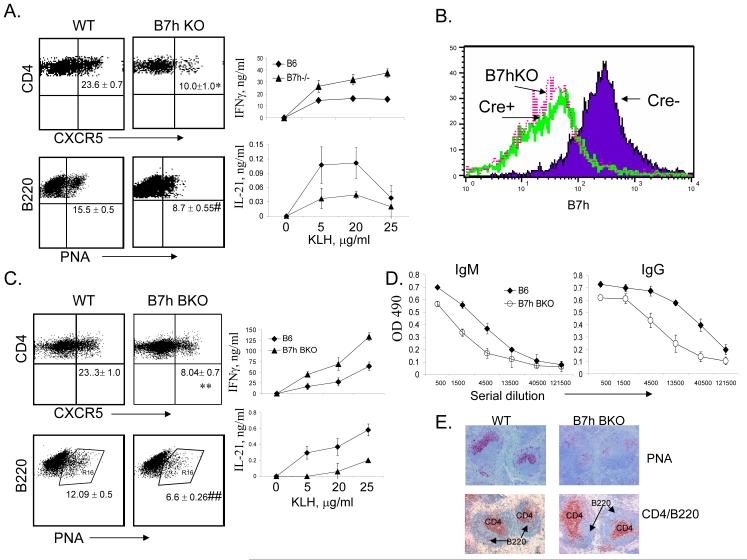
Inducible costimulator (ICOS) is the third member of the CD28 family with an important role in regulation of T-dependant Ab responses and germinal center reactions (Dong and Nurieva, 2003). ICOS was previously shown to be expressed at high levels on human tonsillular CXCR5+ T cells within the light zone of germinal centers and efficiently supported the immunoglobulin production (Breitfeld et al., 2000; Schaerli et al., 2000). In addition, ICOS deficiency in human and mouse resulted in significantly reduced numbers of TFH cell, indicating an essential role of ICOS in the differentiation of CXCR5+ CD4 T cells (Akiba et al., 2005; Bossaller et al., 2006b). Consistently, we also observed reduced percentages of CXCR5+CD4+ cell and decreased expression of IL-21 but not IFNγ in our B7h germline knockout animals (Nurieva et al., 2003b) following KLH immunization (Figure 3A). TFH cells are regarded as regulators of the germinal center reaction by providing help to activated B cells that also express CXCR5. Since B cells constitutively express B7h, we asked whether the generation of TFH cells may require B cell help via engagement of ICOS receptor on T cells. We thus bred mice carrying the B7h conditional flox (f) allele (Nurieva et al., 2003b) with CD19-cre mice (Rickert et al., 1995). In B7h f/f mice carrying the CD19-cre (B7h BKO), in comparison to those without cre expression, there was efficient deletion of the B7h gene in B cells, similar to those in B7h germline knockout mice (Figure 3B). We then immunized B7h BKO mice with KLH and found that absence of B7h in B cells led to a greatly reduced frequency of CXCR5+CD4+ cells (Figure 3C). Similarly, when CD45.1+ OT-II cells were transferred into B7h BKO mice, the numbers of CD45.1+ TFH cells were also greatly reduced following Ova peptide immunization (data not shown). In addition to CXCR5 expression, we found that the expression of IL-21 was also greatly reduced in B7h BKO mice, whereas IFNγ expression was elevated in these mice (Figure 3C). Moreover, PNA+ germinal center B cells were greatly reduced in these animals (Figure 3C and E). KLH-specific IgG production was also reduced (Figure 3D). Overall, these data indicate that B cell expression of B7h is necessary for IL-21 production and for the generation of TFH cells and appropriate antibody responses, indicating that TFH cell differentiation is regulated by B cells.
Figure 3. B7H expressed on B cells is required for generation of TFH cells.
(A) B7h germline knockout mice and their controls (WT, 3 mice per group) were immunized with KLH in CFA. Seven days after the immunization, experimental mice were sacrificed and spleen cells from immunized mice were stimulated in 96-well plates as triplicates with the indicated concentration of KLH. Effector cytokines (IFN-γ and IL-21) were measured after 4 days of treatment. The germinal center B cells and TFH cells were analyzed. P values were calculated with the t-test by comparing the CXCR5+ cells and B220/PNA+ cells between wild-type and B7h KO mice and are indicated as followed: * P < 0.005; #, P < 0.001. Numbers in dot plot quadrants represent the percentages. (B). Splenic B220+ B cells from B7h germline KO (B7hKO), B7h B-cell conditional knockout (B7h BKO, cre+) and the cre- controls were analyzed for B7h expression. (C-E). Wild-type (WT) and B7h BKO (3 mice per group) were immunized with KLH in CFA. Seven days after the immunization, experimental mice were sacrificed and analyzed as in A. P values were calculated with the t-test by comparing the CXCR5+ cells and B220/PNA+ cells between wild-type and B7h BKO mice and are indicated as followed: ** P < 0.001; ##, P < 0.001. (C). (D). Anti-KLH antibodies (IgM and IgG) were measured in the sera by ELISA. The sera from WT and B7h BKO mice were subject to a 3-fold serial dilution, and the concentrations of KLH-specific IgM and IgG were analyzed by ELISA and averaged for each group. (E). GC in the spleens of KLH-immunized WT and B7h BKO mice were identified by PNA staining (brown). T and B cells were identified by staining with anti-CD4 (red) and anti-B220 (blue) Abs. The data represent at least three independent experiments with consistent results.
