Abstract
The Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) is a cAMP dependent chloride channel that is mutated in cystic fibrosis, an inherited disease of high morbidity and mortality. The phosphorylation of its ∼200 amino acid R domain by protein kinase A is obligatory for channel gating under normal conditions. The R domain contains more than ten PKA phosphorylation sites. No individual site is essential but phosphorylation of increasing numbers of sites enables progressively greater channel activity. In spite of numerous studies of the role of the R domain in CFTR regulation, its mechanism of action remains largely unknown. This is because neither its structure nor its interactions with other parts of CFTR have been completely elucidated. Studies have shown that the R domain lacks well-defined secondary structural elements and is an intrinsically disordered region of the channel protein. Here, we have analyzed the disorder pattern and employed computational methods to explore low energy conformations of the R domain. Specific disorder and secondary structure patterns detected suggest the presence of Molecular Recognition Elements (MoREs) that may mediate phosphorylation regulated intra- and inter-domain interactions. Simulations were performed to generate an ensemble of accessible R domain conformations. Although the calculated structures may represent more compact conformers than occur in vivo, their secondary structure propensities are consistent with predictions and published experimental data. Equilibrium simulations of a mimic of a phosphorylated R domain showed that it exhibited an increased radius of gyration. In one possible interpretation of these findings, by changing its size, the globally unstructured R domain may act as an entropic spring to perturb the packing of membrane-spanning sequences that constitute the ion permeability pathway and thereby activate channel gating.
Keywords: CFTR, R domain, phosphorylation, disordered protein, molecular dynamics
Introduction
Cystic fibrosis (CF) is a monogenic disorder characterized by high morbidity and mortality1, 2. CF is caused by mutations in the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR), a cAMP-regulated chloride channel that belongs to the ATP-binding cassette (ABC) transporter family3. CFTR consists of two large transmembrane domains (TMD1 and TMD2), two nucleotide binding domains (NBD1 and NBD2), and a regulatory region (R domain)3. Binding and hydrolysis of ATP at the NBDs gate the channel. Channel opening requires the phosphorylation of the R domain by protein kinase A (PKA)4. The R domain contains multiple phosphorylation sites comprised of either serines or threonines in dibasic or monobasic PKA consensus patterns5. However, the mechanism by which phosphorylation activates the channel remains unknown.
Several experimental studies suggest that the unphosphorylated R domain inhibits CFTR channel function. Phosphorylation and deletion of regions in the R domain, such as residues 708-835 and 768-830, relieve inhibition of channel activity6, 7. However, addition of unphosphorylated R domain fragments to constitutively active channels6, 8, 9 or transferring the R domain to the C-terminus does not inhibit channel function10. These results indicate that the inhibitory function of the R domain is exerted only at its native location within specific distance constraints. In contrast to the inhibitory role of the unphosphorylated R domain, its phosphorylated form has been shown to activate the chloride channel. This was demonstrated experimentally using a deletion construct (denoted as δR CFTR) lacking most of the R domain6, 8, 9. Phosphorylated R domain segments corresponding to regions 645-834, 590-858, and 708-831, stimulated δR CFTR activity. Moreover, translocation of the R domain to the C-terminal end of δR CFTR restored the PKA sensitivity of the channel activity10. However, some studies suggest that the magnitude of the activating effect of the phosphorylated R domain is negligible compared to the release of the inhibitory action based on experiments with severed channels11, 12.
Although the mechanism of activation is not known, the increased negative charge upon phosphorylation has been suggested to contribute to channel activation4. Direct introduction of negative charges by mutation of serines and threonines to either aspartate13 or glutamate14 resulted in activation of CFTR, although the open probability of those constructs was only 40-50 % that of the fully phosphorylated wild type. These experiments did not elucidate the role of specific phosphorylation sites required for activation. However, when increasing numbers of serines were mutated to alanine the level of activation was progressively decreased4, 15 indicating additivity and perhaps redundancy of sites. Fourteen PKA sites in the R domain and one in NBD1 need to be removed to produce a PKA insensitive CFTR5. Sites 660, 700, 737, 768, 795, and 813 were shown to be phosphorylated in vivo4, 16-18. Mutations of sites 660 and 813, near the N- and C- terminal ends of the R domain, to alanine resulted in the largest decrease of stimulated activity suggesting that these two sites may contribute most to activation. Replacement of serine with alanine at position 737 or 768 was reported to produce a channel with higher activity in both the non-phosphorylated and phosphorylated state16, 19-21. These studies came to the indirect conclusion that these two sites are inhibitory in their phosphorylated state. It should be noted that there are also alternative pathways to activate CFTR without phosphorylation, such as stimulation by cyclophilin A, PIP2, and vitamin C22-25.
Several groups investigated structural changes in the R domain to help elucidate its role in CFTR regulation. Slower migration of phosphorylated forms of CFTR or the isolated R domain reflect conformational changes upon phosphorylation4, 16, 26. Phosphorylation dependent conformational changes were also observed by circular dichroism (CD)9, 26 and Fourier Transform Infra Red (FT-IR) spectroscopy27. The study of Ostedgaard et al.9 emphasized that the R domain does not contain well-defined secondary structure elements. These results, observations of high susceptibility to proteolytic digestion, and more directly NMR experiments28 indicate that the R domain is intrinsically disordered. The highly dynamic nature of disordered proteins makes them competent to act as display sites for phosphorylation, molecular recognition partners, and entropic chains (for review see 29, 30). Entropic springs or flexible spacers regulate intramolecular distance as in the case of titin PEVK domain31 or MAP2 projection domain32. It has been observed in the case of proteins regulated by phosphorylation that the amino acid composition around their phosphorylation sites is similar to those observed in disordered regions33. This correlation may apply to the R domain where disorder could help display the phosphorylation sites to both kinases and phosphatases. Disordered proteins likewise contain preformed structural elements, also called molecular recognition elements (MoREs), that promote interaction with binding partners34, 35. The R domain interacts with other parts of CFTR and with other proteins36-38. The disordered property of the R domain is central to its regulatory role by providing a large interaction surface and enabling rapid association and dissociation rates39.
Experimental methods commonly used to study disordered proteins in atomic detail (NMR, H/D exchange, and limited proteolysis39, 40) are limited in their application to CFTR which is a large membrane protein, difficult to express at high levels, purify, and reconstitute in a functional form. Hence, we used computational methods to study the structural properties of the R domain. In other proteins all-atom Monte Carlo simulations have been performed to describe disorder-order transitions coupled to binding41, 42. Unconstrained MD methods also have been used to gain insights into conformational ensembles of disordered polypeptides43-48. Molecular dynamic simulations with time-averaged constraints over many conformers have also been employed, although the large computational cost limits this approach to either short peptides or short time scales49. A completely different technique, TRADES50 (Trajectory Directed Ensemble Sampling), can be used to rapidly sample large conformational space and has been applied successfully to characterize the unfolding of proteins such as the SH3 domain51. However TRADES is a geometrical method, and requires experimental constraints to select populations of physically possible conformers. Since the R domain peptide is large and lacking in constraints, we have employed discrete molecular dynamics (DMD) simulations52, 53 (Ding et al., submitted) and an all-atom force field (Medusa54, 55) to generate an ensemble of three dimensional structures of the R domain at the atomic level.
