Abstract
Although many proteasome inhibitors have been either synthesized or identified from natural sources, the development of more sophisticated, selective proteasome inhibitors is important for a detailed understanding of proteasome function. We have found that antitumor natural product epoxomicin and eponemycin, both of which are linear peptides containing a α,β-epoxyketone pharmacophore, target proteasome for their antitumor activity. Structural studies of the proteasome–epoxomicin complex revealed that the unique specificity of the natural product toward proteasome is due to the α,β-epoxyketone pharmacophore, which forms an unusual six-membered morpholino ring with the amino terminal catalytic Thr-1 of the 20S proteasome. Thus, we believe that a facile synthetic approach for α,β-epoxyketone linear peptides provides a unique opportunity to develop proteasome inhibitors with novel activities. In this chapter, we discuss the detailed synthetic procedure of the α′,β′-epoxyketone natural product epoxomicin and its derivatives.
Introduction
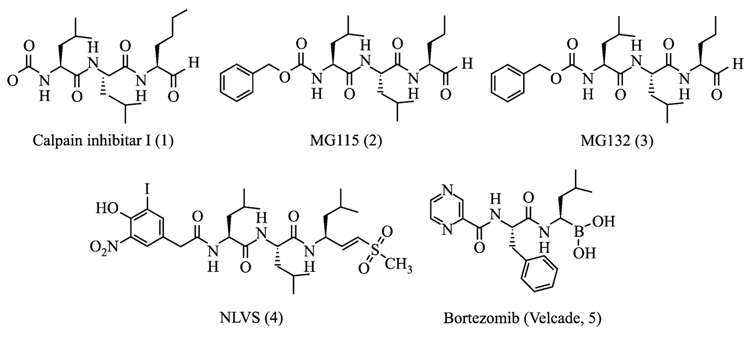
Studies into the chemical synthesis of proteasome inhibitors were initiated during the early 1990s after standard serine/cysteine protease inhibitors were shown to inhibit the 20S proteasome (Figueiredo-Pereira et al., 1994; Orlowski, 1990). Early synthetic efforts were largely focused on the modification of the amino acid sequence of the serine/cysteine protease inhibitors. For example, MG115 and MG132, which were developed by Rock and colleagues (1994) and have been widely used in proteasome biology, are tripeptide aldehydes that share similar peptide backbones and aldehyde pharmacophores with known protease inhibitors such as calpain inhibitor 1 (Fig. 1). Other known peptide-based protease inhibitors possessing different pharmacophores such as vinylketones (Bogyo et al., 1997, 1998) and boronates (Adams, 2002; Adams et al., 1998; Iqbal et al., 1996) have also been developed as proteasome inhibitors (Fig. 1). The major advantage of these peptide backbone–based proteasome inhibitors is their ease of preparation and derivatization, potentially providing an easy access for the development of proteasome inhibitors with novel activities. Although these peptide inhibitors, in general, are cell-permeable potent inhibitors of the 20S proteasome and are still widely used in the study of the role of proteasome in many cellular processes, the cross-reactivity with other proteases remains a major concern for their use as molecular probes in dissecting complex signaling pathways (Kim and Crews, 2003; Kisselev and Goldberg, 2001; Myung et al., 2001a).
FIG. 1.
Proteasome inhibitors derived from other known peptide-based protease inhibitors possessing common pharmacophores.
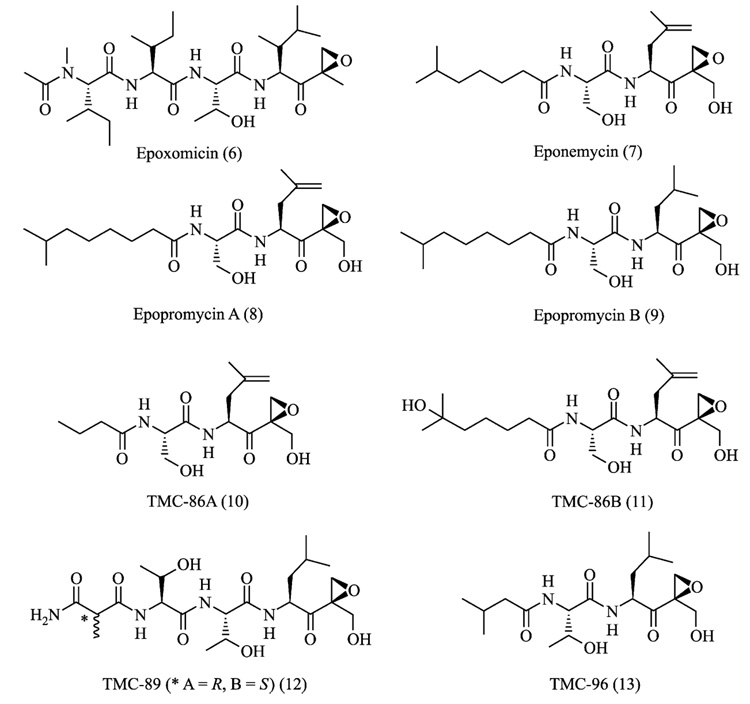
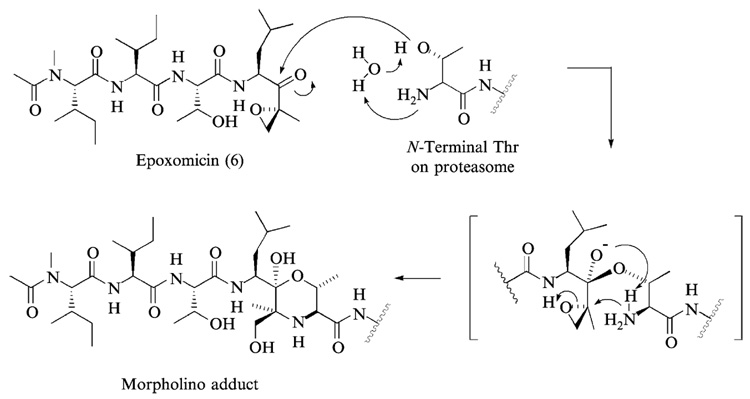
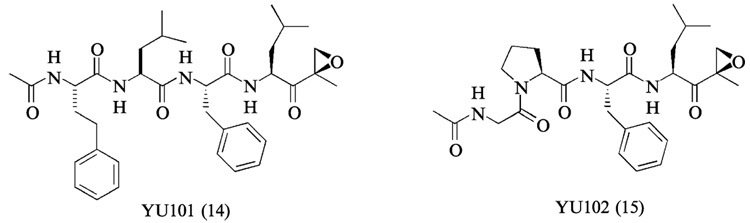
In addition to synthetic approaches, natural products have also provided both peptide and nonpeptide proteasome inhibitors (Kim and Crews, 2003). Examples include epoxomicin (Hanada et al., 1992), eponemycin (Sugawara et al., 1990), lactacystin (Fenteany et al., 1995), TMC-95s (Koguchi et al., 2000a), phepropeptins (Sekizawa et al., 2001), and epigal-locatechin-3-gallate (ECGC) (Nam et al., 2001). Among these, epoxomicin and eponemycin are members of a growing family of α′,β′-epoxyketone natural products having a linear peptide structure (Fig. 2) (Koguchi et al., 1999, 2000b,c; Sugawara et al., 1990; Tsuchiya et al., 1997). We reported the first total synthesis of epoxomicin (Sin et al., 1999), isolated from an unidentified actinomycete strain No.Q996–17, and showed that epoxomicin potently inhibits the 20S proteasome (Meng et al., 1999b) using an affinity reagent–labeled epoxomicin (Meng et al., 1999a; Sin et al., 1998). Interestingly, antitumor natural products epoxomicin and eponemycin are shown to be specific for the proteasome despite their common peptide backbone that resembles structures of known serine/cysteine protease inhibitors (Meng et al., 1999b; Sin et al., 1999). Structural studies of the yeast 20S proteasome complexed with epoxomicin revealed that the unique specificity of epoxomicin is due to the formation of an unusual six-membered morpholino ring between the amino terminal catalytic Thr-1 of the 20S proteasome and the α′,β′-epoxyketone pharmacophore of epoxomicin (Fig. 3) (Groll et al., 2000). The facile synthetic strategy of α′,β′-epoxyketone linear peptides developed through the total synthesis of epoxomicin and the unique specificity of its α′,β′-epoxyketone pharmacophore for proteasome have prompted the development of proteasome inhibitors possessing higher potency or novel inhibitory specificities, such as YU101 (Elofsson et al., 1999) and YU102 (Myung et al., 2001b) (Fig. 4), respectively.