IL-21 and IL-6 are required for generation of TFH cells, which is dependent on STAT3
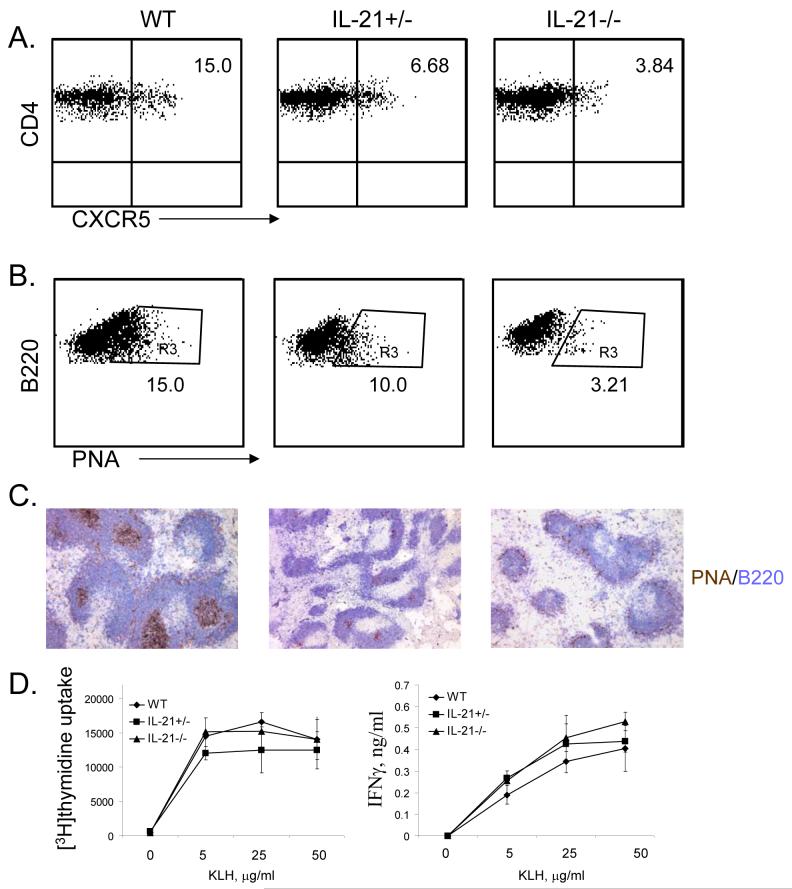
IL-21 has been recently shown to be induced by IL-6 and to autoregulate its own expression during TH17 differentiation (Nurieva et al., 2007a). In addition, TFH T cells produced significantly greatrt amount of IL-21 compared to TH1 and TH2 subsets (Chtanova et al., 2004), and induced the differentiation of autologous B cells into Ig-secreted plasma cells through IL-21 (Bryant et al., 2007). Since IL-21 is also expressed in TFH cells, we assessed if IL-21 is important for TFH cell generation. IL-21+/+, +/- and -/- mice were immunized with KLH and splenic TFH cells were analyzed in these mice. IL-21+/- mice exhibited reduced number of TFH cells, which was further reduced in IL-21-/- mice (Figure 4A). In addition, PNA+ germinal center B cells were also greatly reduced in IL-21-/- mice (Figure 4B-C). In contrast, CD4 T cells from IL-21+/- and -/- mice showed normal proliferation and IFNγ expression after re-stimulation with KLH ex vivo. (Figure 5D). Thus, these results indicate that IL-21 is necessary for TFH cell development.
Figure 4. IL-21 is necessary for TFH cell development.
IL-21+/+, IL-21+/- and IL-21-/- mice (3 mice per group) were immunized subcutaneously with KLH emulsified in CFA. Seven days after the immunization, experimental mice were sacrificed and TFH cells (A) and the germinal center B cells (B) were analyzed. Numbers in dot plot quadrants represent the percentages. Germinal centers were determined by immunohistochemical analysis (C). Spleen cells from immunized mice were stimulated in 96-well plates as triplicates with the indicated concentration of KLH peptide. Proliferation was assayed after 3 days of treatment by adding [3H]thymidine to the culture for the last 8 h. IFN-γ was measured after 4 days of treatment. The experiments were repeated twice with consistent results.
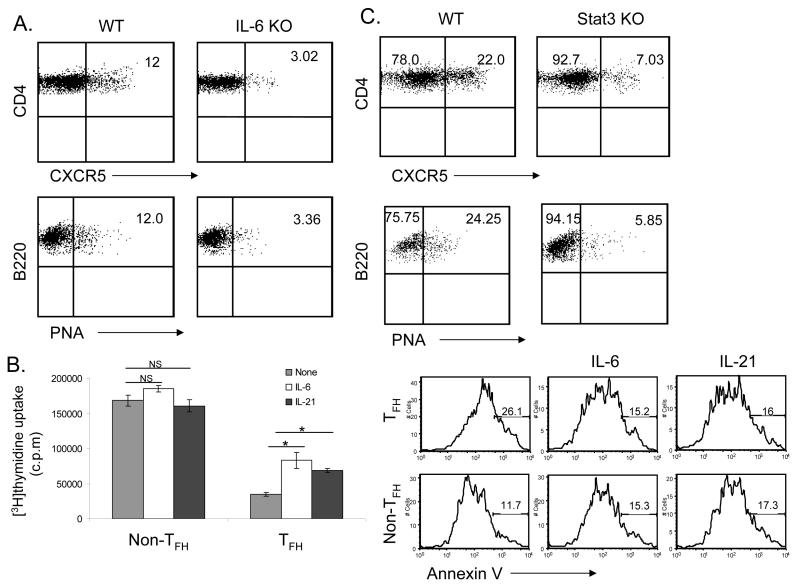
Figure 5. Generation of TFH cells requires IL-6 and STAT3.
