Abstract
Advances in Fourier transform mass spectrometry have made the acquisition of high-resolution and accurate mass measurements routine on a chromatographic time-scale. Here we report an algorithm, Hardklör, for the rapid and robust analysis of high resolution mass spectra acquired in shotgun proteomics experiments. Our algorithm is demonstrated in the analysis of an Escherichia coli enriched membrane fraction. The mass spectrometry data of the respective peptides are acquired by micro-capillary HPLC on an LTQ-Orbitrap mass spectrometer with data-dependent acquisition of MS/MS spectra. Hardklör detects 211,272 total peptide isotope distributions over a two hour analysis (75 min gradient) in only a small fraction of the time required to acquire the data. From these data there are 13,665 distinct, chromatographically persistent peptide isotope distributions. Hardklör is also used to assess the quality of the product ion spectra and finds that more than 11.2% of the MS/MS spectra are composed of fragment ions from multiple different molecular species. Additionally, a method is reported that enzymatically labels N-linked glycosylation sites on proteins, creating a unique isotope signature that can be detected with Hardklör. Using the protein invertase, Hardklör identifies 18O-labeled peptide isotope distributions of four glycosylation sites. The speed and robustness of the algorithm create a versatile tool that can be used in many different areas of mass spectrometry data analysis.
INTRODUCTION
In recent years, mass spectrometry based proteomics has moved toward handling increasingly complicated mixtures. To obtain sufficient peak capacity to characterize complex peptide mixtures, peptides are often separated using multidimensional chromatography to minimize the overlap of analytes of similar m/z entering the mass spectrometer at any one point in time1;2. While effective, this approach is inherently slow, and not particularly amenable to high-throughput analyses. Furthermore, replicate analyses to produce data with appropriate measures of variance for quantitative comparisons are rarely obtained because of the lengthy nature of these experiments. Thus, to handle the increasing demand for proteomics analysis while concurrently performing replicate measurements to obtain statistically meaningful results, the overall throughput for handling mixtures must be increased.
An alternative to multidimensional chromatography is the use of a mass spectrometer with sufficient resolution so that when combined with only a single dimension of chromatographic separation, there is enough peak capacity to handle the complexity of the mixture. Recently, advances in commercially available Fourier transform mass spectrometers have facilitated the routine acquisition of mass spectra at >50,000 resolution, with accurate mass, and on a chromatographic timescale3;4. With these instruments, an incredible wealth of data can be acquired in a single μLC-MS experiment. With these high-resolution data, Smith and coworkers have pioneered the use of accurate monoisotopic mass combined with chromatographic retention time (Accurate Mass and Time tags or AMT tags) to map signals measured by μLC-MS with a database of peptides identified previously by tandem mass spectrometry (MS/MS)5;6. Thus, the throughput of proteomics experiments can be improved dramatically since any peptide identified previously can now be followed in future experiments using only the accurate peptide mass and a normalized chromatographic elution time.
Integral to AMT-based experiments is the robust computational determination of the monoisotopic m/z and charge state for every component in every scan of the μLC-MS run. While simple in principle, this analysis is complicated by 1) many peptides that are present at or near the limit of detection, 2) peptides that have complicated isotope distributions, and 3) overlapping distributions within a complicated mixture. Several computer algorithms have been developed to deconvolute a complex mass spectrum containing overlapping isotope distributions at different charge states to produce a simplified list of monoisotopic masses7;8. Most of these algorithms are either based on, or are a reimplementation of, Horn’s THRASH algorithm9. While powerful, these algorithms are slow relative to the time required to acquire the mass spectrometry data – becoming a bottleneck in interpreting μLC-MS data containing tens of thousands of mass spectra. Furthermore, additional information could potentially be gleaned using the isotope distributions to recognize unique features to target further analyses10;11 as opposed to being restricted to only an averagine model12 for estimating the elemental composition and thus the isotope distribution of a biopolymer.
In this paper we report the development and validation of a new algorithm for the analysis of high resolution μLC-MS data. This algorithm, Hardklör, has similar sensitivity and specificity as alternative approaches for extrapolating monoisotopic mass information from complex datasets, yet is faster and more versatile. Hardklör automates the detection of peptide isotope distributions (PIDs) from high resolution datasets, with improved handling of overlapping and in-phase isotope distributions, and with substantial improvements in speed. The improvement in speed makes possible the identification of unusual isotope distributions and not just those distributions that fit a common averagine12 model. Using this information we can make comparisons between complex datasets, flag MS/MS spectra that may be composed of multiple peptide species, and identify isotope distributions tagged either enzymatically, chemically, or even metabolically to create a unique isotope distribution.
METHODS
Sample Preparation
Escherichia coli membrane sample preparation
E. coli grown to mid log phase in LB media were enriched by centrifugation and lysed in ammonium bicarbonate buffer (pH 7.8) using a French press. The lysate was first spun at low speed (3,000 RPM) and pellet discarded to remove nuclei and unbroken cells. The remaining supernatant was then subjected to a high speed spin to enrich for insoluble membrane vesicles. The pellet was then resolubilized using 0.1% RapiGest in 50 mM ammonium bicarbonate buffer and reduced, alkylated, and digested with trypsin as described previously.13 The resulting peptide mixture was stored at −80 °C until analysis with an LTQ-Orbitrap as described below.
Site-specific isotope labeling of N-linked glycosylation sites
Glycosylated Saccharomyces cerevisiae invertase (Sigma-Aldrich) was suspended in 50 mM ammonium bicarbonate buffer with 0.1% RapiGest (Waters) and H218O (50 APE; Spectra Stable Isotopes), reduced with dithiothreitol (DTT), and alkylated with iodoacetamide (IAA). The invertase was then digested with 5 μl PNGase-F (New England Biolabs) to cleave the N-linked glycosylation and add a unique isotope signature to the respective amino acid residues. The protein was then precipitated with trichloroacetic acid (TCA) to remove the isotope-enriched sample buffer and re-suspended in 200 μL of 50 mM ammonium bicarbonate buffer containing 0.1% RapiGest. The 18O-labeled invertase was then digested by adding trypsin at a 1:100 enzyme/substrate ratio and incubated for four hours with shaking at 37°C. After digestion, 5 M HCl was added to a final concentration of 200 mM and heated at 37°C for 45 minutes to cleave the RapiGest. The insoluble film resulting from the cleavage of RapiGest was removed by centrifugation and the supernatant was stored at −20°C until analyzed with a benchtop LTQ mass spectrometer as described below.
Acquisition of High Resolution Orbitrap Mass Spectrometry Data
Each protein digest (10 μg) was loaded from the autosampler onto a fused-silica capillary column (75-μm i.d.) packed with 15 cm of Luna C18 material (Phenomenex), mounted in the microspray source, and placed inline with a Surveyor MS HPLC and a MicroAS autosampler (ThermoFisher Scientific). The HPLC was operated using two buffer solutions: Buffer A was a mixture of 95% water, 5% acetonitrile, 0.1% formic acid and Buffer B was a mixture of 20% water, 80% acetonitrile, and 0.1% formic acid. The run began with 27 minutes of 95% buffer A during the loading of the sample onto the microcapillary column13 where the HPLC flow was split from 150 μL/min down to ~2 μL/min prior to the autosampler. Following the initial loading of the sample onto the column, the split location was adjusted from prior to the autosampler to a micro-Tee (UpChurch Scientific) immediately upstream of the microcapillary column using the divert valve on the mass spectrometer. The flow through the column was reduced from ~2 μL/min to ~200 nL/min using a split capillary with less restriction than the loading capillary. The peptides were eluted from the column using a 68 minute gradient of 5 to 35% Buffer B, and a 5 minute gradient of 35 to 85% Buffer B. The solvent composition was kept at 85% Buffer B for 2 min. The column was then re-equilibrated with 95% buffer A for 18 minutes, resulting in a total analysis time of 120 min. Mass spectra were acquired using data-dependent acquisition with a single high resolution mass spectrum at 60,000 resolution (at m/z 400) acquired in the Orbitrap mass analyzer in parallel with 5 low resolution MS/MS scans being acquired in the LTQ.
