Abstract
We have recently observed promising success in a mouse model at treating the metabolic disorder phenylketonuria (PKU) with phenylalanine ammonia lyase (PAL) from R. toruloides and A. variabilis. Both molecules, however, required further optimization in order to overcome problems with protease susceptibility, thermal stability and aggregation. We reduced aggregation of the A. variabilis PAL by mutating two surface cysteine residues (C503 and C565) to serines. Here we report the structural and biochemical characterization of the A. variabilis PAL C503S/C565S double mutant. Unlike previously published PAL structures, significant electron density is observed for the two active site loops in the A. variabilis C503S/C565S double mutant, yielding a complete view of the active site. Docking studies and NHS-biotin binding studies support a proposed mechanism in which the amino group of the phenylalanine substrate is attacked directly by the 4-methylidene-imidazole-5-one (MIO) prosthetic group. We propose a helix-to-loop conformational switch in the helices flanking the inner active site loop that regulates accessibility of the active site. Differences in loop stability among PAL homologs may explain the observed variation in enzyme efficiency despite the highly conserved structure of the active site. A. variabilis C503S/C565S PAL is shown to be both more thermally stable and more resistant to proteolytic cleavage than R. toruloides PAL. Additional increases in thermal stability and protease resistance upon ligand binding may be due to enhanced interactions among the residues of the active site, possibly locking the active site structure in place and stabilizing the tetramer. Examination of the A. variabilis C503S/C565S PAL structure combined with analysis of its physical properties provides a structural basis for further engineering of residues that could result in a better therapeutic molecule.
Keywords: phenylketonuria, phenylalanine ammonia lyase, structure, stability, activity
Introduction
Phenylketonuria (PKU, OMIM 261600) is an inherited metabolic disorder where loss or impairment of phenylalanine hydroxylase (PAH, EC 1.14.16.1), an enzyme that metabolizes phenylalanine (L-Phe), leads to a buildup of L-Phe in the body. Excess L-Phe is damaging to tissues and can impair cognitive development, resulting in irreversible intellectual and developmental disabilities.1 Phenylalanine ammonia lyase (PAL, EC 4.3.1.5) is an enzyme that catalyzes the conversion of L-Phe to trans-cinnamic acid (tCA) and ammonia. Deamination of L-Phe is dependent upon a 4-methylidene-imidazole-5-one (MIO) prosthetic group that is produced by the post-translational condensation of a sequential Ala-Ser-Gly triad in the enzyme. The ability of PAL to catalyze the conversion of L-Phe into non-toxic compounds (at the levels generated) in the absence of additional cofactors has led to investigations into its use as a therapeutic agent for the treatment of PKU.2; 3
In rodent models of PKU, PAL has been shown to lower blood L-Phe levels.2; 4; 5 However the pharmacokinetic properties of the enzyme render it unsuitable for therapeutic purposes - it is rapidly cleared from the system through its susceptibility to proteases and it is immunogenic. We have already shown that we can increase in vivo residence times by the covalent addition of polyethylene glycol (PEG) to the surface of the Rhodosporidium toruloides PAL.3; 6 PEGylation of the enzyme presumably increases the in vivo protein half-life by restricting access to epitopes and protease sites. In the PKU mouse model, PEGylated R. toruloides PAL displays lower immunogenicity, higher efficacy and a longer half-life than the unPEGylated enzyme.3; 6
In our continuing search for a more therapeutically effective form of PAL – high catalytic activity and long systemic half-life – we have screened wild-type and mutant PAL proteins from a variety of organisms using the PKU mouse model (manuscript in preparation). We find that in long-term in vivo dosing studies, subcutaneously delivered PEGylated forms of PAL derived from the bacteria Anabaena variabilis display an even higher efficacy and a slower clearance rate than PEGylated R. toruloides PAL. The A. variabilis PAL used in these studies was a C503S/C565S double mutant we designed to eliminate the problems with aggregation displayed by the wild-type A. variabilis protein.7
In order to better understand the differences in the pharmacokinetic behavior of the A. variabilis and R. toruloides PAL enzymes, and to increase our general knowledge of how structure-function relationships affect pharmacokinetic behavior, we determined the crystal structure of the A. variabilis C503S/C565S PAL mutant and analyzed in vitro its kinetic properties as well as its protease and thermal stabilities. We compared these results to the existing data on wild-type R. toruloides PAL and wild-type A. variabilis PAL. Here we present the first structure of a PAL enzyme from any organism where the active site is fully ordered. Previous PAL structures, including the structure of the wild-type A. variabilis PAL,7 contain an active site with two disordered loop regions.3; 7; 8 In our structure, these two regions are ordered, and their structure provides insight into the enzyme’s catalytic mechanism. We believe that the C503S/C565S mutations do not negatively perturb or impair the structure of the active site, as our kinetic measurements show that the C503S/C565S mutations do not significantly alter the activity of the enzyme.
Our analysis of the active site structure provides new insights into the catalytic mechanism of PAL and furthers our understanding of the architecture of proteins in the PAL/histidine ammonia lyase (HAL) enzyme family. Our ability to rationally alter the aggregation properties of a protein, and to compare stability, structure and function across a broad protein sequence space, provides a foundation for a protein engineering program for the design and construction of an effective protein-based treatment for PKU. This work has already enabled us to embark on the first stages of therapeutic development – the execution of animal studies with the potential for moving on to clinical trials using the A. variabilis PAL C503S/C565S mutant – and points the way to future engineering that can be done to further optimize catalytic activity and in vivo enzyme stability.
Results and Discussion
The C503S/C565S double mutation reduces aggregation of A. variabilis PAL
Wild-type A. variabilis PAL has been shown to aggregate upon purification.7 SDS-PAGE of the resulting aggregate in the presence and absence of reducing agent led Mofitt, et al. 7 to suggest that free sulfhydryls may be involved in the aggregation process. In order to identify surface cysteines that may contribute to A. variabilis PAL aggregation, we treated wild-type A. variabilis PAL with N-ethylmaleimide (NEM), an alkylating agent that reacts with the sulfhydryl group of free cysteines. Proteolysis of the modified PAL followed by LC/MS analysis identified residues C503 and C565 as potential surface cysteines. Mutation of both these residues to serine (C503S/C565S) resulted in a protein with reduced aggregation properties as measured by native and denaturing SDS-PAGE as well as HPLC (data not shown). Unlike wild-type A. variabilis PAL, in the absence of reducing agent, the C503S/C565S double mutant migrates as a single, monomer-sized band via SDS-PAGE.
The C503S/C565S double mutation does not alter kinetic behavior of the enzyme
We measured the kinetic properties of the R. toruloides wild-type PAL, the A. variabilis wild-type PAL, and the A. variabilis C503S/C565S PAL double mutant (Table 1). Our measurements for the two wild-type proteins are consistent with previously published measurements of Km and kcat.7; 9; 10 The similarity in the kinetic properties of the wild-type A. variabilis PAL and the C503S/C565S double mutant demonstrate that these stabilizing mutations do not affect the catalytic behavior of the molecule. Thus, we were successful in our attempt to engineer a non-aggregating version of A. variabilis PAL.
Table 1.