IL-6 KO (A) or STAT3 T-cell specific KO mice (C) and their appropriate controls (WT, 3 mice per group) were immunized subcutaneously with KLH emulsified in CFA. Seven days after the immunization, experimental mice were sacrificed and TFH cells and the germinal center B cells were analyzed. Numbers in dot plot quadrants represent the percentages. (B) C57BL/6 mice were immunized with KLH in CFA. 7 days later, CD4+CD44hiCXCR5+ and CD4+CD44hiCXCR5- cells were sorted and restimulated with anti-CD3 and anti-CD28 with or without IL-6 or IL-21 for 48 hours. Proliferation was assayed by adding [3H]-thymidine to the culture for the last 8 h. P values were calculated with the t-test by comparing the CXCR5+ cells proliferation in the absence and in the presence of IL6 (*p<0.005) or in the absence and in the presence of IL21 (*p<0.005). CD4+CD44hiCXCR5+ and CD4+CD44hiCXCR5- cells were restimulated with anti-CD3 and anti-CD28 for 24 hours and stained with Annexin V-FITC. The experiments were performed two times with consistent results.
Since IL-6 induces IL-21 expression, we also tested TFH cell generation in mice lacking IL-6. It has been previously that IL-6 deficient mice showed reduced germinal centers and antigen specific Ig-production (IgG) (Kopf et al., 1998). Compared to the wild-type controls, IL-6-/- mice exhibited greatly reduced numbers of TFH cells and germinal center B cells (Figure 5A), indicating that IL-6 is also necessary for TFH cell generation.
Since TFH cells expressed high levels of IL-21R, IL-6R and IL-6st (Figure 1B), we also examined if IL-6 and IL-21 signaling regulates TFH cells. C57BL/6 mice were immunized with KLH and CD4+CD44hi T cells with or without CXCR5 expression were FACS sorted and restimulated with anti-CD3 and anti-CD28 in the absence or presence of IL-21 or IL-6. Compared to non-TFH cells, CXCR5+ T cells showed a significant reduction in proliferation when activated with anti-CD3 and anti-CD28. Treatment of CXCR5+ T cells with IL-6 or IL-21 significantly enhanced their proliferation (Figure 5B), suggesting that IL-6 and IL-21 preferentially regulate TFH cells. Furthermore, we stained restimulated CXCR5+ and CXCR5- cells with Annexin V. We found that a significant portion of CXCR5+ cells underwent apoptosis compare to CXCR5- (Figure 5B). However, IL-6 or IL-21 treatment reduced apoptosis of TFH cells.
Since both IL-6 and IL-21 signal through STAT3, we analyzed TFH cell generation in STAT3 flox/flox mice (Takeda et al., 1999) bred with CD4-cre mice (Lee et al., 2001). The deletion of STAT3 gene in CD4+ thymocytes was found to be complete (data not shown). When we immunized these mice as well as their controls with KLH, the numbers of CXCR5+ TFH cells were found to be greatly reduced in the absence of STAT3 (Figure 5C). Moreover, STAT3 deficiency in T cells also led to defective germinal center B cell generation (Figure 5C). KLH-specific IgG and IgM production was also reduced in the absence of STAT3 in T cells (Supplementary Figure 5).
TFH cell generation is independent of TH17 differentiation or function
The above results indicate that the IL-6-IL-21 axis and STAT3 transcription factor are necessary for TFH cell generation, which is similar to the regulation of TH17 differentiation. To test whether the development of TFH cells is dependent or independent upon the TH17 lineage, we utilized Rag1-/- mice reconstituted with bone marrow cells from wild-type, ROR α st/st or ROR αst/stROR α-/- mice. We recently reported that the latter mice were completely impaired in TH17 differentiation in vitro and in vivo (Yang et al., 2008b). However, upon KLH immunization, these mice deficient in both RORα and RORγ in lymphocytes developed more CXCR5+ cells in spleen (Figure 6A), indicating that TFH cells can be generated in the absence of TH17 development. Moreover, PNA+ germinal center B cells were not affected in these animals (Supplementary Figure 6B).
Figure 6. Generation of TFH cells is independent of TH17 lineage.
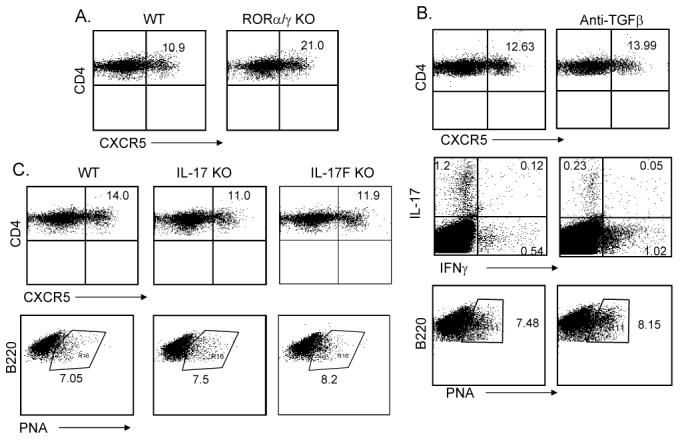
Rag1-/- reconstituted with WT and RORαsg/sg/RORγ KO bone marrow cells (A) or IL-17 KO and IL-17F KO mice and their controls (C) (3 mice per group) were immunized subcutaneously with KLH in CFA. Seven days after the immunization, experimental mice were sacrificed and the TFH cells were determined. Numbers in dot plot quadrants represent the percentages. (B). C57BL/6 mice were immunized with KLH emulsified in CFA with 100 μg of isotype control antibodies or anti-TGFβ blocking Abs (3 mice per group). 7 days later, experimental mice were sacrificed and splenic TFH cells and germinal center B cells were analyzed. Splenocytes were restimulated with KLH for overnight and the production of IL-17 and IFNγ was analyzed in CD4+ gate by intracellular cytokine staining. The results represent one of three individuals with similar results.
