Abstract
TNP-470, the first anti-angiogenic small molecule to enter clinical trials, targets methionine aminopepti-dase-2 (MetAP-2), a metalloprotease that cleaves the N-terminal methionine of proteins. Previously, biochemical binding, in vivo yeast studies, and structural studies of human methionine aminopeptidase-2 bound to TNP-470 and its analogs fumagillin and ovalicin revealed that these compounds exhibit specificity for MetAP-2 over its family member MetAP-1. To further elucidate the nature of this specificity, we developed a yeast-based screen for human MetAP-2 mutations that confer ovalicin resistance. Of the three resistant alleles, A362T appeared in the majority of clones and was found to be the most resistant to the ovalicin class of inhibitors. Alignment of human MetAP-2 with human MetAP-1, which is naturally ovalicin-resistant, revealed that the analogous residue in MetAP-1 is also a threonine. Mutation of this residue to alanine resulted in an ovalicin-sensitive MetAP-1 allele, demonstrating that an alanine at this position is critical for inhibition by ovalicin. These results provide a molecular explanation for the specificity exhibited by this class of anti-angiogenic agents for MetAP-2 over MetAP-1 and may prove useful in the development of additional MetAP-2-specific therapeutic agents.
Fumagillin, a natural product angiogenesis inhibitor, was originally purified from a fungal contaminant of an endothelial cell culture (1). TNP-470, a derivative of the natural product fumagillin, has been shown to be safe and effective in the treatment of solid tumors and arthritis in several animal studies and preclinical trials (1–5). Based on these promising results, TNP-470 entered human clinical trials for the treatment of AIDS-related Kaposi’s sarcoma, metastatic breast cancer, androgen-independent prostate cancer, brain cancer, pediatric solid tumors, lymphomas, acute leukemias, advanced squamous cell cancer of the cervix, and metastatic renal carcinoma (6–9). Because of the clinical potential of TNP-470, it is of great interest to elucidate the mechanism of action of this compound.
The cellular target of fumagillin was purified by natural product affinity chromatography and identified as methionine aminopeptidase-2 (10). In subsequent studies, it was found that methionine aminopeptidase-2 is also the binding protein of TNP-470 and ovalicin, another structural analogue of fumagillin (11). Methionine aminopeptidase-2 is a member of the methionine aminopeptidase family of metal-coordinating proteases whose function is to cleave the N-terminal methionine from nascent proteins. Recently it was shown that manganese is the physiologically relevant cofactor for methionine amino-peptidase-2 (12). Because cleavage of the N-terminal methionine is a process essential to life for both prokaryotes and eukaryotes, the methionine aminopeptidases are highly conserved from bacteria to humans (13, 14). Methionine aminopeptidases (MetAPs)1 are divided into two classes, type I and type II (also known as MetAP-1 and MetAP-2, respectively) (15). In Saccharomyces cerevisiae at least one MetAP is essential for viability, as loss of both MetAPs is lethal (14). Studies on isogenic yeast strains revealed that fumagillin, TNP-470, and ovalicin specifically inhibit yeast MetAP-2 but not yeast MetAP-1 (10, 11).
To investigate the molecular basis of MetAP sensitivity to the fumagillin/TNP-470/ovalicin class of angiogenesis inhibitors, a library of mutant human methionine aminopeptidase-2 alleles was generated and screened for ovalicin resistance in a yeast-based system. After screening more than 106 colonies in three independent screens, we found 3 alleles that were verified to be ovalicin-resistant: A362T, Y444C, and H382Y. These results suggest that the determinants of MetAP-2 ovalicin sensitivity lie not only in the ligand binding pocket (such as Y444C) but also may exert some effect from a distance (such as A362T and H382Y). The A362T allele was present in almost 80% of all ovalicin-resistant yeast colonies obtained from the screen and was found to be the most resistant of the three alleles to ovalicin and its analogues fumagillin and TNP-470. Protein sequence alignment revealed that the analogous residue in human MetAP-1 is a threonine, and interestingly, mutation of this residue to alanine conferred ovalicin sensitivity to MetAP-1. Taken together, these findings imply that A362 is critically important to the sensitivity of human MetAP-2 to the ovalicin class of inhibitors, thus providing a molecular explanation for the specificity of the fumagillin class of compounds for MetAP-2 over MetAP-1.
EXPERIMENTAL PROCEDURES
S. cerevisiae Strains and Growth Media
The BY4741 strain map1::KAN (MATa his3 leu2 met15 ura3) was a gift from the Stanford Genome Technology Center. The W303 haploid strains map1::HIS3 (MATα ade2-1 his3-11, 15 leu2-3, 112 ura3-1 trp1-1) and map2::URA3 (MATα ade2-1 his3-11, 15 leu2-3, 112 ura3-1 trp1-1) were gifts from Dr. Yie-Hwa Chang. Yeast strains were grown either in yeast extract-peptone-dextrose or synthetic minimal medium.
Yeast Transformation
Yeast were transformed according to the lithium acetate method described by Schiestl and Gietz (16), except that twice the prescribed amount of single-stranded DNA was used. Yeast to be screened were plated onto 2× synthetic minimal medium minus leucine agar plates with 10 nM ovalicin and were grown at 30 °C for, 3 days.