Results
Conserved disorder patterns and secondary structure propensities suggest molecular recognition elements within the R domain
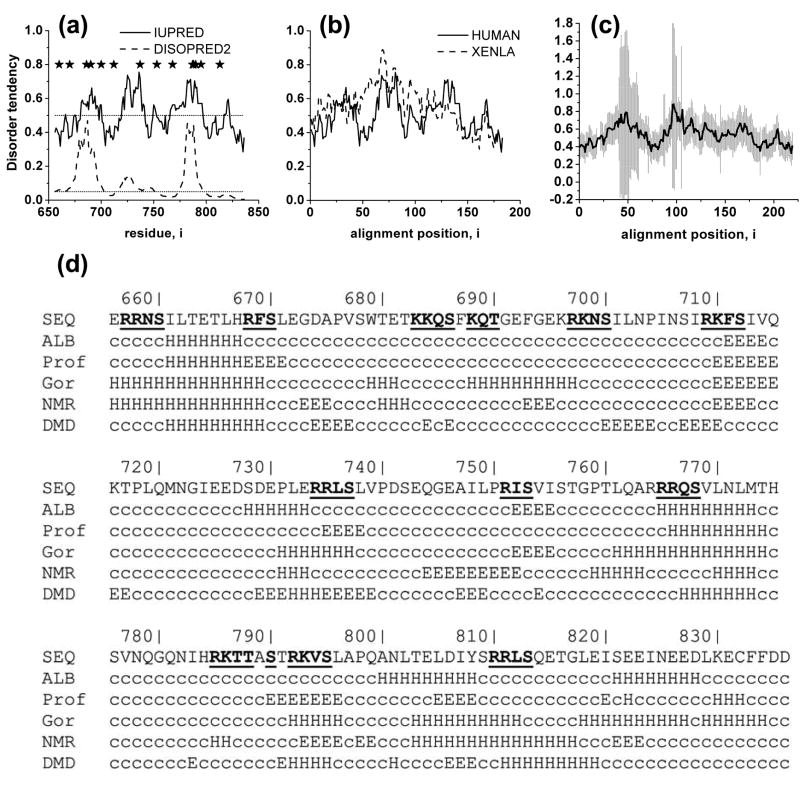
We first mapped disorder-order patterns in the R domain of human CFTR. Two independent computational disorder predictors were employed, the first based on supervised learning algorithms (Disopred256, 57, http://bionif.cs.ucl.ac.uk/disopred) and the second, on estimations of pairwise interaction energies (IUPred58, 59, http://iupred.enzim.hu). Similar patterns of disorder were revealed by the two algorithms (Figure 1(a)) providing some confidence in the validity of the results. Although there are segments of the sequence with distinctly higher and lower degrees of disorder, the magnitudes of the differences between them are relatively small. Furthermore, the regions identified as more ordered still have relatively low propensities for order. Thus overall, the patterns predicted indicate that the whole R domain is intrinsically disordered and highly dynamic, in good agreement with results of CD and NMR experiments9, 28.
Figure 1. Disorder and secondary structure organization of the R domain.
(a) Disorder predictors reveal varying degrees of disorder in the R domain. R domain sequence was submitted to disorder predictors with fundamentally different algorithms (see Methods). Regions with values above the cut-off value (0.5 for IUPred and 0.05 for DISOPRED2) tend to be more disordered, while segments with values below the cut-off are likely ordered parts of the protein. Phosphorylation sites are marked with asterisks. (b) Comparison of disorder propensities of human and Xenopus CFTR using IUPred. (c) Similarity of disorder of R domains from 23 different species (right panel). Average and standard deviation of disorder values at each position in the alignment are calculated. Regions corresponding to amino acids 656-836 in human CFTR are plotted. (d) Secondary structure prediction by GOR4, Prof, and ALB correlate with the structure formed upon binding of a disordered protein or region to its interacting partner. To gain insight into the secondary structure propensities of the R domain its sequence had been submitted to these secondary structure predictors and region 656-836 is depicted. Secondary structural propensities from NMR experiments28 and DMD simulations are extracted from the experimental and simulation data as described in Methods. c: coil, H: helix, E: extended sheet, phosphorylation sites are bold and underlined.
The disorder pattern within the R domain is also found to be conserved among species (Figure 1(b-c)). It is important to emphasize that although the sequence conservation of the R domain is much less than other parts of the protein (amino acid identity between human and Xenopus full length proteins is 79%, while only 58% between the R domains), there is still sufficient conservation to preserve the structural properties of R domains from different species. The disorder patterns of human and Xenopus domains are seen to be very similar (Figure 1(b)) except in one region (between alignment positions 60 and 80). It might be speculated that this difference contributes to the experimentally observed difference in sensitivity to PKA of human and Xenopus CFTR.60 To more extensively assess the degree of conservation of the disorder pattern, a diverse set of R domains from many different species were analyzed. As mammalian sequences are most abundantly represented among cloned CFTR sequences, we did not employ all of them but selected only 23 of a total 48 to reduce bias. Disorder values determined using IUPred across these 23 domains were averaged and plotted in Figure 1(c). The two regions with large error bars (positions 51-70 and 105-110) correspond to insertions in the R domains of marine species (n is only 3 or 4 in these positions). Since most of the phosphorylation sites are located in disordered regions33, it could be that the phosphorylation site spacing determines the disorder pattern. However, this apparently is not the case because there is no such correspondence in other proteins with multiple phosphorylation sites separated by distances similar to those in the R domain. These proteins identified in an earlier study61, exhibit completely different disorder patterns. Thus the conservation of the characteristic periodic positioning of more and less ordered regions in the R domain does imply some level of functional relevance.
Transitions between ordered and disordered segments of disordered proteins often contribute to Molecular Recognition Elements (MoREs) that constitute readily reversible interaction surfaces which recognize and associate with their interaction partners. Although MoREs in other proteins are characterized by much larger differences between the degree of order and disorder in different regions, the possible existence of MoREs within the R domain is suggested by the disorder pattern. However, the presence of MoREs can also be reflected in the secondary structure patterns of proteins. In particular, it has been shown that secondary structure propensities of disordered regions, which are formed upon interaction with binding partners, can be identified computationally by secondary structure predictors35 such as GOR462, ALB63 and PROF64. The R domain secondary structure predictions of these in relation to the phosphorylation site locations are shown in Figure 1(d). Sequences proximal to most of the phosphorylation sites are predicted to form alpha helices while extended strands are predicted around the sites at serines 712 and 753. These are in excellent agreement with most of the predicted alpha-helical propensities (such as those around residues 768, 733-739, 904-814, and 828-835) and the β-sheet propensity of sequences around residues 712 and 753 in the NMR study of Baker and coworkers28. These findings reinforce the suggestion from the disorder patterns that segments of the domain may serve as MoREs, the binding properties of which may be regulated by the phosphorylation state of adjacent sites.