Since TH17 differentiation requires also TGFβ in addition to IL-6 or IL-21, we examined whether TGFβ signaling is required for TFH cell generation. C57BL/6 mice were immunized with KLH in the absence or presence of anti-TGFβ blocking antibody as previously described (Veldhoen et al., 2006b). While IL-17 expression was substantially decreased in anti-TGFβ-treated mice, CXCR5 expression was not (Figure 6B), indicating that TGFβ signaling is not essential for TFH cell generation. Moreover, PNA expression on B cells was not affected either (Figure 6B).
To understand if TFH cell generation requires TH17 function, we also immunized mice deficient in IL-17 or IL-17F. Lack of IL-17 or IL-17F did not significantly reduce the number of TFH cells in spleen or the number of germinal center B cells (Figure 6C, Supplementary Figure 6B). IL-17 or IL-17F is thus not essential in the generation of TFH cells in vivo.
IL-21 in the absence of TGFβ initiates TFH cell differentiation
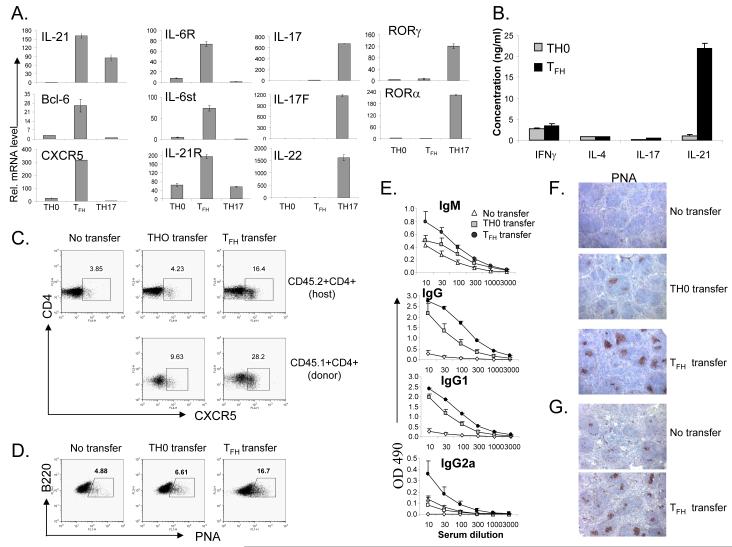
Our results thus far suggest that although IL-6 and IL-21 are required for both TFH and TH17 differentiation, these two subsets appear to have distinct genetic program and differ in their dependency on TGFβ signaling. Since TFH cells have never been generated in vitro, we next assessed whether IL-21 is sufficient to drive TFH cell development in the absence of TGFβ signaling. Naïve OT-II cells were activated by Ova peptide and splenic APC in the absence (neutral condition) or presence of IL-21, TGFβ and antibodies to IL-4 and IFNγ (TH17 condition) or IL-21 plus antibodies to IL-4, IFNγ and TGFβ. 5 days later, the activated T cells were extensively washed and restimulated with anti-CD3 for 4 hours and their gene expression was assessed by real-time RT-PCR. As expected, cells cultured under TH17 condition highly expressed TH17-specific genes, including IL-17, IL-17F, IL-22, RORα and RORγt (Figure 7A). In contrast, T cells treated with IL-21 in the absence of TGFβ signaling upregulated genes that are specifically expressed in TFH cells, including CXCR5, Bcl-6, IL-6R and IL-6st (Figure 7A). They also upregualted IL-21R expression but did not express TH17 genes (Figure 7A). PD-1 was also highly expressed in these cells (data not shown). To confirm this result, cell supernatants were measured 24 hours after restimulation for cytokine secretion by ELISA. IL-21 in the absence of TGFβ signaling did not support IL-17 expression, but the resulting cells expressed high levels of IL-21 (Figure 7B), supporting that IL-21 expression is independent of TGFβ signaling.
Figure 7. IL-21, in the absence of IL-4, IFNγ and TGF-b signaling, generates TFH cells.
A-F, FACS-sorted CD62hiCD44loCD25negCD4+ T cells from CD45.1+ OT-II mice were cultured with irradiated splenic APC plus OVA323-339 peptide under TH0, TFH (IL-21 plus antibodies to IL-4, IFNγ and TGFβ) or TH17 condition for 5 days. After 5 days, CD4 T cells were restimulated with anti-CD3 for 4 hours for real-time PCR analysis (A) or for 24 hours for cytokine measurement by ELISA (B). (C-F). Five days after in vitro differentiation, cells were adoptively transferred into CD45.2+ congenic mice (n=3-4) before the recipient mice were subcutaneously immunized with OVA in IFA. A group of mice that did not receive T cells was used as a control (No transfer). Seven days after immunzation, lymphoid cells from the draining lymph nodes of recipient mice were isolated and TFH cells and germinal center B cells were analyzed (D). Numbers in the boxes represent the percentages. (E). The sera from the recipient mice were subject to a 3-fold serial dilution, and the concentrations of OVA-specific IgM and IgG were analyzed by ELISA and averaged for each group. (F). Germinal center in the spleens of the recipient mice were identified by PNA staining (brown). The results are a representative of multiple mice of two independent experiments with similar results. (G). CXCR5+CD44hi cells were sorted from CD45.1 mice on day 7 after immunization with KLH and transferred to CD45.2 (n=3) recipient mice following immunization with KLH in CFA. A group of mice that did not receive T cells was used as a control (No transfer). Seven days after immunization, germinal center in the spleens of the recipient mice were identified by PNA staining (brown).