Comparison of activity and kinetic constants of wild-type R. toruloides PAL, wild-type A. variabilis PAL and A. variabilis PAL double mutant
| Specific activity (IU/mg) | Km (mM) | kcat (s−1) | kcat/Km (mM−1s−1) | |
|---|---|---|---|---|
| wt R. toruloides PAL | 4.5 | 1.1 | 16 | 15 |
| wt A. variabilis PAL 7 | 1.7 | 0.06 | 4.6 | 77 |
| A. variabilis PAL double mutant | 2.2 | 0.05 | 4.0 | 80 |
pH-dependent activity profile of both A. variabilis C503S/C565S PAL and R. toruloides PAL indicate suitability for therapeutic development
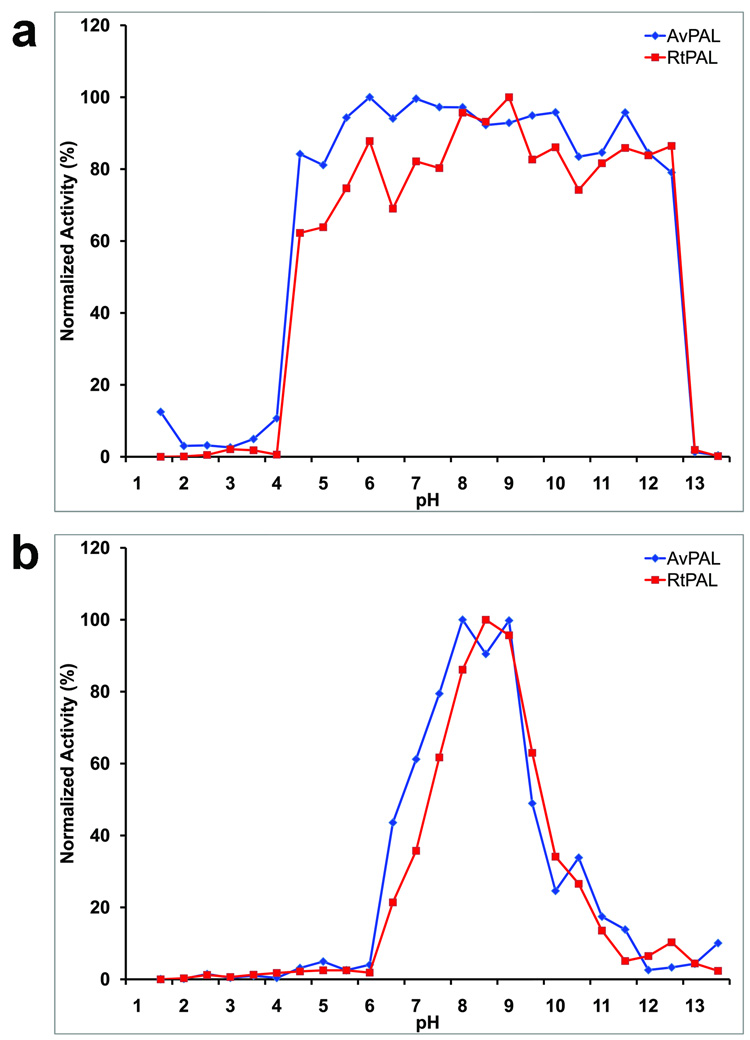
Under standard enzymatic assay condition, the pH-dependent activity profile for the A. variabilis PAL double mutant shows an optimal pH range between 7.5 and 8.5 (Figure 1); R. toruloides PAL has a comparable pH-dependent activity profile, with an optimal pH of 8.0. pH stability studies show that both A. variabilis C503S/C565S PAL and R. toruloides PAL retain their activity after a ten minute exposure to environments in the pH range of 4 to 12 (Figure 1), indicating that they are both stable within a broad pH range. We have found that levels of activity after a ten minute exposure to different pHs is a good indicator for overall pH tolerance of the enzyme. The broad pH range tolerance of A. variabilis C503S/C565S PAL and R. toruloides PAL suggests that these two enzymes could survive the conditions present in the gastro-intestinal tract immediately after a meal and still retain activity. Although the pH in the stomach in the fasting state is ~1.8, it rises to a range of 5.4–6.2 immediately after a meal.11 In addition, recent successful attempts at encapsulating Rhodotorula glutinis PAL demonstrated retention of significant activity12, thus providing an approach to help shield the enzyme from the harsh environment in the stomach. These results, therefore, open the possibility of using A. variabilis C503S/C565S PAL for oral administration in PKU treatment.
Figure 1.
Activity profiles of A. variabilis PAL double mutant and R. toruloides PAL at different pH conditions. (a) pH stability of A. variabilis PAL double mutant and R. toruloides PAL. (b) Optimal pH profiles of A. variabilis PAL and R. toruloides PAL. All data are mean value from three parallel measurements.
Limited proteolysis of A. variabilis C503S/C565S and R. toruloides PAL suggest that the A. variabilis enzyme is more amenable to further engineering for greater protease resistance
We performed limited proteolysis experiments with a variety of proteases to identify candidate sites for future engineering attempts to increase protease resistance. The first two proteases in Table 2 are primarily found in the stomach and provide insight into the potential for oral administration of the protein, whereas the other two enzymes are present in the blood stream and provide insight into the potential for intravenous administration. We found that the A. variabilis C503S/C565S PAL is more resistant to proteases than R. toruloides PAL. The most accessible protease cleavage sites in both the A. variabilis PAL double mutant and R. toruloides PAL are located in the two active site loops, suggesting a feasible way to reduce their protease susceptibility by protein engineering and chemical modification. The protease resistance differences between these two PALs suggest that the A. variabilis PAL double mutant could be more tolerant to rational design than R. toruloides PAL – engineering chymotrypsin resistance into the R. toruloides PAL would require disruption of Y110, a residue that is crucial for catalysis. In contrast, the location of chymotrypsin sensitivity in the A. variabilis enzyme does not overlap with residues known to be critical for catalysis.
Table 2.
Residues shown to be susceptible to protease cleavage in PAL molecules
| Protease | A. variabilis C503S/C565S PAL | R. toruloides PAL |
|---|---|---|
| chymotrypsin | F18 | Y110 (inner loop, highly conserved in ammonia lyases and important for catalysis and binding of substrate) |
| trypsin | R94 (inner loop), R305 (outer loop) | R123 (inner loop) |
| neither are susceptible to Thrombin or FactorX | ||
Thermal stability mediated by a ligand acting as a “keystone” reveals potential targets for engineering more thermally stable therapeutic molecules
We used a fluorescence-based assay 13; 14 to measure thermal stability of the A. variabilis PAL double mutant and R. toruloides PAL, in the presence and absence of ligands. This approach makes use of binding of a hydrophobic fluorescent dye to hydrophobic patches in the protein. As the protein unfolds, more hydrophobic patches are available for dye-binding, resulting in increased fluorescence relative to a standard control. Thermal stabilities are summarized in Table 3. The observed melting temperature (Tm) of the A. variabilis PAL double mutant (78.4 °C) is about 14 °C higher than that of R. toruloides PAL (64.7 °C), indicating a higher global stability which is consistent with previous observations that shorter PAL enzymes are more stable than those with the additional insertion domain 15 (residues 541–655 in R. toruloides PAL). Pilbak et al. used results from molecular dynamics studies to hypothesize that the C-terminal extension destabilizes the enzyme and that this might be important for the rapid responses in the regulation of phenylpropanoid biosynthesis15.
Table 3.
Tm (°C) shift of the A. variabilis PAL double mutant and R. toruloides PAL
| Molecular Species and ligand bound | Tm |
|---|---|
| A. variabilis PAL | 78.4±0.5 |
| + Tyr | 79.0±0.6 |
| + Phe | 85.±0.0 |
| +tCA | 84.6±0.5 |
| R. toruloides PAL | 64.7±0.6 |
| + Tyr | 66.3±0.3 |
| + Phe | 68.1±0.3 |
| +tCA | 64.1±0.3 |
The Tm values and the standard deviation shown in this table are calculated based on four parallel experiments.
The Tm of the A. variabilis PAL double mutant shifts upward by 0.6 °C, 7.1 °C and 6.2°C after pre-incubation with its substrates L-Tyr, L-Phe, and product tCA, respectively, indicating a ligand-binding related increase in thermal stability. Similarly, the Tm of R. toruloides PAL also increases when L-Tyr, L-Phe, and product tCA are present, although the magnitude of the change is less for the R. toruloides enzyme (4 °C versus 7 °C). In contrast to the A. variabilis C503S/C565S PAL, the R. toruloides enzyme is not stabilized by tCA.
We had previously proposed that in R. toruloides PAL, the active site loops are immobilized in a stable conformational state upon L-Tyr binding leading to a reduction in conformational flexibility and an increase in stability.3 This hypothesis implied that differences in active-site loop mobility alone would be the determining factor in measured Tm differences between ligand-bound and unbound enzyme. In retrospect, this proposal does not appear to be sufficient as it suggests that the overall stability of a molecule the size of PAL is determined solely by the conformational state of the active site loops. We now hypothesize that stability of the entire active site, in which five segments from three separate chains converge around the MIO group to form the active site cavity, is an important determinant in the unfolding of the enzyme as temperature is increased. These segments consists of the active site inner loop, 74–96, and the segment 436–458 from one chain, the active site outer loop, 291–311, from a second chain, a loop consisting of residues 394–419 from a third chain, and finally the loop containing the MIO which forms the back wall of the active site. These residues participate in a network of interactions across three domains per active site. Our results appear to indicate that bound ligand provides additional stabilizing interactions in which the ligand acts as a keystone that has to be removed for unfolding and tetramer dissociation to occur. As there are four binding sites, two at each end of the molecule, it is possible that only after these interactions are disrupted will significant unfolding take place. These hypotheses will have to be tested with further experiments to verify the actual unfolding pathway and to better understand the role of the residues in the active site on stability. However, our observations indicate that protein engineering in this region of PAL may result in more thermally stable and thus more therapeutically tractable molecules.