FIG. 2.
α′,β′-Epoxyketone-containing proteasome inhibitors from natural sources.
FIG. 3.
The mechanism of proteasome inhibition by epoxomicin is proposed on the basis of the x-ray structure of yeast 20S proteasome–epoxomicin complex. It is postulated that the unique specificity of epoxomicin is due to the formation of an unusual six-membered morpholino ring between Thr-1 of the catalytic subunit of 20S proteasome and the α′,β′-epoxyketone pharmacophore of epoxomicin.
FIG. 4.
Synthetic α′,β′-epoxyketone proteasome inhibitors designed to target a certain proteasomal proteolytic activity/subunit with a high degree of specificity. YU101 is a chymotrypsin-like activity (CT-L)-selective inhibitor, whereas YU102 is shown to be specific for caspase-like activity.
In this chapter, we discuss the synthesis and characterization of this important class of proteasome inhibitors, focusing on the α′,β′-epoxyketone natural product epoxomicin and its derivatives. It should be noted that the synthetic strategy of epoxomicin described in this chapter can be applied easily to the development of proteasome inhibitors with novel activities.
Synthesis Strategy of α′,β′-Epoxyketone Peptide Inhibitors: Total Synthesis of Epoxomicin
General
There are two main strategies for the chemical synthesis of linear peptides (Fields, 1997; Lloyd-Williams et al., 1997). First, chain elongation in linear synthesis is carried out by repetitive Nα-amino group deprotection and protected amino acid coupling steps. Alternately, convergent synthesis involves the synthesis and coupling of protected peptide segments. Both strategies can be carried out in solution or on a solid support, although solution and solid-phase methods can coexist in convergent synthesis. For the synthesis of epoxomicin, chain elongation on solid support may be a challenging task because of the C-terminus epoxyketone pharmacophore, which lacks a handle for resin attachment. Solution phase chain elongation was also ruled out because of the possibility of decomposition of epoxyketone group during repetitive coupling and deprotection reactions. On the basis of these considerations, a convergent approach was chosen for the total synthesis of epoxomicin (Fig. 5).
FIG. 5.
A convergent approach for the total synthesis of epoxomicin.
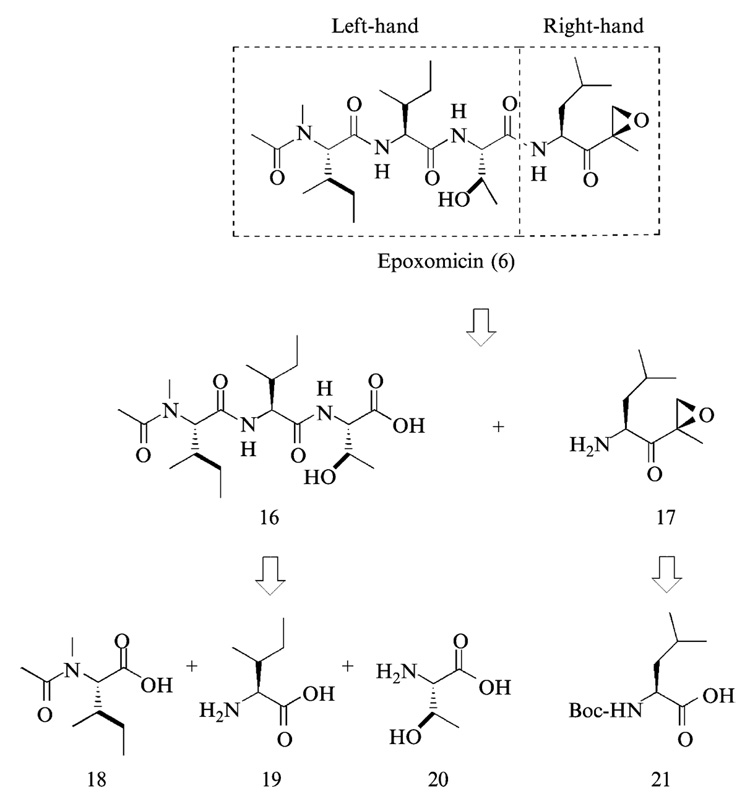
The convergent approach in the synthesis of epoxomicin involves: (1) solution-phase synthesis of the right-hand fragment, (2) solution or solid-phase synthesis of left-hand fragment, (3) solution-phase assembly of complete peptide backbone of epoxomicin, and (4) purification and characterization of the coupled product. After the final coupling, HPLC purification is carried out to separate epoxomicin from the stereoisomeric counterpart generated during the final coupling reaction. Finally, the enzymatic inhibitory activity of epoxomicin was measured using purified bovine proteasome.
Synthesis of the Right-Hand Fragment of Epoxomicin
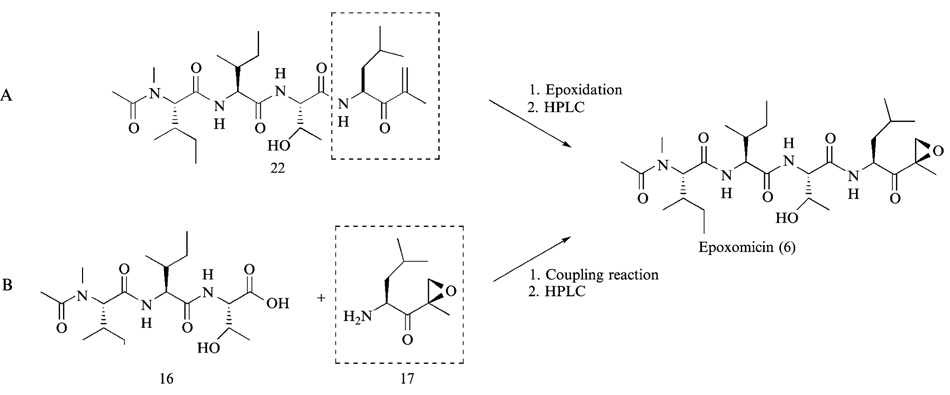
There are two alternative approaches to introduce an epoxy group in the course of synthesis of epoxomicin. First, the epoxidation reaction can be performed after complete assembly of the peptide backbone of epoxomicin, and the resulting desired product is purified from the mixture of epoxomicin and epi-epoxomicin by HPLC (Schmidt and Schmidt, 1994) (Fig. 6A). The second approach relies on the preparation and isolation of stereochemically defined α′,β′-epoxyketone leucine (Bennacer et al., 2003; Dobler, 2001; Hoshi et al., 1993; Iwabuchi et al., 2001; Sin et al., 1998), which is then coupled to the tripeptide left-hand fragment to yield the complete backbone of epoxomicin (Fig. 6B). Because the α′,β′-epoxyketone leucine having the same configuration as that of epoxomicin can be easily chromatographically purified from its stereoisomeric counterpart, the second approach was initially chosen to prepare the right-hand fragment of epoxomicin (Fig. 7).
FIG. 6.
Two potential strategies for the introduction of a α′,β′-epoxyketone group.
FIG. 7.
Synthesis of the right-hand fragment (leucine epoxyketone).