To test the function of these TFH-like cells in vivo, we transferred OT-II cells (CD45.1+) activated under neutral condition or with IL-21 plus blocking antibodies to IFNγ, IL-4 and TGFβ into recipient mice (CD45.2+) followed by immunization with Ova protein. Compared to mice receiving no cells or T cells activated under neutral condition, the recipients of IL-21-treated cells exhibited greatly increased CD45.1+ TFH cells (Figure 7C). Interestingly, host T cells in these mice also had approximately 4 times TFH cells compared to those receiving T cells activated under neutral condition (Figure 7C). The transferred cells also promoted Ova-specific antibody production and germinal center reactions (Figure 7D-F). As a control for the above experiment, we sorted CD45.1+ CXC5+CD44hi cells from KLH immunized mice and transferred them into recipient mice (CD45.2+) followed by immunization with KLH. Similar to in vitro generated TFH cells, in vivo generated CXCR5+ cells also promoted a significant increase in PNA+ germinal center B cells (Figure 7G). These results indicate that IL-21, in the absence of TH1, TH2 and TH17 differentiation, initiates TFH cell development.
Discussion
Following antigenic activation, naïve TH cells differentiate into distinct effector subsets. A fundamental function of TH cells is to “help” humoral immunity. Although strongly implicated in antibody production and germinal center reactions, the ontogeny of TFH cells has been unclear. In the current study, we find that TFH cells are distinct from TH1, TH2 or TH17 cells in their gene expression and developmental regulation. Generation of TFH cells requires IL-21, IL-6 and STAT3.
Although implicated in humoral immunity and antibody-mediated autoimmunity, the regulation of TFH cell development is unclear. ICOS-B7h interaction has been well established in the literature to regulate humoral immunity and germinal center reactions (Dong and Nurieva, 2003). This costimulatory pathway was also found important in generation of TFH cells in mouse (Akiba et al., 2005). Interestingly, ICOS deficiency in human patients also causes a severe reduction of TFH cells (Bossaller et al., 2006a). Conversely, impaired negative regulation of ICOS by Roquin E3 ubiquitin ligase led to increased numbers of CXCR5+ TFH cells and IL-21 hyperproduction (Vinuesa et al., 2005a; Yu et al., 2007). However, the mechanism by which ICOS/B7h regulates TFH cell development has not been clear. In our current study, we find that ICOS-B7h interaction is necessary for IL-21 expression by T cells. ICOS may thus regulate TFH cells through production of IL-21. A recent study by Kim et al has revealed IL-21 regulation by calcium signaling and NFAT factors (Kim et al., 2005). We previously showed that ICOS, together with TcR and CD28, increases the expression of NFATc1 through a PI-3 kinase-Itk-calcium pathway (Nurieva et al., 2007b; Nurieva et al., 2003a). ICOS may act through NFATc1 to regulate IL-21 expression. Since IL-21 also regulates TH17 differentiation, an IL-21 defect may account for the impairment in IL-17 expression in the ICOS-deficient animals (Dong and Nurieva, 2003). Moreover, using a B cell-specific B7h knockout mouse, we show that B7h expression on B cells is required for the generation of TFH cells and IL-21 expression. This result not only substantiates the importance of ICOS-B7h interaction in TFH cell development, but also suggests an important function of B cells in vivo as APCs in the generation or maintenance of TFH cells. Consistent with our findings, Ebert et al previously showed that human B cells regulate TFH cell phenotypes (Ebert et al., 2004).
Previous gene expression analysis has revealed that human TFH cells are distinct from TH1 and TH2 cells (Chtanova et al., 2004; Kim et al., 2004). Our current study also reveals that TFH cells do not produce TH1 or TH2 cytokines. Interestingly, although human TFH cells appear to express IL-10 and CXCL13, a ligand for CXCR5 (Chtanova et al., 2004; Ebert et al., 2004; Kim et al., 2004; Kim et al., 2001), mouse TFH cells do not. What accounts for this species difference would need further investigation. Moreover, we found that naïve T cells differentiate into TFH cells in vivo, which is independent of IFNγ, IL-4 and STAT6. From these data, we conclude that TFH cells develop independent of TH1 and TH2 lineages.
On the other hand, TFH cells share common regulators with TH17 cells. Both subsets express IL-21 and their development depends similarly on IL-6, IL-21 and STAT3. However, TFH cells differ from TH17 cells in the following aspects. First, they are distinct in their gene expression profiles. Second, TFH cells do not produce IL-17, IL-17F or IL-22. Most importantly, TFH cell development does not require RORα or RORγt. Thus, we believe that TFH cells develop independent of the TH17 lineage. TH17 development in mouse is not only mediated by the IL-6-IL-21 axis, but also by TGFβ. We show here that IL-21 can be induced in T cells independent of TGFβ signaling. T cells activated in the presence of IL-21 but in the absence of IL-4, IFNγ and TGFβ signaling produced IL-21 but not IL-4, IFNγ, IL-17, IL-17F or IL-22. Furthermore, these cells acquired expression of CXCR5, Bcl-6, IL-6R and IL-6st, genes that expressed by in vivo generated TFH cells, suggesting that TFH cells may be generated in vitro under the above condition. Moreover, these TFH-like cells generated in vitro preferentially expressed CXCR5 in vivo and functioned to promote humoral immunity, similar to in vivo-generated TFH cells. Furthermore, TGFβ signaling, although required for IL-17 expression in vivo, is not essential for TFH cells, indicating a reciprocal relationship of TFH and TH17 cells. Interestingly, Bcl-6 has recently been shown as a repressor of TGFβ-SMAD signaling (Wang et al., 2008), suggesting that lack of TGFβ signaling may favor TFH cell development. These results further support that TFH cells development is independent of TH1, TH2 and TH17 cells, and IL-21 serves as critical factor for generation of this lineage. Interestingly, in vitro generated TFH cells also enhanced the TFH cell generation in recipient mice, suggesting that IL-21 may function in a paracrine fashion to regulate TFH cell development.
In summary, our current study has extensively characterized the developmental regulation of TFH cells. Our data indicate that TFH cells are distinct in their gene expression and immune function and develop via a pathway that is dependent on IL-21 or IL-6 but independent of TH1, TH2 or TH17 lineages (Supplemental Figure 7). In mice defective in TFH cells, there existed still detectable amounts of antigen-specific antibodies, suggesting that other TH subsets may independently regulate the humoral immunity. This knowledge may help us to find ways to treat antibody-mediated autoimmune diseases.