Simulation of possible R domain 3D structures
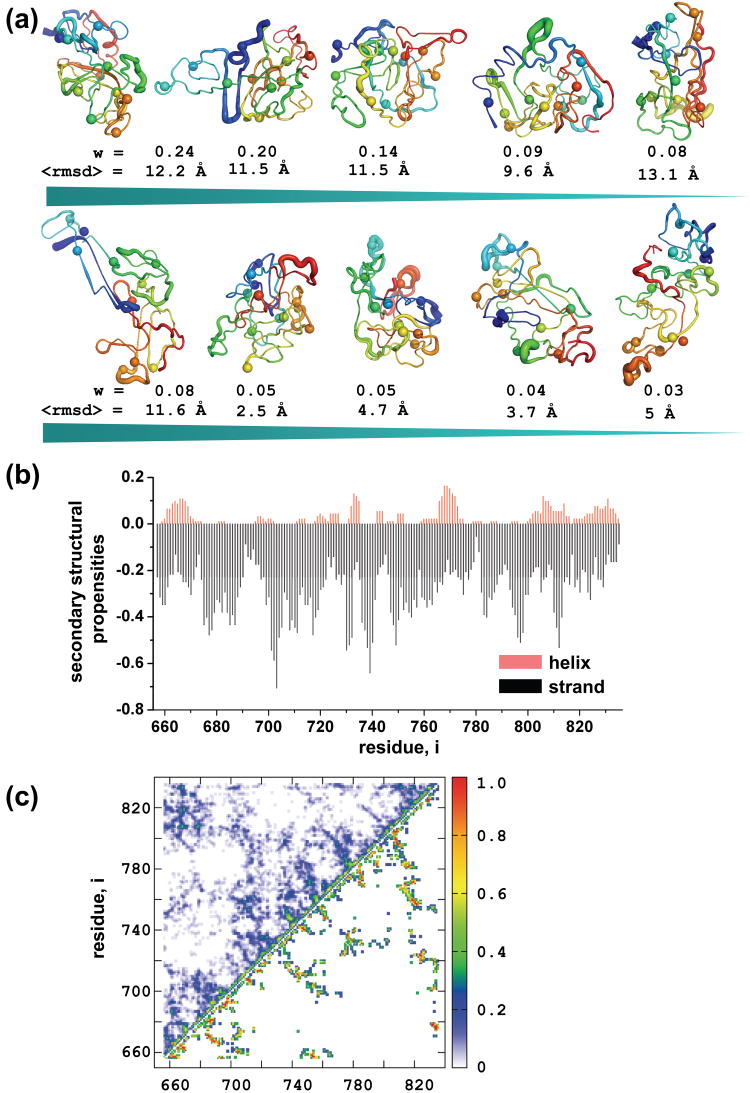
High resolution 3D structural information is not available for either the R domain or the intact CFTR protein. Therefore, to gain further insight into mechanisms whereby the conserved patterns of order, secondary structure, and phosphorylation sites within the globally disordered R domain may control CFTR channel activity, we generated an ensemble of low energy representations using DMD52, 65, 66, a rapid sampling algorithm and an all atom force field called Medusa54, 55, 67. We clustered the decoy set, which consists of low energy members, to identify dominant features of the more ordered R domain conformers within the disordered state ensemble. Shown in Figure 2(a) are the cluster centroids, where backbone thickness is rendered proportional to the average per residue RMSD. Although, the radii of gyration (Rg) do not vary greatly from ∼ 18Å (Table S1), the average intra-cluster RMSD values (Figure 2(a)) indicate that different conformers have been sampled. These conformers appear more compact than what may have been envisaged for such a generally disordered domain. However, there are other examples of collapsed and non-random structures in disordered proteins such as alpha-synuclein68 and polyglutamine proteins69. Other disordered proteins including GccH, tropo M and Nbi6 exhibit compactness typical of a molten globule state70. Thus although the R domain in the whole multidomain CFTR protein could be less compact than our computationally derived low energy conformers, it is important to realize that disorder need not dictate a non-compact configuration. The secondary structure content of the clustered R domain decoys was also analyzed (Figure 2(b)). The alpha helical propensities are very similar to those reveled by NMR data28 (for example peaks at residues 660-770, 737, 768, and 810), supporting the validity of the computed conformers. Although the β strand propensities are more discrepant, there are several possible explanations of this (see Discussion). Because changes in interactions between residues within the R domain may be important to its regulatory role, we also calculated the contact maps of the clusters (Figure 2(c)), which can serve as a guide for mutagenesis to determine positions of intradomain interactions experimentally.
Figure 2. Ensembles of R domain structures.
(a) A diverse pool of structures with low energy is generated using DMD and an all-atom energy function, Medusa. All the decoys are clustered according to pairwise RMSD. Standard deviations of coordinates of all structures in each cluster were calculated and mapped to the centroid structure. Structures are colored blue to red from N- to C-terminus. Phosphorylation sites are represented as spheres. Weights are calculated by dividing the number of decoys in each cluster by the total number of structures (n=92). (b) Secondary structural content of all the decoys were subtracted and population weighted averages were generated for each position. Alpha helical probabilities are plotted in the positive, and the β-sheet propensities in the negative region of the graph. (c) Contact maps of the most (upper diagonal) and the less (lower diagonal) populated clusters. Contacts are counted in all structures in a given cluster and divided by the total number of decoys in that particular cluster.
Phosphorylation increases R domain size
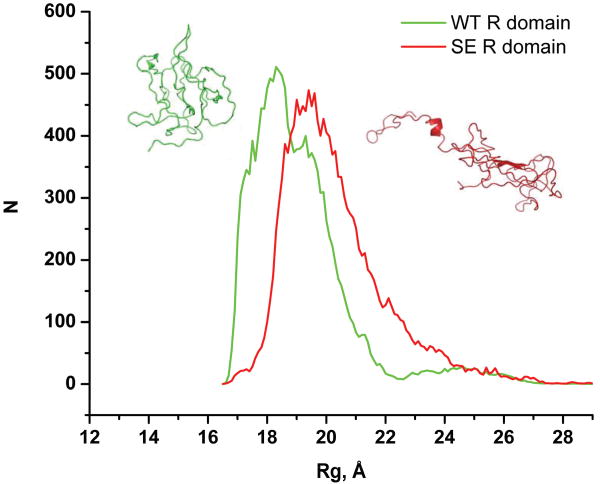
Since our simulations imply that the R domain may be more compact than had been anticipated, we wondered if phosphorylation, as well as altering local secondary structure propensities, might also change the overall size of the domain. Since phosphorylated amino acids cannot be modeled in DMD simulations, we employed serine to glutamate substitutions to mimic phosphorylation as is commonly done experimentally13, 14. As shown in Figure 3, these substitutions do increase Rg values. Although the change in the distribution is relatively small it is statistically significant (Kolmogorov-Smirnov test P value = 0.03). Introduction of carboxylate residues generally serves as only a partial mimic of phosphorylation and S→E substitutions in the R domain of intact CFTR causes far less channel activation than phosphorylation13,14. The influence of phosphorylation on p53 has recently been proposed to occur in a similar manner by promoting open conformers of its transactivation domain47.