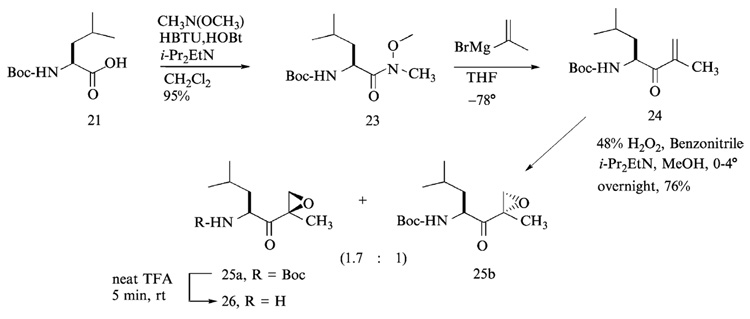
As the first step of the right-hand fragment synthesis, α-tert-butyloxy-carbonyl (Boc)-leucine (Boc-Leu-OH) was coupled to N-methoxy-N-methylamine with O-benzotriazo-1-yl-N,N,N′, N′-tetramethyluronium hexafluorophosphate (HBTU) and 1-hydroxybenzotriazole (HOBt) to yield the Boc-leucine Weinreb amide 23. Readily preparable Weinreb amides (N-methoxy-N-methylamides) (Nahm and Weinreb, 1981) are known to couple in good yields with Grignard and organolithium reagents to produce ketones, and to be reduced with hydrides to afford aldehydes. Fluoren-9-ylmethoxycarbonyl-leucine (Fmoc-Leu-OH), however, cannot be used, because the Fmoc group is unstable under Grignard or organolithium reaction conditions, which are used to introduce the α′,β′-unsaturated ketone 24 in the following step.
Reaction of α-Boc-leucine-Weinreb amide 23 with commercially available propen-2-yl magnesium bromide (Grignard reagent) led to the formation of α-Boc-amino-α′,β′-unsaturated ketone 24 without racemization at low temperature (−78° to room temperature) in tetrahydrofuran (THF). The nucleophilic addition proceeds through stable metal-chelate intermediates that block over-addition. The resulting α-Boc-amino-α′,β′-unsaturated ketone 24 was readily purified by flash column chromatography using hexanes-ethyl acetate system (10:1, v/v) as eluant. Subsequent epoxidation of α-Boc-leucine-α′,β′-unsaturated ketone with alkaline hydrogen peroxide (Schmidt and Schmidt, 1994) in methanol yielded a mixture of epoxide stereoisomers; the ratio of 2-(R)-epoxide 25a to 2-(S)-epoxide 25b was 1.7. The two isomers of leucine epoxyketone were readily separated by flash column chromatography using hexanes-ethyl acetate (5:1, v/v) system. The isomer (2-(R)-epoxide) 25a, which migrates faster than the 2-(S)-epoxide 25b in thin-layer chromatography (TLC, hexanes-ethyl acetate = 5:1, v/v), was found to have the same configuration as that of epoxomicin (Sin et al., 1999). Finally, the Boc group of Boc-Leu-α′,β′-epoxyketone 25a was deprotected by brief treatment (~5 min) with neat trifluoroacetic acid (TFA) without concomitant opening of the epoxide ring. Excess TFA was removed under high vacuum. The resulting TFA salt of leucine-α′,β′-epoxyketone was used without further purification for the final coupling reaction with the left-hand fragment.
Synthesis of the Left-Hand Fragment
The tripeptide left-hand fragment of epoxomicin can be readily prepared either in solution or on a solid support (Sin et al., 1999). Given that peptide synthesis on a solid support has been a subject of many extensive reviews (Fields, 1997), in this chapter we discuss only the solution-phase synthesis.
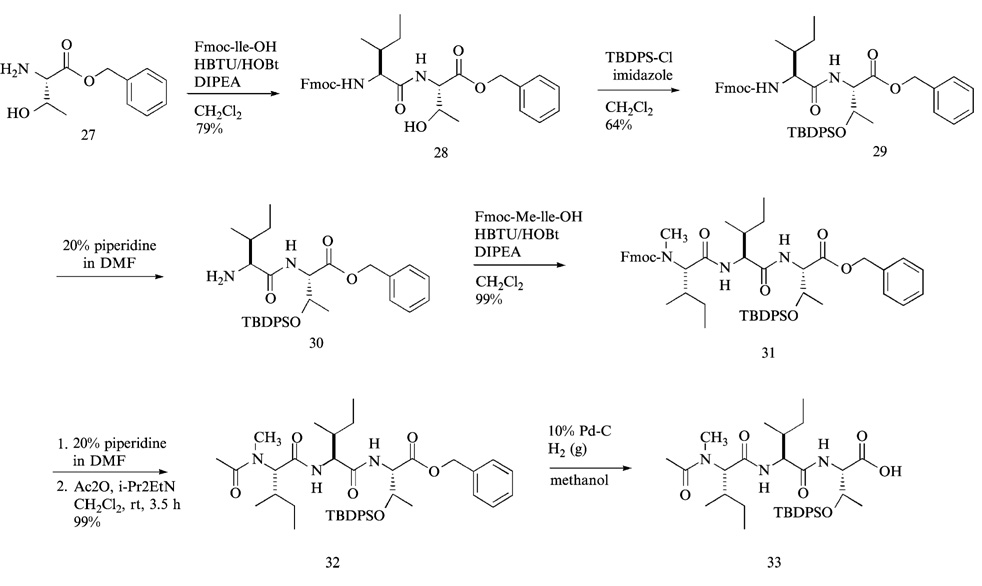
In the first step (Fig. 8), Fmoc-isoleucine-OH was coupled to threonine benzylester 27 with HBTU and HOBt. Although there are many coupling reagents that may be used for peptide bond formation, HBTU/HOBt coupling system was found to be very effective for reducing loss of configuration at the carboxylic acid residue in this coupling reaction. The resulting dipeptide 28 was purified by flash column chromatography using hexanes-ethyl acetate system (1:1, v/v) as eluant. The solubility of the dipeptide 28 was so poor that it was readily precipitated during flash column chromatography, requiring a large volume of elution solvent to redissolve and collect the dipeptide. The protection of threonine hydroxyl group of the dipeptide 28 was then accomplished with tert-butyldiphenylsilylchloride (TBDPSCl) in methylene chloride (CH2Cl2) at room temperature. The same TBDPSCl-protection reaction, however, was less effective in tetrahydrofuran (THF). The advantages of using TBDPS protecting group compared with other commonly used protecting groups such as tert-butyldimethylsilyl (TBDMS) and trimethylsilyl (TMSCl) are: (1) enhanced solubility of TBDPS protected peptides in organic solvents facilitating the following solution-phase reactions and purification process; (2) better stability under acid and base conditions; (3) easy UV detection provided by the two phenyl rings of TBDPS group, thus aiding the HPLC purification of completely assembled TBDPS protected epoxomicin from the stereoisomeric counterpart generated during the final coupling of the left- and right-hand fragments.
FIG. 8.
Preparation of the tripeptide left-hand fragment.
The Fmoc group of TBDPS-protected dipeptide 29 was deprotected using standard protocol (20% piperidine in dimethylformamide, v/v) (Fig. 8). The resulting Fmoc-deprotected dipeptide 30 was purified by flash column chromatography. In this type of Fmoc deprotection reaction, only one fourth or one fifth of the normal quantity of dried silica (SiO2) that is used for routine column chromatography was packed for the purification of the Fmoc-deprotected product. Specifically, after directly loading the Fmoc-deprotected crude product into the silica gel column, fast-migrating Fmoc-adducts, which are shown as bright fluorescent spots under UV, were eluted away using a solvent system (hexanes-ethyl acetate, 1:1, v/v); under this elution condition, the Fmoc-deprotected product 30 was retained in the silica gel column. The retained deprotected dipeptide 30 was then eluted and collected using a different solvent system (CH2Cl2-MeOH, 9:1, v/v).