The tandem mass spectra were searched using SEQUEST14 with no enzyme specificity against a fasta database that consisted of E. coli open reading frames and shuffled decoy sequences of the same length and amino acid composition of the E. coli open reading frames. Peptide identifications were filtered with DTASelect15 using the default parameters with the exception of requiring ΔCN≥0.12 and accepting only fully tryptic peptides.
Acquisition of High Resolution ZoomScans on a Benchtop Linear Ion Trap
Invertase peptides were analyzed by μLC-MS using an LTQ mass spectrometer. The peptides were loaded onto a 75-μm fused-silica capillary packed with Luna 5-μm C18 placed inline with an Agilent 1100 Binary HPLC. Peptides were loaded and eluted from the capillary column as described above for the LTQ-Orbitrap mass spectrometer. The LTQ mass spectrometer was operated using 5 scan events where a full MS scan was followed by the data-dependent acquisition of a ZoomScan and MS/MS spectrum on the most intense signal, followed by a second ZoomScan and MS/MS spectrum on the second most intense signal. Dynamic exclusion was turned on during the entire analysis.
Automated Analysis of High Resolution Orbitrap and ZoomScan Data
Hardklör Algorithm Overview
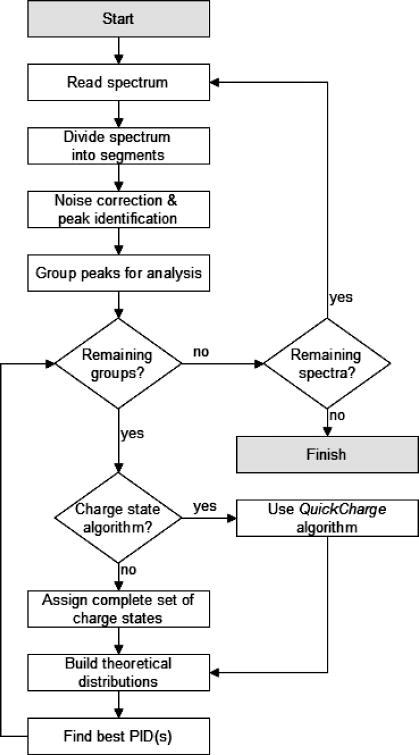
Hardklör is an application written in C++ and compiled with the Visual C++ 6.0 compiler for use on the Microsoft Windows operating system and the GNU GCC compiler for use on GNU/Linux distributions. The algorithm reads ASCII or binary MS1 format files16 and iteratively processes each spectrum within the file. The application outputs a tab-delimited text list of monoisotopic mass, charge state, intensity, and presence/absence of unusual isotope features for each scan. Hardklör follows the general flow shown in Figure 1 and cycles through five main steps in the processing of mass spectrometry data. These steps consist of: 1) Peak Finding, 2) Charge State Estimation, 3) Averagine Modeling and Monoisotopic Mass Prediction, 4) Unusual Peptide Isotope Distribution Detection, and 5) Analysis. Each of these steps will be described in detail below. Information about obtaining Hardklör can be found at http://proteome.gs.washington.edu/software/hardklor/.
Figure 1.
General approach for the analysis of μLC-MS data using Hardklör.
Peak Finding
Hardklör distinguishes signal from background by eliminating signal below a predefined signal-to-noise (S/N) cutoff. To calculate the S/N, Hardklör employs an approach used in the THRASH algorithm9. Briefly, the algorithm assumes that the background signal over a given m/z region will be acquired more frequently than the signal of the isotope peaks. Thus, using a plot of the frequency of each intensity value within the window versus intensity, the background signal (Ib) is estimated by the intensity with the maximum frequency. The noise is calculated by the full-width at half maximum (FWHM) of the smoothed histogram plot and the S/N can be calculated for each signal (Ip) using the equation:
This filter is applied to small, overlapping segments of the entire spectrum. We used segments of 25 m/z, but the size of the filter segments may be specified by the user. The overlapping spectral segments give flexibility when dealing with varying amounts of background noise across the spectrum while minimizing the risk of missing a peak because it resided on the edge of the segment. Peaks shared in the overlap between segments are only considered once in downstream analysis.
A unique aspect of Hardklör is how detected peaks are grouped into potential peptide isotope distributions. Peaks exceeding the S/N threshold (default S/N > 3) are grouped together into potential isotopic distributions using a novel windowing scheme that self-adjusts from the smallest m/z width that includes at least two peaks to a maximum of 5 m/z. Beginning from a window width that encompasses at least two peaks, the window expands to include neighboring peaks until the 5 m/z threshold is reached. To prevent truncation of a valid isotope distribution, the window then contracts back to the largest gap between any two peaks in the group. Peaks that were pushed out of the contracting window are then grouped in a subsequent window and the process is repeated. The resulting groups of peaks are then submitted for charge state determination and correlation with predicted combinations of PIDs.
Charge State Determination
Charge state determination is used in Hardklör to minimize the number of isotope distributions that need to be fit to the group of potential isotope peaks. We have written a routine known as QuickCharge that estimates charge states for each peak by computing the reciprocal of the m/z difference between any two peaks. A potential charge state is computed for every peak by calculating the distance between the respective peak and the other peaks in the same group – often resulting in multiple putative charge states per single isotope peak. While the QuickCharge routine does not return a single correct charge state for each peak, it significantly reduces the total number of combinatorial isotope distributions that need to be fit to each group of peaks in the later steps of the analysis. Alternatively, the user can choose to perform no charge state estimation, and Hardklör will consider all possible isotope distributions within a specified range of charge states. The approach of considering all isotope distributions within a specified range will be referred to as the Complete method.
Averagine Model and Monoisotopic Mass Prediction
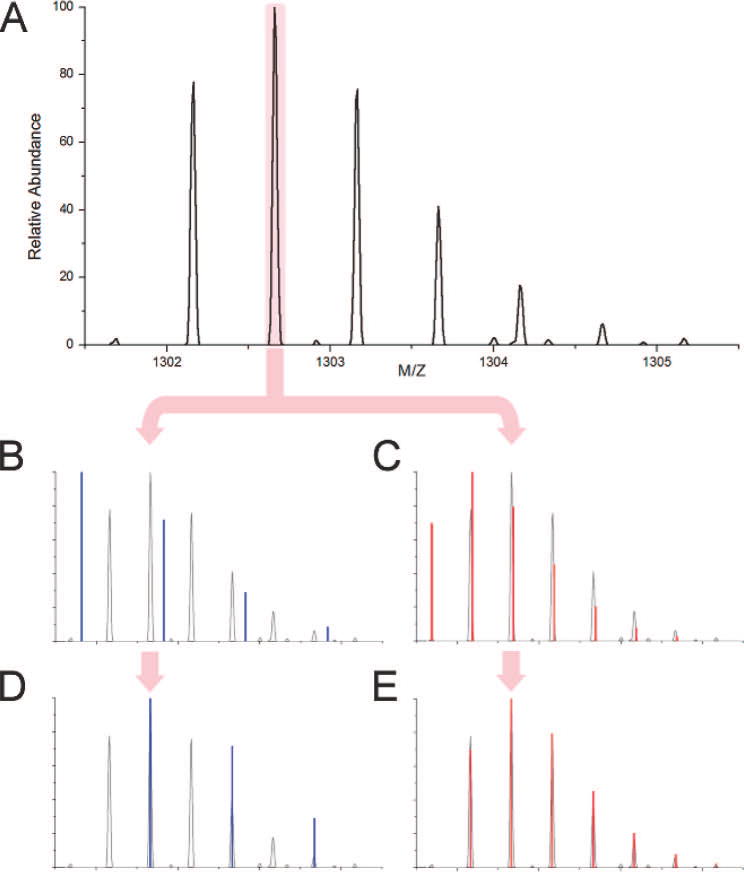
Hardklör uses an averagine12 model to estimate peptide elemental composition. Averagine is the weighted average of the elemental composition of an amino acid found in proteins. The m/z of the base isotope peak is multiplied by the charge-state of the distribution to estimate the peptide mass. Then the mass is divided by the molecular weight of an averagine subunit. The number of averagine subunits estimates the elemental composition of the polypeptide12. Using the elemental composition of the computed poly-averagine molecule, the isotope distribution is calculated with the Mercury algorithm17 to predict an approximate peptide isotope distribution. Because a group of peaks from the experimental data may consist of one or more peptides, the m/z value of each centroided peak is used in combination with the peak’s charge predictions to construct a set of poly-averagine PIDs. The predicted poly-averagine PIDs are aligned to the observed data at the predicted base isotope peaks, and the monoisotopic mass of the predicted PIDs that are accepted, as described below, are used to predict the monoisotopic masses of the observed peptides (Figure 2).