Plasmids and Vector Construction
pSE319 (17) was a gift from Dr. Michael Snyder, and pAD4M (18–20) was a gift from Dr. P. Hieter. YMAP2 was cloned into the SalI sites of pSE319 vector. YMAP2 was amplified by PCR using the following primers: forward, 5′-GCAGTCGACATCGATATCCGAATTTTGTT-3′, and reverse, 5′-TGCGTCGACTCAGTAGTCATCACCTTTCG-3′ (the SalI sites are underlined). Human methionine aminopeptidase-2 was cloned into the SalI and SacI sites of the pAD4M vector. The p415GPD and p416GPD plasmids (21) were a gift from Dr. Susan Baserga. Human methionine aminopeptidase-2 was cloned into the SpeI and XhoI sites of p415GPD and p416GPD. The following primers were used to PCR amplify human methionine aminopeptidase-2: forward, 5′-GGACTAGTATGGCGGGCGTGGAGGAGGTAGC-3′ (the SpeI site is underlined), and reverse, 5′-CCGCTCGAGTTAATAGTCATCTCCTCTGCTGACAACTTCTTTAC-3′ (the XhoI site is underlined). Human methionine aminopeptidase-1 was cloned into the SpeI and XhoI sites of p415GPD. The following primers were used to PCR amplify human methionine aminopeptidase-1: forward, 5′-GGACTAGTATGGCGCTCTTCCAGCGGGCAGG-3′ (the SpeI site is underlined) and reverse, 5′-CCGCTCGAGTTAAAATTGAGACATGAAGTGAGGCCGTGC-3′ (the XhoI site is underlined). All PCR amplifications described above were done with Pfu polymerase (Stratagene) for 35 cycles.
Complementation of YMAP2 with Human MetAP-2
Genomic DNA from the W303 map1::HIS3 MATα strain was isolated with lyticase and the Qiagen Genomic Tip prep kit. The map1::HIS3 locus from the W303 map1::HIS3 MATα strain (22) was PCR-amplified using Tbr (Finnzymes) and the primer pair 5′-CCTATACTATGGCGGAATTCACTCC-3′ (forward) and 5′-GCAAACTGTAGGACGAAGAGC-3′ (reverse). W303 map2::URA3 yeast were transformed with YMAP2 in pSE319 and human methionine aminopeptidase-2 in pAD4M and selected on synthetic minimal medium lacking tryptophan and leucine. This strain was then transformed with the map1::HIS3 PCR product and selected on synthetic minimal medium lacking histidine. The transformants were then grown in medium supplemented with tryptophan and plated onto medium with limiting adenine (23) to develop the sectors.
Gap Repair and Error-prone PCR
p415GPD with insert human methionine aminopeptidase-2 was cut internally with BsaBI and PstI enzymes to create a gapped vector. Human methionine aminopeptidase-2 was PCR-amplified using the following primers: forward, 5′-GGACTAGTATGGCGGGCGTGGAGGAGGTAGC-3′, and reverse, 5′-CCGCTCGAGTTAATAGTCATCTCCTCTGCTGACAACTTCTTTAC-3′. The human methionine aminopeptidase-2 was PCR-amplified with Taq polymerase (Roche Applied Science) in 4 separate reactions in which one of the dNTPs was 40 µM and the remaining three were 200 µM. The reactions were then pooled (24). For the gap repair in each screen, 500 ng of gapped vector was transformed with 2.5 µg of pooled mutagenized human methionine aminopeptidase-2 PCR products (25) into the BY4741 map1::KAN MATa yeast strain.
Isolation of Plasmids from Yeast and DNA Sequencing
Human methionine aminopeptidase-2 plasmids were rescued from yeast by the glass bead lysis procedure (26). The DNA isolated from the yeast was then transformed into competent DH5α bacteria, and individual colonies were picked and mini-prepped. DNA was sequenced by the Keck Foundation Biotechnology Resource Laboratory at Yale. Sequence analysis was performed with DNASTAR Seqman.
Dot Titrations and Yeast Quantitation Assay
Yeast colonies were inoculated into minimal medium lacking leucine and grown overnight at 30 °C with shaking. A600 was measured, and the yeast were diluted to 0.200 absorbance units, then serially diluted 10-fold to 0.00002. Using a 1–10-µl multichannel pipettor, 3-µl aliquots of diluted yeast were plated onto extract-peptone-dextrose agar plates with various concentrations of ovalicin, fumagillin, and TNP-470. Plates were then incubated at 30 °C for 2–7 days and then photographed. In the yeast quantitation assay, 3-µl aliquots of yeast diluted to A600 = 0.200 were plated onto extract-peptone-dextrose agar plates with various concentrations of ovalicin, fumagillin, and TNP-470. Plates were then incubated at 30 °C for 2–4 days, and the yeast colonies were scraped from the plate and resuspended in 1 ml of double-distilled H2O. The amount of yeast was quantitated spectrophotometrically at 600 nm, and % growth was determined by the following formula.
| (Eq. 1) |
Site-directed Mutagenesis
Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit by Stratagene (La Jolla, CA). Synthetic oligonucleotide primers were used to PCR amplify the supercoiled double-stranded DNA template plasmid. Primers complementary to opposite strands of the vector were extended using Pfu Turbo polymerase (Stratagene) for 18 cycles. The resulting PCR product was digested with DpnI to eliminate parental DNA and transformed into DH5α Escherichia coli. The template for each of the following primer pairs was wild type human MetAP-2 in the p415GPD vector: A362T forward, 5′-ggaaggagaagtatatAcaattgaaacctttgg-3′, and reverse, 5′-ccaaaggtttcaattgTatatacttctccttcc-3′; Y444C forward, 5′-gcattgtagatccatGtccaccattatgtg-3′, and reverse, 5′-cacataatggtggaCatggatctacaatgc-3′; H382Y forward, 5′-gatatggaatgttcaTattacatgaaaaattttg-3′, and reverse, 5′-caaaatttttcatgtaatAtgaacattccatatc-3′. The template for the following primer pair was wild type human MetAP-1 in the p415GPD vector: T334A forward, 5′-ggccatgtatttGcaattgagccaatg-3′, and reverse, 5′-cattggctcaattgCaaatacatggcc-3′.