Structure of the A. variabilis PAL double mutant
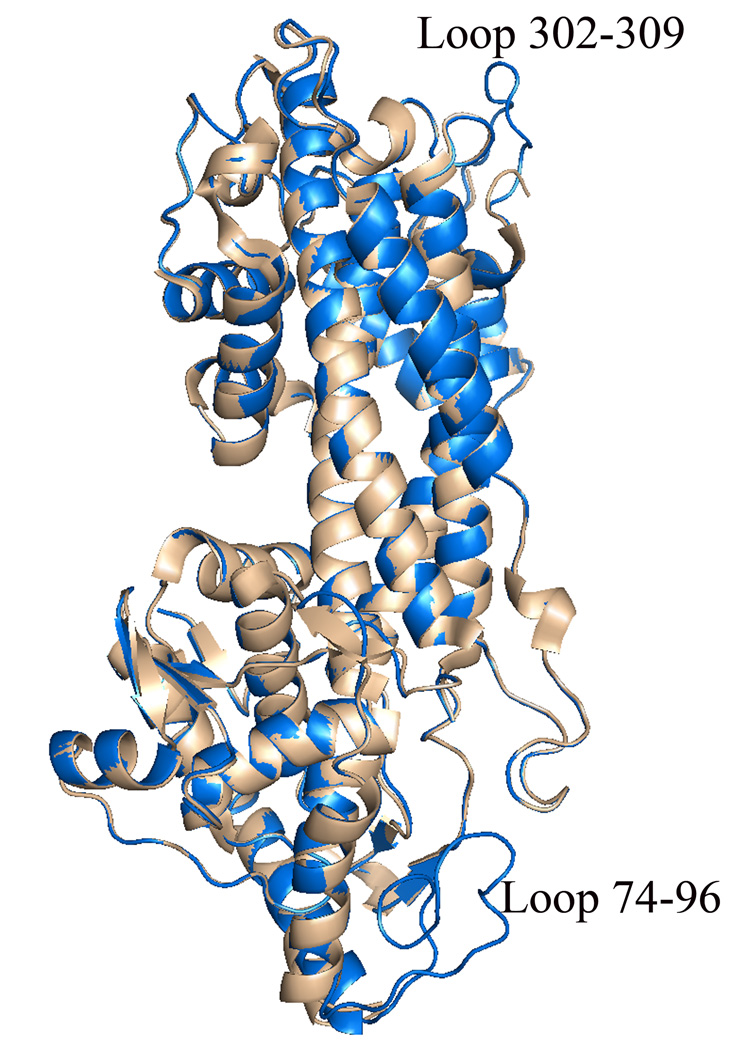
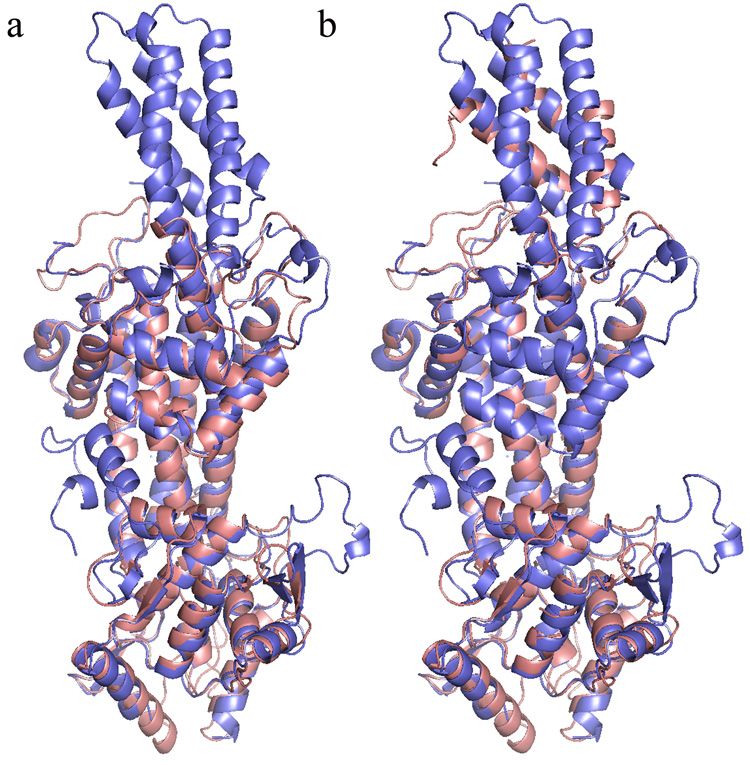
The overall structure of the A. variabilis PAL C503S/C565S double mutant is similar to the wild-type structure (Figure 2).7 Superposition of Cα coordinates of all residues in the wild-type tetramer with those in the double mutant showed an overall rmsd of 0.604 Å. Significant deviations of up to 5.8 Å are observed between residues in segments immediately adjacent to the inner active-site loop (residues 74–96) and residues 302–309 of the outer active-site loop (residues 281–314). In the crystal structure of the wild-type enzyme, these loops were observed to be disordered; in the double mutant crystal structure, significant density was observed for these loops allowing the construction of a complete model for the A. variabilis PAL active site. Thus, this study provides the first ever complete picture of the PAL active site.
Figure 2.
Superimposed monomer structures of wild-type A. variabilis PAL (light brown) and the A. variabilis PAL C503S/C563S double mutant (blue). Highlighted are the two loops that are observed in an ordered conformation only in the A. variabilis PAL double mutant structure.
The A. variabilis PAL double mutant formed crystals where the asymmetric unit (asu) contains two dimers, requiring a two-fold symmetry operation to generate a complete tetrameric enzyme. This is in contrast to the wild-type structure, where the crystallographic asu is a complete tetramer. This difference in crystal packing may play an important role in crystallizing a PAL molecule with an ordered active site. As in the wild-type structure, the first 25 residues of the double mutant are not visible.
The PAL active site
Previous structures of the PAL active site (R. toruloides,3 A. variabilis and Nostoc punctiforme,7 R. toruloides,8 Petroselinum crispum16) show an open mouthed cavity with the electrophile MIO at its back wall (Figure 3). In five of these six structures (structures from two crystal forms are reported in ref. 8), the inner and outer active site loops were disordered and in the sixth, that of parsley (P. crispum) PAL,16 the inner loop is drawn away from the cavity with Tyr110 (the equivalent of Tyr78 in A. variabilis PAL) pointing towards solvent regions. In the parsley PAL structure, a DL-dithiothreitol (DTT) molecule is covalently linked to the MIO methylidene carbon, potentially stabilizing the active site in the loop-out conformation. In the structurally and functionally similar H89F mutant of tyrosine ammonia lyase (TAL) from Rhodobacter sphaeroides, which has a shifted substrate preference for L-Phe instead of L-Tyr, these loops are also ordered.
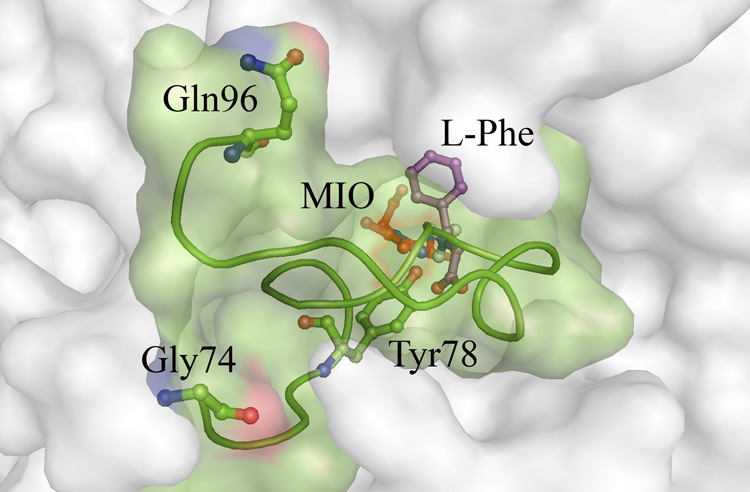
Figure 3.