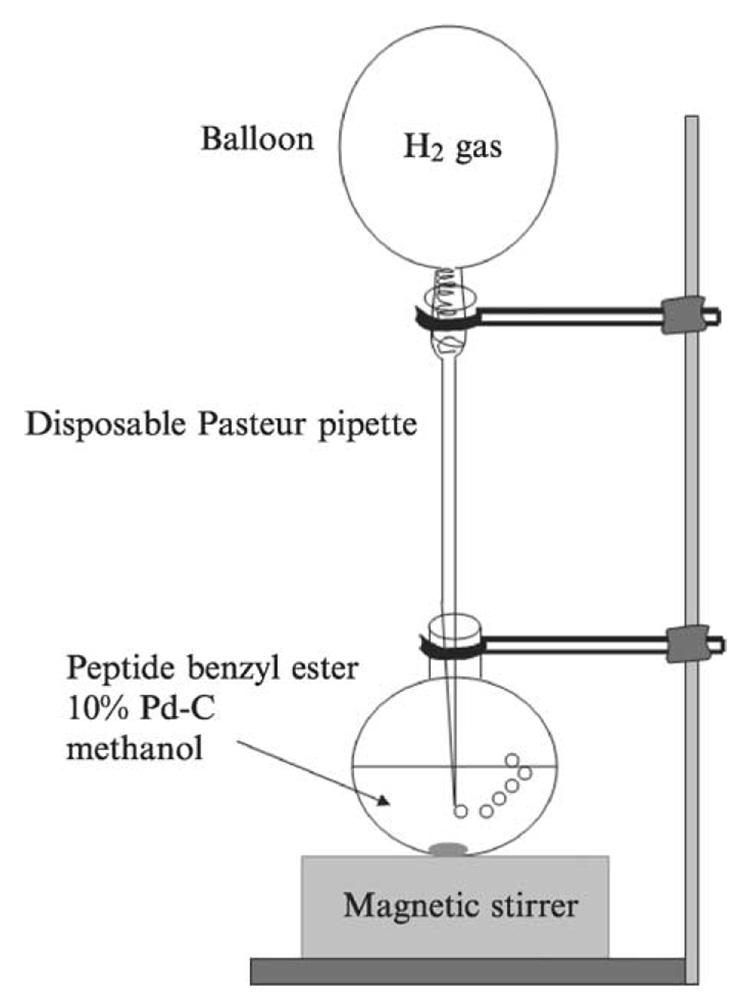
Next, Fmoc-N-methyl-isoleucine was coupled to the deprotected dipeptide 30 with HBTU/HOBt to yield the tripeptide 31, which was easily purified by flash column chromatography (hexanes-ethyl acetate, 1:1, v/v). After the Fmoc-protected tripeptide 31 was treated with 20% piperidine in DMF (v/v) for 20 min at room temperature, DMF and piperidine were removed under high vacuum. Without further purification, the crude product was mixed with excess acetic anhydride in pyridine for 1 h at room temperature. The resulting N-acetylated tripeptide 32 was purified by flash column chromatography (hexanes-ethyl acetate, 1:1, v/v). Finally, the C-terminus benzyl group of the left-hand fragment 32 was deprotected by catalytic hydrogenolysis mediated by 10% activated palladium-charcoal in methanol under hydrogen gas atmosphere. In this reaction, instead of performing hydrogenolysis under high pressure, a stream of hydrogen gas was directly applied to the reaction mixture providing hydrogen bubbles into the solution (Fig. 9). The benzyl protecting group was readily removed in nearly quantitative yield, and the reaction mixture was filtered through a coarse, Celite-packed, fritted-glass filter to afford the TBDPS-protected left-hand fragment 33, which was coupled to the right-hand fragment without further purification for the final coupling with the right-hand fragment.
FIG. 9.
Schematic of hydrogenation reaction, in which hydrogen gas is provided to the reaction mixture through a stream of hydrogen gas.
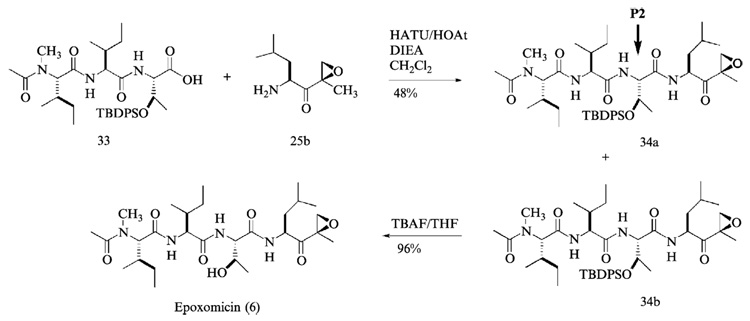
Assembly of Complete Epoxomicin Peptide Backbone
The final coupling reaction between the right- and left-hand fragments of epoxomicin was performed with O-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) and 1-hydroxy-7-azabenzotriazol (HOAt) to give TBDPS protected epoxomicin 34b (Fig. 10). Unlike the coupling reactions performed during the synthesis of left-hand fragment, HATU was used for the final assembly of epoxomicin peptide backbone to enhance the efficiency of coupling reaction. HATU was developed by Carpino (Albericio and Carpino, 1997; Carpino, 1993) and shown to be particularly effective with hindered couplings. After overnight stirring at room temperature, the resulting TBDPS protected epoxomicin 34b was initially purified by flash column chromatography (hexanes-ethyl acetate, 1:1, v/v) to verify the successful assembly of complete peptide backbone of epoxomicin by 1H NMR. 1H NMR spectra analysis showed the complete assembly of epoxomicin peptide backbone and the presence of minor stereoisomer 34a (5–15% of the assembled product), the formation of which during the amide bond formation is well documented (Carpino, 1993, 1997). It is assumed that the loss of configuration occurs at the carboxylic acid residue of P2 amino acid during the final coupling reaction (Fig. 10). It was difficult to purify the TBDPS protected epoxomicin 34b from the isomer 34a using standard column chromatography, but it was readily separated by normal-phase HPLC (hexanes-isopropanol, linear gradient, hexanes 100% to 50%). In normal-phase HPLC, the TBDPS-protected epoxomicin 34b displayed a longer retention time compared with its stereoisomeric counterpart 34a. Finally, treatment of the TBDPS protected epoxomicin with tetrabutylammonium fluoride (TBAF) gave epoxomicin (Fig. 10), which was purified by silica gel column chromatography (CH2Cl2-MeOH, 98:2, v/v), followed by reverse-phase HPLC.
FIG. 10.
The final assembly of epoxomicin peptide backbone and preparation of epoxomicin.
Purification of Epoxomicin by HPLC
Epoxomicin was purified by RP-HPLC (Rainin Dynamax) system composed of two solvent delivery pumps (Model SD200) and variable wavelength detector (Model UV-1) set at 214 nm. The column used was an YMC-Pack ODS-AM, 250 mm × 20 mm, 5-µm particles size and 120 Å pore size (Waters, Milford, MA) coupled to a guard column ODS-AM (10 mm × 10 mm) with same stationary phase specifications. The separation was carried out with a linear gradient of solvent A (water) into solvent B (MeOH): 65% of B to 80% of B over 90 min, run at 5.0 ml/min, room temperature. Epoxomicin was eluted at 49 min and the peak was collected as seven fractions of nearly 1 ml each.
Chemical Characterization of Epoxomicin
Analytical HPLC traces of collected fractions were carried out in a Waters Separation Module 2795 coupled to a Waters 2795 Photodiode Array Detector (set at 214 nm) and to a Micromass ZQ 2000 Electrospray Mass detector (cone voltage = +20V). A linear gradient of solvent A (water) in solvent B (MeOH), 30% of B to 90% of B in 30 min was run at 0.2 ml/min, at 25°. The analytical column used was an XTerra MS C18 (4.6 mm × 50 mm, 2.5-µm particle size, and 80 Å pore size).