Sequence Analysis
Protein sequence alignments were performed with the DNAssist 2.0 computer program (DNAssist cc, South Africa).
RESULTS
Human Methionine Aminopeptidase-2 Complements the Function of Yeast Methionine Aminopeptidase-2
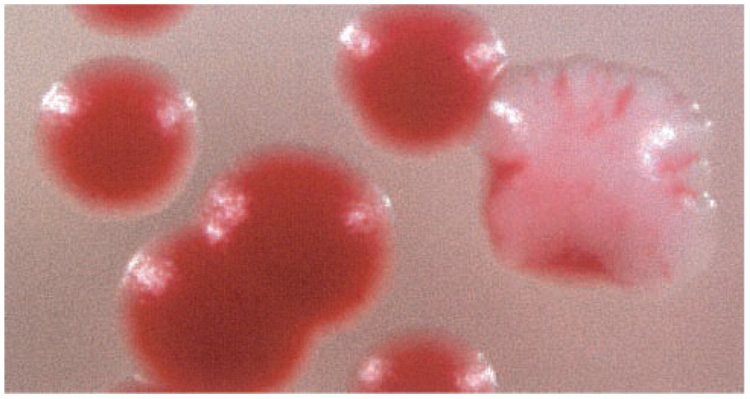
Because of the nature of gap repair as a method for creating a library of mutant MetAP-2 alleles, it was undesirable to screen yeast MetAP-2 because it preferentially recombines with the yeast genome. It was, therefore, desirable to screen MetAP-2 from a different species. Alignment of human and yeast methionine aminopeptidase-2 reveals that these proteins share a modest 54% amino acid sequence identity and 69% similarity. A demonstration of the functionality of the human MetAP-2 gene in S. cerevisiae was necessary to perform a fruitful screen; therefore, human MetAP-2 was tested for its ability to complement the function of yeast MetAP-2 or YMAP2. Because loss of both MetAPs in yeast is lethal, the overall strategy was to rescue a ΔMAP1ΔMAP2 yeast strain with human MetAP-2. The MetAP null strain, containing the ade2-1 mutation, was constructed and rescued with episomal human MetAP-2 and YMAP2. Growth of the resulting strain under non-selective conditions led to loss of the YMAP2 gene, and surviving yeast were viable due to the presence of functional human MetAP-2. The ade2-1 mutation in the MetAP null strain causes the buildup of adenine precursor, which colors the yeast red. However, the plasmid into which YMAP2 is cloned also contains an ade2-1 suppressor gene and eliminates the red color when present. As shown in Fig. 1, the yeast are either completely red or have sectored, indicating loss of the YMAP2 plasmid and evidence that human MetAP-2 is sufficient for yeast viability in the absence of YMAP1 and YMAP2. Further evidence for the functionality of human MetAP-2 in yeast was provided by its ability to confer a growth advantage to ΔMAP1 yeast in the presence of fumagillin (data not shown).
FIG. 1. Human methionine aminopeptidase-2 complements the function of yeast MAP2.
ΔMAP2 yeast were co-transformed with the human MetAP-2 pAD4M and YMAP2 pSE319 vectors. Yeast MAP1 was knocked out by transformation of the map1::HIS3 locus. The resulting strain was then grown in medium supplemented with tryptophan to promote segregation of the pSE319 plasmid and development of red sectors. Sectors or colonies that are red are yeast rescued by human MetAP-2.
Given that fumagillin, TNP-470, and ovalicin all target MetAP-2, it was important to determine whether these three analogues were equivalently effective in inhibition of yeast growth. Although wild type yeast are resistant to fumagillin, yeast with a deleted MetAP-1 gene (ΔMAP1) cannot grow in the presence of fumagillin. Comparison of the growth of human MetAP-2-overexpressing ΔMAP1 yeast in response to the fumagillin analogues showed that ovalicin was the most potent, followed by fumagillin and TNP-470 (Supplemental Fig. 1). Therefore ovalicin was the inhibitor of choice for the screening procedure.