Figure 2.
Illustration of base isotope peak alignment. Hardklör selects a peak (A) and constructs two averagine models at charge states of +1 (B) and +2 (C). The averagine models are aligned to the base isotope peak and a correlation score is computed (D and E). The predicted peptide isotope distribution that matches the measured isotope distribution with the highest similarity is accepted (E).
A key component to Hardklör is the ability to detect unusual PIDs that deviate from the averagine model. These differences may include the presence of atoms other than C, H, O, N or S and/or non-natural isotope abundances. A user-defined list of possible differences is used to create variations to the averagine model. Either the standard averagine model or one of the variants is accepted based on the best similarity score to the observed data.
Combinatorial Analysis of Peptide Isotope Distribution Predictions
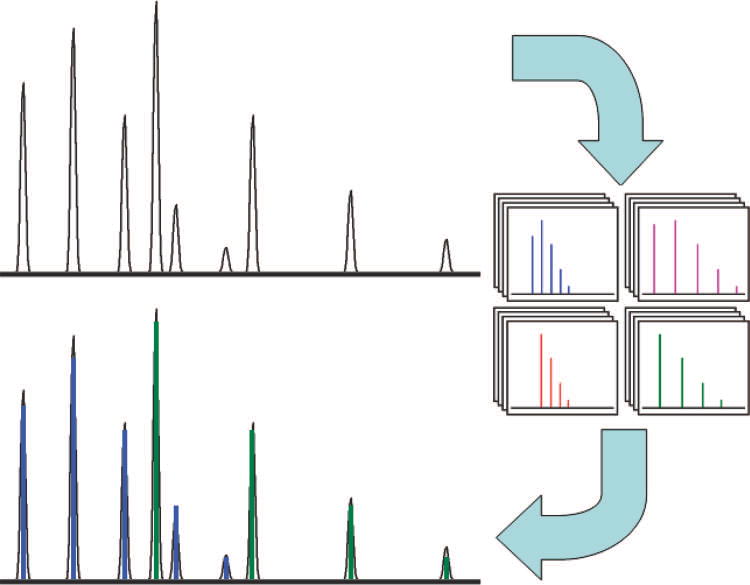
Each group of peaks has multiple estimated PIDs: one PID based upon each peak, and variations for each of these PIDs including different charge states and averagine model variants. The estimated PIDs are compared to the observed data using the dot-product similarity metric18. To account for multiple peptide isotope distributions in the group of peaks, each predicted PID is considered alone and in combination with one or more other peptide predictions (Figure 3). No two variants of the same peptide prediction are ever considered together. When combining predicted PIDs, the intensity of each predicted PID is scaled to the intensity of the experimentally derived data using the base isotope peak of the predicted distribution and its respective peak in the experimentally measured spectrum. If two or more predicted PIDs share an isotope peak, then the contribution to the individual peaks from the respective PIDs, scaled to the individual base isotope peaks, are summed. If the intensity of a base isotope peak of a predicted PID is shared by a peak from previously combined predicted PIDs, then the scaled intensity of the subject PID is the intensity of the respective observed peak minus the intensity of the respective peak of the combined PIDs. Thus, the contribution of any one predicted isotope distribution may differ relative to the other PIDs in the combination; however, the relative abundance of all the peaks within a model remains the same. The PIDs are combined in order of base isotope peak intensity from least intense to most intense.
Figure 3.
Combinatorial analysis of peptide isotope distributions with Hardklör. A list of averagine models is created for each peak in the observed distribution. Each averagine model is scored alone and in combination with the other models. The averagine model distribution or combination of distributions that has the closest similarity to the observed distribution is accepted.
To minimize over-fitting the data, the combination with the fewest PID predictions to exceed the user-defined dot-product threshold is accepted. To make this process as efficient as possible, each single PID is analyzed first. If the threshold is not exceeded, all possible combinations of two PIDs are analyzed next. This process is repeated, increasing the number of combined PIDs, until either the maximum number of combinations specified by the user is reached or the dot-product threshold is exceeded. Hardklör will always iterate through all combinations of predicted PIDs at each depth (i.e. the number of combined distributions). Because all combinations at any depth are considered before terminating the routine, the order in which the combinations are analyzed does not affect the results. Once the dot-product score threshold has been exceeded, the PID or PIDs with the highest score is accepted.
RESULTS
Hardklör was tested using an enriched membrane fraction from E. coli. Peptides formed from the digestion of this sample by trypsin were analyzed by μLC-MS on a hybrid LTQ-Orbitrap mass spectrometer. The high-resolution mass spectra acquired in the electrostatic Fourier transform mass analyzer of the Orbitrap were processed using Hardklör. Within each group of peaks, Hardklör was configured to map up to three different peptide isotope distributions with charge states ranging from +1 to +3, with a dot-product threshold of 0.90. The software then returned a predicted monoisotopic mass and charge state for each peptide isotope distribution from each scan. These predictions were further supported by observing the same peptide isotope distribution in at least three of four consecutive scans. These PIDs were considered to be a set of persistent distributions and were separated from spurious incorrect distributions.
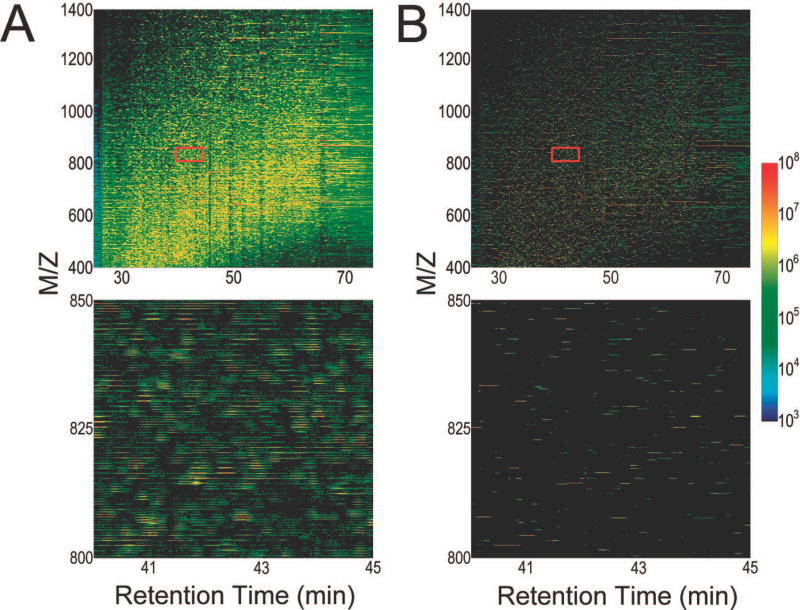
Figure 4 illustrates the detection of the monoisotopic m/z from the raw LTQ-Orbitrap μLC-MS data using Hardklör. The spectra acquired during the entire run are represented as a two dimensional image with time on the x-axis, m/z on the y-axis, and the log normalized intensity as a heat map color scale. The image of the raw data displays all peaks and is complicated by chemical noise and by the complexity of the isotope distributions. The image of the same data processed by Hardklör displays only the predicted monoisotopic peaks. Even over the relatively narrow m/z and time range that is displayed in the figure, Hardklör is able to extract hundreds of PIDs and reduce the unwieldy raw data (Figure 4A) to a very simplified output (Figure 4B). When applied over the entire μLC-MS datafile, Hardklör can detect hundreds of thousands of PIDs.