Epoxomicin eluted at 19.8 min [observed m/z = 555.2 (M + H)+, 577.6 (M + Na)+]. Fractions showing purity ≥98% were grouped together and vacuum-dried with no heat until only water remained. Samples were then lyophilized. Epoxomicin was further characterized by 1H NMR and 13C NMR (Meng et al., 1999b; Sin et al., 1999).
Development Strategy of Proteasome Inhibitors with Novel Activities
General
The emergence of the ubiquitin-proteasome pathway as a major player in many important biological processes has prompted many synthetic efforts in proteasome inhibitor development (Kim and Crews, 2003; Myung et al., 2001a). In addition, systematic natural product screening has also provided a number of novel proteasome inhibitors (Kim and Crews, 2003). Thus far, most of these proteasome inhibitors have been shown to target the chymotrypsin-like (CT-L) activity. Although the CT-L activity of the proteasome has been shown to be largely responsible for the proteolytic function of the proteasome in vivo and in vitro (Myung et al., 2001b), the contribution of other major activities remains to be determined. Therefore, a novel class of inhibitors that target individual catalytic subunit/activity of proteasome may be required to dissect the role of each catalytic subunit.
Exposure of cells to stimuli such as IFN-γ, TNF-α, and LPS induces the synthesis of certain catalytic subunits (respectively, LMP2, MECL-1, and LMP7) that together are incorporated into alternative proteasome form (Kloetzel, 2001). This isoform, known as the immunoproteasome, has an enhanced capacity to generate peptides bearing hydrophobic and basic amino acids at their C-termini and a reduced capacity to produce peptides bearing acidic residues at their C-terminus (Kloetzel, 2001; Rock and Goldberg, 1999). Consequently, the spectrum of the produced peptides is shifted toward peptides that associate with MHC class I molecules with increased affinity (Fruh et al., 1994), implicating a major role of immunoproteasome in antigen presentation. Furthermore, it has been suggested that the immunoproteasome may be involved in some pathological processes, such as diabetes and autoimmune diseases (Casp et al., 2003). However, the exact role of the immunoproteasome remains unclear, caused in large part by the lack of appropriate molecular probes. Given that currently available proteasome inhibitors target both the constitutive proteasome and the immunoproteasome, development of immunoproteasome-specific inhibitors may be useful in the dissection of the role of immunoproteasomes.
Lessons from SAR Studies on the Natural Products Epoxomicin and Dihydroeponemycin
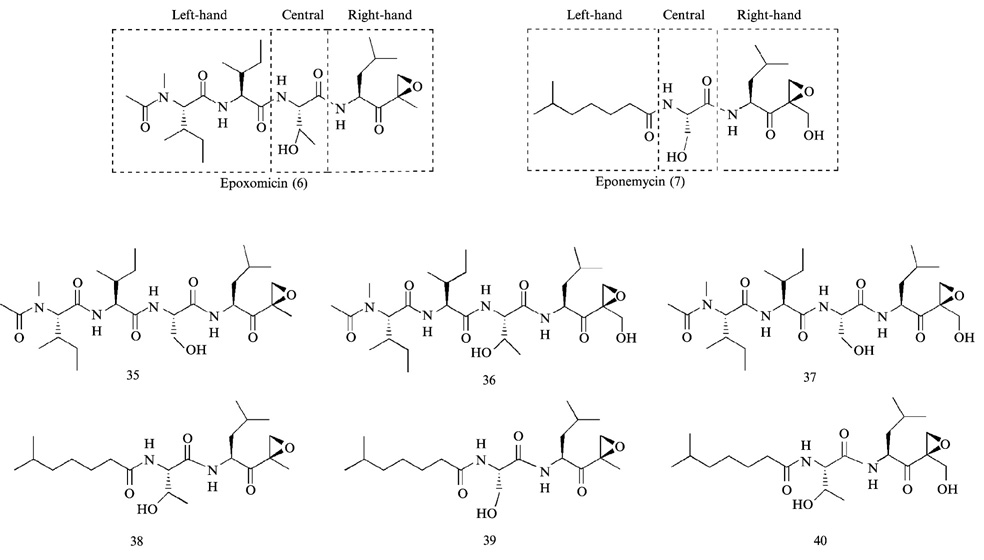
Although antitumor natural products epoxomicin (Meng et al., 1999b; Sin et al., 1999) and eponemycin (Meng et al., 1999a) have been shown to target proteasome, they markedly differ in proteasome subunit binding specificity and rates of proteasome inhibition. To understand such differences and develop proteasome inhibitors with novel activities, a combinatorial approach was taken to investigate the structure-activity relationship (SAR) of these compounds (Kim et al., 1999). Although both are members of the α′,β′-epoxyketone linear peptide natural product family, there are some structural differences in their left-, central, and right-hand fragments (Fig. 11) that may cause the different subunit binding specificity and inhibitory potency between two natural products. To this end, epoxomicin/dihydroeponemycin chimerae and their biotinylated counterparts were synthesized to correlate these structural features with proteasome inhibitory activity and their specific subunit labeling (Fig. 11). Kinetic data of epoxomicin/dihydroeponemycin chimerae for proteasome activities showed that α′,β′-epoxyketone tetrapeptide inhibitors (compounds 35–37) generally display 300–500 fold greater inhibition for the CT-L activity compared with that of isooctanoic-containing tripeptide inhibitors (compounds 38–40) (Kim et al., 1999).
FIG. 11.
Epoxomicin/eponemycin chimerae were prepared by a random combination of left- and right-hand and central fragments.
On the other hand, the α′,β′-epoxyketone tetrapeptide inhibitors (compounds 35–37), regardless of changes in the central and right-hand fragments, displayed the same proteasome subunit binding pattern as epoxomicin, which predominantly binds LMP7/X and MECL1/Z (Meng et al., 1999b), whereas tripeptide inhibitors (compounds 38–40) that possess isooctanoic residue at the N-terminus displayed the same labeling pattern as that of dihydroeponemycin, which predominantly labels immunoproteasome subunit LMP2 and, to a lesser extent, LMP7/X (Meng et al., 1999a). Taken together, SAR studies on eponemycin/dihydroepoxomicin show that (1) α′,β′-epoxyketone tetrapeptide inhibitors generally possess a higher inhibitory activity for the proteasome than tripeptide counterparts; (2) α′,β′-epoxyketone tripeptide inhibitors containing the isooctanoic residue at the N-terminus, regardless of changes in the central and right-hand fragments, display greater specificity toward immunoproteasome over the constitutive proteasome. It is expected that the information on SAR studies will provide a basis for development of more potent proteasome inhibitors. In addition, the insights gained from SAR studies may facilitate the development of proteasome inhibitors with a high degree of immunoproteasome specificity.
Preparation of Epoxomicin/Dihydroeponemycin Chimerae
All the chimerae were prepared following the procedure used for the synthesis of epoxomicin with the exception of the right-hand fragment of dihydroeponemycin. The right-hand fragment of dihydroeponemycin was synthesized by a slight modification of a previously reported procedure (Hoshi et al., 1993; Schmidt and Schmidt, 1994). Recently, similar procedures for the preparation of the right-hand fragment have also been reported (Bennacer et al., 2003; Dobler, 2001; Iwabuchi et al., 2001). Given that a convergent approach that is applied for the preparation of epoxomicin/dihydroeponemycin chimerae is essentially the same as that described for epoxomicin, we only describe the total synthesis of dihydroeponemycin.