Figure 3. R domain size is increased by phosphorylation-mimicking S→E substitutions.
Radius of gyration of the WT and S→E R domain are collected from trajectories of equilibrium simulations. Histograms had been generated and Gaussian fitting is performed to determine the peaks of the WT (18.2 Å, 19.1 Å) and the S→E (19.7 Å, 22.1 Å) distributions. Kolmogorov-Smirnov test is used to verify the difference between the two distributions (P-value = 0.03). A more closed conformation of the WT domain is depicted on the left (green), and a more open conformation of the S→E R domain on the right (red).
Distance constraint model of R domain function
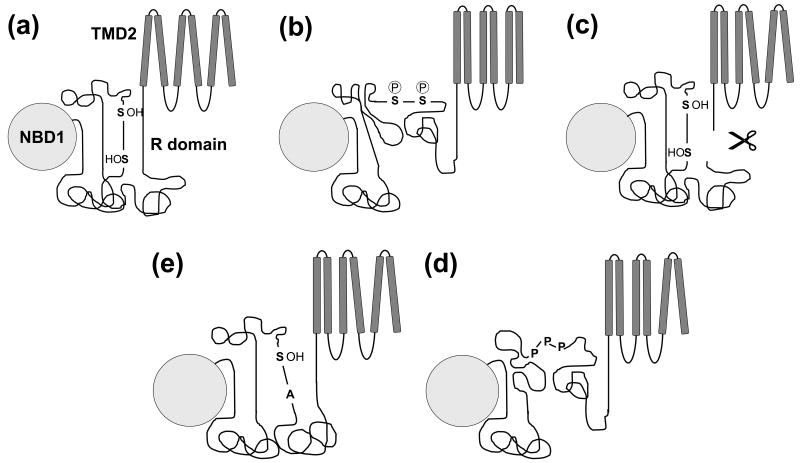
The disordered nature of the R domain has fostered a notion that it may be a loosely-formed, segmented region of the protein in which phosphorylation-induced modulation of local elements adjacent multiple phosphorylation sites alters their interactions with sites elsewhere in CFTR. The data of Baker et al.28 support such a mode of action. However, the overall mechanism is still unclear. Our computational findings that the R domain has the potential to form a fairly compact structure whose size may be increased by phosphorylation as well as published electrophysiological data11 are consistent with a role of the domain as a distance constraint between the two halves of CFTR which it separates. These spatial changes are postulated to alter the packing of membrane-spanning helices or their susceptibility to the impact of ATP binding to the NBDs. We hypothesize that the regions with higher order tendency are most likely to form interactions with other parts of CFTR to inhibit channel opening in the non-phosphorylated state (Figure 4(a)). Upon activation, the phosphorylation at serine and threonine residues by PKA has several consequences. First of all, those amino acids gain negative charge from the phosphate moiety. Phosphorylation may also affect the structure by changing the secondary structure of some regions, which eventually leads to reorganization of intramolecular interactions. Ostedgaard et al.9 showed that in the presence of TMAO, an osmolyte promoting protein folding, there was an increase in alpha helical content in the non-phosphorylated R domain but not in the phosphorylated domain. This observation suggests decreased helical propensity in the phosphorylated polypeptide as originally shown by Dulhanty et al.26. A recent NMR study also demonstrated a decreased helical tendency upon phosphorylation of the R domain28. The most likely outcome of all of these changes is the extension and increased mobility of the R domain, which in turn may cause the reorientation of the transmembrane helices (Figure 4(b)). Other activation events (Figure 4(c)-(e)) may similarly change the distances between the cytoplasmic parts of the transmembrane domains and lead to the rearrangement of transmembrane helices (see below).
Figure 4. Working model for R domain function.
(a) In the non-phosphorylated state, R domain regions with higher ordered tendency form intramolecular interactions with the R domain itself and NBD1, maintaining a short distance between the two ends of the R domain inhibiting channel gating. Serines and threonines are important in the stabilization of this state. (b) Upon phosphorylation there is a net gain in negative charge and conformational changes within the R domain. All of these alterations cause reorientation of helices and signal transduction from the transmembrane to the nucleotide binding domains to promote gating. (c) Severing CFTR C-terminal of the R domain abolishes the distance constraint imposed by the R domain and leads to increased activation coupled to reorganization of helix packing. (d) The slightly increased activity of unphosphorylated Ser→Ala CFTR mutants may be caused by losing the stabilization provided by the hydroxyl amino acid, Serine. The alanine substitutions result in a more extended conformation of the R domain and therefore chloride channel activation. (e) The three prolines responsible for CFTR activation by PPIase are located in a region with higher ordered propensity. The basis of this alternative R domain activation pathway might be the weakened interaction between that region and other parts of the R domain or NBD1 after isomerization.
Discussion
The R domain product of the largest exon in the CFTR gene is the most unique feature of the ion channel, distinguishing it from all other known ABC transporters. There is strong evidence that its role is to provide tight quantitative control of the level of channel gating activity since in its absence the channel gates continuously, precluding its crucial regulated physiological function. The molecular mechanism whereby this control is exerted is not yet understood. In this study we focused on the contribution of the disorder-order pattern of local segments within the globally disordered domain as well as on the properties and response to phosphorylation of computationally derived low energy conformers of the domain.
With respect to the first aspect, the relatively lower sequence identity among R domains of different species compared to other domains of CFTR is apparently sufficient to provide conservation of structural organization including the disorder-order pattern. This conservation is also reflected in the distribution of secondary structure elements predicted either from sequence or a computed low energy conformer ensemble. Both the more ordered and the more structured segments are characteristic of molecular recognition elements34, 35 and hence are likely to participate in interactions with sites elsewhere in the protein. Their conformation and binding properties may be regulated by phosphorylation of adjacent phosphorylation sites similar to that which occurs in many other disordered proteins, such as the Kinase-Inducible Domain of CREB61, the cyclin-dependent kinase inhibitor Sic162, and the transcription factor Ets-163. In these cases phosphorylation promotes ordering and facilitates binding, while in the R domain, phosphorylation is reported to decrease alpha helical propensity and to disrupt many of its interactions with other parts of the molecule28.
We aimed to characterize the disordered state ensemble of the R domain by generation of low energy conformers of this polypeptide computationally. To compare members of our ensemble with experiments, we compared the secondary structure distribution within the ensemble to secondary structural propensities from NMR studies28. The location and probability of the fluctuating α-helices are consistent with the experiments (Figure 1(d)). The propensity to form strands in the DMD simulations (Figure 2(b)) are much larger compared to the NMR experiments, however, it is in good agreement with an earlier CD study indicating much higher β-sheet propensity (30%) compared to the helical propensity (10%)26. In the latter measurements a significant portion of this helical content arises from the C-terminal part of the NBD1, which was a part of the construct used in the CD experiments. Moreover, the Medusa force field has not shown bias towards strands in any previous simulations. In fact, when we applied cutoff values for the average of the helical and strand content (0.06 and -0.4 for helices and strands, respectively), the extracted secondary structural pattern of the R domain is very similar to both the propensities from the NMR experiments and the positions of helices and strands located by the predictors (Figure 1(d)). In spite of this, some properties of the low energy conformers such as their apparent compactness may not entirely reflect the R domain structure under physiological conditions. This compactness also raises the question of the accessibility of the phosphorylation sites to kinases and phosphatases. However a somewhat compact R domain does not necessarily limit its dynamics which is probably sufficient to expose the phosphorylation sites. Furthermore, phosphorylation of exposed sites may make other, more buried sites accessible to kinases, as suggested by the different rates of phosphorylation of different sites16. Additionally, cellular and macromolecular crowding conditions may affect the compactness and secondary structure conformation of disordered proteins, as suggested by in vivo experiments73, 74.