Screening and Discovery of Ovalicin-resistant Mutations of Human Methionine Aminopeptidase-2
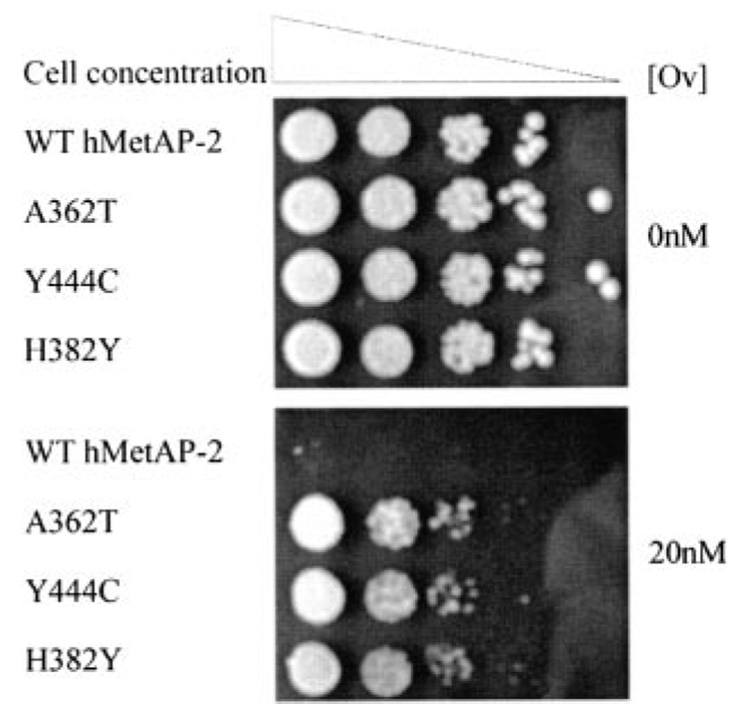
To determine the optimal concentration of ovalicin for screening, ΔMAP1 yeast expressing wild type human MetAP-2 were plated in the presence of various ovalicin concentrations. These pilot studies revealed that no colonies grew above 10 nM ovalicin. Mutant alleles of human methionine aminopeptidase-2 were generated by error-prone PCR and co-transformed with gapped p415GPD yeast expression vector into ΔMAP1 yeast to simultaneously create and screen a library of mutant human MetAP-2 alleles. These yeast were then plated directly onto plates of minimal medium lacking leucine with 10 nM ovalicin. Plasmids were rescued from the colonies that grew in the presence of 10 nM ovalicin, and the identities of the alleles was determined by sequencing. As shown in Table I, the screen was performed independently three times, generating greater than 106 colonies. Of the 23 colonies confirmed to be resistant, 18 of them had the A362T mutation, 3 had the Y444C mutation, and 2 had the H382Y mutation. To confirm that these mutations were the cause of ovalicin resistance, site-directed mutagenesis was used to generate single point mutations. Yeast expressing these alleles were dot-titrated onto plates containing 20 nM ovalicin. As shown in Fig. 2, there was significant growth of the mutants in comparison with the wild type in the presence of 20 nM ovalicin. To determine whether these alleles were also resistant to other ovalicin analogs, yeast expressing wild type or mutant MetAP-2 alleles were dot-titrated in the presence of fumagillin and TNP-470. All three alleles found in the screen were resistant to TNP-470, but only A362T and H382Y were found to be resistant to fumagillin (Supplemental Fig. 2). To exclude the possibility that these alleles cause hyperproliferation, doubling times were measured for the strains expressing wild type, A362T, Y444C, and H382Y alleles. No significant difference in doubling times was observed (Supplemental Fig. 3).
TABLE I.
Tabulation of screen results
| Screen No. |
Total | |||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| No. colonies screened | 105 | 5 × 105 | 5 × 105 | 1.1 × 106 |
| Total No. OvR colonies/screen | 6 | 9 | 8 | 23 |
| No. colonies with A362T | 4 | 7 | 7 | 18 |
| No. colonies with Y444C | 1 | 1 | 1 | 3 |
| No. colonies with H382Y | 1 | 1 | 0 | 2 |
OvR, ovalicin resistance.
FIG. 2. Dot titrations of ovalicin-resistant human MetAP-2 alleles.
ΔMAP1 yeast were transformed with either wild type (WT) or ovalicin (Ov)-resistant alleles of human MetAP-2 (hMetAP-2). At 20 nm ovalicin the alleles found in the screen confer significant ovalicin resistance to ΔMAP1 yeast. Plates were photographed after a 6-day incubation at 30 °C.
Quantitation of Yeast Growth in the Presence of Ovalicin, Fumagillin, TNP-470
To quantitatively determine the relative resistance of the mutants to wild type human MetAP-2 and to each other, yeast growth in the presence of ovalicin, fumagillin, and TNP-470 was measured spectrophotometrically. Ovalicin was tested at concentrations of 1, 5, 10, 15, 20, and 30 nM. Significant growth differences between yeast expressing wild type or mutant MetAP-2 began appearing at 10 nM and were the most dramatic at 20 nM ovalicin (Table II). At this concentration, A362T exhibited ~63-fold more growth than wild type followed by Y444C at 33.5 and H382Y at 19. Also of note is the fact that A362T is about twice as resistant as Y444C and three times as resistant as H382Y in the presence of 20 nM ovalicin.
TABLE II.