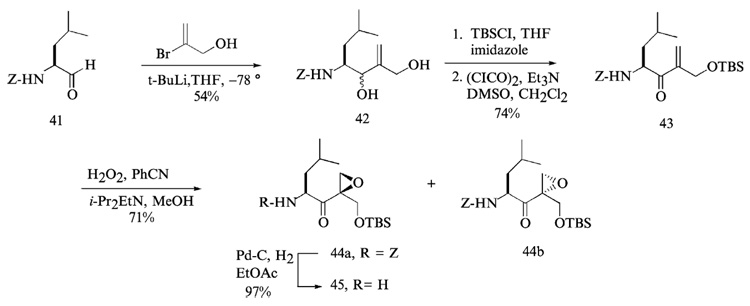
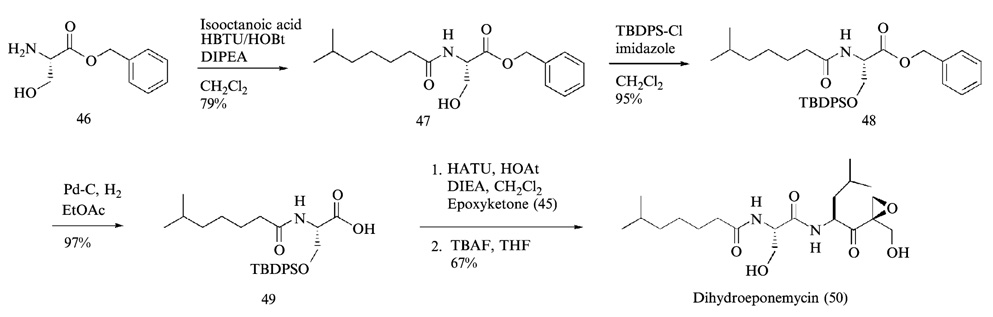
The synthesis of the right-hand fragment of dihydroeponemycin began with the addition of vinyl lithium derived from treatment of 2-bromo-1-hydroxy-2-propene (Hoshi et al., 1993) with t-butyl lithium (Corey and Widiger, 1975) to leucine aldehyde 41, which afforded a 1:1 inseparable mixture of diol 42 (Fig. 12). The leucine aldehyde was prepared by the reduction of leucine Weinreb amide with LiAlH4 in anhydrous ether. Selective protection of the primary alcohol of 42 with t-butyldimethylsilyl (TBS) group and a Swern oxidation of the remaining secondary alcohol yielded the α,β-unsaturated ketone 43. Epoxidation of 43 with hydrogen peroxide (Schmidt and Schmidt, 1994) afforded two epoxyketone isomers 44a and 44b as a 1.2:1 mixture that were readily separated by flash column chromatography using an elution system (hexanes-ethyl acetate = 5:1, v/v). The isomer (2-(R)-epoxide) 44a, which migrates faster than the 2-(S)-epoxide 44b in thin-layer chromatography (TLC), was found to have the same configuration as that of eponemycin epoxide. Final removal of the benzyloxycarbonyl (Z) protecting group of 44a through a catalytic hydrogenation reaction (Hoshi et al., 1993) gave leucine epoxyketone 45 in nearly quantitative yield. The left-hand fragment was prepared by coupling of isooctanoic acid with serine benzylester using HBTU, followed by protection of the hydroxyl side chain of serine residue with TBDPS group and hydrogenolysis of the benzyl protecting group yielded TBDPS-protected left-hand fragment 49 (Fig. 13). The final coupling reaction between epoxyketone 45 and dipeptide 49 was performed with HATU, followed by removal of the TBDPS group and normal-phase HPLC (hexanes-isopropanol, linear gradient, hexanes 100–50%) to yield dihydroeponemycin 50.
FIG. 12.
Preparation of the epoxyketone residue of eponemycin.
FIG. 13.
The final assembly of eponemycin peptide backbone, which yielded dihydroeponemycin by TBDPS deprotection from serine side chain.
Development of Activity-Specific α′,β′-Epoxyketone Proteasome Inhibitors
CT-L Activity-Specific α′,β′-Epoxyketone Inhibitors
On the basis of the information gathered from SAR studies on epoxomicin/dihydroeponemycin, α′,β′-epoxyketone tetrapeptide was chosen as a lead compound in the development of CT-L activity-specific proteasome inhibitors. To develop highly potent CT-L specific α′,β′-epoxyketone proteasome inhibitors, a combinatorial positional scanning approach with a variety of hydrophobic amino acids, such as alanine, leucine, phenylalanine, homophenylalanine, 3-(1-naphthyl)-alanine, and p-benzoylphenylalanine, was applied to find the optimal amino acids at each position (Elofsson et al., 1999). After the synthetic strategy of epoxomicin described previously, peptide α′,β′-epoxyketones were assembled from left-hand fragments, prepared by standard solid-phase synthesis, and the leucine α′,β′-epoxyketone. The coupling reactions were carried out with HATU/HOAt and diisopropylethylamine (DIEA) in DMF. Unlike the final coupling in epoxomicin synthesis, in which CH2Cl2 was used as a solvent, DMF was used because of the poor solubility of the left-hand fragments. It seems that a slightly higher level of stereoisomer (5–30%) is formed in DMF conditions compared with CH2Cl2 environment. After reverse-phase HPLC purification, the resulting α′,β′-epoxyketone tetrapeptides were tested for inhibition of three catalytic activities of the 20S proteasome. Once amino acids that display the best CT-L activity inhibition at each position were identified, the optimized inhibitor with optimized amino acids at each position was prepared, tested for CT-L activity inhibition to yield the most potent CT-L activity selective inhibitor (YU101, Fig. 4) to date (Elofsson et al., 1999).
Purification of YU101
Impure YU101 was purified by RP-HPLC (for HPLC system and column specifications see the purification procedure for epoxomicin). A linear gradient of solvent A into B (water and methanol, respectively) consisting of 70% of B to 85% of B in 90 min, run at 5 ml/min, room temperature. YU101, which was eluted at 62.5 min, was collected as seven fractions of nearly 2 ml each.
Chemical Characterization
LC-MS analysis of collected fractions were performed in a Waters Separation Module 2795 coupled to a Waters 2795 Photodiode Array Detector (set at 214 nm), and to a Micromass ZQ 2000 Electrospray Mass detector (cone voltage = +20V). A linear gradient of solvent A (water) in solvent B (MeOH), 5% of B to 98% of B in 30 min was run at 0.2 ml/min, 25°. The analytical column used was the same as the one described for epoxomicin.
YU101 was eluted at 10.3 min [observed m/z = 635.5 (M + H)+, 658.4 (M + Na)+]. Fractions showing purity ≥98% were grouped together and vacuum-dried with no heat. YU101 was also confirmed by 1H NMR.
Development of Caspase-Like Activity-Specific α′,β′-Epoxyketone Peptide Inhibitors
α′,β′-Epoxyketone-based caspase-like activity-specific proteasome inhibitors were also developed using a combinatorial positional scanning approach (Myung et al., 2001b). Initial efforts involved peptidyl α′,β′-epoxyketones with glutamic acid at the P1 position. It was reasoned that the presence of Glu at the P1 position of peptide inhibitors alone would suffice to render higher selectivity toward the caspase-like activity. However, α′,β′-epoxyketone peptide inhibitors with Glu at the P1 position did not display any significant selectivity for the caspase-like activity (Myung et al., 2001b). To screen an optimum P1 amino acid residue for the caspase-like activity inhibition, α′,β′-epoxyketones with various natural and unnatural amino acid residues (i.e., Leu, Ala, Val, Phe, norleucine, cyclohexyl, and tert-butyl) in place of Glu at the P1 position were prepared, which were then coupled to dipeptides containing the serine at the C-terminus and isooctanoic group at the N-terminus. Among these, α′,β′-epoxyketone tetrapeptide inhibitors with Leu at the P1 position provided the best caspase-like selectivity. In this regard, incorporation of leucine at the P1 position in an N-terminus protected peptidyl aldehyde proteasome inhibitor (i.e., Z-GPFL-CHO) has previously been shown to inhibit competitively the CASPASE-LIKE activity with a ~13-fold selectivity over the CT-L activity (Vinitsky et al., 1994).