Solvent accessible surface of the A. variabilis PAL double mutant around the active site showing the inner active site loop 74–96 (shown in green colored ribbon and surface representation) acting as a door to the cavity. MIO is shown in red, Tyr78 in green, and a docked Phe molecule in magenta. Analysis with CastP shows that the active site cavity in this structure does not have a solvent accessible opening.
We were able to locate the active site cavities of the PAL double mutant using the protein cavity searching program, CastP,17 showing that they are completely sequestered from solvent. Residues from the inner loop act like a lid, closing off the active site in a manner similar to the corresponding loop in the active site structures of HAL (PDB: 1B8F) and the TAL H89F mutant (PDB: 2O78). Clearly this loop must be displaced or disordered for substrate to enter and product to exit the active site cavity. Given that the length and stability of this loop varies in PAL from different organisms, it seems likely that the efficiency of opening and closing of the active site cavity, as well as the stability of the closed conformation relative to the open one will influence Km and kcat. For example, in the R. toruloides PAL structure, which is assumed to have more flexible active site loops, the Km is significantly higher than that in the A. variabilis PAL double mutant, which appears to have a less flexible active site loop. It would be interesting to pursue further studies to investigate if there is indeed a correlation between Km and loop flexibility. A similar search should be made for the case of kcat to see if a higher kcat indicates higher degree of flexibility in these loops.
Superposition of the Cα atoms from the various PAL homolog tetramers to the structure of the A. variabilis PAL double mutant provides a clear picture of a highly conserved active site
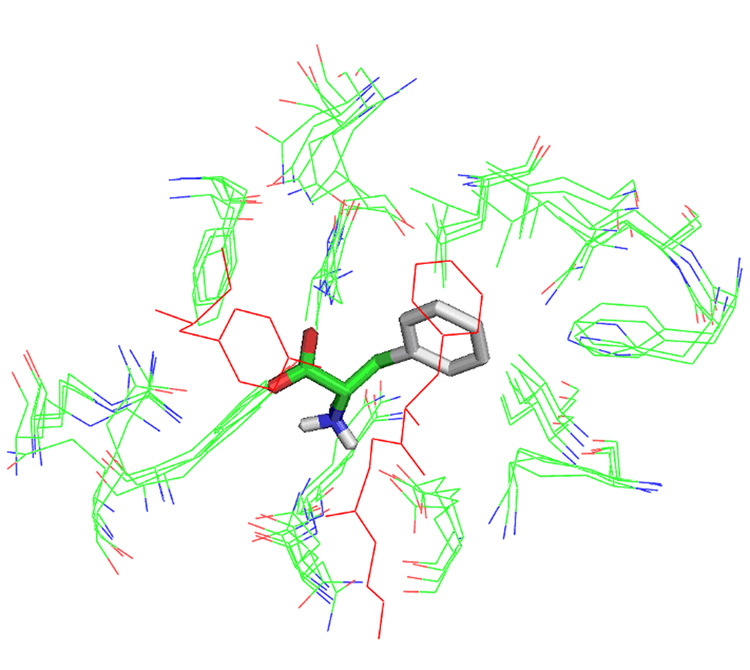
We used AutoDock18 to dock L-Phe into the A. variabilis double mutant PAL structure. The docked L-Phe molecule was used as a center to view the active sites from the different PAL homologs after structural alignment, and we identified residues from all these crystal structure that have atoms within 5.0 Å of atoms of the docked L-Phe (Figure 4). Equivalent residues in the cavity localize quite closely to each other, with their side chains pointing in the same direction. Exceptions include Tyr78 and Phe84 in the plant PAL structure (PDB: 1W27) which are part of the inner loop and which point away from the cavity. The location of equivalent residues from all active sites examined are close enough to each other that they could be considered as a distribution of allowable conformations in solution as normally observed in NMR structure determination studies. Also noteworthy is the location of His137 of R. toruloides PAL (PDB: 1Y2M, 1T6J, 1T6P) which is coincident with Phe107 of A. variabilis PAL, as well as the H89F mutation in TAL. These appear to be involved in an edge-on interaction with the docked L-Phe substrate ring. Leu108 in A. variabilis PAL, which corresponds to Gln138 in R. toruloides PAL and a leucine in the other PAL structures, appears to be in a good position to interact with the ring of the docked L-Phe in a charge-pi type of interaction. It would be interesting to see if activity is retained or improved if further modification in the TAL H89F mutant is carried out in which the TAL Leu90 is replaced with glutamine, thus producing an environment similar to R. toruloides PAL.
Figure 4.
Superposition of residues in published PAL active sites (PDB code 1Y2M, 1W27, 1T6J, 1T6P, 2NYN and 2NYF) with atoms within 5.0 Å of a Phe substrate molecule docked using AutoDock shows a highly conserved active site. Residues shown in red are from the active site loop 74–96 of the A. variabilis PAL double mutant.
Equivalent atoms in residues within the inner loop of the A. variabilis PAL double mutant and the TAL H89F mutant, the only two views of the loop for an enzyme with significant PAL activity, superimpose quite well with each other. In the H89F TAL structure, the tyrosine equivalent to Tyr314 in A. variabilis C503S/C565S PAL is offset by less than 1.0 Å and with its hydroxyl group within interacting distance of the amine group of the docked Phe substrate. The aromatic rings in corresponding tyrosines in the other ammonia lyase structures are co-planar with the A. variabilis C503S/C565S PAL. The hydroxyl groups of these tyrosine residues are also within 3.0 Å of the methylidene carbon of the MIO. Finally, all the MIO groups superimpose with their methylidene group pointing towards the Cβ of the L-Phe substrate. Clearly, the active site of ammonia lyases contains high structural conservation. This suggests that kinetics parameters such as Km and kcat (Table 4) are not solely determined by the structure or chemistry of the residues lining the active site.
Table 4.
Crystallographic statistics for the A. variabilis PAL (C503S/C564S) double mutant
| Data collection | |
|---|---|
| Wavelength (Å) | 0.9183 |
| Resolution range (Å) | 134.84-1.87 |
| No. of observations | 421,119 |
| No. of unique observations | 122,951 |
| R unmerge (%) | 11 |
| I/σ(I) | 9.0 |
| Space group | C2 |
| Cell dimensions (a,b,c,β) | 195.1, 78.1, 156.8, 120.4 |
| Refinement | |
| Resolution range (Å) | 134.84-2.2 |
| Reflections | 90,926 |
| Completeness (%) | 92.56 (89.9) |
| R work (%) | 19.6 |
| R free (%) | 27.3 |
| R.m.s. deviations | |
| Bond length (Å) | 0.017 |
| Bond angles (°) | 1.556 |
| Average B of overall | 25.82 |
Unassigned electron density within the A. variabilis C503S/C565S PAL active site is consistent with the presence of tCA
We observed unassigned electron density approximately 1.7 Å away from the methylidene carbon of the prosthetic MIO in the A. variabilis C503S/C565S PAL active site. It is possible that this unidentified molecule contributes to the stabilization of the disordered loops in the A. variabilis PAL double mutant, in a manner similar to the proposed DTT-mediated stabilization of Tyr110 in the loop-out conformation in the parsley PAL structure.16 TAL structures (PDB: 2O6Y, 2O7B, 2O7D, 2O78, 2O7F, and 2O7E) which also have ordered active site loops also show extra electron density (an ammonium group) near the MIO, although a covalently attached amine at the MIO in the R. toruloides crystal structures does not seem to have stabilized the structure of the active site in these molecules. The recently published structure of tyrosine ammonia mutase also reported the existence of unexpected density which they assumed was from a reaction between the MIO group and β-mercaptoethanol that was used in sample preparation.19 Regardless of whether the extra electron density in the active site contributes to stabilization of the active site, the presence of covalent modifications and stable excess electron density within this region highlight the reactive nature of the electrophile within the active site cavity.