Quantitation of growth of yeast expressing mutant MetAP-2 in the presence of ovalicin (Ov), fumagillin (Fg) and TNP-470
| MetAP-2 allele | -Fold growth over wild type at 20 nm Ov | -Fold growth over wild type at 50 nm Fg | -Fold growth over wild type at 750 nm TNP |
|---|---|---|---|
| A362T | 63.35 | 53.29 | 5.33 |
| Y444C | 33.55 | No growth | 4.32 |
| H382Y | 19.18 | 18.95 | 1.84 |
Fumagillin was tested at concentrations of 1, 5, 10, 20, 35, 50, 100, 250, and 500 nM. Growth differences between yeast expressing wild type or mutant MetAP-2 in the presence of fumagillin were observed over a larger range than with ovalicin (between 10 and 500 nM), with the maximal difference occurring at 50 nM. Table II summarizes the growth of strains expressing wild type and mutant human MetAP-2 in the presence of 50 nM fumagillin. A362T exhibits about a 53-fold increase in growth relative to wild type and H382Y displays a 19-fold increase. The A362T allele is more than twice as resistant as H382Y and continues to grow up to concentrations of 500 nM. Consistent with the previous dot titration observations, allele Y444C is not resistant to fumagillin. Finally, TNP-470 was tested at concentrations of 50, 100, 200, 300, 400, 500, 750, and 1000 nM. The concentrations are much higher due to our previous observation that TNP-470 is much less potent in arresting growth of S. cerevisiae than either ovalicin or fumagillin. The growth differences between wild type and mutant MetAP-2 are most clear at 750 nM, with A362T displaying greater than 5-fold growth over wild type followed by Y444C at about 4-fold and H382Y at almost 2-fold (Table II).
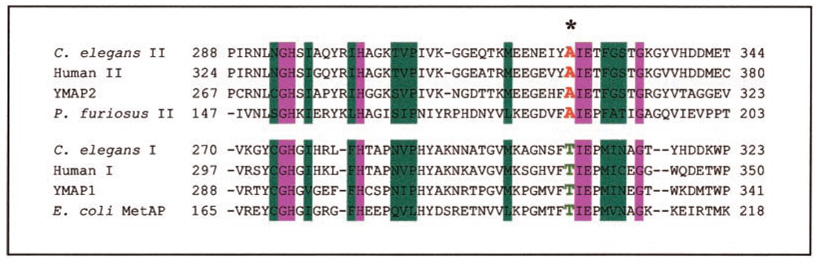
Because of the superior resistance of A362T to ovalicin, fumagillin, and TNP-470 and its abundant appearance in all three screens, this allele warranted further study. Alignment of MetAP-2 protein sequences demonstrated that alanine 362 of human MetAP-2 is completely conserved. In addition, alignment of MetAP-1 and MetAP-2 amino acid sequences revealed that a threonine residue at the corresponding position to A362 is completely conserved in MetAP-1 (Fig. 3). The fact that the A362T ovalicin-resistant allele of MetAP-2 has a threonine at this position led to the hypothesis that this threonine residue in wild type MetAP-1 might be responsible for its lack of ovalicin sensitivity.
FIG. 3. Further characterization of the A362T allele.
Alignment of the amino acid sequences of various MetAP-2 isoforms shows complete conservation of the alanine residue (as denoted by the asterisk). Alignment of MetAP-2 with MetAP-1 reveals that the analogous residue in MetAP-1 is a completely conserved threonine. Residues highlighted in blue are similar; residues highlighted in pink are identical. C. elegans, Caenorhabditis elegans; P. furiosus; Pyrococcus furiosus.
Generation of an Ovalicin-sensitive Human Methionine Aminopeptidase-1 Allele
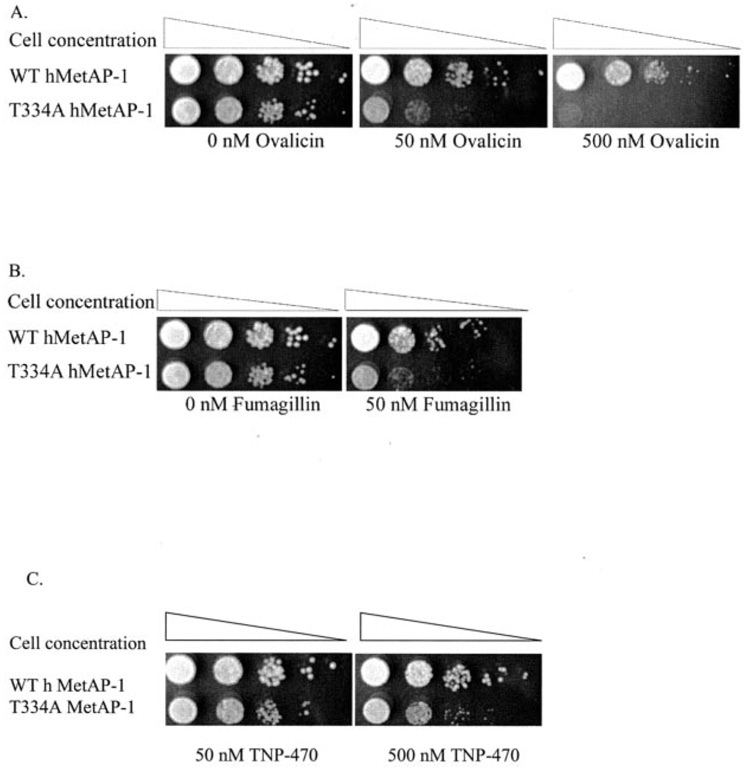
Based on sequence analysis with the MetAP-2 amino acid sequences, it was hypothesized that mutation of threonine 334 to alanine might render the MetAP-1 enzyme sensitive to ovalicin. Given that human MetAP-1 complements yeast MAP1 function (27), wild type and T334A human MetAP-1 were transformed into ΔMAP1 yeast and compared for ovalicin sensitivity. Interestingly, this mutation indeed renders human MetAP-1 sensitive to ovalicin (Fig. 4, panel A). The sensitivity of the T334A allele is not due to reduced in vivo function, as yeast strains expressing wild type and mutant human MetAP-1 do not differ significantly in their doubling times (Supplemental Fig. 4).