Crucial to the understanding of the regulatory function is the mapping of R domain interaction sites within the domain and elsewhere in the molecule. The segments in the R domain biased towards α-helices or β-sheets may interact with sites in R and/or in other domains. Interactions within the R domain are suggested by non-uniform R2 relaxation rates in NMR experiments28, which indicate restricted mobility by contact formation within and between the dynamic secondary structural elements. The interactions with other elements, corresponding to the ones also identified by NMR studies, were detected in in vitro experiments in which only R and NBD1 were present.28 Since phosphorylation by PKA can promote association between NBD1 and NBD2 in the whole CFTR protein71, the R/NBD1 interactions may be a part of the regulatory mechanism. However, channel gating is not entirely dependent on the state of the interface between NBDs as it can occur in the complete absence of NBD272, 73. Therefore, we postulate that R domain phosphorylation may more directly influence channel gating by acting on the MSDs which constitute the channel pore.
Because we found also that reasonably compact low energy R conformers, generated computationally, are increased in size on “pseudophosphorylation” by substituting carboxylate residues at PKA sites (Figure 3), a change in the configuration of transmembrane helices (TMs) within the MSDs may occur due to a change in the overall distance between sites on either side of the enlarged R domain. Such a mechanism would be conceptually analogous to the activation mechanism of multi-drug resistance (MDR) proteins, which are active transporter members of the ABC family. Activation (drug-stimulated ATPase activity) of MDR proteins is initiated by the interaction of hydrophobic drugs with sites formed by transmembrane helices74-78. Drug binding results in rearrangement of transmembrane helices79-81, which in turn signals the nucleotide binding domains to stimulate the enzymatic cycle. In our model of CFTR, similar changes in helix packing would be required for activation, with the R domain playing a similar role as the hydrophobic drug.
Activation modes alternative to phosphorylation are also consistent with this model. First, a single peptide cleavage between the C-terminal end of the R domain and TMD2 increases channel activity (Figure 4(c))11, 12. Second, the addition of an R domain polypeptide to phosphorylated CFTR is not inhibitory6, 8, 9. Third, relocation of the R domain to the CFTR C-terminus also has no inhibitory influence10. Two of the PKA phosphorylation sites at serines 737 and 768 have been reported to be inhibitory rather than stimulatory16, 19-21. It may be postulated that these mutations destabilize important intramolecular interactions involving the two hydroxyl amino acids (Figure 4(d)). The region between 737 and 768 is predicted to be a more highly ordered part of the domain and probably contributes strongly to the inhibitory role of the unphosphorylated R domain via interactions that keep it compact and less dynamic. Cyclophilin A, a peptidyl-prolyl isomerase acts on three prolines between amino acids 737 and 768 activating CFTR without phosphorylation by PKA23, 24. This alternative mode of CFTR activation agrees well with our model, in which the inhibitory intramolecular interactions in this region are destabilized by the isomerization of prolines 740, 750, and 759 (Figure 4(e)).
Specific aspects of this model including changes in distances between residues in TMs will become testable experimentally in conjunction with a high resolution structural model of CFTR which we have generated recently82 as well as structural changes observed by electron crystallography (R.C. Ford, unpublished observations).
Methods
Disorder predictions
Many collected predictors are available at http://www.disprot.org/predictors.php with application of different algorithms for recognition of disordered region of proteins. In order to gain higher confidence of prediction, we selected two predictors based on two different approaches. Disopred2 (http://bioinf.cs.ucl.ac.uk/disopred) implements support vector machines and is trained on a non-redundant set of the PDB database56, 57. The cutoff value is 0.05, which is the false-hit rate. The other server, IUPred (http://iupred.enzim.hu), assesses the capacity of polypeptides to form stabilizing contacts by estimation of total pairwise interaction energies of residues58, 59. In IUPred the energy values are transformed into a probabilistic score ranging from 0 (complete order) to 1 (complete disorder). Residues with a score above 0.5 can be regarded as disordered. Values higher than the cut-off (0.05 and 0.5 for DISOPRED2 and IUPred, respectively) indicate regions that have higher disorder propensity. Both prediction methods give similar results of alternating high and low disorder tendencies (Figure 1). Since IUPred performs extremely rapidly compared to other predictors and accepts sequences with arbitrary length, we used it mainly in our study. Alignments to compare disorder tendencies in different proteins were generated by clustalw83. The following CFTR entries were used from the UniProt database84 (http://www.ebi.ac.uk/uniprot): A0M8T4_FELCA, CFTR_HUMAN, CFTR_HYLLE, CFTR_CALMO, CFTR_SAIBB, CFTR_CARPS, CFTR_MICMU, CFTR_BOVIN, CFTR_SHEEP, CFTR_PIG, CFTR_RABIT, A4D7T4_MACEU, CFTR_DIDMA, CFTR_ORNAN, CFTR_MOUSE, Q2IBD3_RAT, A0M8U4_CHICK, CFTR_XENLA, CFTR_SQUAC, Q9IAR8_SALSA, Q9W750_SALSA, O73677_FUNHE, A0M8W1_DANRE. Locally written scripts were used to calculate the average and standard deviation at each position in the alignments. Alignments were edited and colored in Jalview85.
Secondary structure prediction
Three different predictors already assessed on disordered proteins35 were used. GOR462 (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) uses a simple statistical algorithm, while ALB63 (http://i2o.protres.ru/alb/alb.cgi) estimates short and long range interactions between residues based on physicochemical properties of amino acids. Prof64 (http://www.aber.ac.uk/∼phiwww/prof) is a more advanced method employing a neural network algorithm using evolutionary information.