Figure 4.
Two-dimensional images of peptide isotope distributions before and after Hardklör analysis. (A) Images of the complexity of raw data from which peptide isotope distributions are derived. The red rectangle in the top image shows the region that is enlarged in the bottom image. (B) Hardklör has reduced the data to monoisotopic m/z values. The lengths of the lines represent persistence over multiple scans, one of the criteria for validating the isotope distributions, and the log normalized intensity is expressed using a heat map color scheme.
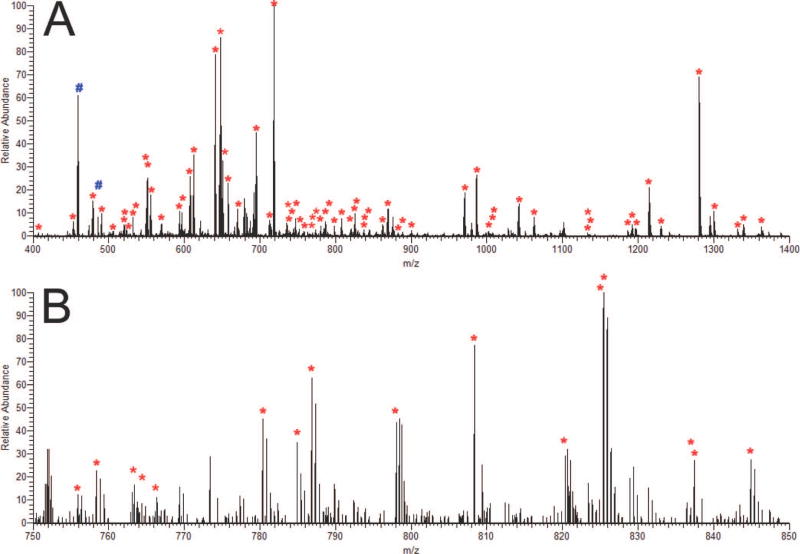
Figure 5 shows a typical example of Hardklör results from a single high resolution Orbitrap scan. From this single scan, Hardklör correctly identified the monoisotopic mass and charge state of 71 PIDs (Figure 5A). These data span a dynamic range of >3 orders of magnitude – a large range for a single scan of data this complicated. Within this scan, there are several overlapping isotope distributions (e.g. m/z range 824–828), as there are with most scans, that are correctly computed. Also, within this scan are two clearly detectable signals (indicated by # signs) that are not labeled by Hardklör. After additional analysis (discussed below), Hardklör correctly detects and assigns these PIDs a charge state of +4. While Hardklör can handle peptides (and proteins) of any charge state, the user settings limited the analysis to +1, +2, and +3 charge states to minimize the overall analysis time.
Figure 5.
Example of Hardklor PIDs identified in a single scan. (A) Red stars (*) indicate the monoisotopic masses of PIDs with charge states of +1, +2, or +3. Blue pound signs (#) are some examples of visually obvious PIDs that were skipped because their charge states were outside the user-specified parameters (e.g. +4) (B) Enlarged region from 750 to 850 m/z.
For the entire μLC-MS datafile, Hardklör found 211,272 PIDs (FDR < 1.0%; estimated using a decoy averagine model with 50% atom percent excess 15N) from a total of 4,376 Orbitrap spectra acquired over a 2 hour HPLC gradient. These total PIDs were further reduced to 13,665 chromatographically persistent PIDs as described above. By grouping PIDs into those that persist chromatographically over time, we reduce the likelihood that any one detected PID could be a result of a spurious random event and minimize redundancy by grouping isotope distributions that belong to a single molecular species.
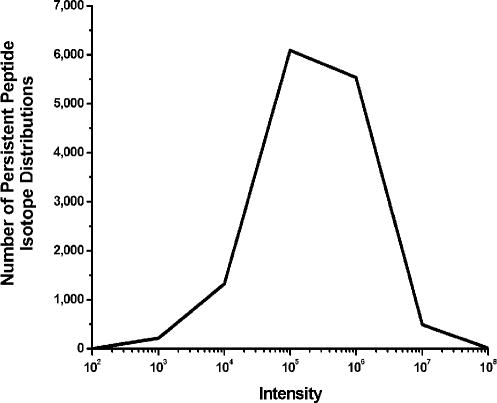
Figure 6 shows the dynamic range of Hardklör among the persistent PIDs. The log-scaled maximum intensities of the PIDs are on the x-axis, and the number of PIDs is on the y-axis. While the majority of the PIDs have an intensity of 105 counts, Hardklör can identify distributions with intensities ranging from 103 to greater than 107 counts. Even for a short 75 minute chromatographic separation, the LTQ-Orbitrap combined with Hardklör can detect features spanning a dynamic range of 5-orders of magnitude. This large dynamic range illustrates that Hardklör is not limited to identification of PIDs for only the most abundant proteins in a cell. When combined with an instrument like the Orbitrap, Hardklör can characterize peptides in a very short analysis time that span a dynamic range that is normally only feasible with lengthy multidimensional separations2;19.
Figure 6.
Dynamic range of persistent peptide isotope distributions identified using Hardklör. Isotope distributions were automatically detected over an intensity range that spans ~103 to ~107 counts on the LTQ-Orbitrap.
To assess the performance of different methods for PID identification, comparisons were made between five different methods for charge state determination. The five methods implemented are the Complete method, our QuickCharge algorithm, and the previously reported Patterson, FFT, and combined Patterson-FFT algorithms20. The Complete method does not apply a charge state algorithm per se but instead assigns all possible charge states (+1, +2, or +3) to each peak’s m/z value. In contrast to using a charge state algorithm, when the Complete method was used, a poly-averagine distribution was calculated for all charge states, and the distribution with the best dot-product similarity score was accepted. The Patterson, FFT, and Patterson-FFT algorithms were expanded so that, as with the QuickCharge algorithm, they provided a set of estimated charge states that is a subset of the Complete method. Incorrect charge states from each method should score poorly because the spacing of the isotope peaks and the number of averagine subunits in the calculated isotope distribution will be incorrect. Thus, by applying an additional layer of computation to reduce the number of charge states, and therefore PIDs to analyze, we hope to increase the overall speed of analysis.
Table 1 summarizes the performance of each charge state method by comparing the number of persistent PIDs and the computation time at different score thresholds. Each analysis was performed on an Intel Xeon 2.4GHz processor. For three different charge states, the use of the Patterson, FFT, and Patterson-FFT algorithms increased the computation time beyond any benefit gained by reducing downstream computation; however, the QuickCharge algorithm performed fast enough to reduce the computation time to approximately 1/3 of the time necessary to compute the Complete method. At a similarity score threshold of 0.90, the Patterson and FFT algorithms produced more persistent PIDs than the Complete method, yet they did not increase the fraction of PIDs that matched the SEQUEST results. Although the QuickCharge algorithm had the fewest persistent PIDs, it maintained a roughly equivalent fraction of PIDs that matched the SEQUEST results as well as the Complete method. Thus, our QuickCharge algorithm fulfills its purpose in providing an increase in speed without a significant loss in performance.