FIG. 4. Discovery of an ovalicin-sensitive human methionine aminopeptidase-1 allele.
Wild type (WT) and T334A human MetAP-1 alleles were expressed in ΔMAP1 yeast, and a dot titration was subsequently performed. Shown is substitution of threonine 334 with alanine renders human MetAP-1 (hMetAP-1) sensitive to ovalicin (A), fumagillin (B), and TNP-470 (C). Plates were photographed after a 2-day incubation at 30 °C.
All of the structural evidence to date suggests that the ovalicin analogue, fumagillin, is not easily accommodated by MetAP-1 due to the narrow specificity pocket (28). It was hypothesized that the bulk of the fumagillin hydrocarbon tail would prohibit its entry to the narrow MetAP-1 pocket. To test this hypothesis, yeast expressing wild type human MetAP-1 and T334A MetAP-1 were compared for sensitivity in the presence of fumagillin. Fig. 4, panel B, shows that although wild type MetAP-1 is resistant, T334A retains its sensitive phenotype when grown in the presence of fumagillin. Therefore we conclude that the hydrocarbon tail of fumagillin does not play a role in MetAP-1 resistance. To determine whether the sensitivity of T334A extends to the analog TNP-470, growth of yeast expressing wild type or T334A MetAP-1 was compared in the presence of TNP-470. The T334A allele was also sensitive to TNP-470 (Fig. 4C).
To quantitatively determine the relative resistance of wild type and T334A human MetAP-1, growth of yeast expressing these alleles was measured spectrophotometrically. As shown in Table III, yeast expressing the wild type allele of human MetAP-1 are ~60-fold more resistant to ovalicin and fumagillin than yeast expressing the T334A allele. In the presence of TNP-470 there is 2.5-fold difference in growth between strains expressing wild type and T334A. This is consistent with our previous studies on MetAP-2 alleles in the presence of TNP-470 and its poor efficacy in arresting yeast growth.
TABLE III.
Quantitation of growth of yeast expressing T334A allele of MetAP-1 in the presence of ovalicin, fumagillin, and TNP-470
| Drug, concentration of maximum effect | -Fold decrease in growth relative to wild type |
|---|---|
| Ovalicin, 100 nm | 59.2 |
| Fumagillin, 500 nm | 58.8 |
| TNP-470, 1 µm | 2.5 |
DISCUSSION
Although much has been learned about the structure and function of the metal-coordinating and catalytic residues of the MetAPs, the molecular basis for the specificity of the fumagillin class of angiogenesis inhibitors for MetAP-2 has remained elusive. Through the identification and characterization of ovalicin-resistant alleles of MetAP-2, we were able to further study the interaction between ovalicin and MetAP-2 and pinpoint the most critical residues underlying this specificity. The conclusions drawn from these results are strongly supported by 1) the reproducibility of obtaining these mutants among independent screens, 2) the conservation of key residues among MetAPs, and 3) the biological relevance of these mutations due to the fact that they were selected in vivo.
The alleles found in this screen offer some interesting insight into the manner in which the ovalicin class of compounds inhibits MetAP-2. According to Liu et al. (28) tyrosine 444 of human MetAP-2 provides hydrophobic contacts to fumagillin, which may serve to stabilize the drug in the active site. Although Y444C is resistant to both ovalicin and TNP-470, it was a surprise to discover that Y444C was not resistant to fumagillin. This suggests that although tyrosine 444 is critical to ovalicin and TNP-470 stabilization in the active site, this residue is not critical for fumagillin. It is important to note that this tyrosine is conserved; thus, the provision of such hydrophobic contacts to ovalicin is conserved among MetAP-2 enzymes as well. Our surprise finding that Y444C is not resistant to fumagillin suggests that fumagillin forms critical hydrophobic contacts with residues other than 444. These hydrophobic contacts would most likely occur between MetAP-2 and the long hydrocarbon tail present in fumagillin that is lacking in both ovalicin and TNP-470. In contrast to the insight gained from the Y444C mutation, the mechanism of H382Y ovalicin resistance is elusive. Previously, superimposition of the crystal structures of human MetAP-2 and E. coli MetAP (a Type I MetAP) revealed that His-382 of human MetAP-2 corresponds in space to Tyr-65 of E. coli MetAP (29). Protein sequence alignment of the type I MetAPs revealed that Tyr-65 of E. coli MetAP corresponds to a phenylalanine in human, yeast, and worm MetAP-1. However, the importance of an imidazole versus a phenyl group at position 382 for ovalicin sensitivity remains unknown.