Generating structural ensemble of the R domain
A diverse and large pool of structures was generated from the linear R domain peptide (residues 656-836) using a dynamic sampling algorithm, discrete molecular dynamics (DMD)54, 66, 86, 87. DMD provides rapid and efficient sampling of conformational space compared to conventional molecular dynamics. We used Medusa, an all-atom force field54, 55. Detailed description of the energy terms and the parametrization can be found in our recent work54. The search for low energy conformers was performed by replica exchange87 DMD simulations54, 66, 86, 87 in two steps. Replica exchange was employed to overcome energy barriers between local minima and to efficiently search for states with low energy. In the ideal case the simulation temperatures of replicas cover a wide range, for ordered proteins both below and above the Tm, but these types of simulations take a long time with a polypeptide of the size of the R domain. To circumvent this problem, simulations were performed in two rounds. In the first round, we sampled accessible conformers of the R domain in a higher temperature range (0.5-0.78 ε/kb). Then we constructed a preliminary decoy set composed of structures with low potential energy values (< -450 ε). Then each decoy in the preliminary set was subjected to a second round replica exchange-DMD with exchange temperatures in a lower range (0.3-0.58 ε/kb). Our final decoy set consists of 92 low-energy structures (with energy less than or equal to -750 ε), which were relaxed in a final step of equilibrium simulation at low temperature (0.2 ε/kb). The sampled low-energy conformers allowed determination of the characteristic properties of the more ordered R domain conformers within the disordered state ensemble.
Equilibrium simulations of WT and S→E R domain
An R domain mutant with S→E substitutions (S 660, 670, 686, 690, 700, 712, 737, 753, 768, 787, 788, 790, 795, 813 E) was generated by flexible backbone redesign using Medusa54, 55. First equilibrium simulations were performed at various temperatures on a few structures to determine the melting profile of both the WT and the SE R domain. No discrete Tm was detected, which is characteristic of disordered proteins indicating the lack of cooperative folding (data not shown). Then long equilibrium simulations at temperature 0.55 ε/kb were carried out with one structure from each cluster. Structures and their properties were analyzed as described below.
Analysis of clusters of R domain structures
To find the major conformations accessed by the R domain, decoys were grouped into clusters using the k-means algorithm88, a simple unsupervised learning algorithm to solve clustering problems. It clusters objects into k partitions based on attributes, which are assumed to form a vector space. The k-means algorithm attempts to find centers in the data and minimize the total intra-cluster variance. The distance (i.e. similarity) metric between two structures was defined as the Cα root-mean-square-deviation (RMSD). Clustering was done using MatLab (MathWorks, Inc.) code. Weight of clusters was defined as the ratio of the number of elements in a cluster and the total number of structures. In order to demonstrate the main properties of clusters, standard deviations of atomic coordinates were calculated and projected onto the centroid. The values were inserted into the B-factor column in the pdb file of the centroid structure and displayed by PyMol (http://www.pymol.org).
We generated contact maps by defining a contact between two residues if their heavy atoms are within a distance of 5 Å. Contacts at each position were counted for each structure in a cluster, normalized, and plotted (Figure 2(c)). For each residue, the numbers of contacts formed in the structures within a cluster were counted, divided by the total number of structures in the given cluster, and plotted.
Secondary structure was assigned using a method proposed by Srinivasan and Rose89. Occurrence of each residue in the four types of secondary structural elements (coil, α-helix, β-sheet, turn) was counted in the decoy set of each cluster. The values were averaged and population weighted, then α-helix and β-sheet propensity values were plotted. To generate a string representing the secondary structure content, cutoff values 0.06 and -0.4 were used for helices and strands, respectively. The secondary structure string for NMR experiments was extracted from Figure 2(a) in Baker et al.28 applying a cutoff of 0.15 for both helices and strands.
Supplementary Material
Acknowledgments
We wish to thank Péter Tompa and Zsuzsa Dosztányi (Institute of Enzymology, Hungary) for valuable discussions on intrinsically disordered proteins and Feng Ding for help in using DMD. This work was supported by the NIH and partially by the Cystic Fibrosis Foundation Grant DOKHOL07I0 (to N. V. D.) and a Marie Curie Fellowship (to T.H.; MTKD-CT-2006-042794).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boucher RC. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur Respir J. 2004;23:146–58. doi: 10.1183/09031936.03.00057003. [DOI] [PubMed] [Google Scholar]
- 2.Riordan JR. Assembly of functional CFTR chloride channels. Annu Rev Physiol. 2005;67:701–18. doi: 10.1146/annurev.physiol.67.032003.154107. [DOI] [PubMed] [Google Scholar]
- 3.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–73. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 4.Cheng SH, Rich DP, Marshall J, Gregory RJ, Welsh MJ, Smith AE. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell. 1991;66:1027–36. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- 5.Seibert FS, Chang XB, Aleksandrov AA, Clarke DM, Hanrahan JW, Riordan JR. Influence of phosphorylation by protein kinase A on CFTR at the cell surface and endoplasmic reticulum. Biochim Biophys Acta. 1999;1461:275–83. doi: 10.1016/s0005-2736(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 6.Ma J, Zhao J, Drumm ML, Xie J, Davis PB. Function of the R domain in the cystic fibrosis transmembrane conductance regulator chloride channel. J Biol Chem. 1997;272:28133–41. doi: 10.1074/jbc.272.44.28133. [DOI] [PubMed] [Google Scholar]
- 7.Vankeerberghen A, Lin W, Jaspers M, Cuppens H, Nilius B, Cassiman JJ. Functional characterization of the CFTR R domain using CFTR/MDR1 hybrid and deletion constructs. Biochemistry. 1999;38:14988–98. doi: 10.1021/bi991520d. [DOI] [PubMed] [Google Scholar]
- 8.Winter MC, Welsh MJ. Stimulation of CFTR activity by its phosphorylated R domain. Nature. 1997;389:294–6. doi: 10.1038/38514. [DOI] [PubMed] [Google Scholar]
- 9.Ostedgaard LS, Baldursson O, Vermeer DW, Welsh MJ, Robertson AD. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc Natl Acad Sci U S A. 2000;97:5657–62. doi: 10.1073/pnas.100588797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldursson O, Ostedgaard LS, Rokhlina T, Cotten JF, Welsh MJ. Cystic fibrosis transmembrane conductance regulator Cl- channels with R domain deletions and translocations show phosphorylation-dependent and -independent activity. J Biol Chem. 2001;276:1904–10. doi: 10.1074/jbc.M006934200. [DOI] [PubMed] [Google Scholar]
- 11.Csanady L, Chan KW, Seto-Young D, Kopsco DC, Nairn AC, Gadsby DC. Severed channels probe regulation of gating of cystic fibrosis transmembrane conductance regulator by its cytoplasmic domains. J Gen Physiol. 2000;116:477–500. doi: 10.1085/jgp.116.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csanady L, Chan KW, Nairn AC, Gadsby DC. Functional roles of nonconserved structural segments in CFTR's NH2-terminal nucleotide binding domain. J Gen Physiol. 2005;125:43–55. doi: 10.1085/jgp.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rich DP, Berger HA, Cheng SH, Travis SM, Saxena M, Smith AE, Welsh MJ. Regulation of the cystic fibrosis transmembrane conductance regulator Cl- channel by negative charge in the R domain. J Biol Chem. 1993;268:20259–67. [PubMed] [Google Scholar]