Table 1.
Comparison of the resulting output from five different charge state methods
| Score Threshold(Dot Product) | Total Peptide Predictions1 | Persistent Peptide Distributions2 | False Persistent Peptide Distributions3 | Matches to SEQUEST/DTASelect Results (out of 1530)4 | Approximate Computation Time (minutes) |
|---|---|---|---|---|---|
| Complete | |||||
| 0.1 | 257596 | 16136 | 1213 | 1184 | 87 |
| 0.5 | 251228 | 15881 | 1070 | 1184 | 92 |
| 0.9 | 247604 | 15854 | 36 | 1207 | 203 |
| 0.95 | 213558 | 14103 | 2 | 1196 | 262 |
| 0.99 | 81268 | 6172 | 0 | 946 | 303 |
| Patterson | |||||
| 0.1 | 288850 | 17631 | 1815 | 1182 | 2824 |
| 0.5 | 281504 | 17213 | 1622 | 1182 | 2481 |
| 0.9 | 266592 | 16463 | 45 | 1192 | 2676 |
| 0.95 | 221789 | 14439 | 1 | 1187 | 2734 |
| 0.99 | 81017 | 6082 | 0 | 917 | 2764 |
| FFT | |||||
| 0.1 | 287753 | 17551 | 1846 | 1193 | 191 |
| 0.5 | 280163 | 17117 | 1643 | 1192 | 190 |
| 0.9 | 262630 | 16367 | 44 | 1201 | 258 |
| 0.95 | 217886 | 14163 | 1 | 1187 | 296 |
| 0.99 | 80439 | 6095 | 0 | 920 | 317 |
| FFT and Patterson Combination | |||||
| 0.1 | 277614 | 17232 | 1805 | 1184 | 2714 |
| 0.5 | 269227 | 16702 | 1610 | 1183 | 2472 |
| 0.9 | 242895 | 15329 | 45 | 1177 | 2755 |
| 0.95 | 199360 | 13087 | 1 | 1167 | 2537 |
| 0.99 | 81017 | 7203 | 0 | 917 | 2764 |
| QuickCharge | |||||
| 0.1 | 242881 | 15923 | 1150 | 1186 | 14 |
| 0.5 | 236776 | 15602 | 1028 | 1186 | 14 |
| 0.9 | 211272 | 13665 | 36 | 1200 | 15 |
| 0.95 | 178895 | 11981 | 2 | 1182 | 15 |
| 0.99 | 71228 | 5680 | 0 | 926 | 15 |
Total peptide predictions are the absolute number of peptide isotope distributions (PID) that exceeded the dot-product threshold (cosθ).
Peptides of the same monoisotopic mass and charge state that are observed across consecutive scans are defined as persistent peptide isotope distributions. The threshold for being listed as a persistent peptide distribution is described in the methods.
False persistent peptide distributions are determined using a decoy averagine model with 50% atom percent excess 15N.
Peptide sequences were identified for the persistent peptide distributions if they had the same monoisotopic mass, charge state, and retention time as peptides identified from the MS/MS analysis using SEQUEST and DTASelect.
To assess the sensitivity and specificity of each charge state method, we used a nonstandard isotope composition for nitrogen (50% atom percent excess 15N) to estimate the chance of obtaining a random match to an incorrect isotope distribution. Because the nitrogen isotope composition used in the E. coli growth medium was of natural abundance, the number of PIDs returned with this alternative isotopic composition could be used as a proxy measure of the number of false discoveries returned by Hardklör (Table 1, Column 4). The FFT, Patterson, and Patterson-FFT algorithms showed minimal improvement over the Complete method and QuickCharge algorithm. The QuickCharge algorithm had significant reduction in computational overhead when compared to the other charge-state algorithms evaluated. Thus, the QuickCharge algorithm provides a fast, yet robust, method for PID feature detection.
A unique feature of Hardklör over alternative approaches to PID feature detection7;9;21 is how it handles overlapping isotope distributions. Hardklör deconvolutes multiple overlapping peptide distributions by considering multiple combinations of PIDs. This combinatorial approach evaluates the similarity of each combined set of averagine models against the peaks in the experimentally measured distribution. Each poly-averagine distribution and its combination with one or more of the other poly-averagine distributions in the set are scored until a combination exceeding a preset similarity threshold is found. In this manner, accurate PIDs can be obtained even for overlapping distributions.
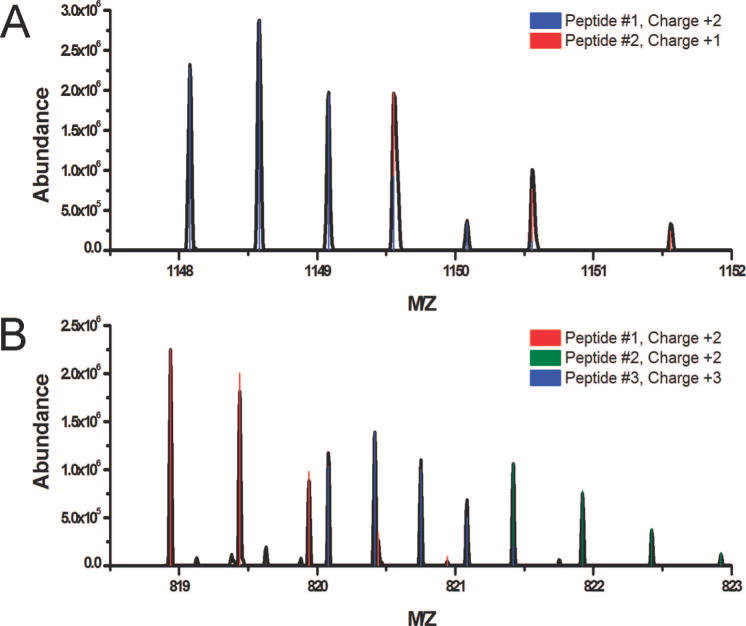
Figure 7 illustrates Hardklör’s ability to deconvolute overlapping PIDs. The distributions show isotope peaks from one peptide nested between the isotope peaks of a second peptide. Some isotope peaks from different peptides share the same m/z and each contributes to the abundance of the peak. Hardklör can deconvolute two PIDs that share peaks, even when the PIDs have different charge states (Figure 7A). Additionally, Figure 7B illustrates Hardklör’s deconvolution of three PIDs over a narrow 4 m/z range with nested isotope peaks, shared isotope peaks, and charge states of +2 and +3. Hardklör’s accurate deconvolution of complex spectra allows the mining of rich samples instead of constraining analysis to simplified mixtures and only the most abundant peptides.
Figure 7.
Identification of overlapping peptide isotope distributions. (A) Deconvolution of two peptide isotope distributions of different charge states that share two peaks. (B) Deconvolution of three peptide isotope distributions over a 4 m/z spectral segment. Peptides were identified with different charge states, overlapping distributions (peptides #1 and #3), and shared peaks between distributions (peptides #2 and #3).