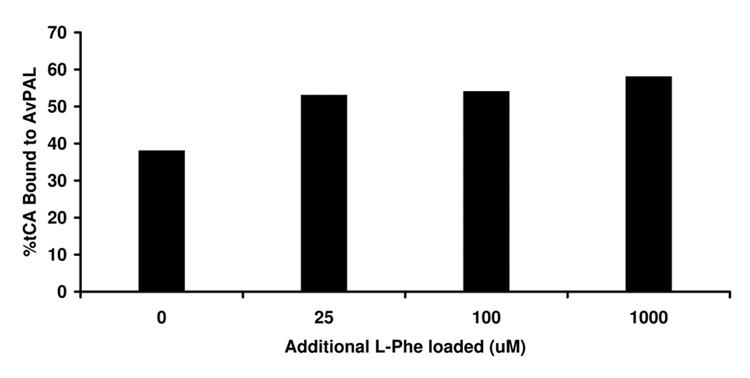
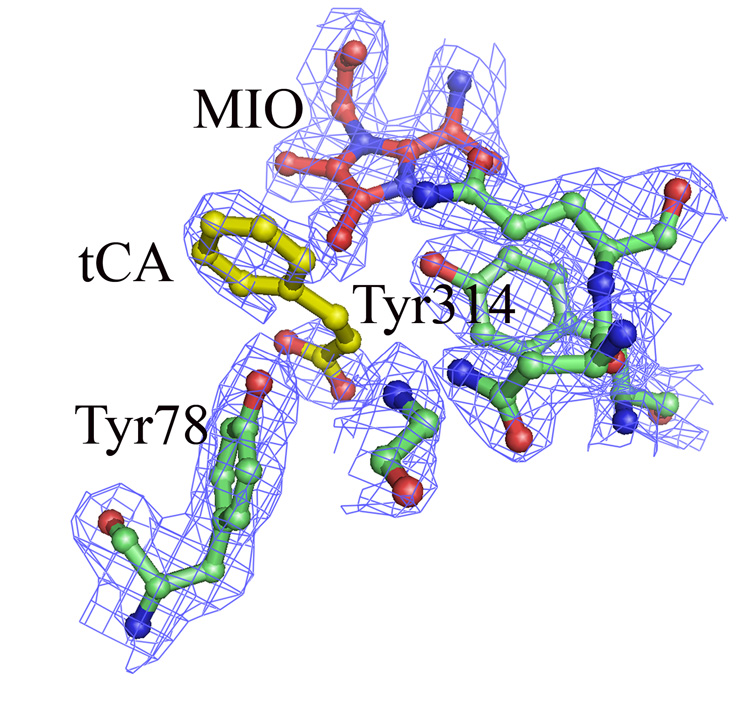
We believe that the extra electron density we see in the active site of the A. variabilis double mutant is from tCA product that remains bound to the enzyme during purification. When we measured the kinetic properties of A. variabilis C503S/C565S PAL, we were surprised to discover that we could only recover half of the tCA product that we had measured spectroscopically (Figure 5), leading us to suspect that a portion of the tCA remains tightly bound in the active site. This tightly bound tCA was only recoverable upon addition of L-Phe. We modeled tCA into the extra density in active site of the A. variabilis PAL double mutant monomer B (Figure 6) and found an excellent fit, especially in the phenyl ring region. Moreover, the position of the modeled tCA is consistent with the positions of coumarate in the TAL-product complex structure,20 and that in the tCA docking study (see below). As in the TAL-product complex structure, the position of tCA in this A. variabilis PAL double mutant does not directly support any of the proposed enzyme mechanisms, unless one assumes that the observed conformation of the side chains of the active site residues represent a product-bound enzyme conformation, and that either on or prior to L-Phe binding, these residues adopt a different conformation.
Figure 5.
Percentage tCA bound to the A. variabilis PAL double mutant as a function of L-Phe loaded. The indicated concentration of Phe was incubated in the presence of 13 µM A. variabilis PAL double mutant for a sufficient period of time to convert all substrate to product. Samples were then desalted to remove excess L-Phe and unbound tCA and injected on a C4-RP column for analysis. Results appear to show that two PAL active sites remain occupied with the product tCA which is not easily dislodged.
Figure 6.
Close-up view of active site residues that have direct interaction with the MIO and modeled tCA in the A. variabilis PAL double mutant monomer B, the only monomer that has clear electron density for the phenyl ring of tCA. Extra electron density are also observed in the other monomers, but none could be modeled as a ring. The electron density map (2Fo-Fc) is contoured at 1σ level
It is unclear why, unlike the other PAL structures including that of the wild-type A. variabilis PAL, the A. variabilis C503S/C565S PAL structure contains a fully ordered active site. It is possible that the putative tCA present in the active site helps order the external loops, although why it is only seen in the double mutant but not the wild-type structure is unclear. Other likely possibilities include differences in sample handling, differences in the crystallographic asu, and stabilization of global protein structure from the C503S and C565S mutations. Further experimentation would need to be done in order to fully understand if any or all of these factors contribute to stabilization of the active site.
Docking studies position tCA in the active site in a manner similar to the TAL H89F co-crystal structure
Further characterization of the active site was carried out by molecular docking studies, probing the region around the active site with L-Phe, L-Tyr, and tCA. Using AutoDock’s 18 novel global-local evolutionary algorithm, we searched for sites with the lowest free-energy of binding between the ligand and the enzyme. All three ligands show binding sites clustered around the MIO in a location and orientation similar to that observed for tCA in the H89F TAL co-crystal structure 20. Positioning of tCA in our docking studies is consistent with our models of tCA as the source of the observed extra electron density in the active site (Figure 6). Docking results show that the docked molecule bisects the active site cavity into a region containing aromatic and hydrophobic residues adjacent to the aromatic ring and a region containing charged and hydrophilic residues adjacent to the hydrophilic tail of L-Phe. Lastly, the Cβ of L-Phe is within 3 Å of the methylidene group of the MIO. Both Tyr78 and Tyr314 are within hydrogen bonding distance of the L-Phe, with Tyr78 interacting with the carboxylate group and Tyr314 to the amine group. The carboxylate group of the substrate is also within hydrogen-bonding distance of the guadininium group of Arg317 from an adjacent monomer. Thus, the picture that emerges from these docking studies replicates what was experimentally observed in the crystal structure of TAL complexes.
The structure of the A. variabilis C503S/C565S active site is most consistent with an enzyme mechanism that proceeds through a carbanion intermediate rather than a carbocation intermediate
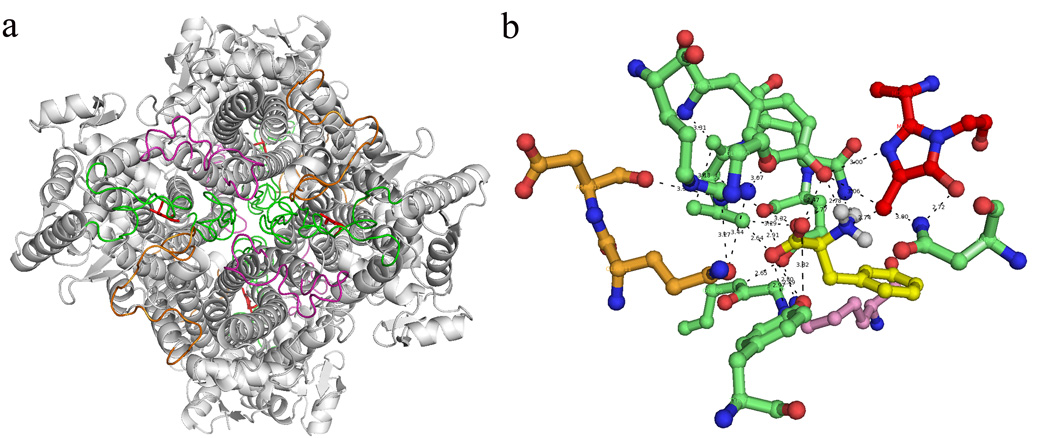
Consistent with the similarities between the structure presented here and the partially modeled active site presented in Calabrese, et al.,8 our structure supports an enzyme mechanism that occurs via a carbanion intermediate and not a Friedel-Crafts-type mechanism that occurs via a carbocation intermediate (see Scheme 1 and Scheme 2 in ref. 8). In this reaction scheme, the methylidene of the MIO group reacts with the NH2 of the substrate. As pointed out by Louie et al.,20 a rotation by the substrate from its observed position will be required to bring the amino group of the substrate into close proximity to the methylidene group of the MIO. Interestingly, preliminary studies using AutoDock’s facility to allow flexibility in receptor side chains shows that movement of just one residue, Arg317 of an adjacent monomer, is enough to produce an energy landscape in which the L-Phe amino group moves from being at a distance of 4.42 Å to 2.74 Å (Figure 7). Additionally, in the A. variabilis PAL double mutant crystal structure Tyr314 is 2.78 Å from the methylidene carbon, which is close enough to act as a general acid to generate product ammonia. Finally, in the flexible docking run, Tyr78 is 2.94 Å from the Cβ of the Phe substrate; this close proximity to this group will allow Tyr78 to act as a general base, abstracting a proton from the L-Phe methylene carbon.