- 14.Aleksandrov AA, Chang X, Aleksandrov L, Riordan JR. The non-hydrolytic pathway of cystic fibrosis transmembrane conductance regulator ion channel gating. J Physiol. 2000;528(Pt 2):259–65. doi: 10.1111/j.1469-7793.2000.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang XB, Tabcharani JA, Hou YX, Jensen TJ, Kartner N, Alon N, Hanrahan JW, Riordan JR. Protein kinase A (PKA) still activates CFTR chloride channel after mutagenesis of all 10 PKA consensus phosphorylation sites. J Biol Chem. 1993;268:11304–11. [PubMed] [Google Scholar]
- 16.Csanady L, Seto-Young D, Chan KW, Cenciarelli C, Angel BB, Qin J, McLachlin DT, Krutchinsky AN, Chait BT, Nairn AC, Gadsby DC. Preferential phosphorylation of R-domain Serine 768 dampens activation of CFTR channels by PKA. J Gen Physiol. 2005;125:171–86. doi: 10.1085/jgp.200409076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picciotto MR, Cohn JA, Bertuzzi G, Greengard P, Nairn AC. Phosphorylation of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1992;267:12742–52. [PubMed] [Google Scholar]
- 18.Cohn JA, Nairn AC, Marino CR, Melhus O, Kole J. Characterization of the cystic fibrosis transmembrane conductance regulator in a colonocyte cell line. Proc Natl Acad Sci U S A. 1992;89:2340–4. doi: 10.1073/pnas.89.6.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson DJ, Strong TV, Mansoura MK, Wood DL, Smith SS, Collins FS, Dawson DC. CFTR activation: additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am J Physiol. 1997;273:L127–33. doi: 10.1152/ajplung.1997.273.1.L127. [DOI] [PubMed] [Google Scholar]
- 20.Baldursson O, Berger HA, Welsh MJ. Contribution of R domain phosphoserines to the function of CFTR studied in Fischer rat thyroid epithelia. Am J Physiol Lung Cell Mol Physiol. 2000;279:L835–41. doi: 10.1152/ajplung.2000.279.5.L835. [DOI] [PubMed] [Google Scholar]
- 21.Vais H, Zhang R, Reenstra WW. Dibasic phosphorylation sites in the R domain of CFTR have stimulatory and inhibitory effects on channel activation. Am J Physiol Cell Physiol. 2004;287:C737–45. doi: 10.1152/ajpcell.00504.2003. [DOI] [PubMed] [Google Scholar]
- 22.Himmel B, Nagel G. Protein kinase-independent activation of CFTR by phosphatidylinositol phosphates. EMBO Rep. 2004;5:85–90. doi: 10.1038/sj.embor.7400034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie J, Zhao J, Davis PB, Ma J. Conformation, independent of charge, in the R domain affects cystic fibrosis transmembrane conductance regulator channel openings. Biophys J. 2000;78:1293–305. doi: 10.1016/S0006-3495(00)76685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, Xie J, Ma J, Davis PB. Intermolecular interaction between R domains of cystic fibrosis transmembrane conductance regulator. Am J Respir Cell Mol Biol. 2004;30:242–8. doi: 10.1165/rcmb.2002-0108OC. [DOI] [PubMed] [Google Scholar]
- 25.Fischer H, Schwarzer C, Illek B. Vitamin C controls the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci U S A. 2004;101:3691–6. doi: 10.1073/pnas.0308393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dulhanty AM, Riordan JR. Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1994;33:4072–9. doi: 10.1021/bi00179a036. [DOI] [PubMed] [Google Scholar]
- 27.Grimard V, Li C, Ramjeesingh M, Bear CE, Goormaghtigh E, Ruysschaert JM. Phosphorylation-induced conformational changes of cystic fibrosis transmembrane conductance regulator monitored by attenuated total reflection-Fourier transform IR spectroscopy and fluorescence spectroscopy. J Biol Chem. 2004;279:5528–36. doi: 10.1074/jbc.M311014200. [DOI] [PubMed] [Google Scholar]
- 28.Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–82. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 30.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–33. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 31.Ma K, Forbes JG, Gutierrez-Cruz G, Wang K. Titin as a giant scaffold for integrating stress and Src homology domain 3-mediated signaling pathways: the clustering of novel overlap ligand motifs in the elastic PEVK segment. J Biol Chem. 2006;281:27539–56. doi: 10.1074/jbc.M604525200. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay R, Hoh JH. AFM force measurements on microtubule-associated proteins: the projection domain exerts a long-range repulsive force. FEBS Lett. 2001;505:374–8. doi: 10.1016/s0014-5793(01)02844-7. [DOI] [PubMed] [Google Scholar]
- 33.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–49. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–70. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 35.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–26. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, He Z, O'Shaughnessy TJ, Rux J, Reenstra WW. Domain-domain associations in cystic fibrosis transmembrane conductance regulator. Am J Physiol Cell Physiol. 2002;282:C1170–80. doi: 10.1152/ajpcell.00337.2001. [DOI] [PubMed] [Google Scholar]
- 37.Wei L, Vankeerberghen A, Cuppens H, Cassiman JJ, Droogmans G, Nilius B. The C-terminal part of the R-domain, but not the PDZ binding motif, of CFTR is involved in interaction with Ca(2+)-activated Cl- channels. Pflugers Arch. 2001;442:280–5. doi: 10.1007/s004240100531. [DOI] [PubMed] [Google Scholar]
- 38.Ko SB, Zeng W, Dorwart MR, Luo X, Kim KH, Millen L, Goto H, Naruse S, Soyombo A, Thomas PJ, Muallem S. Gating of CFTR by the STAS domain of SLC26 transporters. Nat Cell Biol. 2004;6:343–50. doi: 10.1038/ncb1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–56. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bracken C, Iakoucheva LM, Romero PR, Dunker AK. Combining prediction, computation and experiment for the characterization of protein disorder. Curr Opin Struct Biol. 2004;14:570–6. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding. Proc Natl Acad Sci U S A. 2003;100:5148–53. doi: 10.1073/pnas.0531373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verkhivker GM. Protein conformational transitions coupled to binding in molecular recognition of unstructured proteins: hierarchy of structural loss from all-atom Monte Carlo simulations of p27Kip1 unfolding-unbinding and structural determinants of the binding mechanism. Biopolymers. 2004;75:420–33. doi: 10.1002/bip.20149. [DOI] [PubMed] [Google Scholar]
- 43.Wong KB, Clarke J, Bond CJ, Neira JL, Freund SM, Fersht AR, Daggett V. Towards a complete description of the structural and dynamic properties of the denatured state of barnase and the role of residual structure in folding. J Mol Biol. 2000;296:1257–82. doi: 10.1006/jmbi.2000.3523. [DOI] [PubMed] [Google Scholar]
- 44.Wen EZ, Luo R. Interplay of secondary structures and side-chain contacts in the denatured state of BBA1. J Chem Phys. 2004;121:2412–21. doi: 10.1063/1.1768151. [DOI] [PubMed] [Google Scholar]
- 45.Mayor U, Guydosh NR, Johnson CM, Grossmann JG, Sato S, Jas GS, Freund SM, Alonso DO, Daggett V, Fersht AR. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–7. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 46.Marianayagam NJ, Jackson SE. The folding pathway of ubiquitin from all-atom molecular dynamics simulations. Biophys Chem. 2004;111:159–71. doi: 10.1016/j.bpc.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Lowry DF, Stancik A, Shrestha RM, Daughdrill GW. Modeling the accessible conformations of the intrinsically unstructured transactivation domain of p53. Proteins. 2007 doi: 10.1002/prot.21721. [DOI] [PubMed] [Google Scholar]