Because Hardklör can handle the deconvolution of multiple overlapping PIDs, it can be used to evaluate the frequency at which an MS/MS spectrum contains multiple molecular species. Database searching algorithms generally assume that each MS/MS spectrum contains only a single component and will likely fail to provide an adequate interpretation for a spectrum with more than one component at similar signal intensities. The MS/MS spectra in this LTQ-Orbitrap dataset were obtained using data-dependent acquisition by isolating and activating a 3 m/z isolation window around the selected precursor ion. The 3 m/z isolation widths of the preceding scans of the MS/MS spectra were compared to the Hardklör results. A total of 9,567 MS/MS spectra were acquired over the 2 hour analysis, of which there were 1,530 unique MS/MS peptide spectrum matches (PSMs) when searched with SEQUEST and filtered using DTASelect (FDR < 1.0%; assessed using a shuffled decoy database). These 1,530 PSMs were mapped to 278 non-redundant proteins. Hardklör identified a single PID in the 3 m/z window of the preceding Orbitrap survey scan for 1,086 (71.0%) of the PSMs. However, a surprisingly large number of the MS/MS spectra that were successfully mapped to peptide sequences contained two and three PIDs: 144 (9.4%) and 15 (1.0%), respectively. A likely explanation for why SEQUEST was still able to find a PSM even for MS/MS spectra containing multiple molecular species is that one peptide provided the most dominant signal. Interestingly, Hardklör did not find any PIDs in the Orbitrap data from which 285 (18.6%) of the PSMs were obtained. The relatively large number of missing PIDs cannot be explained solely by false positive peptide identifications by SEQUEST, because the FDR was estimated to be less than 1% using a shuffled decoy database. Instead, the lack of an apparent PID was often because the intensity of the PID was insufficient to score well with Hardklör. While an MS/MS spectrum can be triggered from a single spurious signal in the spectrum, Hardklör requires an entire isotope distribution of sufficient S/N and accuracy to obtain a PID.
The analysis of the Hardklör results can also be expanded to include all 9,567 MS/MS spectra. From these data, the number of MS/MS spectra that Hardklör found zero, one, two, and three PIDs in the 3 m/z isolation widow from the Orbitrap data was 2,805 (29.3%), 5,693 (59.5%), 947 (9.9%) and 122 (1.3%), respectively. When a SEQUEST search was performed on only the MS/MS spectra with zero or one Hardklör PID and filtered using DTASelect, 1,387 unique PSMs were obtained (FDR < 1.0%; assessed using a shuffled decoy database). These 1,387 PSMs were mapped to 263 non-redundant proteins. While only a fraction of the acquired MS/MS spectra were ultimately matched to peptide sequences, elimination of the 11.2% of the total spectra containing multiple species that cannot be handled appropriately by database search algorithms still yielded 94.6% of the protein identifications made when using the entire datafile.
The number of Hardklör persistent PIDs matching the database search results was similar regardless of the approach used to derive the charge-state (Table 1). The QuickCharge algorithm performed competitively with alternative charge state determination methods at score thresholds greater than 0.90. The 1,530 unique MS/MS PSMs were compared to the list of monoisotopic masses and charge states of the Hardklör results obtained from the high resolution Orbitrap scans. If a result from both lists had the same charge state, the same monoisotopic mass within 10 ppm, and the same approximate retention time, then the two results were considered a match. Thus, the 1,200 chromatographically persistent PIDs that matched MS/MS identifications using SEQUEST accounted for 78.4% of the total peptides identified. While at first glance the fraction of the Hardklör-detected PIDs that matched SEQUEST results (8.8%) might seem surprisingly low, this value is similar to the 5–10% value usually reported for the fraction of total MS/MS spectra that are identified by database searching.
At the resolution that these experiments were performed, the Orbitrap mass spectrometer was able to resolve PIDs with charge states greater than +3. Thus, the analysis was performed a second time using the QuickCharge algorithm on charge states from +1 to +5 with a score threshold of 0.90. Hardklör identified 219,941 PIDs, 14,070 persistent PIDs, and 1,202 matches to the SEQUEST results. In total, 501 and 130 of the total persistent PIDs were identified as charge states +4 and +5 respectively. The two additional matches to the SEQUEST results come from better deconvolution of overlapping PIDs at +4 or +5 charge states with those of +1, +2, or +3. Addition of the two extra charge states to the Hardklör analysis only added one minute to the computation time when using the QuickCharge algorithm.
To assess our ability to identify peptides containing unusual isotope distributions, we developed a method to specifically tag asparagine residues containing an N-linked glycosylation with a non-endogenous mixture of 16O and 18O. Glycosylated asparagine residues of the S. cerevisiae protein invertase were labeled with 16O:18O mix using the enzyme PNGase-F. PNGase-F catalytically cleaves the N-acetylglucosamine-asparagine amide bond, converting the asparagine to aspartic acid through the incorporation of oxygen from water in the solution. Invertase was digested with PNGase-F in 1:1 mixture of unlabeled water and H218O (50% APE) to label the sites of glycosylation. The isotope distributions were then measured using zoom-scans acquired over a 10 m/z window using data-dependent acquisition on an LTQ ion trap mass spectrometer.
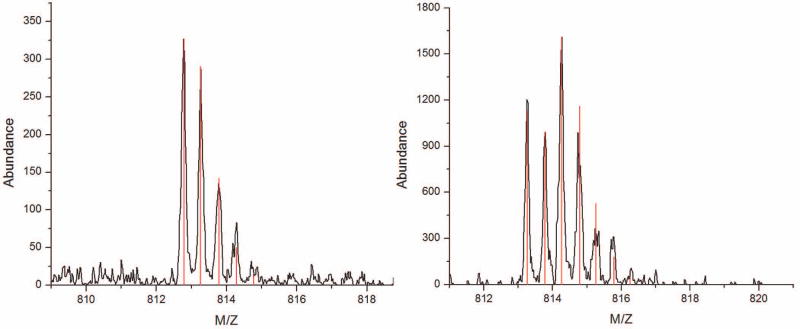
If a peptide contained an N-linked glycosylation site, then the carboxylic acid of the resulting aspartic acid was labeled with a mixture of 18O-enriched and natural abundance oxygen at a ratio of approximately 1:1. The mixture of 16O:18O labeling created a unique shape to the peptide distribution that can be predicted by the summation of the 18O-labeled and unlabeled distributions weighted by the APE (Figure 8). Peptide isotope distributions containing the unique shape were identified from the zoom-scan mass spectra using Hardklör and the correct site of glycosylation was validated from the MS/MS spectra.
Figure 8.
Example of isotope labeling a glycosylated peptide. (A) Isotope distribution for the non-glycosylated form of the invertase peptide, AEPILNISNAGPWSR. (B) Isotope distribution of the glycosylated form of the same peptide after digestion with PNGase-F in the presence of H218O at 50% APE. The monoisotopic mass (first peak) is slightly heavier because of the conversion of an asparagine to aspartic acid. The distinctive pattern of peak heights is a result of a single 18O present in 50% of the peptide molecules.
Seventeen unique invertase peptides were identified from the MS/MS spectra, of which four of these PSMs should contain sites of glycosylation in agreement with previous studies22. Analysis of the peptide isotope distributions with Hardklör correctly assigned the monoisotopic mass, charge state, and presence or absence of 18O-labeling to 16 of the invertase peptides. The single exception was a case where Hardklör made no determination because the isotope distribution within the respective zoom-scan spectrum was of insufficient signal-to-noise. Hardklör correctly identified 18O-labeled PIDs for the four peptides containing sites of glycosylation. One peptide (AEPILNISNAGPWSR) was identified from the MS/MS spectra to have both glycosylated and non-glycosylated forms. The unglycosylated peptide contained an asparagine residue with natural abundance isotopes, whereas the glycosylated peptide was present as an aspartic acid (after PNGase-F treatment) with a 50% enrichment of a single 18O atom (Figure 8). Thus, Hardklör can discriminate between labeled and unlabeled peptides using a simple and efficient enzymatic labeling strategy, even when the modified peptide is present at low stoichiometry.