Figure 7.
Loops from three different monomers located around each active site of A. variabilis PAL double mutant. (a) The bottom view of A. variabilis PAL with loop 74–96 (monomer A, green), 436–458 (monomer A, green), 291–311 (monomer B, orange) and 394–419 (monomer C, magenta) are highlighted. (b) Networking among residues from three monomers in each active site with MIO (red) and docked Phe (yellow).
The ability of Tyr78 to act as a general base is further supported by our finding that Tyr78 can react with NHS-biotin to form a covalent Tyr78-biotin adduct (data not shown). Given that the pKa for tyrosine is normally 10, under our reaction conditions Tyr78 should be protonated and therefore nonreactive with NHS-biotin. However, we find that upon NHS-biotin treatment, a significant portion of the Tyr78 becomes biotinylated, indicating that this active-site residue is deprotonated. Biotinylation at Tyr78 inactivates the enzyme (data not shown), but activity is slowly regained over time in a manner consistent with resumption of activity upon hydrolysis of the Tyr78-biotin bond. The ability for Tyr78 to act as a general base is further support for an enzyme mechanism that proceeds through a carbanion intermediate. How Tyr78 is deprotonated is unclear, although the backbone amide of an invariant Gly85 lies within 2.78 Å of Tyr78 OH and can potentially abstract the proton from the Tyr78 hydroxyl group.
The inner loop may control access to the active site by undergoing a conformational switch, making it a potential target for engineering a better therapeutic molecule, between helix and loop that could control access to the active site
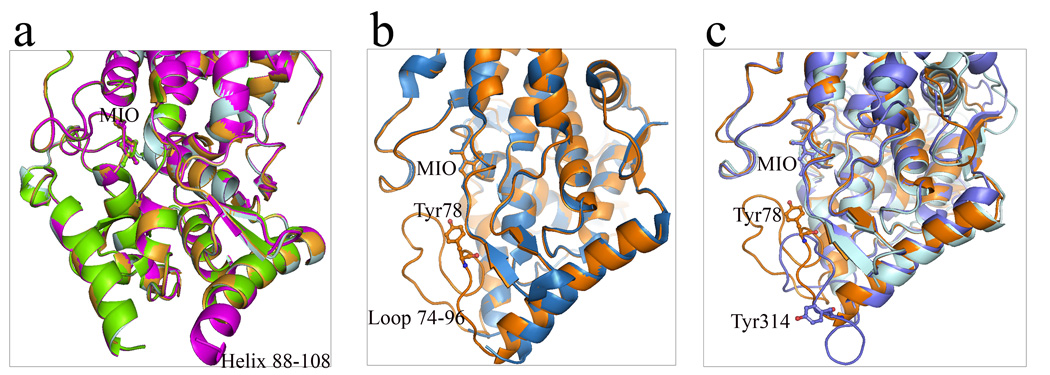
Careful examination of the superposition of PAL structures shows an interesting variability in the position of ordered residues at both ends of the helices adjacent to the inner active site loop. In the A chain of the R. toruloides PAL structure (PDB: 1Y2M), the helix starting at residue 88 terminates at residue 102 and is disordered after residue 103. In the B chain of this structure, the same helix starts at residue 88 but terminates at residue 108 and is disordered after residue 109; thus, two additional turns of the helix are observed in this chain. In chains C and D of this structure the helix ends at residue 103 and is disordered after residue 104 (Figure 8a). In another R. toruloides PAL structure (PDB: 1T6P), the helix flanking the C-terminal end of the inner loop starts at residue 125 in chain A, while in the B chain it starts at residue 121 with the first ordered residue at 119. In the wild-type A. variabilis PAL, the equivalent helix at the N-terminus of the loop ends at residue 72 with loop disorder starting after residue 74. In the A. variabilis PAL double mutant, this helix ends at residue 73 with the rest of the loop in a coil conformation until residue 96 followed by a helix starting at residue 97 (Figure 8b). In the wild-type A. variabilis PAL, the loop is disordered until residue 92 and is helical starting at residue 94. Similar variability in the position of helix termini can be found in other PAL structures. We did not observe any correlation in the thermal vibration parameters in the residues along these helices indicating that this variability is not due to a moving helix but is more likely the result of helix unwinding, rewinding, or transitioning from helix to coil and/or coil to helix. Thus, there is evidence for a conformational switch at the ends of the inner loop similar to that observed in a number of other structures. We suggest that in the A. variabilis PAL double mutant, the helix at residues 125–134 uncoils from Phe131 to Gln134 and the helix from residues 139–143 becomes disordered, perhaps to allow access to or egress from the active site. If these residues do behave as a conformational switch affecting access to the enzyme active site, then factors which affect the stability and fluidity of these structures should affect the kinetic properties of this enzyme, making these residues potential targets for future protein engineering attempts.
Figure 8.
Helix-loop conformational switch of aromatic ammonia lyases. (a) Superimposed monomer A, B, C and D of R. toruloides PAL (1Y2M) shows helix-loop switch in helix 88–108 region in monomer B. Monomer B: magenta; monomer A: pale cyan; monomer C: bright orange; monomer D: chartreuse. (b) Superimposed monomers of wild-type A. variabilis PAL (sky blue) and its double mutant (orange) presents disordered and well-ordered conformations of residues 75–91 region. (c) Three conformational forms of the active site loop 74–91 in A. variabilis PAL (loop-in position, orange) and its corresponding region in P. crispum PAL (loop-out position, slate) and R. toruloides PAL (disordered, cyan). In all figures, MIO, Tyr78 in A. variabilis PAL and Tyr110 in P. crispum PAL are shown in sticks.
Structural alignment of eukaryotic and prokaryotic PAL C-terminal domains suggest gene duplication event
One common feature of prokaryotic aromatic ammonia lyases that has been characterized is the absence of the insertion domain that exists in the longer eukaryotic yeast R. toruloides PAL (residues 541–655) and plant P. crispum PAL (residues 527–648). Previous studies suggested that phosphorylation of the insertion domain could possibly play a regulatory role by modulating substrate access to the active site.16; 21 An additional role of this insertion domain was proposed by Pilbak and colleagues, who hypothesized that this insertion domain may be an important factor in the destabilization of eukaryotic PAL as a response to a regulation signal eventually leading to a decrease in its lifetime.15 However, to date, there is no clear experimental evidence to support either hypothesis.
In an attempt to understand the relationship between the C-terminal domains of eukaryotic and prokaryotic PALs and to identify its possible functional role, we superimposed the A. variabilis PAL double mutant structure with the R. toruloides PAL structure using the program FATCAT.22 A rigid superposition of the two structures (rmsd 1.73 Å) clearly shows that the A. variabilis PAL double mutant has no corresponding region to the insertion domain present in yeast R. toruloides PAL (Figure 9a). However, using the option to flexibly align structures through the introduction of conformational changes, we found that adding a twist to the very end region of the C-terminus (residues 498–563) of the A. variabilis PAL double mutant structure aligns that segment with a portion of the insertion domain of R. toruloides PAL structure (residues 546–612) with an overall rmsd of 2.12 Å (Figure 9b). In contrast, rigid alignment shows that this region of the A. variabilis PAL double mutant (residues 498–563) overlapped with the very end of C-terminus of R. toruloides PAL (residues 652–716). This leads to the possibility that the insertion domain may have arisen by gene duplication, fusion and followed by divergence to optimize their structural biological roles. Although this analysis provides some clues as to the possible origins of this domain, its functional role remains a mystery.
Figure 9.
Structural alignment of R. toruloides PAL (blue) and the A. variabilis PAL double mutant (salmon) monomer. (a) Rigid alignment. The C-terminal residues 498–563 in the A. variabilis PAL double mutant are aligned with C-terminal residues 652–716 in R. toruloides PAL (b) Flexible alignment. The C-terminal region of residues 498–563 in the A. variabilis PAL double mutant makes a twist and is overlapped with the region of residues 546–612, which is a part of the insertion domain in R. toruloides PAL.