- 48.Olson KE, Narayanaswami P, Vise PD, Lowry DF, Wold MS, Daughdrill GW. Secondary structure and dynamics of an intrinsically unstructured linker domain. J Biomol Struct Dyn. 2005;23:113–24. doi: 10.1080/07391102.2005.10507052. [DOI] [PubMed] [Google Scholar]
- 49.Makowska J, Rodziewicz-Motowidlo S, Baginska K, Vila JA, Liwo A, Chmurzynski L, Scheraga HA. Polyproline II conformation is one of many local conformational states and is not an overall conformation of unfolded peptides and proteins. Proc Natl Acad Sci U S A. 2006;103:1744–9. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman HJ, Hogue CW. A fast method to sample real protein conformational space. Proteins. 2000;39:112–31. [PubMed] [Google Scholar]
- 51.Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol. 2001;308:1011–32. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 52.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–87. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 53.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Identifying the protein folding nucleus using molecular dynamics. J Mol Biol. 2000;296:1183–8. doi: 10.1006/jmbi.1999.3534. [DOI] [PubMed] [Google Scholar]
- 54.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007;15:1567–76. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 56.Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53 6:573–8. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- 57.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–45. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 58.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–39. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 59.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Altenberg GA, Reuss L. Mechanism of activation of Xenopus CFTR by stimulation of PKC. Am J Physiol Cell Physiol. 2004;287:C1256–63. doi: 10.1152/ajpcell.00229.2004. [DOI] [PubMed] [Google Scholar]
- 61.Hegedus T, Riordan JR. Search for proteins with similarity to the CFTR R domain using an optimized RDBMS solution, mBioSQL. Central European Journal of Biology. 2006;1:29–42. [Google Scholar]
- 62.Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 63.Ptitsyn OB, Finkelstein AV. Theory of protein secondary structure and algorithm of its prediction. Biopolymers. 1983;22:15–25. doi: 10.1002/bip.360220105. [DOI] [PubMed] [Google Scholar]
- 64.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci U S A. 1993;90:7558–62. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding F, Buldyrev SV, Dokholyan NV. Folding Trp-cage to NMR resolution native structure using a coarse-grained protein model. Biophys J. 2005;88:147–55. doi: 10.1529/biophysj.104.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding F, Dokholyan NV. Simple but predictive protein models. Trends Biotechnol. 2005;23:450–5. doi: 10.1016/j.tibtech.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4:466–7. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 68.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127:476–7. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 69.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci U S A. 2006;103:16764–9. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Csizmok V, Szollosi E, Friedrich P, Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol Cell Proteomics. 2006;5:265–73. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.Mense M, Vergani P, White DM, Altberg G, Nairn AC, Gadsby DC. In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. Embo J. 2006;25:4728–39. doi: 10.1038/sj.emboj.7601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, Gentzsch M, Aleksandrov A, Balch WE, Riordan JR. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol. 2007;365:981–94. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 73.Wang W, Bernard K, Li G, Kirk KL. Curcumin opens cystic fibrosis transmembrane conductance regulator channels by a novel mechanism that requires neither ATP binding nor dimerization of the nucleotide-binding domains. J Biol Chem. 2007;282:4533–44. doi: 10.1074/jbc.M609942200. [DOI] [PubMed] [Google Scholar]
- 74.Ferry D, Boer R, Callaghan R, Ulrich WR. Localization of the 1,4-dihydropyridine drug acceptor of P-glycoprotein to a cytoplasmic domain using a permanently charged derivative N-methyl dexniguldipine. Int J Clin Pharmacol Ther. 2000;38:130–40. doi: 10.5414/cpp38130. [DOI] [PubMed] [Google Scholar]
- 75.Lugo MR, Sharom FJ. Interaction of LDS-751 and rhodamine 123 with P-glycoprotein: evidence for simultaneous binding of both drugs. Biochemistry. 2005;44:14020–9. doi: 10.1021/bi0511179. [DOI] [PubMed] [Google Scholar]
- 76.Qu Q, Sharom FJ. Proximity of bound Hoechst 33342 to the ATPase catalytic sites places the drug binding site of P-glycoprotein within the cytoplasmic membrane leaflet. Biochemistry. 2002;41:4744–52. doi: 10.1021/bi0120897. [DOI] [PubMed] [Google Scholar]
- 77.Shapiro AB, Ling V. Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur J Biochem. 1997;250:122–9. doi: 10.1111/j.1432-1033.1997.00122.x. [DOI] [PubMed] [Google Scholar]
- 78.Shapiro AB, Ling V. Transport of LDS-751 from the cytoplasmic leaflet of the plasma membrane by the rhodamine-123-selective site of P-glycoprotein. Eur J Biochem. 1998;254:181–8. doi: 10.1046/j.1432-1327.1998.2540181.x. [DOI] [PubMed] [Google Scholar]
- 79.Loo TW, Bartlett MC, Clarke DM. Substrate-induced conformational changes in the transmembrane segments of human P-glycoprotein. Direct evidence for the substrate-induced fit mechanism for drug binding. J Biol Chem. 2003;278:13603–6. doi: 10.1074/jbc.C300073200. [DOI] [PubMed] [Google Scholar]
- 80.Loo TW, Clarke DM. Drug-stimulated ATPase activity of human P-glycoprotein requires movement between transmembrane segments 6 and 12. J Biol Chem. 1997;272:20986–9. doi: 10.1074/jbc.272.34.20986. [DOI] [PubMed] [Google Scholar]
- 81.Loo TW, Clarke DM. The packing of the transmembrane segments of human multidrug resistance P-glycoprotein is revealed by disulfide cross-linking analysis. J Biol Chem. 2000;275:5253–6. doi: 10.1074/jbc.275.8.5253. [DOI] [PubMed] [Google Scholar]
- 82.Serohijos AW, Hegedus T, Aleksandrov A, He L, Cui L, Dokholyan NV, Riordan JR. Phenylalanine 508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0800254105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 84.Consortium TU. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–7. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–7. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 86.Zhou YQ, Karplus M, Wichert JM, Hall CK. Equilibrium thermodynamics of homopolymers and clusters: Molecular dynamics and Monte Carlo simulations of systems with square-well interactions. Journal of Chemical Physics. 1997;107:10691–10798. [Google Scholar]
- 87.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical physics letters. 1999;314:141–51. [Google Scholar]
- 88.MacQueen JB. Some Methods for classification and Analysis of Multivariate Observations. Proceedings of 5-th Berkeley Symposium on Mathematical Statistics and Probability; University of California Press; 1967. pp. 281–297. [Google Scholar]
- 89.Srinivasan R, Rose GD. A physical basis for protein secondary structure. Proc Natl Acad Sci U S A. 1999;96:14258–63. doi: 10.1073/pnas.96.25.14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.