Methods
Mice
IL-4-, IFNγ- and IL-6-deficient mice on C57BL/6 background and STAT6 and STAT4-deficient mice on BALB/c background were purchased from Jackson Laboratories and C57BL/6, B6.SJL (CD45.1) and BALB/c mice were used as controls. RORαst/st, RORαst/st RORγ-/- and wild-type bone marrow chimeras were generated as described (Yang et al., 2008b). Stat3 fl/fl mice (Takeda et al., 1999) were bred with CD4-Cre mice provided by Dr. Christopher Wilson (Lee et al., 2001). IL-21 knockout mice on 129×C57BL/6 F1 mixed background were obtained from NIH Mutant Mouse Regional Resource Centers (MMRRC) (Nurieva et al., 2007a). B7h BKO mice were created by breeding B7H flox mice (Nurieva et al., 2003b) with CD19-cre mice (Rickert et al., 1995). IL-17- and IL-17F- mice were recently generated in our lab (Yang et al., 2008a). Mice were housed in the SPF animal facility at M. D. Anderson Cancer Center and the animal experiments were performed at the age of 6-10 weeks using protocols approved by Institutional Animal Care and Use Committee.
T cell differentiation
Differentiation of OT-II cells in Figure 1A-B was performed as previously described (Chung et al., 2006). CD62LhiCD44loCD25negCD4+ T cells were isolated and cultured with irradiated splenic APC plus OVA323-339 peptide (10 μg/ml) (TH0 condition) and in the presence of polarizing reagents as the following for 5 days: 10 μg/ml of anti-IL-4 (11B11) and 10 ng/ml of IL-12 for TH1; 10 μg/ml of anti-IFNγ (XMG 1.2) and 10 ng/ml of IL-4 for TH2; 30 ng/ml of IL-6, 5 ng/ml of TGFβ, 50 ng/ml IL-23, 10 μg/ml of anti-IL-4 and 10 μg/ml of anti-IFNγ for TH17. In Figure 7A, the cytokine stimuli for TH17 differentiation were 100 ng/ml of IL-21, 5 ng/ml of TGFβ, and 10 μg/ml of anti-IL-4 and 10 μg/ml of anti-IFNγ, and for generation of TFH cells were 50 ng/ml of IL-21, 10 μg/ml anti-IFNγ, 10 μg/ml anti-IL-4 and 20 μg/ml anti-TGF-β (1D11) neutralizing Abs. IL-4, IL-6, IL-12 and TGFβ were purchased from Peprotech. IL-21, IL-23 and anti-TGF-β (1D11) neutralizing Abs were purchased from R&D. To characterized the in vitro-differentiated CD4 T cells under these conditions, these cells were restimulated with plate-bound anti-CD3 Ab (5 μg/ml) for 4hours for real-time PCR analysis, or for 24 hours for cytokine measurement by ELISA.
Keyhole Limpet Hemocyanin (KLH) Immunization
Various strains of mice (6-8 wk old; three per group) were immunized with KLH (0.5 mg/ml) emulsified in CFA (0.5 mg/ml) at the base of the tail (100 μl each mouse). In Figure 6 for local blockade of TGF-β, 100 μg anti-TGFβ (1D11) was included in the emulsion; the control group received 100 μg isotype control antibodies. Seven days after immunization, these mice were sacrificed and analyzed individually. The germinal center B cells were determined by staining with FITC-labeled PNA (Pharmingen) and PerCP-labeled anti-B220 mAb (Pharmingen). The TFH cell induction was determined by staining with PerCP-labeled anti-CD4 mAb (Pharmingen) and biotinylated anti-CXCR5 mAb (Pharmingen), followed by APC-labeled streptavidin (Jackson ImmunoResearch Laboratories, Inc.). In some experiments, sera from immunized mice were collected, and antigen-specific IgM, and IgG antibodies were measured by using ELISA. Briefly, serum samples were added in a 3-fold serial dilution onto plates precoated with 10 μg/ml KLH or Ova protein. Antigen-specific antibodies were detected with biotinylated goat antimouse IgM or rat anti-mouse IgG antibodies (Southern Biotechnology Associates). To analyze the role of B7h in regulation of T cell responses in vivo (Fig. 3A and C), spleen cells from KLH-immunized mice were stimulated in 96-well plates as triplicates with or without KLH. Effector cytokines (IFN-γ and IL-21) were analyzed 4 days later by ELISA (Pharmingen). In Figure 6 B, spleen cells from immunized mice were restimulated with 50 μg KLH for 24 h. In the final 5 h, Golgi-stop (BD Bioscience) was added and IL-17- and IFNγ-producing cells were analyzed using a BD CytoFix/CytoPerm intracellular staining kit (BD Bioscience).
Statistical analysis of microarray data
The DNA microarray analysis was carried out at the Institute for Systems Biology microarray core facility using Affymetrix Mouse 430 2.0 chips. The total RNA samples were labeled according to manufacturer’s instruction using One-Cycle Target Labeling method, which consists of oligo-dT primed cDNA synthesis followed by in vitro transcription that incorporates biotinylated nucleotides. The microarray data were normalized using GCRMA (Zhijin et al., 2004). We then selected the genes whose expression levels were changed across the TH1, TH2, TH17, and TFH cells using a False Discovery Rate (FDR) estimation method (Storey and Tibshirani, 2001): 1) one-way ANOVA was performed to compute unadjusted p-value for the four classes of cells; 2) the number of non-differentially expressed probe-sets (m0) was estimated as 2 × the number of the probe-sets with p-value>0.5 (m0= 25088); 3) the expected number of false positives under the complement null hypothesis E(V0) was estimated for a given ANOVA F-statistic value by performing 500 times of resampling; and 4) FDR was finally estimated as m0/m × E(V0)/R where m is the total number of the probe-sets (m=45037) and R is the number of the genes being selected using the given ANOVA F-statistic value (FDR estimated for various F-statistic value is shown in Supplementary Figure 2). Then, the expression levels of 8350 probe-sets showing differential expressions among the four types of cells (FDR = 0.1, corresponding to an unadjusted p-value = 0.033 from one-way ANOVA) were used for hierarchical clustering (Euclidian distance metric and Ward Minimum Variance Linkage) and PCA.