Biochemical and structural characterization of the A. variabilis PAL C503S/C565S double mutant has led to the development of testable new hypotheses on the enzyme’s mechanism and demonstrates how aspects of the enzyme’s structure potentially affect its mechanism. Our structural and biochemical characterization of this enzyme elucidate regions of the enzyme that are likely targets for future optimization and engineering efforts for the development of a therapeutic compound to treat PKU. Based on docking studies as well as NHS-biotin binding studies, we now propose that Tyr314/Tyr78 act as general acid/base extracting and donating protons during catalysis. Furthermore, these data and analysis support an enzyme mechanism described first by Peterkofsky,23 and later by Calebrese,8 where the prosthetic MIO group reacts with the NH2 of the substrate L-Phe. Our biochemical study results show that the A. variabilis PAL double mutant is more thermally stable and more resistant to proteases than R. toruloides PAL, possibly due to more rigid or constrained active site loops and/or the existence of the insertion domain in R. toruloides PAL. Of particular therapeutic importance, ligands that bind to the enzyme such as L-Phe, L-Tyr, and tCA lead to a more thermally stable molecule, possibly through the creation of a kinetic barrier that helps to stabilize the tetramer with the ligand serving the role of a “keystone”. Such stabilization is more pronounced on the A. variabilis PAL double mutant than on R. toruloides PAL, which could be due to the higher substrate affinity of the A. variabilis PAL double mutant. The proposed conformational switch in the inner active site loop and its role in determining enzyme kinetics, if successfully validated by follow-on studies, should contribute significantly to attempts at engineering an enzyme more suitable for use as a therapeutic.
Materials and Methods
Wild-type A. variabilis PAL surface cysteine identification
Final concentration of 10 mM DTT was added to A. variabilis PAL (0.1 M Tris, pH 7.5 containing 140 mM NaCl) to remove potential surface cysteine adducts and intermolecular disulfides. Following 30 min incubation at 37 °C, the freshly reduced A. variabilis PAL was desalted to remove excess DTT (0.1 M potassium phosphate, pH 7.0). N-ethylmaleimide (NEM) was immediately added to the desalted-reduced A. variabilis PAL at a final concentration of 10 mM to modify surface cysteines (free sulfhydryls). The reaction mixture was incubated for 30 min at 37 °C, and excess NEM was removed by a desalting column after the reaction was completed. The NEM modified A. variabilis PAL was concentrated by precipitation (tCA/Acetone). The protein pellet was denatured in 8 M urea, reduced by DTT (20 mM for 30 min at 37 .C), followed by alkylation (40 mM iodoacetamide (IAM) for 30 min at 37 °C) in order to block the remaining free sulfhydryls that were either buried or in intramolecular disulfides. After that, the NEM/IAM modified A. variabilis PAL was digested by trypsin and the digested fragments were assayed by LC/MS to map surface cysteines, and buried or partially buried cysteines, by mass change of each fragment compared to unmodified A. variabilis PAL. The mass gain from each NEM and IAM modification is 124 Da and 57 Da per cysteine, respectively.
Molecular cloning and mutagenesis
The full-length A. variabilis PAL gene was cloned into the same pIBX vector used for R. toruloides PAL.2 Point mutations C503S and C565S were introduced in A. variabilis PAL by a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The full-length R. toruloides PAL expression construct has been described previously.2 Planned future use of the protein as a therapeutic precluded the use of affinity tags for expression and purification purposes.
Protein expression, purification and activity assay
The non-tagged selenomethionine-A. variabilis PAL double mutant was expressed in the methionine-auxotroph E. coli B834 (DE3) (Novagen) host cell using selenomethionine-containing medium. The cell culture was grown at 37 °C to an OD600nm of 0.8, and was then induced by 1 mM IPTG at 20 °C for 14 hr. The cell pellet was lysed in buffer [100 mM Tris (pH 7.8), 10 mM NaCl, 1 mM MgCl2, 5% glycerol, 3 mM methionine, 0.01% β-mercaptoethanol, EDTA-free protease inhibitor cocktail (Roche diagnostics GmbH, Mannheim, Germany)] and was then sonicated. After ultracentrifugation, the soluble extract was applied onto a Poros HQ (Applied Biosystems, Foster City, CA) column equilibrated with 25 mM Tris, pH 8.0 and flow-through was collected. Ammonium sulfate was added in the flow-through from the HQ column to a final concentration of 1.45 M followed by a centrifugation and filtration step. The supernatant was applied onto a Poros HP (Applied Biosystems) column equilibrated with 25 mM Tris, 1.45 M ammonium sulfate, pH 8.0, and was eluted using 25 mM Tris, pH 8.0 with a 20 column volume gradient. The A. variabilis PAL fraction was diluted using 25 mM Tris, pH 8.0 to a final 50× volume, and then was applied onto a second HQ column using 25 mM Tris, pH 8.0 as running buffer and 25 mM Tris, 250 mM NaCl, pH 8.0 as elution buffer, using a 20 column volume gradient. The final active A. variabilis PAL double mutant fraction was concentrated to 12 mg/mL with a buffer exchange into 25 mM Tris, pH 8.0, 150 mM NaCl for crystallization trials.
The PAL activity assay was started by the addition of 50 µL protein sample into 950 µL 100 mM Tris, pH 8.5, with 22.5 mM Phe. The formation of tCA was monitored by optical absorption at 290 nm at 25 °C and activity was calculated in a standard fashion 24.
Crystallization, data collection and structure determination
The selenomethionine-A. variabilis PAL double mutant crystals were grown at 25 °C in 96-well plate by sitting drop vapor diffusion. The reservoir solution contained 70 µL 12–20% PEG1500 and 100 mM SPG (succinic acid, sodium dihydrogen phosphate, glycine), pH 7.0. The drops were made with 0.3 µL protein sample and 0.3 µL reservoir solutions. A. variabilis PAL double mutant crystals were cryo-protected in 25% glucose and mother liquor before being flash-cooled in liquid nitrogen. Diffraction data sets were collected at the SSRL beam line 11-1. The crystals belong to space group C2 with four monomers per asymmetric unit and a solvent content of 45.5%. Diffraction data were indexed, integrated, and scaled using HKL2000 and Scalepack.25 Data collection and refinement statistics are given in Table 4. The structure of the A. variabilis PAL double mutant was determined by molecular replacement by program Phaser26 and used the wild-type A. variabilis PAL monomer structure as a search model (2NYN). Model rebuilding was carried out with the program Coot.27; 28 REFMAC29; 30 was used for final model refinement.
Limited proteolysis and protease cleavage site mapping
The trypsin and chymotrypsin limited digestion of R. toruloides PAL and cleavage sites mapping has been previously reported.3 In this study, the reaction temperature was 37 °C. Limited proteolysis was carried out by incubating the purified A. variabilis PAL double mutant with trypsin (40 µg/mL); chymotrypsin (40 µg/mL) in 0.1 M Tris, 150 mM NaCl, pH 8.5, at 37 °C; 0.01U thrombin; or 0.2U factor Xa in 50 mM Tris pH 8.0, 100 mM NaCl and 2 mM CaCl2. At various time points, a digested sample was withdrawn for activity assay and SDS-PAGE analysis. The two major protein fragments obtained from proteolytic digestion were blotted onto FVDF membrane and analyzed by protein sequencing via standard methods.
Thermal stability assay
All thermal stability studies were performed using an iCycler iQ real-time PCR instrument. Temperature-dependent Sypro orange binding was measured at a 0.5 °C/min scan rate from 20 °C to 95 °C. The unfolding temperature Tm was defined as the midpoint temperature of the protein-unfolding transition and determined by program MyiQ. All samples contained 5 µL 1:250 diluted sypro orange and were prepared by same buffer (50 mM Tris, 150 mM NaCl, pH 8.0) to a final volume of 50 µL. There was 2 µg of protein in all protein-containing samples. All samples were divided into groups and the samples in each group are listed below. Group 1: sypro orange only, Group 2: A. variabilis PAL double mutant only, A. variabilis PAL double mutant plus 5 mM tyrosine; plus 5 mM phenylalanine; plus 5 mM tCA. The samples in Groups 3 are equivalent to the ones in groups 2 with R. toruloides PAL substituted for the A. variabilis PAL double mutant. Samples containing tyrosine or phenylalanine were prepared by incubation of the protein and the corresponding amino acids for 10 min.
pH stability assay and pH profile
The pH stability assay and optimal pH profile study were carried out by incubating the A. variabilis PAL (0.07 mg/mL) in different buffers with pH ranging from 1–13 in 0.5 unit increments for 10 min. For the pH stability studies, the samples were then subjected to a standard activity assay, measured at pH 8.5. For the pH activity profile studies, the activity assays were carried out in buffers at the corresponding pH.