Interestingly, α′,β′-epoxyketone inhibitors with Pro-Phe-Leu at the P3-P1 positions display no significant inhibition for the T-L activity, regardless of the nature of residues at the P4 position. Expanding on this finding, therefore, it was decided to derivatize the N-terminus of Pro-Phe-Leu-α′,β′-epoxyketone inhibitors to explore the importance of length, steric bulk, and protecting group at the P4 position for the caspase-like activity inhibition. The results from these studies showed that the presence of a bulky aromatic protecting group in place of an acetyl group at the amino terminus provides a rather stronger inhibition toward the CT-L activity, thus making them less caspase-like selective. On the basis of these findings, several α′,β′-epoxyketone peptides possessing a smaller amino-terminal group (i.e., acetyl) instead of a bigger group (i.e., benzyloxycarbonyl group) were prepared and tested for their inhibitory activity.
To this end, one compound (YU102, Fig. 4) showed the highest selectivity toward the caspase-like activity with ~50-fold higher values of kobs/[I] for inhibition of the caspase-like activity than the CT-L activity (Myung et al., 2001b); 8 µM YU102 was found to inhibit only the caspase-like activity but not the CT-L activity at 8 µM concentration. Up to 90% of the caspase-like activity was found to be inhibited under the assay conditions. Against T-L activity, YU102 is a very poor inhibitor; even at concentrations of 100–150 µM, it displayed no significant inhibition of the T-L activity. Moreover, YU102 showed a time-dependent inhibition, indicating the irreversible modification of the catalytic Thr-1 of the proteasome. Therefore, a greater than 90% inhibition of the caspase-like activity under these conditions suggests that the caspase-like subunits were at least 90% saturated irreversibly with YU102. The CT-L activity of the 20S proteasome, however, was not affected. This results show that near quantitative occupancy of the caspase-like sites with YU102 did not trigger inhibition off the CT-L activity. YU102 was applied to probe the role of different catalytic subunits in cell-based protein degradation assays in living cells (Myung et al., 2001b), revealing that selective caspase-like activity inhibition is not sufficient to inhibit total protein degradation.
Enzyme Kinetic Assays
Purification of 20S Proteasome
20S proteasome was purified from bovine reticulocyte lysates by batch DE-52 binding, DEAE Sephacel chromatography, gel filtration on Sephacryl S-300, and chromatography on hydroxyapatite. This procedure is as previously described (Elofsson et al., 1999; Myung et al., 2001b), with the exception that bovine reticulocytes were used as a starting source.
To evaluate the rates of proteolytic inactivation by epoxomicin and YU101, Kassociation(Kobs/[I]), values were determined by use of fluorogenic peptide substrates over a range of inhibitor concentrations.
Epoxomicin and YU101 inhibition of the chymotrypsin-like catalytic activity of the 20S proteasome complex was determined as follows: Inhibitors solubilized in DMSO were mixed with a final concentration of 5 µM of the fluorogenic peptide substrate Suc-LLVY-AMC and assay buffer (20 mM Tris·HCl, pH 8.0, 0.5 mM EDTA/0.035% SDS) in a 96-well plate. Inhibitors concentrations were adjusted so that the final DMSO concentration would not exceed 1%. Hydrolysis was initiated by the addition of bovine red blood cell 20S proteasome to a final volume of 100 µl/well, and the reaction was followed by fluorescence (360 nm excitation/460 nm detection) using a multilable plate-reader Wallac Victor2, set at 25°. Reactions were allowed to proceed for 50 min, and fluorescence data were collected every 10 sec. Fluorescence was quantified as arbitrary units, and progression curves were plotted for each reaction as a function of time. Kobs/[I] values were obtained using KALEIDOGRAPH software by nonlinear least-squares fit of the data to the following equation: fluorescence = νSt + [(νO − νS)/Kobs][1 − exp (− Kobst)], where νO and νS are the initial and final velocities respectively, and Kobs is the reaction rate constant. Kassociation = Kobs/[I] (M−1s−1).
The range of inhibitors final concentrations tested were 40–100 nM for epoxomicin and 5–12 nM for YU101. Bovine erythrocyte 20S proteasome (2.5 mg/ml) was diluted 1:500.
YU101 most potently inhibits the chymotrypsin-like activity of the 20S proteasome with a Kassociation = 310,000 ± 19,000 (M−1s−1), CV = 60%. The chymotrypsin-like inhibitory activity of epoxomicin was Kassociation = 20,000 ± 4,600 (M−1s−1), CV = 24%.
For YU102, peptide-AMC (10 µM Suc-LLVY-AMC, 10 µM Z-LLE-AMC, or 20 µM Boc LRR-AMC) and 20S proteasome were added to assay buffer (20 mM Tris·HCl, pH 8.0, and 0.5 mM EDTA). For Suc-LLVY and Z-LLE-AMC assays, 0.035% (w/v) SDS was added to the assay buffer. After the steady state of hydrolysis for each substrate was established, an inhibitor was added to the assay buffer containing substrate and enzyme in a Dynex™ 96-well plate at room temperature. Release of fluorescent 7-amino-4-methylcoumarin (AMC) was measured using a Cytofluor spectrofluorometer with an excitation wavelength of 360 nm, and kinetic data were processed as described previously.
Summary
Given the complex proteolytic activities associated with the proteasome and poorly understood biological role of each catalytic subunit in many important signaling pathways, there are unmet needs for more sophisticated, selective proteasome inhibitors to dissect proteasome function. Here we describe strategies for developing α′,β′-epoxyketone peptide-based proteasome inhibitors: (1) a general approach for the synthesis of α′,β′-epoxyketone peptides that was developed through the total synthesis of epoxomicin; (2) SAR studies on epoxomicin/dihydroeponemycin, potentially shedding light on a means to design catalytic subunit- or immunoproteasome-specific α′,β′-epoxyketone peptides; (3) development of highly potent, CT-L activity-specific α′,β′-epoxyketone peptides; and (4) development of caspase-like activity-specific α′,β′-epoxyketone inhibitors. Fortunately, all of the reactions for the synthesis of α′,β′-epoxyketone peptides can readily be carried out in solutions (CH2Cl2 or DMF solvents) with good to excellent yields and easily repeatable. In conclusion, our studies have shown that derivatization of α′,β′-epoxyketone peptide proteasome inhibitors at positions P1–P4 can be easily accomplished to provide novel proteasome-specific, subunit-selective small molecule inhibitors.