Because of the fact that the A362T mutation occurred in the majority of the resistant colonies isolated in the screen and was the most resistant to ovalicin/fumagillin/TNP-470, this allele was extensively characterized. In the absence of structural determination of the A362T mutant human MetAP-2 protein, the mechanism by which this allele functions is currently speculative. One possibility is that mutation of 362 from an alanine to a larger threonine residue may push histidine 331 into the space where histidine 339 resides after ovalicin binding (28) and as such could prevent entry of the drug into the binding site. To test this hypothesis we attempted to reverse the ovalicin resistance phenotype of the A362T allele by constructing a double mutant (A362T,H339A) to relieve potential steric hindrance between 339 and 331. Unfortunately we were not able to confirm this model because the double mutant allele was not functional (data not shown), suggesting a critical role for His-339 in catalysis. This finding was supported by Griffith et al. (30), who found that the H339N mutant allele lost almost all catalytic activity. Another possible explanation for resistance of the A362T mutant is that threonine 362 may simply force histidine 331 directly into the space where ovalicin would reside when bound. Either of these two hypotheses could account for the prevention of ovalicin binding and the lack of inactivation of the peptidase activity of MetAP-2.
Given the similarity in enzymatic function of the MetAPs, the specificity for MetAP-2 over MetAP-1 of the fumagillin class of angiogenesis inhibitors is quite remarkable. Previously, in an attempt to explain this specificity, the structures of E. coli MetAP, a type I MetAP (31), and human MetAP-2 were compared (28). This comparison suggested that the active sites of Type I and Type II MetAPs are very similar except for the fact that the catalytic histidine in E. coli MetAP appears sufficiently distant from fumagillin so as not to form a bond, thus suggesting a reason for the difference in specificity. However, support for the ability of fumagillin to interact with MetAP-1 comes from a study by Lowther et al. (32), which showed that fumagillin covalently modifies the active site histidine in E. coli MetAP in vitro, albeit at significantly higher concentrations (2.4–2.7 mm ovalicin or fumagillin) than needed to inhibit the T334A allele of human MetAP-1 (20 nm).
A potential application of the discovery of ovalicin-resistant alleles is to answer definitively the question of whether MetAP-2 mediates the cytostatic response of endothelial cells to the fumagillin class of angiogenesis inhibitors. Although there is much evidence to support this hypothesis, it has not been proven conclusively. These resistant alleles may also have the potential to explain differential responses of humans to TNP-470 in clinical trials. Such varied responses may be due to differential progression of the cancer among subjects and possibly also to genetic predisposition. A study of angiogenesis in inbred mouse strains demonstrates that there is angiogenic heterogeneity in mice (33). Interestingly, one of the strains in the study exhibited resistance to TNP-470 in the corneal micropocket assay. If the genetic basis for response to angiogenesis inhibitors is shared by humans, perhaps a resistant allele of MetAP-2 that is functional but blocks drug efficacy (such as A362T) could be responsible in some of the cases that are not responsive to TNP-470 therapy.
The discovery of ovalicin-resistant MetAP-2 alleles and an ovalicin-sensitive MetAP-1 allele suggests that the geometry of the active sites of these enzymes is far more similar than had originally been presumed and that, remarkably, this specificity is due to a single amino acid difference between the two enzymes. Based on the mutant phenotypes, sequence alignments, and information obtained from the crystal structures, it appears that an alanine residue at this crucial position is the determining factor for ovalicin interaction and inhibition of the MetAP enzymes. Our work highlights the similarities of the active sites of these enzymes while providing an explanation for their sensitivity to ovalicin in vivo. Determination of the crystal structures of A362T human MetAP-2 and T334A human MetAP-1 would elucidate how these mutations affect the movement of amino acids in the binding pocket in response to ovalicin binding. Such studies would enable structure-based drug development of new MetAP-1 and MetAP-2 inhibitors, which is of considerable interest both from a chemical synthesis perspective (34–37) as well as a clinical one. Taken together, our findings offer the strongest evidence to date of the key structural elements of the MetAPs that define specificity of inhibition by fumagillin and its analogues.
Footnotes
This work was supported by the National Institutes of Health Grant RO1 CA83049.
The on-line version of this article (available at http://www.jbc.org) contains Supplemental Figs. 1–4.
The abbreviation used is: MetAP, methionine aminopeptidase.