DISCUSSION
Unique Aspects of Hardklör for the Detection of Peptide Isotope Distributions
The combinatorial approach for the analysis of PIDs used by Hardklör provides a powerful method for identifying multiple overlapping peptide distributions, including those of mixed charged states. This combinatorial approach has a distinct advantage over alternative methods that first try to identify a single isotope distribution from the overlapping mixture, subtract the signal from that predicted distribution, and then proceed by identifying additional distributions. In using a subtractive method, an incorrect PID prediction at the beginning can result in a cascade of erroneous subsequent PID predictions, because the incorrect signal will be subtracted from the combination of isotope distributions. The downside of a combinatorial approach is that it can create a computational bottleneck because the number of combinations to analyze (C) is represented by the following equation:
where v is the number of variations in the predicted distributions and p is the number of predicted distributions to analyze. The number of combinations to be analyzed exponentially increases with the number of variations to compute for each of the predicted distributions. For example, if we have five predicted distributions to analyze using the Complete method, each with three charge state variants (+1, +2, or +3), the total number of combinations to analyze all possibilities is a reasonable 1,023. However, if the number of predicted distributions were doubled to 10, the number of combinations explodes to 1,048,575. A simple solution is to limit the depth of combinations, i.e. do not consider combinations of more than three predicted distributions. This bottleneck can be further minimized by reducing the number of variations and predicted distributions in the combinatorial set. The QuickCharge algorithm with Hardklör is a simple single-pass, inverse distance computation of charge states that dramatically reduces the complexity of the combinatorial set by reducing the number of charge state variations, and thus considerably improves the speed of analysis. In some cases the number of combinatorial distributions considered is reduced if, for any predicted distribution, the QuickCharge algorithm can eliminate all potential charge states. While the QuickCharge algorithm is simplistic, it performs well when compared to other common charge state algorithms in correctly identifying PIDs, and dramatically outperforms them in terms of computational speed. Because of this improvement in speed, Hardklör is capable of analyzing shotgun proteomics data from an LTQ-Orbitrap mass spectrometer in significantly less time than it takes to acquire the data and using only a single processor with humble specifications.
A novel feature of Hardklör’s isotope distribution analysis is the ability to identify unusual isotope distributions. Hardklör is able to differentiate PIDs that include uncommon atoms or isotopic enrichments that deviate from a standard averagine model. We have demonstrated identification of 18O-label peptides, but Hardklör’s differential analysis can be extended to include other atoms, molecules, or levels of enrichment. This capability makes Hardklör a powerful tool for molecular profiling when combined with a specific labeling approach to target specific classes of molecules.
Application to Peptide Identification and Comparative Experiments
One of the primary uses of Hardklör in our laboratory is to make comparisons between experiments. These comparisons can either be qualitative comparisons to previous runs where the PIDs have been identified using tandem mass spectrometry or quantitative comparisons in PID abundance. The qualitative aspects of this application bear a strong resemblance to AMT tag experiments. By using normalized chromatographic elution times combined with the detection of high resolution and accurate monoisotopic mass measurements, peptides can be identified from a database of previously identified isotope distributions23;24. The dynamic range of intensities of PIDs that we are able to detect with Hardklör using a one-dimensional 2 hour analysis on an LTQ-Orbitrap is greater than that reported previously for the detection of peptides using data-dependent MS/MS with a multidimensional separation of ~24 hours2;19. By combining Hardklör with high resolution mass spectrometry data, which improves the overall peak capacity in the μLC-MS analysis, peptides can be identified using high speed separations that either make MS/MS acquisition impractical or the characterization of the MS/MS spectra impossible because of the overlapping PIDs. Once a database of identified persistent PIDs has been acquired using extensive fractionation and data-dependent acquisition of tandem mass spectra, future analyses can be used to characterize the unfractionated mixture using only normalized elution time and accurate mass from the deconvolution of multiple overlapping PIDs.
As mentioned above, Hardklör also facilitates quantitative comparisons between multiple different samples. In this case, PIDs are not being mapped to a database of previously identified features, but are used to compare the intensities between analyses. Because of the high mass measurement accuracy of Fourier transform mass spectrometers, even PIDs that deviate somewhat in retention time can be associated with persistent PIDs between different samples by only imposing crude restrictions for similarity in retention time. Thus, we believe that because of Hardklör’s speed and ability to handle overlapping mixtures of high complexity, it will become an essential tool in increasing the overall throughput of comparative proteomics measurements.
Use of Hardklör for Improving MS/MS Identifications
Hardklör can also be used to improve MS/MS PSMs. One method of estimating the false discovery rate of PSMs is to perform the search on a database containing both real protein sequences and random amino acid sequences, known as decoys25;26. Because false PSMs are expected to hit the decoys at the same frequency as real peptides, parameters can be adjusted to minimize decoy hits and thus improve the confidence of peptide identification; however, using very stringent parameters can also increase elimination of true PSMs — i.e. false negatives. Other labs have reported the use of high mass measurement accuracy of the peptide precursor ion to minimize false positives while maintaining a large percentage of true positive peptide identifications27;28. An under-appreciated step in this process is the accurate determination of the monoisotopic mass of PIDs from large molecular weight peptides. At a molecular weight above approximately 2,000 Da, the monoisotopic mass will no longer be the most intense isotope peak. Hardklör can accurately identify the monoisotopic mass of PIDs to within the mass accuracy of the mass spectrometer. Thus, by eliminating peptide identifications with monoisotopic mass outside the Orbitrap mass measurement accuracy, it is possible to validate PSMs and eliminate false positives without having to resort to overly stringent database filtering parameters.
High resolution mass spectrometers enable increased throughput relative to low resolution instruments because a loss in peak capacity of the chromatographic separation can be made up by the improved resolution of the mass analyzer. However, assuming the MS/MS precursor isolation window remains unchanged, the compressed chromatographic separation increases the number of MS/MS spectra containing fragment ions from multiple precursor ions. We have shown that in the analysis of the E. coli fraction presented here, greater than 12% of the acquired MS/MS spectra contained more than one molecular species. Because Hardklör is able to identify overlapping PIDs, we are able to flag MS/MS spectra containing multiple components prior to database searching. By flagging MS/MS spectra prior to database searching, these spectra can either be triaged to minimize computational resources spent on spectra that are unlikely to provide a PSM or redirected to software specifically designed to interpret fragmentation spectra containing multiple components29.
Hardklör’s capability to identify unusual PIDs can be used to improve database searching efficiency. The most common approach to handling MS/MS spectra derived from peptides containing modified amino acid residues is to perform differential modification searches30. A single differential modification on an amino acid requires the algorithm to consider that amino acid with and without a change in mass every time it is encountered in the database. Thus each modification adds an exponential increase in search time on an already computationally expensive process.
When combined with a strategy to specifically tag peptides to create a unique isotope distribution, Hardklör can separate MS/MS spectra into distinct groups so that customized differential modification searches can be performed on only a small number of spectra. We have shown how Hardklör can be used to identify peptides that contain N-linked glycosylation, but this approach can be expanded to the analysis of any chemical moiety that can be selectively tagged to produce a unique isotope signature. An excellent example is the selective alkylation of cysteine residues with an isotope distribution encoded tag (IDEnT) to constrain search parameters in the identification of peptides10. While IDEnT labeling is a powerful tool, its widespread adoption has been hindered by the lack of available software to automatically detect the peptides containing cysteine tagged residues. Hardklör can be used to automatically identify residue-specific chemical modifications and is not limited to IDEnT labeling. Additionally, Hardklör can identify peptides containing endogenous post-translational modifications such as chloro- and bromo-tyrosine31;32 (data not shown). Thus, the availability of Hardklör creates a flexible platform for scientists to design experiments to specifically target peptides containing modifications (either in vivo or in vitro) of interest.
Application to Real-Time “Intelligent” Data-Dependent Acquisition
The current approach to data-dependent acquisition is to select precursor ions from the most intense signal and to work down in intensity. This approach limits MS/MS spectra acquisition to the most intense peptides and peptides of low intensity in the context of much more intense peptides may elute from the column before they are ever selected for MS/MS. As a result, many interesting classes of molecules may be ignored.