References
- Adams J. Development of the proteasome inhibitor PS-341. Oncologist. 2002;7:9–16. doi: 10.1634/theoncologist.7-1-9. [DOI] [PubMed] [Google Scholar]
- Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Potent and selective inhibitors of the proteasome: Dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- Albericio F, Carpino LA. Coupling reagents and activation. Methods Enzymol. 1997;289:104–126. doi: 10.1016/s0076-6879(97)89046-5. [DOI] [PubMed] [Google Scholar]
- Bennacer B, Rivalle C, Grierson D. A new route for the total synthesis of 6,7-dihydroeponemycin. Eur. J. Org. Chem. 2003;23:4569–4574. [Google Scholar]
- Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogyo M, Shin S, McMaster JS, Ploegh HL. Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chem. Biol. 1998;5:307–320. doi: 10.1016/s1074-5521(98)90169-7. [DOI] [PubMed] [Google Scholar]
- Carpino LA. 1-Hydroxy-7-azabenzotriazole. An efficient peptide coupling additive. J. Am. Chem. Soc. 1993;115:4397–4398. [Google Scholar]
- Casp CB, She JX, McCormack WT. Genes of the LMP/TAP cluster are associated with the human autoimmune disease vitiligo. Genes Immun. 2003;4:492–499. doi: 10.1038/sj.gene.6364016. [DOI] [PubMed] [Google Scholar]
- Corey EJ, Widiger GN. J. Org. Chem. 1975;40:2975–2976. [Google Scholar]
- Dobler MR. Total synthesis of (+)-epopromycin B and its analogues—studies on the inhibition of cellulose biosynthesis. Tetrahedron Lett. 2001;42:215–218. [Google Scholar]
- Elofsson M, Splittgerber U, Myung J, Mohan R, Crews CM. Towards subunit-specific proteasome inhibitors: Synthesis and evaluation of peptide α′,β′-epoxyketones. Chem. Biol. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Fields GB. Methods in Enzymology. New York: Academic Press; 1997. [Google Scholar]
- Figueiredo-Pereira ME, Banik N, Wilk S. Comparison of the effect of calpain inhibitors on two extralysosomal proteinases: The multicatalytic proteinase complex and m-calpain. J. Neurochem. 1994;62:1989–1994. doi: 10.1046/j.1471-4159.1994.62051989.x. [DOI] [PubMed] [Google Scholar]
- Fruh K, Gossen M, Wang K, Bujard H, Peterson PA, Yang Y. Displacement of housekeeping proteasome subunits by MHC-encoded LMPs: A newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO J. 1994;13:3236–3244. doi: 10.1002/j.1460-2075.1994.tb06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Kim KB, Kairies N, Huber R, Crews CM. Crystal structure of epoxomicin: 20s proteasome reveals a molecular basis for selectivity of α′,β′-epoxyketone proteasome inhibitors. J. Am. Chem. Soc. 2000;122:1237–1238. [Google Scholar]
- Hanada M, Sugawara K, Kaneta K, Toda S, Nishiyama Y, Tomita K, Yamamoto H, Konishi M, Oki T. Epoxomicin, a new antitumor agent of microbial origin. J. Antibiot. (Tokyo) 1992;45:1746–1752. doi: 10.7164/antibiotics.45.1746. [DOI] [PubMed] [Google Scholar]
- Hoshi H, Ohnuma T, Aburaki S, Konishi M, Oki T. A total synthesis of 6,7-dihydroeponemycin and determination of stereochemistry of the epoxide ring. Tetrahedron Lett. 1993;34:1047–1050. [Google Scholar]
- Iqbal M, Chatterjee S, Kauer JC, Mallamo JP, Messina PA, Reiboldt A, Siman R. Potent a-ketocarbonyl and boronic ester derived inhibitors of proteasome. Bioorg. Med. Chem. Lett. 1996;6:287–290. [Google Scholar]
- Iwabuchi Y, Sugihara T, Esumi T, Hatakeyama S. An enantio-and stereocontrolled route to epopromycin B via cinchona alkaloid-catalyzed Baylis-Hillman reaction. Tetrahedron Lett. 2001;42:7867–7871. [Google Scholar]
- Kim KB, Crews CM. Natural product and synthetic proteasome inhibitors. In: Adams J, editor. Cancer Drug Discovery and Development: Proteasome Inhibitors in Cancer Therapy. Totowa, NJ: Humana Press Inc; 2003. pp. 47–63. [Google Scholar]
- Kim KB, Myung J, Sin N, Crews CM. Proteasome inhibition by the natural products epoxomicin and dihydroeponemycin: Insights into specificity and potency. Bioorg. Med. Chem. Lett. 1999;9:3335–3340. doi: 10.1016/s0960-894x(99)00612-5. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- Koguchi Y, Kohno J, Nishio M, Takahashi K, Okuda T, Ohnuki T, Komatsubara S. TMC-95A, B, C, and D, novel proteasome inhibitors produced by Apiospora montagnei Sacc. TC 1093. Taxonomy, production, isolation, and biological activities. J. Antibiot. (Tokyo) 2000a;53:105–109. doi: 10.7164/antibiotics.53.105. [DOI] [PubMed] [Google Scholar]
- Koguchi Y, Kohno J, Suzuki S, Nishio M, Takahashi K, Ohnuki T, Komatsubara S. TMC-86A, B and TMC-96, new proteasome inhibitors from Streptomyces sp. TC 1084 and Saccharothrix sp. TC 1094. I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. (Tokyo) 1999;52:1069–1076. doi: 10.7164/antibiotics.52.1069. [DOI] [PubMed] [Google Scholar]
- Koguchi Y, Kohno J, Suzuki S, Nishio M, Takahashi K, Ohnuki T, Komatsubara S. TMC-86A, B and TMC-96, new proteasome inhibitors from Streptomyces sp. TC 1084 and Saccharothrix sp. TC 1094. II. Physico-chemical properties and structure determination. J. Antibiot (Tokyo) 2000b;53:63–65. doi: 10.7164/antibiotics.53.63. [DOI] [PubMed] [Google Scholar]
- Koguchi Y, Nishio M, Suzuki S, Takahashi K, Ohnuki T, Komatsubara S. TMC-89A and B, new proteasome inhibitors from Streptomyces sp. TC 1087. J. Antibiot. (Tokyo) 2000c;53:967–972. doi: 10.7164/antibiotics.53.967. [DOI] [PubMed] [Google Scholar]
- Lloyd-Williams P, Albericio F, Giralt E. Chemical Approaches to the Synthesis of Peptides and Proteins. Boca Raton, Florida: CRC Press; 1997. [Google Scholar]
- Meng L, Kwok BH, Sin N, Crews CM. Eponemycin exerts its antitumor effect through the inhibition of proteasome function. Cancer Res. 1999a;59:2798–2801. [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo anti-inflammatory activity. Proc. Natl. Acad. Sci. USA. 1999b;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 2001a;21:245–273. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J, Kim KB, Lindsten K, Dantuma NP, Crews CM. Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Mol. Cell. 2001b;7:411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]
- Nahm S, Weinreb SM. N-methoxy-N-methylamides as effective acylating agents. Tetrahedron Lett. 1981;22:3815–3818. [Google Scholar]
- Nam S, Smith DM, Dou QP. Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J. Biol. Chem. 2001;276:13322–13330. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- Orlowski M. The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochemistry. 1990;29:10289–10297. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Schmidt U, Schmidt U. The total synthesis of eponemycin. Synthesis. 1994;3:300–304. [Google Scholar]
- Sekizawa R, Momose I, Kinoshita N, Naganawa H, Hamada M, Muraoka Y, Iinuma H, Takeuchi T. Isolation and structural determination of phepropeptins A, B, C, and D, new proteasome inhibitors, produced by Streptomyces sp. J. Antibiot. (Tokyo) 2001;54:874–881. doi: 10.7164/antibiotics.54.874. [DOI] [PubMed] [Google Scholar]
- Sin N, Kim KB, Elofsson M, Meng L, Auth H, Kwok BH, Crews CM. Total synthesis of the potent proteasome inhibitor epoxomicin: A useful tool for understanding proteasome biology. Bioorg. Med. Chem. Lett. 1999;9:2283–2288. doi: 10.1016/s0960-894x(99)00376-5. [DOI] [PubMed] [Google Scholar]
- Sin N, Meng L, Auth H, Crews CM. Eponemycin analogues: Syntheses and use as probes of angiogenesis. Bioorg. Med. Chem. 1998;6:1209–1217. doi: 10.1016/s0968-0896(98)00089-3. [DOI] [PubMed] [Google Scholar]
- Sugawara K, Hatori M, Nishiyama Y, Tomita K, Kamei H, Konishi M, Oki T. Eponemycin, a new antibiotic active against B16 melanoma. I. Production, isolation, structure and biological activity. J. Antibiot. (Tokyo) 1990;43:8–18. doi: 10.7164/antibiotics.43.8. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Kobayashi S, Nishikiori T, Nakagawa T, Tatsuta K. Epopromycins, novel cell wall synthesis inhibitors of plant protoplast produced by Streptomyces sp. NK04000. J. Antibiot. (Tokyo) 1997;50:261–263. [PubMed] [Google Scholar]
- Vinitsky A, Cardozo C, Sepp-Lorenzino L, Michaud C, Orlowski M. Inhibition of the proteolytic activity of the multicatalytic proteinase complex (proteasome) by substrate-related peptidyl aldehydes. J. Biol. Chem. 1994;269:29860–29866. [PubMed] [Google Scholar]