Adoptive transfer study
CD4+ T cells from OT-II mice (CD45.2) were intravenously transferred into C57BL/6 (CD45.1+) mice (3×106 cells/mouse) (3 groups; 3 mice per group). 2 groups of recipient mice were immunized subcutaneously with 100 μg Ova protein emulsified in CFA and treated with a 300 μg of control rat Ig or anti-IFNγ and anti-IL-4 mAbs at the time of immunization (day 0) and on days 2 and 4. Seven days after the immunization, experimental mice were sacrificed and splenic CD45.1+ and CD45.2+ CD4 cells were stained with biotinylated anti-CXCR5 mAb, followed by APC-labeled streptavidin. In Figure 7C-E, FACS-sorted naïve (CD4+CD62LhiCD44-CD25-) T cells from CD45.1+ OT-II mice were activated under TH0 or TFH condition, washed and intravenously transferred into C57BL/6 (CD45.2+) mice (4×106 cells/mouse) and the recipient mice were subcutaneously immunized with 100 μg OVA protein emulsified in IFA. A group of C57BL/6 mice that did not receive T cells was used as a control (No transfer). In Fig. 7G, CXCR5+CD44hi cells were sorted from B6.SJL (CD45.1) mice immunized with KLH. These cells were transferred into C57BL/6 (CD45.2+) mice (5×106 cells/mouse) (3 mice per group). Second group did not receive cells. All mice were immunized subcutaneously with 1000 μg KLH. Seven days after immunzation, lymphoid cells from the draining lymph nodes of the recipient mice were isolated and stained with FITC-labeled anti-CD45.1 mAb and PerCP-labeled anti-CD4 mAb plus biotinylated anti-CXCR5 mAb, followed by APC-labeled streptavidin, or stained with FITC-labeled PNA and PerCP-labeled anti-B220 mAb.
Quantitative real-time PCR
Total RNA was prepared from T cells using TriZol regent (Invitrogen). cDNA were synthesized using Superscript reverse transcriptase and oligo(dT) primers (Invitrogen) and gene expression was examined with a Bio-Rad iCycler Optical System using iQ™ SYBR green real-time PCR kit (Bio-Rad Laboratories, Inc.). The data were normalized to β-actin reference. The following primer pair for Bcl-6, IL-6R, IL-6st, and CXCR5 was used: Bcl6 forward: CACACCCGTCCATCATTGAA, reverse: TGTCCTCACGGTGCCTTTTT; IL6R forward: GGTGGCCCAGTACCAATGC, reverse: GGACCTGGACCACGTGCT; CXCR5 forward: ACTCCTTACCACAGTGCACCTT, reverse: GGAAACGGGAGGTGAACCA; IL-6st forward: ATT TGT GTG CTG AAG GAG GC, reverse: AAA GGA CAG GAT GTT GCA GG. The primers for IL-21, IL-17, IL-17F, IL-4, IFNγ, RORγ, RORα, T-bet, GATA3, and β-actin were previously described (Yang et al., 2007).
Immunohistochemical analysis
Fresh mouse spleen tissues were embedded in OCT and frozen with isopentane in Histobath. Tissue blocks were sliced 6 μm with cryotome. Slides were fixed with acetone cold. Purified anti-mouse CD4 or biotin labeled anti-mouse PNA were applied as primary Ab following with biotinylated anti-rat secondary Ab and avidin-peroxidase complex reagent. Novared was used as substrate. For double staining, biotin-labeled B220 was applied and following with avidin-aklinphosphotase complex reagent. Vector blue was used as substrate. CD4 and B220 are from BD Pharmagen. PNA and other reagent are from Vector Laboratary. Slides for PNA staining were count stained with Hematoxlin.