The speed of Hardklör should facilitate real-time calculations during the acquisition of mass spectrometry data. Instead of selecting only the most intense precursors for MS/MS spectra, Hardklör could be used for data-dependent selection or elimination of PIDs with specific features. For example, Hardklör could be used to direct MS/MS spectra acquisition to regions that contain only a single PID and to eliminate regions with overlapping isotope distributions. Because most co-eluting peptides have some small variation in their elution times, Hardklör could be used to acquire MS/MS spectra at points where the two precursor ions do not overlap, potentially leading to two PSMs instead of only one or none. Furthermore, Hardklör could be used to target the acquisition of MS/MS spectra of peptides containing a uniquely labeled isotope distribution as described above. Integral to the development of Hardklör has been the provision of a modular and flexible platform for the analysis of high resolution shotgun proteomics data. This flexibility makes possible the integration of Hardklör with other software, including instrument data-systems, with only minor modifications.
CONCLUSION
The Hardklör algorithm is a robust tool with many applications in the analysis of high resolution mass spectra. We have demonstrated the algorithm’s ability to rapidly detect the monoisotopic mass of peptides in the context of a very complex mixture. Using a combinatorial approach, we demonstrate the deconvolution of overlapping PIDs. Although our combinatorial approach to deconvolution trades the speed of subtractive methods for a more robust analysis, the use of the relatively simple QuickCharge method to reduce the number of charge state combinations improves the overall speed of the algorithm almost 100-fold over previously reported approaches. We have demonstrated the identification of peptides containing an unusual isotope distribution using high resolution scans on a benchtop ion-trap mass spectrometer. While the identification of unusual isotope distributions was demonstrated by enzymatic cleavage of N-linked glycosylation in the presence of 18O-enriched water, Hardklör can be used to detect virtually any unique isotope distribution feature. We also demonstrated Hardklör’s use in determining whether MS/MS spectra were acquired from an m/z window that contained multiple PIDs. Using these data can provide a measure of the quality of the product ion scans because algorithms used to interpret these spectra assume one spectrum equals one peptide sequence. These capabilities make Hardklör a useful tool for the analysis of μLC-MS data from high resolution mass spectrometers.
Acknowledgments
The authors would like to acknowledge Adele Blackler and Christine Wu from the University of Colorado Health Sciences Center for preparing the E. coli membrane enriched fraction. We are indebted to ThermoFisher Scientific for providing time on the LTQ-Orbitrap to perform these experiments. We also acknowledge Xianhua Yi, Barbara Frewen, Gennifer Merrihew and Aaron Klammer for aiding in the experiments and for critical reading of the manuscript. The authors would like to thank Alan Rockwood for providing source code for the Mercury isotope distribution algorithm. Financial support for this work was provided in part from National Institutes of Health grants R01-DK069386 and P41-RR011823 and an unrestricted research grant from Merck Research Laboratories. MRH and GLF gratefully acknowledge training grant support from T32-HG00035.
References
- 1.Link AJ, Eng JK, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Nat Biotechnol. 1999;17:676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 2.Washburn MP, Wolters D, Yates JR., III Nat Biotechnol. 2001;19:242–47. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 3.Syka JE, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. J Proteome Res. 2004;3:621–26. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 4.Hardman M, Makarov AA. Anal Chem. 2003;75:1699–705. doi: 10.1021/ac0258047. [DOI] [PubMed] [Google Scholar]
- 5.Conrads TP, Anderson GA, Veenstra TD, Pasa-Tolic L, Smith RD. Anal Chem. 2000;72:3349–54. doi: 10.1021/ac0002386. [DOI] [PubMed] [Google Scholar]
- 6.Strittmatter EF, Ferguson PL, Tang K, Smith RD. J Am Soc Mass Spectrom. 2003;14:980–91. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 7.Kaur P, O'Connor PB. J Am Soc Mass Spectrom. 2006;17:459–68. doi: 10.1016/j.jasms.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Du P, Angeletti RH. Anal Chem. 2006;78:3385–92. doi: 10.1021/ac052212q. [DOI] [PubMed] [Google Scholar]
- 9.Horn DM, Zubarev RA, McLafferty FW. J Am Soc Mass Spectrom. 2000;11:320–32. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 10.Goodlett DR, Bruce JE, Anderson GA, Rist B, Pasa-Tolic L, Fiehn O, Smith RD, Aebersold R. Anal Chem. 2000;72:1112–18. doi: 10.1021/ac9913210. [DOI] [PubMed] [Google Scholar]
- 11.MacCoss MJ, Wu CC, Matthews DE, Yates JR., III Anal Chem. 2005;77:7646–53. doi: 10.1021/ac0508393. [DOI] [PubMed] [Google Scholar]
- 12.Senko MW, Beu SC, McLafferty FW. J Am Soc Mass Spectrom. 1995;6:229–33. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 13.Klammer AA, MacCoss MJ. J Proteome Res. 2006;5:695–700. doi: 10.1021/pr050315j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eng JK, McCormack AL, Yates JR., III J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 15.Tabb DL, McDonald WH, Yates JR., III J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., III Rapid Commun Mass Spectrom. 2004;18:2162–68. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 17.Rockwood AL, Vanorden SL, Smith RD. Anal Chem. 1995;67:2699–704. [Google Scholar]
- 18.Alfassi ZB, Boger Z, Ronen Y. Statistical Treatment of Analytical Data. CRC Press; Boca Raton: 2005. [Google Scholar]
- 19.Wolters DA, Washburn MP, Yates JR., III Anal Chem. 2001;73:5683–90. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 20.Senko MW, Beu SC, McLafferty FW. J Am Soc Mass Spectrom. 1995;6:52–56. doi: 10.1016/1044-0305(94)00091-D. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Hines W, Adamec J, Asara JM, Naylor S, Regnier FE. J Am Soc Mass Spectrom. 2005;16:1181–91. doi: 10.1016/j.jasms.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Reddy VA, Johnson RS, Biemann K, Williams RS, Ziegler FD, Trimble RB, Maley F. J Biol Chem. 1988;263:6978–85. [PubMed] [Google Scholar]
- 23.Pasa-Tolic L, Masselon C, Barry RC, Shen Y, Smith RD. Biotechniques. 2004;37:621–33. 636. doi: 10.2144/04374RV01. [DOI] [PubMed] [Google Scholar]
- 24.Strittmatter EF, Ferguson PL, Tang K, Smith RD. J Am Soc Mass Spectrom. 2003;14:980–91. doi: 10.1016/S1044-0305(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 25.Moore RE, Young MK, Lee TD. J Am Soc Mass Spectrom. 2002;13:378–86. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 26.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 27.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. Nat Biotechnol. 2006;24:1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 28.Pilch B, Mann M. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang N, Li XJ, Ye M, Pan S, Schwikowski B, Aebersold R. Proteomics. 2005;5:4096–106. doi: 10.1002/pmic.200401260. [DOI] [PubMed] [Google Scholar]
- 30.Yates JR, III, Eng JK, McCormack AL, Schieltz DM. Anal Chem. 1995;67:1426–36. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 31.Bergt C, Fu X, Huq NP, Kao J, Heinecke JW. J Biol Chem. 2004;279:7856–66. doi: 10.1074/jbc.M309046200. [DOI] [PubMed] [Google Scholar]
- 32.Gaut JP, Yeh GC, Tran HD, Byun J, Henderson JP, Richter GM, Brennan ML, Lusis AJ, Belaaouaj A, Hotchkiss RS, Heinecke JW. Proc Natl Acad Sci USA. 2001;98:11961–66. doi: 10.1073/pnas.211190298. [DOI] [PMC free article] [PubMed] [Google Scholar]