REFERENCES
- 1.Ingber D, Fujita T, Kishimoto S, Sudo K, Kanamaru T, Brem H, Folkman J. Nature. 1990;348:555–557. doi: 10.1038/348555a0. [DOI] [PubMed] [Google Scholar]
- 2.Kusaka M, Sudo K, Fujita T, Marui S, Itoh F, Ingber D, Folkman J. Biochem. Biophys. Res. Commun. 1991;174:1070–1076. doi: 10.1016/0006-291x(91)91529-l. [DOI] [PubMed] [Google Scholar]
- 3.Peacock DJ, Banquerigo ML, Brahn E. Cell. Immunol. 1995;160:178–184. doi: 10.1016/0008-8749(95)80025-e. [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly MS, Brem H, Folkman J. J. Pediatr. Surg. 1995;30:325–330. doi: 10.1016/0022-3468(95)90583-9. [DOI] [PubMed] [Google Scholar]
- 5.Shusterman S, Grupp SA, Barr R, Carpentieri D, Zhao H, Maris JM. Clin. Cancer Res. 2001;7:977–984. [PubMed] [Google Scholar]
- 6.Dezube BJ, Von Roenn JH, Holden-Wiltse J, Cheung TW, Remick SC, Cooley TP, Moore J, Sommadossi JP, Shriver SL, Suckow CW, Gill PS. J. Clin. Oncol. 1998;16:1444–1449. doi: 10.1200/JCO.1998.16.4.1444. [DOI] [PubMed] [Google Scholar]
- 7.Kruger EA, Figg WD. Expert Opin. Investig. Drugs. 2000;9:1383–1396. doi: 10.1517/13543784.9.6.1383. [DOI] [PubMed] [Google Scholar]
- 8.Kudelka AP, Levy T, Verschraegen CF, Edwards CL, Piamsomboon S, Termrungruanglert W, Freedman RS, Kaplan AL, Kieback DG, Meyers C, Jaeckle KA, Loyer E, Steger M, Mavligit G, Killian A, Tang RA, Gutterman JU, Kavanagh JJ. Clin. Cancer Res. 1997;3:1501–1505. [PubMed] [Google Scholar]
- 9.Stadler WM, Kuzel T, Shapiro C, Sosman J, Clark J, Vogelzang NJ. J. Clin. Oncol. 1999;17:2541–2545. doi: 10.1200/JCO.1999.17.8.2541. [DOI] [PubMed] [Google Scholar]
- 10.Sin N, Meng L, Wang MQ, Wen JJ, Bornmann WG, Crews CM. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith EC, Su Z, Turk BE, Chen S, Chang YH, Wu Z, Biemann K, Liu JO. Chem. Biol. 1997;4:461–471. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Sheppard GS, Lou P, Kawai M, Park C, Egan DA, Schneider A, Bouska J, Lesniewski R, Henkin J. Biochemistry. 2003;42:5035–5042. doi: 10.1021/bi020670c. [DOI] [PubMed] [Google Scholar]
- 13.Chang SY, McGary EC, Chang S. J. Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Chang YH. Proc. Natl. Acad. Sci. U. S. A. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arfin SM, Kendall RL, Hall L, Weaver LH, Stewart AE, Matthews BW, Bradshaw RA. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiestl RH, Gietz RD. Curr. Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 17.Hieter P, Pridmore D, Hegemann JH, Thomas M, Davis RW, Philippsen P. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 18.Colicelli J, Birchmeier C, Michaeli T, O’Neill K, Riggs M, Wigler M. Proc. Natl. Acad. Sci. U. S. A. 1989;86:3599–3603. doi: 10.1073/pnas.86.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballester R, Michaeli T, Ferguson K, Xu HP, McCormick F, Wigler M. Cell. 1989;59:681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- 20.Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O’Connell P, Cawthon RM, Innis MA, McCormick F. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 21.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 22.Chang YH, Teichert U, Smith JA. J. Biol. Chem. 1992;267:8007–8011. [PubMed] [Google Scholar]
- 23.Hieter P, Mann C, Snyder M, Davis RW. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 24.Weisman R, Finkelstein S, Choder M. J. Biol. Chem. 2001;276:24736–24742. doi: 10.1074/jbc.M102090200. [DOI] [PubMed] [Google Scholar]
- 25.Muhlrad D, Hunter R, Parker R. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 26.Fink G, Guthrie C. Methods Enzymol.: Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego, CA: Academic Press; 1991. pp. 322–323. [Google Scholar]
- 27.Dummitt B, Fei Y, Chang YH. Protein Pept. Lett. 2002;9:295–303. doi: 10.2174/0929866023408607. [DOI] [PubMed] [Google Scholar]
- 28.Liu S, Widom J, Kemp CW, Crews CM, Clardy J. Science. 1998;282:1324–1327. doi: 10.1126/science.282.5392.1324. [DOI] [PubMed] [Google Scholar]
- 29.Lowther WT, Matthews BW. Biochim. Biophys. Acta. 2000;1477:157–167. doi: 10.1016/s0167-4838(99)00271-x. [DOI] [PubMed] [Google Scholar]
- 30.Griffith EC, Su Z, Niwayama S, Ramsay CA, Chang YH, Liu JO. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15183–15188. doi: 10.1073/pnas.95.26.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roderick SL, Matthews BW. Biochemistry. 1993;32:3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- 32.Lowther WT, McMillen DA, Orville AM, Matthews BW. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12153–12157. doi: 10.1073/pnas.95.21.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohan RM, Fernandez A, Udagawa T, Yuan J, D’Amato RJ. FASEB J. 2000;14:871–876. doi: 10.1096/fasebj.14.7.871. [DOI] [PubMed] [Google Scholar]
- 34.Kwon JY, Jeong HW, Kim HK, Kang KH, Chang YH, Bae KS, Choi JD, Lee UC, Son KH, Kwon BM. J. Antibiot. (Tokyo) 2000;53:799–806. doi: 10.7164/antibiotics.53.799. [DOI] [PubMed] [Google Scholar]
- 35.Han CK, Ahn SK, Choi NS, Hong RK, Moon SK, Chun HS, Lee SJ, Kim JW, Hong CI, Kim D, Yoon JH, No KT. Bioorg. Med. Chem. Lett. 2000;10:39–43. doi: 10.1016/s0960-894x(99)00577-6. [DOI] [PubMed] [Google Scholar]
- 36.Baldwin JE, Bulger PG, Marquez R. Tetrahedron. 2002;58:5411–5452. [Google Scholar]
- 37.Son KH, Kwon JY, Jeong HW, Kim HK, Kim CJ, Chang YH, Choi JD, Kwon BM. Bioorg. Med. Chem. 2002;10:185–188. doi: 10.1016/s0968-0896(01)00268-1. [DOI] [PubMed] [Google Scholar]