Supplementary Material
Acknowledgements
We thank B. Marzolf in Institute for Systems Biology for his assistance in microarray analysis, Drs. Robert Rickert and Robert Carter for the CD19-cre mice, Dr. Chris Wilson for the CD4-cre mice, Dr. Yue Wang for advice and the Dong lab members for their help. The work is supported by research grants from NIH (to CD), an Intramural Research Program of the NIEHS, NIH (to AMJ) and MD Anderson Cancer Center and the Gillson Longenbaugh Foundation (to SSW). RN is a recipient of a Scientist Development Grant from the American Heart Association. CD is a Trust Fellow of the MD Anderson Cancer Center, a Cancer Research Institute Investigator and an American Lung Association Career Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiba H, Takeda K, Kojima Y, Usui Y, Harada N, Yamazaki T, Ma J, Tezuka K, Yagita H, Okumura K. The Role of ICOS in the CXCR5+ Follicular B Helper T Cell Maintenance In Vivo. J Immunol. 2005;175:2340–2348. doi: 10.4049/jimmunol.175.4.2340. [DOI] [PubMed] [Google Scholar]
- Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- Bossaller L, Burger J, Draeger R, Grimbacher B, Knoth R, Plebani A, Durandy A, Baumann U, Schlesier M, Welcher AA, et al. ICOS Deficiency Is Associated with a Severe Reduction of CXCR5+CD4 Germinal Center Th Cells. J Immunol. 2006a;177:4927–4932. doi: 10.4049/jimmunol.177.7.4927. [DOI] [PubMed] [Google Scholar]
- Bossaller L, Burger J, Draeger R, Grimbacher B, Knoth R, Plebani A, Durandy A, Baumann U, Schlesier M, Welcher AA, et al. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J Immunol. 2006b;177:4927–4932. doi: 10.4049/jimmunol.177.7.4927. [DOI] [PubMed] [Google Scholar]
- Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, de Waal Malefyt R, Tangye SG. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol. 2007;179:8180–8190. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T Follicular Helper Cells Express a Distinctive Transcriptional Profile, Reflecting Their Role as Non-Th1/Th2 Effector Cells That Provide Help for B Cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- Chung Y, Yang X, Chang SH, Ma L, Tian Q, Dong C. Expression and regulation of IL-22 in the IL-17-producing CD4+ T lymphocytes. Cell Res. 2006;16:902–907. doi: 10.1038/sj.cr.7310106. [DOI] [PubMed] [Google Scholar]
- Dong C. Diversification of T-helper-cell lineages: finding the family root of IL-17-producing cells. Nat Rev Immunol. 2006;6:329–334. doi: 10.1038/nri1807. [DOI] [PubMed] [Google Scholar]
- Dong C, Flavell RA. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res. 2000;2:179–188. doi: 10.1186/ar85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Nurieva RI. Regulation of immune and autoimmune responses by ICOS. J Autoimmun. 2003;21:255–260. doi: 10.1016/s0896-8411(03)00119-7. [DOI] [PubMed] [Google Scholar]
- Ebert Lisa M., Horn Michael P., Lang Alois B., Moser B. B cells alter the phenotype and function of follicular-homing CXCR5+ T cells. Eur J Immunol. 2004;34:3562–3571. doi: 10.1002/eji.200425478. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- Haynes NM, Allen CDC, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in Follicular Th Cell Positioning and Appearance of a Programmed Cell Death Gene-1High Germinal Center-Associated Subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Junt T, Fink K, Forster R, Senn B, Lipp M, Muramatsu M, Zinkernagel RM, Ludewig B, Hengartner H. CXCR5-Dependent Seeding of Follicular Niches by B and Th Cells Augments Antiviral B Cell Responses. J Immunol. 2005;175:7109–7116. doi: 10.4049/jimmunol.175.11.7109. [DOI] [PubMed] [Google Scholar]
- Kim CH, Lim HW, Kim JR, Rott L, Hillsamer P, Butcher EC. Unique gene expression program of human germinal center T helper cells. Blood. 2004;104:1952–1960. doi: 10.1182/blood-2004-03-1206. [DOI] [PubMed] [Google Scholar]
- Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-P, Korn LL, Gamero AM, Leonard WJ. Calcium-dependent Activation of Interleukin-21 Gene Expression in T Cells. J Biol Chem. 2005;280:25291–25297. doi: 10.1074/jbc.M501459200. [DOI] [PubMed] [Google Scholar]
- Kopf M, Herren S, Wiles MV, Pepys MB, Kosco-Vilbois MH. Interleukin 6 influences germinal center development and antibody production via a contribution of C3 complement component. J Exp Med. 1998;188:1895–1906. doi: 10.1084/jem.188.10.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 Signaling via STAT5 Constrains T Helper 17 Cell Generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, Flavell RA. T Cell-Produced Transforming Growth Factor-[beta]1 Controls T Cell Tolerance and Regulates Th1- and Th17-Cell Differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007a;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chuvpilo S, Wieder ED, Elkon KB, Locksley R, Serfling E, Dong C. A Costimulation-Initiated Signaling Pathway Regulates NFATc1 Transcription in T Lymphocytes. J Immunol. 2007b;179:1096–1103. doi: 10.4049/jimmunol.179.2.1096. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Duong J, Kishikawa H, Dianzani U, Rojo JM, Ho I, Flavell RA, Dong C. Transcriptional regulation of th2 differentiation by inducible costimulator. Immunity. 2003a;18:801–811. doi: 10.1016/s1074-7613(03)00144-4. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Mai XM, Forbush K, Bevan MJ, Dong C. B7h is required for T cell activation, differentiation, and effector function. Proc Natl Acad Sci U S A. 2003b;100:14163–14168. doi: 10.1073/pnas.2335041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–36. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-l cell development in CD19-deficient mice. Nature. 1995;376:352–355. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, Leonard WJ. Interleukin-21: Basic Biology and Implications for Cancer and Autoimmunity. Annu Rev Immunol. 2008;26 doi: 10.1146/annurev.immunol.26.021607.090316. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Estimating the Positive False Discovery Rate Under Dependence, with Applications to DNA Microarrays. Vol. 2001. Department of Statistics, Stanford University; 2001. [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 Activity and Development of Chronic Enterocolitis in Mice Devoid of Stat3 in Macrophages and Neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006a;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat Immunol. 2006b;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005a;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Tangye SG, Moser B, Mackay CR. FOLLICULAR B HELPER T CELLS IN ANTIBODY RESPONSES AND AUTOIMMUNITY. Nature Reviews Immunology Nat Rev Immunol. 2005b;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- Wang D, Long J, Dai F, Liang M, Feng X-H, Lin X. BCL6 Represses Smad Signaling in Transforming Growth Factor-B Resistance. Cancer Research. 2008 doi: 10.1158/0008-5472.CAN-07-0008. [DOI] [PubMed] [Google Scholar]
- Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang Y, Schluns KS, Broaddus RR, Zhu Z, Dong C. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008a doi: 10.1084/jem.20071978. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T Helper 17 Lineage Differentiation Is Programmed by Orphan Nuclear Receptors ROR[alpha] and ROR[gamma] Immunity. 2008b;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- Zhijin W, Rafael AI, Robert G, Francisco M-M, Forrest S. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.