NHS (N-hydroxysuccinimide)-ester modifications
All NHS-ester labeling studies were performed according to the general procedure as outlined in the Pierce catalog using NHS-Biotin. Reactions were quenched with a 10× molar excess of Tris, pH 7.5 and split into two microcentrifuge tubes, one to be stored at 4 °C and the other to be stored at −80 °C. Specific activity determinations were made immediately after the NHS modification (T=0) and 2 weeks later. Prior to tryptic digestion, samples were denatured in the presence of 8 M Urea. A 10 µL aliquot of the denatured samples (~100 µg) was digested in 50 mM Tris, pH 8.2 containing 25 µg/ml trypsin (100 µL final volume) and incubated overnight (~16 hr) at 37 °C. Digests were then reduced by the addition of DTT (20 mM final) and incubated an additional hour at 37 °C. The reaction was quenched by the addition of 2 µL of 15% formate and injected onto a C18 reverse-phase column equipped with an ESI-MSD. Unmodified and biotin-labeled peptides were quantified by mass spectroscopy using a differential peptide mapping technique and compared to the specific activity data for the same sample set.
Determination of bound tCA
A 0.6 mL volume of A. variabilis PAL (6.4 mg/mL) in 10 mM Tris, pH 7.5 containing 210 mM NaCl were incubated in the presence of 0, 25, 100 or 1000 µM Phe (final concentration) for 1 hr at 37 °C. The samples were stored overnight at 4 °C. The next morning, samples were desalted in TBS buffer. The protein concentration after gel filtration for all samples was 0.8 mg/mL as determined by the extinction coefficient at 280 nm for A. variabilis PAL (1 mg/mL = 0.75 absorbance). A 100 µL aliquot was injected onto a C4-RP column. tCA and protein was eluted from the column using an increasing acetonitrile/water gradient containing 0.05% TFA. A tCA standard curve was run to determine the concentration of tCA bound to protein. tCA was monitored by absorbance at 280 nm and quantified by measuring the area under the curve.
ACKNOWLEDGEMENTS
We are grateful to Dr. Gye Won Han, Dr. Ellen Chien, Dr. Kumar Saikatendu and Dr. Joe Ng for invaluable advice on structure determination and the thermal stability study, and Dr. Anand Kolatkar and Professor Art Olson’s laboratory for assistance with Autodock. Jovylyn Gatchalian assisted in carrying out a number of the experiments described in this paper. We also wish to thank Angela Walker and A. Pia Abola for assistance with manuscript preparation. This research was supported in part by U01 NS051353, A.G. was supported by the Tia Piziali Fellowship for PKU Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein Data Bank accession codes
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank and are available under accession code 3CZO.
REFERENCES
- 1.Scriver CR. In: The Metabolic and Molecular Bases of Inherited Disease. 8th edit. Scriver CR, editor. McGraw-Hill: Professional Publishing; 2000. [Google Scholar]
- 2.Sarkissian CN, Shao Z, Blain F, Peevers R, Su H, Heft R, Chang TM, Scriver CR. A different approach to treatment of phenylketonuria: phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proc Natl Acad Sci U S A. 1999;96:2339–2344. doi: 10.1073/pnas.96.5.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Gamez A, Sarkissian CN, Straub M, Patch MG, Han GW, Striepeke S, Fitzpatrick P, Scriver CR, Stevens RC. Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria. Mol Genet Metab. 2005;86:134–140. doi: 10.1016/j.ymgme.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Hoskins JA, Jack G, Wade HE, Peiris RJ, Wright EC, Starr DJ, Stern J. Enzymatic control of phenylalanine intake in phenylketonuria. Lancet. 1980;1:392–394. doi: 10.1016/s0140-6736(80)90944-7. [DOI] [PubMed] [Google Scholar]
- 5.Ambrus CM, Anthone S, Horvath C, Kalghatgi K, Lele AS, Eapen G, Ambrus JL, Ryan AJ, Li P. Extracorporeal enzyme reactors for depletion of phenylalanine in phenylketonuria. Ann Intern Med. 1987;106:531–537. doi: 10.7326/0003-4819-106-4-531. [DOI] [PubMed] [Google Scholar]
- 6.Gamez A, Sarkissian CN, Wang L, Kim W, Straub M, Patch MG, Chen L, Striepeke S, Fitzpatrick P, Lemontt JF, O'Neill C, Scriver CR, Stevens RC. Development of pegylated forms of recombinant Rhodosporidium toruloides phenylalanine ammonia-lyase for the treatment of classical phenylketonuria. Mol Ther. 2005;11:986–989. doi: 10.1016/j.ymthe.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Moffitt MC, Louie GV, Bowman ME, Pence J, Noel JP, Moore BS. Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization. Biochemistry. 2007;46:1004–1012. doi: 10.1021/bi061774g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabrese JC, Jordan DB, Boodhoo A, Sariaslani S, Vannelli T. Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry. 2004;43:11403–11416. doi: 10.1021/bi049053+. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki Watanabe S, Iturbe-Chinas GH-VF, Lopes-Munguia A. Phenylalanine ammonia lyase from Sporidiobolus pararoseus and Rhodosporidium toruloides: application for phenylalanine and tyrosine deamination. World Journal of Microbiology and Biotechnology. 1992;8:406–410. doi: 10.1007/BF01198755. [DOI] [PubMed] [Google Scholar]
- 10.Ogata K, Uchiyama K, Yamada H, Tochikura Metabolism of aromatic amino acids in microorganisms. Part II. Properties of phenylalanine ammonia lyase of Rhodotorula. Agricultural and Biological Chemistry. 1967;31:600–606. [Google Scholar]
- 11.Tyssandier V, Reboul E, Dumas JF, Bouteloup-Demange C, Armand M, Marcand J, Sallas M, Borel P. Processing of vegetable-borne carotenoids in the human stomach and duodenum. Am J Physiol Gastrointest Liver Physiol. 2003;284:G913–G923. doi: 10.1152/ajpgi.00410.2002. [DOI] [PubMed] [Google Scholar]
- 12.Shah RM, D'Mello AP. Strategies to maximize the encapsulation efficiency of phenylalanine ammonia lyase in microcapsules. Int J Pharm. 2008 doi: 10.1016/j.ijpharm.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 13.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 14.Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 15.Pilbak S, Tomin A, Retey J, Poppe L. The essential tyrosine-containing loop conformation and the role of the C-terminal multi-helix region in eukaryotic phenylalanine ammonia-lyases. FEBS J. 2006;273:1004–1019. doi: 10.1111/j.1742-4658.2006.05127.x. [DOI] [PubMed] [Google Scholar]
- 16.Ritter H, Schulz GE. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell. 2004;16:3426–3436. doi: 10.1105/tpc.104.025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 19.Christianson CV, Montavon TJ, Van Lanen SG, Shen B, Bruner SD. The structure of L-tyrosine 2,3-aminomutase from the C-1027 enediyne antitumor antibiotic biosynthetic pathway. Biochemistry. 2007;46:7205–7214. doi: 10.1021/bi7003685. [DOI] [PubMed] [Google Scholar]
- 20.Louie GV, Bowman ME, Moffitt MC, Baiga TJ, Moore BS, Noel JP. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem Biol. 2006;13:1327–1338. doi: 10.1016/j.chembiol.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allwood EG, Davies DR, Gerrish C, Ellis BE, Bolwell GP. Phosphorylation of phenylalanine ammonia-lyase: evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 1999;457:47–52. doi: 10.1016/s0014-5793(99)00998-9. [DOI] [PubMed] [Google Scholar]
- 22.Ye Y, Godzik A. Database searching by flexible protein structure alignment. Protein Sci. 2004;13:1841–1850. doi: 10.1110/ps.03602304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterkofsky A. The mechanism of action of histidase: amino-enzyme formation and partial reactions. J Biol Chem. 1962;237:787–795. [PubMed] [Google Scholar]
- 24.Zucker M. Induction of phenylalanine ammonia-lyase in Xanthium leaf disks. Photosynthetic requirement and effect of daylength. Plant Physiol. 1969;44:912–922. doi: 10.1104/pp.44.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov G, Vagin A, Dodson E. Proc. Daresbury Study Weekend. 1996 [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]